Abstract
Healthcare-associated infections are a leading cause of morbidity and mortality worldwide. Treatment is increasingly complicated by the escalating incidence of antimicrobial resistance. Among drug-resistant pathogens, carbapenem-resistant Acinetobacter baumannii (CRAb) is of increasing concern because of the limited applicable therapies and its expanding global distribution in developed countries and newly industrialized countries. Therefore, a rapid detection method that can be used even in resource-poor countries is urgently required to control this global public health threat. Conventional techniques, such as bacterial culture and polymerase chain reaction (PCR), are insufficient to combat this threat because they are time-consuming and laborious. In this study, we developed a loop-mediated isothermal amplification (LAMP) method for detecting bla OXA-23-positive CRAb, the most prevalent form of CRAb in Asia, especially in Thailand, and confirmed its efficacy as a surveillance tool in a clinical setting. Clinical samples of sputum and rectal swabs were collected from patients in a hospital in Bangkok and used for LAMP assays. After boiling and centrifugation, the supernatants were used directly in the assay. In parallel, a culture method was used for comparison purposes to evaluate the specificity and sensitivity of LAMP. As a first step, a total of 120 sputum samples were collected. The sensitivity of LAMP was 88.6% (39/44), and its specificity was 92.1% (70/76) using the culture method as the “gold standard”. When surveillance samples including sputum and rectal swabs were analyzed with the LAMP assay, its sensitivity was 100.0%. This method enables the direct analysis of clinical specimens and provides results within 40 minutes of sample collection, making it a useful tool for surveillance even in resource-poor countries.
Healthcare-associated infections are a leading cause of morbidity and mortality worldwide. Treatment is increasingly complicated by the escalating incidence of antimicrobial resistance. Among drug-resistant pathogens, carbapenem-resistant Acinetobacter baumannii (CRAb) is of increasing concern because of the limited applicable therapies and its expanding global distribution in developed countries and newly industrialized countries. Therefore, a rapid detection method that can be used even in resource-poor countries is urgently required to control this global public health threat. Conventional techniques, such as bacterial culture and polymerase chain reaction (PCR), are insufficient to combat this threat because they are time-consuming and laborious. In this study, we developed a loop-mediated isothermal amplification (LAMP) method for detecting bla OXA-23-positive CRAb, the most prevalent form of CRAb in Asia, especially in Thailand, and confirmed its efficacy as a surveillance tool in a clinical setting. Clinical samples of sputum and rectal swabs were collected from patients in a hospital in Bangkok and used for LAMP assays. After boiling and centrifugation, the supernatants were used directly in the assay. In parallel, a culture method was used for comparison purposes to evaluate the specificity and sensitivity of LAMP. As a first step, a total of 120 sputum samples were collected. The sensitivity of LAMP was 88.6% (39/44), and its specificity was 92.1% (70/76) using the culture method as the “gold standard”. When surveillance samples including sputum and rectal swabs were analyzed with the LAMP assay, its sensitivity was 100.0%. This method enables the direct analysis of clinical specimens and provides results within 40 minutes of sample collection, making it a useful tool for surveillance even in resource-poor countries.
Introduction
Acinetobacter baumannii is a gram-negative bacterium that causes nosocomial infections, such as ventilator-associated pneumonia, bacteremia, and urinary tract infections, particularly in immune-compromised patients [1]. This organism has a remarkable capacity to acquire mechanisms that confer resistance to various types of antibiotics [2]. Among these resistant bacteria, carbapenem-resistant A. baumannii (CRAb) is becoming a major concern in clinical settings because this pathogen is associated with high morbidity and mortality as well as longer hospital stays [3]. Recent studies have reported high rates of resistance worldwide, particularly in Asia-Pacific countries [4]. This issue is especially critical in developing countries where this organism spreads unnoticed and appropriate examinations cannot be performed because of a lack of resources. An easy, rapid, and sensitive detection system is required for proper therapy and isolation precautions [5, 6].
To establish a method that can be widely used in cost-limited situations, we selected LAMP (Loop-mediated isothermal amplification), which is a DNA amplification method that was developed in 2000 [7]. LAMP quickly, specifically, and efficiently amplifies DNA under isothermic conditions. The LAMP method is a valuable tool for the rapid diagnosis of infectious diseases in hospitals [8]. In this study, because CRAb isolates in Asia mostly possess bla OXA-23 [4, 9–12], we established a novel method for detecting bla OXA-23-positive CRAb directly from clinical specimens using LAMP (CRAb-LAMP). We also confirmed that this tool could be easily used as a surveillance tool in a hospital: its ease and low cost permit its use in resource-poor countries, and its detection capability for bla OXA-23 alone is adequate for surveillance.
Materials and Methods
Genotyping of CRAb isolates in Thailand
In 2011 clinical isolates of CRAb were collected from various wards and an outpatient department of Ramathibodi Hospital and Phranungklao Hospital in Bangkok in 2011. The sources of specimens were perianal swabs, wounds swabs, tracheal suction and urine, and collected. Duplicate samples were not obtained from one patient. These isolates were analyzed by conventional biochemical testing [13] and MALDI-TOF mass spectrometry (Bruker, Leipzig, Germany) [14] for bacterial identification. The Sensititre (Trek, West Sussex, UK) with the THANF panel of antimicrobial agents was used for the susceptibility tests. The antimicrobial sensitivity testing results (minimum inhibitory concentrations, MICs) were interpreted and reported according to breakpoints published in the CLSI document M100-S23.
The bla genes associated with drug resistance were investigated via PCR assays (S1 Table) [11, 12, 15–17]. The PCR mixture (30 μl) contained 0.2 mM of each dNTP, 0.5 μM of each primer, 3 μl of 10× Ex Taq buffer, 2.5 U of Ex Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan), and 1 μl of template DNA that was extracted with a QIAamp DNA mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol. PCR was performed with a thermal cycler (Bio-Rad, Hercules, CA, USA) for 40 cycles. Each cycle consisted of 30 seconds at 95°C, 30 seconds at 55°C, and 45 seconds at 72°C. The amplified products were resolved by agarose gel electrophoresis and visualized with ethidium bromide staining under UV illumination.
LAMP assay for CRAb isolates
A total of 113 bacterial strains, including A. baumannii, were used to examine the CRAb-specific reliability of the LAMP reaction, as shown in S2 Table [18–22]. The CRAb strains that were tested included the Thai isolates shown in Table 1, which represent different genotypes, and isolates from Japan, Korea, and Taiwan. We used a diverse collection of clinical strains to evaluate the ability of our LAMP assay to detect various bacterial clones.
Table 1. Genotypes of carbapenem-resistant A. baumannii (CRAb) isolates from hospital patients in Bangkok.
| Type number | OXA -10 | OXA -23 | OXA -24 | OXA -51 | OXA -58 | TEM | IMP | VIM | VEB1 | ARR2 | CMLA | Total isolates |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | + | - | + | - | - | - | - | - | - | - | 1 |
| 2 | - | + | - | + | - | - | - | - | - | + | - | 2 |
| 3 | + | - | - | + | - | - | - | - | - | - | - | 1 |
| 4 | + | - | - | + | - | - | - | - | - | + | - | 1 |
| 5 | + | - | - | + | - | - | - | - | - | + | + | 2 |
| 6 | + | - | - | + | - | + | - | - | - | - | - | 1 |
| 7 | + | - | - | + | - | + | - | - | - | + | - | 1 |
| 8 | + | - | - | + | - | + | - | - | - | + | + | 2 |
| 9 | + | - | - | + | + | - | - | - | + | + | + | 2 |
| 10 | + | + | - | + | - | - | - | - | - | - | - | 6 |
| 11 | + | + | - | + | - | - | - | - | - | + | - | 14 |
| 12 | + | + | - | + | - | - | - | - | - | + | + | 4 |
| 13 | + | + | - | + | - | - | - | - | + | + | + | 2 |
| 14 | + | + | - | + | - | + | - | - | - | - | - | 34 |
| 15 | + | + | - | + | - | + | - | - | - | + | - | 83 |
| 16 | + | + | - | + | - | + | - | - | - | + | + | 16 |
| 17 | + | + | - | + | + | + | - | - | - | + | - | 1 |
| Total a | 170 | 163 | 0 | 173 | 3 | 138 | 0 | 0 | 4 | 140 | 28 | 173 |
| % b | 98.2 | 94.2 | 0 | 100 | 1.7 | 79.8 | 0 | 0 | 2.3 | 80.9 | 16.2 | 100 |
Totala: Total number of isolates with each resistance gene
%b: The proportion of isolates with each resistance gene
A set of primers targeting bla OXA-23 was designed using Primer Explorer V4 software (Net Laboratory, Kanagawa, Japan) based on the genome sequence of A. baumannii AB0057 (NC_011585), as shown in Table 2 [23]. The primer set targeting the 16S-23S rRNA gene intergenic spacer (ITS) sequence that was used for A. baumannii identification and as an internal control for the LAMP reaction has been previously described [24]. The reaction mixture (25 μl) consisted of 40 pM each FIP and BIP, 5 pM each F3 and B3, 20 pM each LF and LB, 1.4 mM dNTPs, 10× Bst reaction buffer, and 8 U of Bst DNA polymerase (Nippon Gene, Osaka, Japan). The mixture was incubated at 60–65°C for 20–60 minutes. The results were evaluated by visual inspection because the LAMP reaction generates insoluble magnesium pyrophosphate as the amplification proceeds [25]. A Loop amp real-time turbidimeter (LA-320; Teramecs, Kyoto, Japan) was also used to confirm the amplification by monitoring the turbidity in the reaction tube in real time according to the OD650.
Table 2. LAMP primers used for bla oxa-23 and the ITS sequence.
| Target gene | Nucleotide sequence (5′-3′) | Reference | |
|---|---|---|---|
| bla OXA-23 | OXA23 F3 | GAAGCCATGAAGCTTTCTG | This study |
| OXA23 B3 | GTATGTGCTAATTGGGAAACA | ||
| OXA23 FIP | ACCGAAACCAATACGTTTTACTTCTCAGTCCCAGTCTATCAGGA | ||
| OXA23 BIP | CTGAAATTGGACAGCAGGTTGACTCTACCTCTTGAATAGGCG | ||
| OXA23 LF | TTTTGCATGAGATCAAGACCGA | ||
| OXA23 LB | CTGGTTGGTAGGACCATTAAAGGTT | ||
| 16S-23S rRNA gene intergenic spacer | ITS-F3 | CGGTAATTAGTGTGATCTGAC | [12] |
| ITS-B3 | CATTTCAGTTTAGAGCACTGT | ||
| ITS-FIP | TTGCTTAACCTAAACTCTTGAGTGAGAAGACACATTAACTCATTAACAGA | ||
| ITS-BIP | AGCAAATTAACTGAATCAAGCGTTTACTTAAGCACCGTACAGC | ||
| ITS-LF | AATTTATTTCAGACTCAATTTTGCCAA | ||
| ITS-LB | TGGTATGTGAATTTAGATTGAA |
F3: outer forward primer; B3: backward inner primer; LF/LB: loop primersouter backward primer; FIP: forward inner primer; BIP:
Ethics Statements
Ethical approval for the collection of patient specimens was obtained from the Ethics Committee of Osaka University Graduate School of Medicine and the Ethical Clearance Committee on Human Rights Related to Research Involving Human Subjects Faculty of Medicine Ramathibodi Hospital, Mahidol University. Because sample collection was performed as part of routine clinical work, informed consent was omitted as the both Institutional Review Boards approved. Instead, the patients were informed of the research procedure and wavering rights via a poster in the hospital.
LAMP assay using clinical samples
All of the clinical samples were collected at the Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand from March-June. 2013. Sputum specimens were collected randomly to investigate the efficacy of CRAb-LAMP. As surveillance samples the sputum specimens (intubated patients only) and rectal swabs were obtained in ICUs at the Faculty of Medicine Ramathibodi Hospital
After examination using a conventional culture method, each sample was stored at -20°C until its use in the LAMP assay. Sputazyme (Kyokuto, Tokyo, Japan) was added to 200–500 μl of sputum to reduce the viscosity. The rectal swabs were steeped in 500 μl of saline to dissolve the components. The sputum and rectal swab samples were boiled for 5 minutes and subsequently centrifuged for 10 minutes at 6,600 g. The supernatants were used as templates for LAMP and PCR reaction. Simultaneously, the DNA in the supernatant was extracted using a QIAamp DNA mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol and used for LAMP. The same samples were also used for the PCR assay and to compare those results with the LAMP results.
Statistical Analysis
Statistical analyses of sensitivity and specificity were analyzed with GraphPad Prism software (GraphPad Software, La Jolla, CA, USA).
Results
Genotyping of clinical isolates of CRAb in Thailand
CRAb was mainly manifested by plasmid-encoded beta-lactamases (OXA-23, OXA-24, and OXA-58) categorized as OXA enzymes [1, 9, 26]. This prevalence differed based on the region, and OXA-23-producing strains are highly prevalent in the Asia-Pacific region including Thailand [9–12]. In order to confirm this knowledge and identify a target gene that could be used to detect CRAb via LAMP a total of 173 isolates of CRAb were collected in Bangkok. PCR amplification analyses of the genes associated with carbapenem resistance were performed (Table 1). bla OXA-23 was identified in most of the isolates (94.2%) as previously described [9–12]. However, bla OXA-24 and bla OXA-58, which are frequently reported carbapenemase genes in A. baumannii samples from Europe and the United States [1, 9, 26], were rarely detected in Thailand. bla OXA-51 was identified in all of the isolates; however every A. baumannii strain possesses a chromosomally encoded OXA-51 irrespective of its resistance to carbapenem, and cannot be used in CRAb-detection [9, 27]. Although bla OXA-10 was found in most of the isolates, OXA-10 was unable to hydrolyze carbapenem and was not an appropriate target for the detection of CRAb [28]. Overall, most CRAb isolates can be detected by amplifying bla OXA-23; thus, bla OXA-23 was selected as a LAMP target for the detection of CRAb in this study.
Establishment of LAMP targeting CRAb carrying bla OXA-23
LAMP is becoming increasingly popular as an in vitro diagnostic method because it amplifies DNA quickly, efficiently, and highly specifically under isothermic conditions [7, 29]. To identify A. baumannii, a previously reported assay for the ITS sequence [12] was performed. Because a LAMP assay for bla OXA-23 has not yet been published, we designed several sets of LAMP primers in advance. To determine the best primer set and the optimal reaction conditions, a LAMP assay was performed on extracted bacterial DNA at temperatures ranging from 62°C to 67°C. The results were evaluated with a turbidimeter, as shown in Fig 1. One successful primer set (Table 2) amplified the target gene, bla OXA-23, within 20 minutes, and the negative control remained transparent after 60 minutes of incubation. Because the reaction at 65°C produced optimal results (Fig 1), we incubated the samples at 65°C for 20 minutes in all subsequent reactions.
Fig 1. Real-time turbidity assays under various conditions using a turbidimeter.
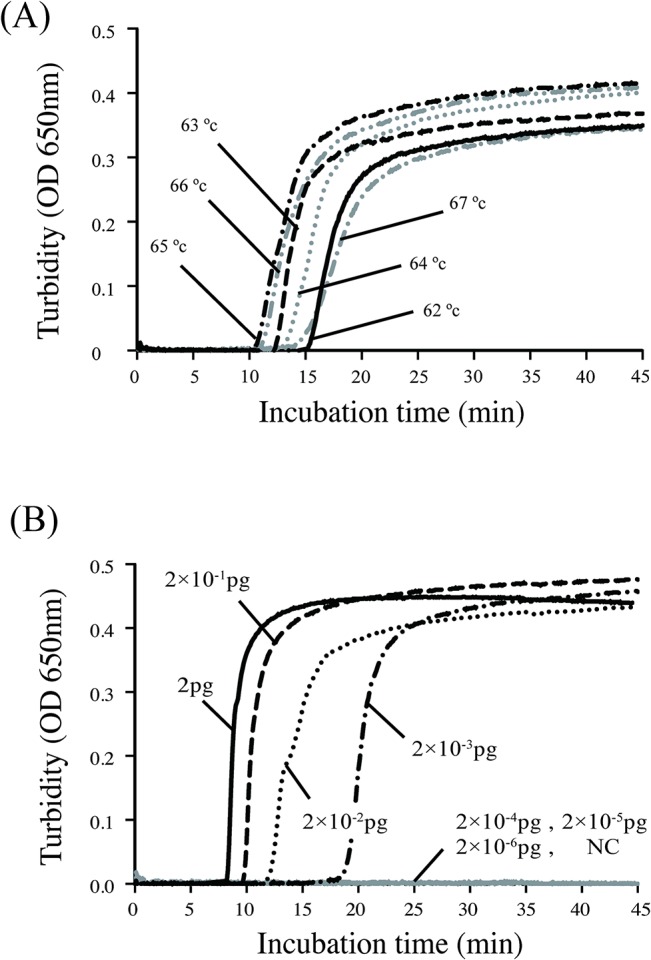
(A) To determine the optimal reaction conditions, a LAMP assay was performed on extracted bacterial DNA at temperatures ranging from 62°C to 67°C. At 65°C, the reaction finished within the shortest period of time, and the negative control remained transparent after 60 minutes of incubation. (B) To determine the detection limit, the extracted DNA templates were serially diluted 10 times (from 2 pg to 2×10−6 pg) and used in the LAMP assay. The turbidity was evaluated with a turbidimeter every 5 minutes.
To evaluate the specificity of the LAMP assay for A. baumannii and CRAb, 113 bacterial strains were used (S2 Table). All of the A. baumannii isolates were positive for ITS, and all of the CRAb isolates carrying bla OXA-23 were positive for bla OXA-23. The results of the LAMP assay matched the PCR results, and LAMP demonstrated 100% sensitivity and 100% specificity for targeting ITS and bla OXA-23.
To determine the detection limit, the extracted DNA templates were serially diluted 10 times (from 2 pg to 2×10−6 pg) and used in the LAMP assay. The turbidity of the LAMP samples was evaluated using a turbidimeter and by visual inspection every 5 minutes (Fig 1). Within 20 minutes, the turbidity of the reaction containing 2×10−3 pg of DNA had increased enough for visual inspection; this result indicated that the LAMP assay detected less than 10 CFU per assay as calculated by the amount of DNA extracted from bacterial culture with known CFU. In contrast, conventional PCR can detect 2×10−1 pg of DNA using the same templates as the LAMP assay. This result indicated that our LAMP assay was 100 times more sensitive than conventional PCR.
LAMP assay using clinical samples
We used clinical specimens to examine the feasibility, sensitivity, and specificity of the LAMP assay. To confirm the presence of CRAb in sputum specimens, LAMP assays for ITS and bla OXA-23 were performed simultaneously, and the samples that were positive for both ITS and bla OXA-23 were considered CRAb-positive. We refer to this LAMP method as “CRAb-LAMP”. A total of 120 samples were subsequently collected for this study, of which 44 tested positive for CRAb using the culture method. In our CRAb-LAMP assay, 39 samples were considered CRAb-positive with a sensitivity of 88.6% (95% CI: 75.4–96.2%) and a specificity of 92.1% (95% CI: 83.6–97.1%) (Table 3). When sputum specimens were used directly as templates for PCR, no amplicon was obtained due to inhibitory substances present in samples such as blood and sputum [30]. When DNA extracted from the sputum specimens was used as a template for CRAb-LAMP, the sensitivity was 88.6% (95% CI: 75.4–96.2%), the specificity was 90.8% (95% CI: 81.9–96.2%) (Table 3), and the results matched those of the fresh sputum specimens with the exception of one sample (ITS:119/120 and bla OXA-23:120/120, (accord/total)). This result indicated that LAMP was not affected by interfering substances. When the same DNA samples were directly used in a PCR assay, the sensitivity was 61.3% (95% CI: 45.4–75.6%), and the specificity was 96.0% (95% CI: 73.5–97.9%) for CRAb compared with a culture-based method.
Table 3. Sensitivity and specificity of the CRAb-LAMP assay.
| Culture | Sensitivity | Specificity | PPVa | NPVb | PLRc | ||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||||
| Clinical Samples | Sputum Directly | Positive | 39 | 6 | 88.6 | 92.1 | 86.7 | 94.5 | 11.2 |
| Negative | 5 | 70 | (75.4–96.2) | (83.6–97.0) | (73.2–95.0) | (85.1–97.8) | |||
| Extracted DNA | Positive | 39 | 7 | 88.6 | 90.8 | 84.8 | 93.2 | 9.63 | |
| Negative | 5 | 69 | (75.4–96.2) | (81.4–96.2) | (71.1–93.7) | (84.9–97.8) | |||
PPVa: positive predictive value;NPVb: negative predictive value; PLRc: positive likelihood ratio
LAMP assay as a surveillance tool
We next applied surveillance samples to CRAb-LAMP to investigate this method as a surveillance tool. Rectal swab samples were also used as surveillance samples because A. baumannii colonizes the gastrointestinal tract as well as the oropharynx [31]. After examination using the conventional culture method, each sample was used in a LAMP assay, and the results were compared. If either sample of a patient tested positive, we defined the patient as positive. A total of 108 patients participated in this study, resulting in a sensitivity of 100.0% and a specificity of 83.3% (Table 4). CRAb-LAMP was able to completely identify reservoirs regardless of the type of sample used (S3 Table)
Table 4. Sensitivity and specificity of the Surveillance LAMP compared with culture.
| Culture | Sensitivity | Specificity | PPVa | NPVb | PLRc | |||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||
| LAMP | Positive | 30 | 13 | 100 | 83.3 | 69.7 | 100 | 6.00 |
| Negative | 0 | 65 | (88.4–100) | (73.2–90.8) | (53.9–82.8) | (94.5–100) | ||
PPVa: positive predictive value; NPVb: negative predictive value; PLRc: positive likelihood ratio
Discussion
A. baumannii is one of the most problematic pathogens in clinical settings worldwide, primarily because it readily manifests resistance to many different classes of antibiotics [1, 2, 32]. An easy, rapid, and sensitive detection system for CRAb is required. Although classical culture methods have been used to detect CRAb, these require at least 2 days to determine the antibiotic spectrum. Moreover, the sensitivity of the culture method is insufficient to detect patients colonized with CRAb [33]. To solve these problems, we developed “CRAb-LAMP”, which can be used to detect CRAb more easily and rapidly
To identify specific gene candidates, we have genotyped CRAb isolates from Thailand and successfully identified bla OXA-23. These epidemiological data for CRAb isolates were obtained from just two hospitals; however, the results were similar to previously published data [9–12] and we established LAMP method for bla OXA-23. Our assay method was not able to detect CRAb in every instance, as a few isolates lacked bla OXA-23; however, most CRAb isolates can be detected by amplifying bla OXA-23 due to its high prevalence among CRAb isolates in Thailand [9–12]. The culture-based method was used as the “gold standard”, thus there were some “false-positive” samples; however, these samples may indeed be “true-positives” as a result of the low sensitivity of the culture method [33] and the high sensitivity of the LAMP method. To verify this presumption, we subjected a pair of samples that were obtained from the same patient on different days to both the culture method and CRAb-LAMP. The culture method detected positivity only in the latter sample, whereas LAMP detected positivity in both samples (unpublished data). Therefore, CRAb-LAMP was able to detect CRAb with higher sensitivity.
Three recent reports have described the detection of A. baumannii via LAMP [16, 34, 35]. None of these studies used clinical samples directly or mentions the utility of LAMP as a surveillance tool. Two of these studies could only detect A. baumannii and required more time for sample preparation. In clinical settings, clonal spreading is an important concern [1, 36], and we observed that detection of one target gene is sufficient to perform infection control measures. Therefore, our method is expected to be clinically useful in detecting CRAb. The three most popular genotypes of CRAb are bla OXA-23, which prevails in Asia, and bla OXA-24 and bla OXA-58, which are prevalent in Europe and the United States [1, 9, 27]. By designing primers for these target genes, we can detect different genotypes of CRAb easily. The sensitivity and specificity of our CRAb-LAMP method using clinical samples is equivalent to those of currently available diagnostic kits for diseases such as tuberculosis and pathogens such as mycoplasma.
Conclusion
The CRAb-LAMP assay requires 10 minutes to prepare the template and the reaction master mix, and 20 minutes for the reaction. No special equipment, such as a thermal cycler, is required. The speed of this assay facilitates the early treatment and isolation of CRAb-positive patients. Therefore, our method can facilitate the detection of CRAb and provide a useful tool for hospitals in resource-poor countries, especially in the event of an outbreak.
Supporting Information
To analyze the genotypes of CRAb, the bla genes associated with drug resistance were investigated via PCR assays shown in this table.
(DOCX)
A total of 113 bacterial strains shown in this table were used to examine the CRAb-specific reliability of the LAMP reaction.
(DOCX)
CRAb-LAMP was performed as a surveillance tool. When either of the samples was used, the sensitivity and specificity showed all most the same level regardless of the type of sample used.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by JSPS KAKENHI Grant Number 24406022 for KT [http://www.jsps.go.jp/english/e-grants/index.html]. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008. 21: 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev.1996. 9: 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latibeaudiere R, Rosa R, Laowansiri P, Arheart K, Namias N, Munoz-Price LS. Surveillance cultures growing Carbapenem-Resistant Acinetobacter baumannii Predict the Development of Clinical Infections: a Cohort Study. Clin Infect Dis. 2014: in press. [DOI] [PubMed]
- 4. Lemos EV, de la Hoz FP, Einarson TR, McGhan WF, Quevedo E, Castañeda C, et al. Carbapenem resistance and mortality in patients with Acinetobacter baumannii infection: systematic review and meta-analysis. Clin Microbiol Infect. 2013. 20: 416–23. 10.1111/1469-0691.12363 [DOI] [PubMed] [Google Scholar]
- 5. Erbay A, Idil A, Gozel MG, Mumcuoglu I, Balaban N. Impact of early appropriate antimicrobial therapy on survival in Acinetobacter baumannii bloodstream infections. Int J Antimicrob Agents. 2009. 34: 575–579. 10.1016/j.ijantimicag.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 6. Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013. 57: 1237–1245. 10.1093/cid/cit498 [DOI] [PubMed] [Google Scholar]
- 7. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000. 28: E63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okada K, Chantaroj S, Taniguchi T, Suzuki Y, Roobthaisong A, Puiprom O, et al. A rapid, simple, and sensitive loop-mediated isothermal amplification method to detect toxigenic Vibrio cholerae in rectal swab samples. Diagn Microbiol Infect Dis. 2010. 66: 135–139. 10.1016/j.diagmicrobio.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 9. Evans BA, Amyes SG. OXA beta-lactamases. Clin Microbiol Rev. 2014. 27: 241–263 10.1128/CMR.00117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niumsup PR, Boonkerd N, Tansawai U, Tiloklurs M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn J Infect Dis. 2009. 62: 152–154. [PubMed] [Google Scholar]
- 11. Thapa B, Tribuddharat C, Srifuengfung S, Dhiraputra C. High prevalence of bla OXA-23 in oligoclonal carbapenem-resistant Acinetobacter baumannii from Siriraj Hospital, Mahidol University, Bangkok, Thailand. Southeast Asian J Trop Med Public Health.2010. 41: 625–635. [PubMed] [Google Scholar]
- 12.Thapa B. Ph.D. Thesis. University of Mahidol, Bangkok, Thailand. 2009
- 13. Gerner-Smidt P, Tjernberg I, Ursing J. Reliability of phenotypic tests for identification of Acinetobacter species. J Clin Microbio. 1991. 29: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alvarez-Buylla A, Culebras E, Picazo JJ. Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect Genet Evol. 2012. 12: 345–349. 10.1016/j.meegid.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 15. Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005. 49: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Afzal-Shah M, Woodford N, Livermore DM. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii . Antimicrob Agents Chemother. 2001. 45: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marti S, Sanchez-Cespedes J, Blasco MD, Ruiz M, Espinal P, Alba V, et al. Characterization of the carbapenem-hydrolyzing oxacillinase oxa-58 in an Acinetobacter genospecies 3 clinical isolate. Antimicrob Agents Chemother. 2008. 52: 2955–2958. 10.1128/AAC.00072-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouyama Y, Harada S, Ishii Y, Saga T, Yoshizumi A, Tateda K, et al. Molecular characterization of carbapenem-non-susceptible Acinetobacter spp. in Japan: predominance of multidrug-resistant Acinetobacter baumannii clonal complex 92 and IMP-type metallo-beta-lactamase-producing non-baumannii Acinetobacter species. J Infect Chemother. 2012. 18: 522–528. 10.1007/s10156-012-0374-y [DOI] [PubMed] [Google Scholar]
- 19. Mitsuda T, Kuroki H, Ishikawa N, Imagawa T, Ito S, Miyamae T, et al. Molecular epidemiological study of Haemophilus influenzae serotype b strains obtained from children with meningitis in Japan. J Clin Microbio. 1999. 37: 2548–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romero-Steiner S, Fernandez J, Biltoft C, Wohl ME, Sanchez J, Feris J, et al. Functional antibody activity elicited by fractional doses of Haemophilus influenzae type b conjugate vaccine (polyribosylribitol phosphate-tetanus toxoid conjugate). Clin Diagn Lab Immunol. 2001. 8: 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogunniyi AD, LeMessurier KS, Graham RM, Watt JM, Briles DE, Stroeher UH, et al. Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. Infect Immun. 2007. 75: 1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus . Antimicrob Agents Chemother. 2004. 48: 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii . J Bacteriol. 2008. 190: 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soo PC, Tseng CC, Ling SR, Liou ML, Liu CC, Chao HJ, et al. Rapid and sensitive detection of Acinetobacter baumannii using loop-mediated isothermal amplification. J Microbiol Methods. 2013. 92: 197–200. 10.1016/j.mimet.2012.11.020 [DOI] [PubMed] [Google Scholar]
- 25. Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004. 59: 145–157. [DOI] [PubMed] [Google Scholar]
- 26. Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii . Drug Resist Updat. 2012. 15: 237–247. 10.1016/j.drup.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 27. Evans BA, Hamouda A, Towner KJ, Amyes SG. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii . Clin Microbiol Infect. 2008. 14: 268–275. 10.1111/j.1469-0691.2007.01919.x [DOI] [PubMed] [Google Scholar]
- 28. De Luca F, Benvenuti M, Carboni F, Pozzi C, Rossolini GM, Mangani S, et al. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D beta-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci U S A. 2011. 108: 18424–18429. 10.1073/pnas.1110530108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother. 2013. 19: 404–411. 10.1007/s10156-013-0590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trombley Hall A, McKay Zovanyi A, Christensen DR, Koehler JW, Devins Minogue T. Evaluation of inhibitor-resistant real-time PCR methods for diagnostics in clinical and environmental samples. PLOS ONE. 2013. 8: e73845 10.1371/journal.pone.0073845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baraibar J, Correa H, Mariscal D, Gallego M, Valles J, Rello J. Risk factors for infection by Acinetobacter baumannii in intubated patients with nosocomial pneumonia. Chest. 1997. 112: 1050–1054. [DOI] [PubMed] [Google Scholar]
- 32. Dijkshoorn L, Nemec A, Seifert H An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii . Nat Rev Microbiol. 2007. 5: 939–951. [DOI] [PubMed] [Google Scholar]
- 33. Marchaim D, Navon-Venezia S, Leavitt A, Chmelnitsky I, Schwaber MJ, Carmeli Y. Molecular and epidemiologic study of polyclonal outbreaks of multidrug-resistant Acinetobacter baumannii infection in an Israeli hospital. Infect Control Hosp Epidemiol. 2007. 28: 945–950. [DOI] [PubMed] [Google Scholar]
- 34. Wang Q, Zhou Y, Li S, Zhuo C, Xu S, Huang L, et al. Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii . PLOS ONE. 2013. 8: e66406 10.1371/journal.pone.0066406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vergara A, Zboromyrska Y, Mosqueda N, Morosini MI, Garcia-Fernandez S, Roca I, et al. Evaluation of a loop-mediated isothermal amplification-based methodology to detect carbapenemase carriage in Acinetobacter clinical isolates. Antimicrob Agents Chemother. 2014. 58: 7538–7540. 10.1128/AAC.03870-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peleg AY, Franklin C, Bell JM, Spelman DW. Emergence of carbapenem resistance in Acinetobacter baumannii recovered from blood cultures in Australia. Infect Control Hosp Epidemiol. 2006. 27: 759–761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To analyze the genotypes of CRAb, the bla genes associated with drug resistance were investigated via PCR assays shown in this table.
(DOCX)
A total of 113 bacterial strains shown in this table were used to examine the CRAb-specific reliability of the LAMP reaction.
(DOCX)
CRAb-LAMP was performed as a surveillance tool. When either of the samples was used, the sensitivity and specificity showed all most the same level regardless of the type of sample used.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


