Abstract
Technological advances in experimental neuroscience are generating vast quantities of data, from the dynamics of single molecules to the structure and activity patterns of large networks of neurons. How do we make sense of these voluminous, complex, disparate and often incomplete data? How do we find general principles in the morass of detail? Computational models are invaluable and necessary in this task and yield insights that cannot otherwise be obtained. However, building and interpreting good computational models is a substantial challenge, especially so in the era of large datasets. Fitting detailed models to experimental data is difficult and often requires onerous assumptions, while more loosely constrained conceptual models that explore broad hypotheses and principles can yield more useful insights.
Introduction
By nature, experimental biologists collect and revere data, including the myriad details that characterize the particular system they are studying. At the same time, as the onslaught of data increases, it is clear that we need tools that allow us to crisply extract understanding from the data that we can now generate. How do we find the general principles hiding among the details? And how do we understand which details are critical features of a process, and which details can be approximated or ignored while still permitting insight into an important biological question? Intelligent model building coupled to disciplined data analyses will be required to progress from data collection to understanding.
Computational models differ in their objectives, limitations and requirements. Conceptual models examine the consequences of broad assumptions. These kinds of models are useful for conducting rigorous thought experiments: one might ask how noise impacts latency in a forced choice between multiple alternatives [1], or how network topology determines the fusion and rivalry of visual percepts [2]. While conceptual models must be constrained by data in the sense that they cannot violate known facts about the world, they do not strive to assimilate or reproduce detailed experimental measurements. Phenomenological data-driven models aim to capture details of empirically observed data in a parsimonious way. For example, reduced models of single neurons [3,4] can often capture the behavior of neurons, but with simplified dynamics and few parameters. These kinds of models are useful for understanding ‘higher level’ functions of a neural system, be it a dendrite, a neuron or a neural circuit [5**] that, in the appropriate context, are independent of low-level details. Used carefully, they can tell us biologically relevant things about how nervous systems work without needing to constrain large numbers of parameters. Detailed data-driven or “realistic” models attempt to assimilate as much experimental data as are available and account for detailed observations at the same time. Successful examples might include detailed structural models of ion channels that capture voltage-sensing and channel gating [6], or carefully parameterized models of biochemical signaling cascades underlying long-term potentiation [7]. With notable exceptions, models of this kind are often the least satisfying, as they can be most compromised by what hasn’t been measured or characterized [8**].
How should we approach computational modelling in the era of ‘big data’? The non-linear and dynamic nature of biological systems is a key obstacle for building detailed models [8**,9**] even when large amounts of data are available. For example, even well-characterized neural circuits such as crustacean CPGs that have full connectivity diagrams have not, to date, been successfully modelled in a level of detail that incorporates all of what is known about the synaptic physiology, intrinsic properties and circuit architecture [10]. As a consequence, there is still a big role for conceptual models that tell investigators what kinds of processes may underlie the data [11], or, more importantly, what potential mechanisms one should rule out [12,13*].
Relating data to models
The Hodgkin-Huxley [14] model stands almost alone in its level of impact and in the way it achieved a more-or-less complete fit of the data. In hindsight their success came from extraordinarily good biological intuition about how action potentials are generated and a clever choice of experimental preparation. Their model revealed fundamental principles of how a ubiquitous phenomenon – the spike, or action potential – resulted from few processes, namely two voltage-dependent membrane currents mediated by separate ionic species.
By contrast, the success of subsequent attempts to fit and model the biophysics of more complex neuronal conductances, neurons and circuits has been less dramatic – although insight into the roles of specific currents in neuronal dynamics has certainly been achieved [6,14,15,16*,17,18]. Understanding why this is the case requires investigators to step back and view the problem in a general setting. Biological systems are assembled from many component enzymes, signaling molecules and cellular structures. Modelling these components and their interactions produces complex nonlinear dynamical systems with multiple parameters for each component. For example, even if one specifies quite rigidly the desired output of a neuronal network, the underlying parameters that can give rise to these properties is weakly constrained as multiple solutions to neuronal and network dynamics are found [19,20]. Subsequent work, informed by this general finding, explored families of models with parameters scattered over plausible ranges [21,22,23,24*]. Although these studies abandoned the idea of finding unique fits to data, they nonetheless revealed important principles about how specific combinations of conductances contribute to neuronal and network behavior [22,23], and how temperature-robust neuronal function might emerge in cold-blooded animals that experience significant changes in temperature [21,24*].
There are fundamental reasons why it is challenging to fit large numbers of parameters in biological models [9**,25]. First, the models are typically nonlinear, so the relation between the parameters and the output can be complicated and many-valued. Averages of measured parameters can give rise to non-observed behavior [26] and models can be exquisitely sensitive to measured parameters [27,28,29,30]. The value of averaging as a means of combating experimental noise might thus be obviated by the possibility that the average values are not valid parameter combinations themselves. Second, biological systems have degenerate pathways and components, meaning that properties and functions of structurally distinct components overlap. While this confers robustness to the systems themselves, it means that models can be remarkably insensitive to many combinations of parameters [5**,21,22,23,27,29,30,31]. This ‘sloppy’ property of biological systems is well-documented in systems biology [8**] and neuroscientists may benefit from a wider appreciation of the tribulations and successes of model building in this sister field [32].
Sloppiness (Figure 1) means that models with large numbers of parameters exhibit relatively few sensitive directions in local regions of parameter space, although these directions are not generically aligned with parameter axes. Instead, the sensitive (and insensitive) directions are comprised of mixtures of parameters (Figure 1c), meaning that performance of a detailed model will be severely compromised by poor measurement, or ignorance of even a single parameter [8**]. A recent, elegant modelling study of oculomotor integration [5**] revealed a handful of sensitive directions in the high-dimensional parameter space of a complex neuronal circuit model (Figure 1d). The model permitted fresh insight into the trade-offs between structural and functional properties of a circuit and did so by constraining model behavior rather than measured parameters. As this study illustrates, useful insight into circuit function can be obtained from phenomenological matching of the overall model behavior to experimental data, provided the non-sloppy, or ‘stiff’, parameter combinations are identified [33].
Figure 1.
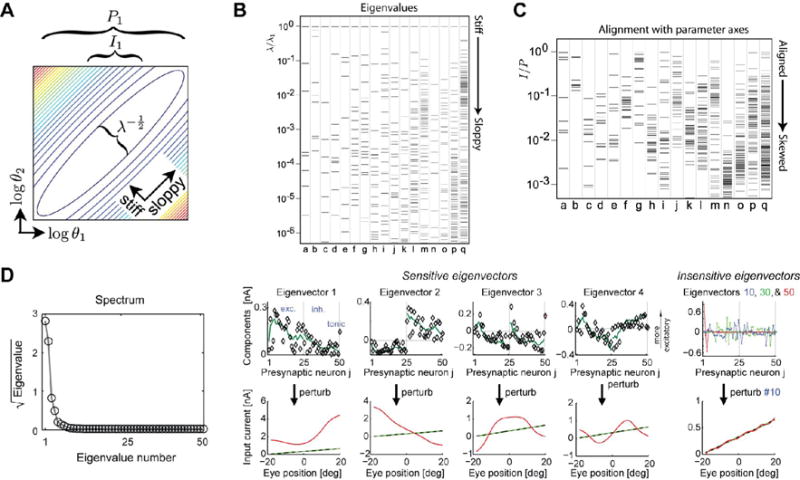
In high-dimensional biological models there are often many parameter combinations that can co-vary without significant effect on the behavior of the model, known as sloppy parameter combinations [8**]. (A) Elliptical level sets in the deviation of model output from a nominal value (computed from the Hessian, or second derivative) shows a direction in which parameter variation does not change model behavior (sloppy direction) and an orthogonal direction in which the model is sensitive (stiff). The major and minor axes of these ellipses (and thus the relative sensitivity to the stiff/sloppy directions) are determined by the eigenvalues, λi, and the projections of these onto the parameter axes (θ1, θ2) parameter axes are denoted by P1 and I1, respectively. (B) Eigenvalues computed for 17 different systems biology models [8**], including detailed models of circadian transcriptional circuits and yeast metabolism (a–q) are spread evenly across many orders of magnitude. Only the first few eigenvectors have significant effects on the behavior of the model, thus only a few parameter directions determine model behavior. (C) The alignment of the ‘error ellipsoids’ I/P relative to model parameters shows that most eigenvectors tend to be composed of many underlying parameters (tend to be skewed). Thus, while there are relatively few stiff directions in parameter space that change model behavior, these directions usually have contributions from many experimentally measurable parameters. (D, left) A computational model of an oculomotor circuit [5**] shows similar sensitivity to only four or five parameter combinations (D, right) to the systems biology models (A–C). The sensitive directions, projected onto the underlying parameter axes (presynaptic input weights) have substantial contributions from all parameters. Figures (A–C) reproduced from [8**], (D) Reproduced from [5**].
A third reason for the difficulty of the ‘fitting problem’ arises because biological systems are intrinsically variable [34]. This variability is well-appreciated in the context of single neuron parameters, where neurons with highly stereotyped properties exhibit surprisingly large variability in their membrane conductance expression [20,35,36,37,38]. High variability is present wherever one looks, whether it is the synaptic connectivity of well-defined neural circuits [39,40,41,42] or the behavior of entire animals [43]. As a consequence, the number of valid, distinct parameter sets – should they be accessible – can equal the number of biological repeats of an experiment. This kind of variability is not noise; it represents genuinely different parameter combinations that the biological system has found. For this reason, understanding the regulatory logic of the nervous system is of fundamental importance [44**].
In an age when increasingly voluminous and complex datasets are demanding interpretation, these fundamental model-fitting problems are sobering. However, there are direct means of taming these difficulties by exploiting the resolution and high-dimensionality of the data themselves. An elegant analysis of the requirements for fitting a multicompartment model [31] showed that if one could access, at high temporal resolution, the membrane voltage of each compartment in a neuron, then one can recover the densities of multiple voltage-gated conductances – providing the identity and kinetics of the conductances are known. At the time this study was published, such measurements seemed impractical. Nearly ten years later, we are on the verge of being able to make such measurements thanks to new molecular tools and improved microscopy.
Advances in statistical methods and fitting algorithms are accompanying advances in data collection. Many of these exploit fast computers and numerical methods such as Monte Carlo sampling to solve complex statistical inference problems, such as inferring synaptic inputs from noisy physiological traces [45,46*]. Knowledge of the general properties of the system permits ill-posed problems to be regularized, allowing noisy or incomplete data to yield informative measurements [47*,48]. Statistical inference has other important roles aside from making biological parameter values accessible. Oftentimes, inference can be performed in a way that incorporates important assumptions – such as the presence of interneuronal connections in a network – thus embedding a modelling question in the data analysis task. Such statistical modelling approaches can yield valuable hidden information, such as how common noise sources may explain population activity in the retina [49**] and how the statistics of complex multiunit activity can encode aversive and appetitive taste [50,51*](Figure 2).
Figure 2.
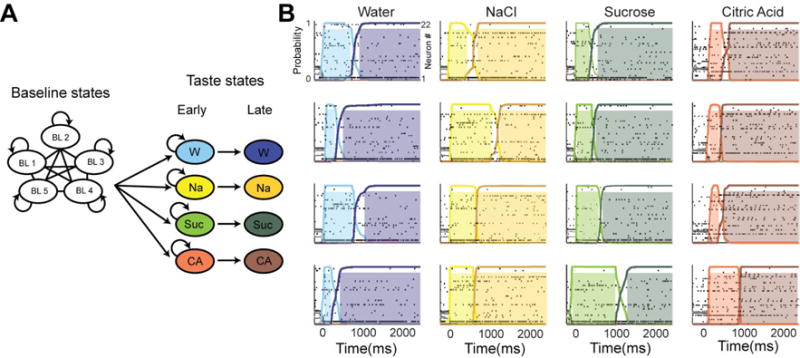
A Markov Models describing the statistics of transitions in multiunit network activity in sensory (taste) cortex [50,51*] during delivery of one of four tastes (water – W, salt – Na, sugar – Suc, acid – CA). (A) Baseline activity before tastes were presented was disorganized: all transitions are possible. After taste presentation the networks entered one state in a probabilistic way, determined by the stimulus. Each state is characterized by distinct combinations of neural firing patterns (raster plots in (B)). The network can remain in the early state or advance to the late state. (B) Spiking data across four trials illustrates how the same network of neurons leaves a baseline state to enter an early state, which entirely depends on the stimulus, then advances to the late state after a certain amount of time. Each color represents a state that can be distinguished from all other states by the network firing activity. Thus given only spiking data the taste stimulus can be inferred based on the state of the network. An important feature of this statistical model is that the variable latencies of discrimination ‘decision’ events is evident, something that is lost if activity is averaged over trials. Figure reproduced from [50].
Alternative strategies for fitting data, including evolutionary algorithms [52,53] and dynamic state estimation [29] have also been developed to exploit multiple, time-series measurements. In spite of the sophistication of current data analysis techniques and the increasing richness and quality of data, any model that is constrained by data is only as sound as the necessary assumptions upon which it rests: even incorrect models can fit the data.
Conceptual models as tools for explaining data and asking “what if?”
The mammalian prefrontal cortex (PFC) is one of the most complex and mysterious structures in neuroscience. Single-unit activity from tens to hundreds of neurons reveals a diverse and puzzling array of activity profiles during behavioral tasks, with no obvious relation to external variables. Faced with a snapshot of data from a miniscule and only loosely identified population of neurons, a recent study was nonetheless successful in shedding light on how behavioral output can be represented in this brain region [54**](Figure 3). The role of the computational model in this study was not to fit and explain the data in painstaking detail – far too many unknowns exist for this to be practical even if the fitting problem could be solved. Instead, the authors appealed to the general properties of an abstract, recurrent neural network to explain ‘how’ such a structure could represent the external world in its internal state. In spite of the gulf between the unknown and complex properties of the PFC and the simpler and more abstract nature of the model, a striking agreement was evident in the way population activity evolved during a decision.
Figure 3.
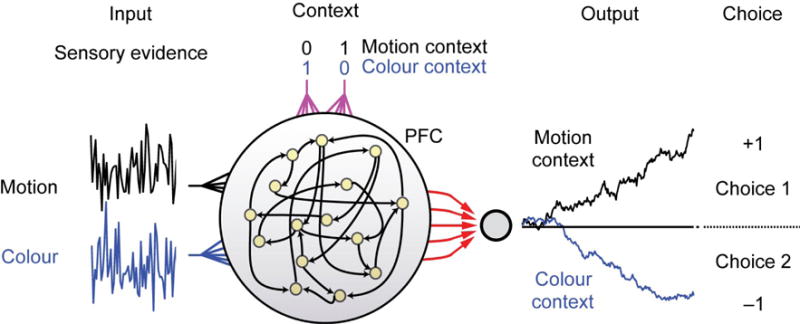
A conceptual/phenomenological model [66] of recurrent networks such as the pre-frontal cortex (PFC) can account for the observed data even when precise understanding of the underlying, anatomy and detailed mechanisms is lacking. The experimental task involves a monkey looking at moving, colored dots and reporting the perceived direction of movement, or the color, depending on a context cue. Here, the same physiological data regarding the color and motion are fed into the network, along with a context cue, and the model reliably selects the correct choice. Thus without knowing the precise molecular/network mechanisms of the PFC the authors were still able to postulate how such a network might work and create testable hypotheses. Figure reproduced from [54**].
Similarly, a wealth of neurophysiological and behavioral data is emerging from models of motor sequence learning and navigation. For example, the brain structures involved in bird song learning are still being mapped and characterized. Nonetheless, deep insights into the nature of reinforcement learning [55] and temporal sequence learning [56**] have emerged from modelling studies that focused on conceptual, rather than detailed features of experimental data. Similarly, the power of C elegans in linking circuit dynamics to behavior was recently demonstrated in a combined experimental and modelling study of chemotaxis [57*]. Notably, this study used phenomenological models to characterize single neuron dynamics that informed a behavioral model of active sensing.
Conceptual models are not confined to ‘high level’ neurophysiological phenomena such as decision making and learning. Low level, mechanistic phenomena such as how protein synthesis impinges on synaptic plasticity can be studied using computational models without attempting to parameterize every molecule involved. A recent study by O’Donnell and Sejnowski [58*] shows that memory generalization can emerge from diffusion of plasticity proteins in dendritic trees. Similarly, a coarse model of activity dependent ion channel regulation has recently helped explain physiologically important expression patterns in the mRNA of ion channels in identified neurons, while accounting for cell-to-cell variability [44**,59]. Building more realistic and detailed molecular models is becoming more feasible as imaging and subcellular biochemistry are providing more data to constrain these models [60], but there will always be a role for conceptual models – especially in gaining intuition and in situations where data-fitting is impractical for reasons we have already discussed.
A skeptic might worry that conceptual models can be adjusted ad-hoc, or post-hoc, to agree with data and thus be consistent with any finding. If this were the case, conceptual models would only make vacuous statements about the world and not generate new understanding. However, many conceptual models can be falsified, and can stimulate important, fruitful research programs in experimental neuroscience. For example, the oscillatory interference model of grid cell formation was proposed very soon after the discovery of grid cells [61]. The power of the oscillatory interference model was that it used a simple mechanism – interference – and combined it with a well-documented phenomenon – theta oscillations – to account for a puzzling observation. However, recent work [62*], motivated by tension between this model and a rival theory, the continuous attractor model [63], found compelling evidence for the latter. It is important to note how much has been learned in the wake of these modelling attempts, irrespective of whether they are correct. Deeper understanding of intrinsic cellular properties, network dynamics and robustness of alternative coding schemes [64] have all descended from simple conceptual models.
Exploring an artificial model universe comes with its own risks. If exploration is done without reality-checking assumptions, it is easy to fall into the trap of building irrelevant models. There are infinitely many models consistent with any one piece of experimental data, so it is important to avoid just-so explanations that can arise when a model spuriously matches an observed phenomenon. Well-conceived models rest on underlying principles that ensure the model does not only work under idiosyncratic circumstances. Sometimes this can be done formally; for example, physiological models of central pattern generating neurons and networks can be reduced to the underlying family of dynamical systems, permitting an understanding of intrinsic neuronal dynamics and network interactions that is model-independent [4,65]. In other cases, strong biological intuition and close contact with the experimentalist, or experimental preparation can combat fragile or spurious modelling results.
All experimentalists have, on occasion, seen a piece of new data and said, “Of course!” There is a sense of recognition that comes from seeing the answer to a previously puzzling question. The best computational models are equally illuminating: an idea or a principle is revealed and recognized as part of the path to understanding a biological conundrum. Principled model building will be ever more important in the era of big data, as it is only principled model construction and evaluation that will allow us to understand which details are important for what functions of the brain.
Computational models will prove increasingly useful for understanding large datasets
Substantial challenges exist for fitting detailed models to data
Conceptual and phenomenological models are often more useful than detailed models
Acknowledgments
This work was funder by NIH grant MH 46742 and the Charles A King Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* = of special interest
** = of outstanding interest
- 1.Krajbich I, Rangel A. Multialternative drift-diffusion model predicts the relationship between visual fixations and choice in value-based decisions. Proc Natl Acad Sci U S A. 2011;108:13852–13857. doi: 10.1073/pnas.1101328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diekman CO, Golubitsky M. Network symmetry and binocular rivalry experiments. J Math Neurosci. 2014;4:12. doi: 10.1186/2190-8567-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brette R, Gerstner W. Adaptive exponential integrate-and-fire model as an effective description of neuronal activity. J Neurophysiol. 2005;94:3637–3642. doi: 10.1152/jn.00686.2005. [DOI] [PubMed] [Google Scholar]
- 4.Franci A, Drion G, Seutin V, Sepulchre R. A balance equation determines a switch in neuronal excitability. PLoS Comput Biol. 2013;9:e1003040. doi: 10.1371/journal.pcbi.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Fisher D, Olasagasti I, Tank DW, Aksay ER, Goldman MS. A modeling framework for deriving the structural and functional architecture of a short-term memory microcircuit. Neuron. 2013;79:987–1000. doi: 10.1016/j.neuron.2013.06.041. Fisher et al. note that separately fitting parameters to a single value can be overly constraining and parameter-space explorations can be infeasible as dimensionality increases. Instead, the authors collectively fit a model of occulomotor integration to experimental data. They report that this technique is viable in recovering structural and functional connectivity in a model network and that only a handful of dominant parameter combinations determines overall behavior, while the majority of directions in parameter space are unconstrained. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 7.Bhalla US. Molecular computation in neurons: a modeling perspective. Curr Opin Neurobiol. 2014;25:31–37. doi: 10.1016/j.conb.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 8**.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. This study analyses 17 biochemical models from the systems biology literature and reveals that in all cases models depend on their parameters in a ‘sloppy’ way that means many parameter combinations do not significantly affect model behavior. On the other hand, the few sensitive directions in parameter space that do influence model behavior span most of the available parameters. Thus uncertainty in only a few experimentally-determined parameters can swamp precision in others and lead to poor recapitulation of overall behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Transtrum MK, Machta BB, Sethna JP. Why are nonlinear fits to data so challenging? Phys Rev Lett. 2010;104:060201. doi: 10.1103/PhysRevLett.104.060201. This study examines the difficulties in fitting nonlinear parameters. It outlines the ways a model might be stuck in a local optimum, or along a parameter edge, and discusses strategies to avoid such pitfalls. [DOI] [PubMed] [Google Scholar]
- 10.Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri R, Bernacchia A, Wang XJ. A diversity of localized timescales in network activity. Elife. 2014;3:e01239. doi: 10.7554/eLife.01239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs AL, Fridman G, Douglas RM, Alam NM, Latham PE, Prusky GT, Nirenberg S. Ruling out and ruling in neural codes. Proc Natl Acad Sci U S A. 2009;106:5936–5941. doi: 10.1073/pnas.0900573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Goodman DF, Benichoux V, Brette R. Decoding neural responses to temporal cues for sound localization. eLife. 2013;2:e01312. doi: 10.7554/eLife.01312. This study draws comparisons between two theories of interaural time differences that underlie sound localization. The authors examine the effects that factors such as stimulus spectrum and background noise have on the decoder models and, in doing so, reveal that one is demonstrably more efficient. This study highlights how distinct conceptual models can be evaluated with respect to experimental data and how models allow relevant neurophysiological questions to be explored in ways that are prohibitive experimentally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor JA, Stevens CF. Prediction of repetitive firing behaviour from voltage clamp data on an isolated neurone soma. J Physiol. 1971;213:31–53. doi: 10.1113/jphysiol.1971.sp009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Almog M, Korngreen A. A quantitative description of dendritic conductances and its application to dendritic excitation in layer 5 pyramidal neurons. J Neurosci. 2014;34:182–196. doi: 10.1523/JNEUROSCI.2896-13.2014. Using dual-electrode recordings from the soma and apical dendrite the authors show that heterogeneous Ca2+ conductance gradients found experimentally can be mapped onto model pyramidal neurons. The resulting anisotropy allows the model to recapitulate several experimental findings that were previously difficult to reproduce, including dendritic spikes and their activation with backpropogating action potentials. Key parameters of the model were found using genetic algorithms and experimental perturbations with pharmacological blockers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouwens NW, Wilson RI. Signal propagation in Drosophila central neurons. J Neurosci. 2009;29:6239–6249. doi: 10.1523/JNEUROSCI.0764-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mease RA, Famulare M, Gjorgjieva J, Moody WJ, Fairhall AL. Emergence of adaptive computation by single neurons in the developing cortex. J Neurosci. 2013;33:12154–12170. doi: 10.1523/JNEUROSCI.3263-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci. 2004;7:1345–1352. doi: 10.1038/nn1352. [DOI] [PubMed] [Google Scholar]
- 20.Marder E, Taylor AL. Multiple models to capture the variability in biological neurons and networks. Nat Neurosci. 2011;14:133–138. doi: 10.1038/nn.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caplan JS, Williams AH, Marder E. Many parameter sets in a multicompartment model oscillator are robust to temperature perturbations. J Neurosci. 2014;34:4963–4975. doi: 10.1523/JNEUROSCI.0280-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doloc-Mihu A, Calabrese RL. Identifying crucial parameter correlations maintaining bursting activity. PLoS Comput Biol. 2014;10:e1003678. doi: 10.1371/journal.pcbi.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor AL, Goaillard JM, Marder E. How multiple conductances determine electrophysiological properties in a multicompartment model. J Neurosci. 2009;29:5573–5586. doi: 10.1523/JNEUROSCI.4438-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Roemschied FA, Eberhard MJ, Schleimer JH, Ronacher B, Schreiber S. Cell-intrinsic mechanisms of temperature compensation in a grasshopper sensory receptor neuron. eLife. 2014;3:e02078. doi: 10.7554/eLife.02078. Roemschied et al. investigate temperature sensitivity of the biophysical properties of cricket auditory neurons. They model biophysical channel properties of specific ion channels and find that the neurons can maintain consistent spiking activity without excessive metabolic cost. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lillacci G, Khammash M. Parameter estimation and model selection in computational biology. PLoS Comput Biol. 2010;6:e1000696. doi: 10.1371/journal.pcbi.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golowasch J, Goldman MS, Abbott LF, Marder E. Failure of averaging in the construction of a conductance-based neuron model. J Neurophysiol. 2002;87:1129–1131. doi: 10.1152/jn.00412.2001. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez GJ, O’Leary T, Marder E. Multiple mechanisms switch an electrically coupled, synaptically inhibited neuron between competing rhythmic oscillators. Neuron. 2013;77:845–858. doi: 10.1016/j.neuron.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marin B, Barnett WH, Doloc-Mihu A, Calabrese RL, Cymbalyuk GS. High prevalence of multistability of rest states and bursting in a database of a model neuron. PLoS Comput Biol. 2013;9:e1002930. doi: 10.1371/journal.pcbi.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meliza CD, Kostuk M, Huang H, Nogaret A, Margoliash D, Abarbanel HD. Estimating parameters and predicting membrane voltages with conductance-based neuron models. Biol Cybern. 2014;108:495–516. doi: 10.1007/s00422-014-0615-5. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez GJ, Marder E. Rectifying electrical synapses can affect the influence of synaptic modulation on output pattern robustness. J Neurosci. 2013;33:13238–13248. doi: 10.1523/JNEUROSCI.0937-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huys QJ, Ahrens MB, Paninski L. Efficient estimation of detailed single-neuron models. J Neurophysiol. 2006;96:872–890. doi: 10.1152/jn.00079.2006. [DOI] [PubMed] [Google Scholar]
- 32.De Schutter E. Why are computational neuroscience and systems biology so separate? PLoS Comput Biol. 2008;4:e1000078. doi: 10.1371/journal.pcbi.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutenkunst RN, Casey FP, Waterfall JJ, Myers CR, Sethna JP. Extracting falsifiable predictions from sloppy models. Ann N Y Acad Sci. 2007;1115:203–211. doi: 10.1196/annals.1407.003. [DOI] [PubMed] [Google Scholar]
- 34.Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- 35.Schulz DJ, Goaillard JM, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- 36.Golowasch J, Abbott LF, Marder E. Activity-dependent regulation of potassium currents in an identified neuron of the stomatogastric ganglion of the crab Cancer borealis. J Neurosci. 1999(19):RC33. doi: 10.1523/JNEUROSCI.19-20-j0004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swensen AM, Bean BP. Robustness of burst firing in dissociated purkinje neurons with acute or long-term reductions in sodium conductance. J Neurosci. 2005;25:3509–3520. doi: 10.1523/JNEUROSCI.3929-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amendola J, Woodhouse A, Martin-Eauclaire MF, Goaillard JM. Ca2+/cAMP-sensitive covariation of IA and IH voltage dependences tunes rebound firing in dopaminergic neurons. J Neurosci. 2012;32:2166–2181. doi: 10.1523/JNEUROSCI.5297-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caron SJ, Ruta V, Abbott LF, Axel R. Random convergence of olfactory inputs in the Drosophila mushroom body. Nature. 2013;497:113–117. doi: 10.1038/nature12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roffman RC, Norris BJ, Calabrese RL. Animal-to-animal variability of connection strength in the leech heartbeat central pattern generator. J Neurophysiol. 2012;107:1681–1693. doi: 10.1152/jn.00903.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norris BJ, Wenning A, Wright TM, Calabrese RL. Constancy and variability in the output of a central pattern generator. J Neurosci. 2011;31:4663–4674. doi: 10.1523/JNEUROSCI.5072-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goaillard JM, Taylor AL, Schulz DJ, Marder E. Functional consequences of animal-to-animal variation in circuit parameters. Nat Neurosci. 2009;12:1424–1430. doi: 10.1038/nn.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, Priebe CE, Zlatic M. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science. 2014;344:386–392. doi: 10.1126/science.1250298. [DOI] [PubMed] [Google Scholar]
- 44**.O’Leary T, Williams AH, Franci A, Marder E. Cell types, network homeostasis, and pathological compensation from a biologically plausible ion channel expression model. Neuron. 2014;82:809–821. doi: 10.1016/j.neuron.2014.04.002. In this study, O’Leary et al. examine how regulation rules allow a neuron to synthesize and degrade ion channels and ion channel mRNAs while maintaining electrical activity. Using only a single physiological variable, intracellular [Ca2+], the model can generate populations of neurons with stable properties but variable underlying parameters, consistent with experimental observations. The model illustrates important conceptual issues, such as how robustness to certain perturbations is accompanied by sensitivity to others, and how ‘homeostasis’ can fail even if the underlying compensation mechanism is intact. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huys QJ, Paninski L. Smoothing of, and parameter estimation from, noisy biophysical recordings. PLoS Comput Biol. 2009;5:e1000379. doi: 10.1371/journal.pcbi.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Paninski L, Vidne M, DePasquale B, Ferreira DG. Inferring synaptic inputs given a noisy voltage trace via sequential Monte Carlo methods. J Comput Neurosci. 2012;33:1–19. doi: 10.1007/s10827-011-0371-7. Using a statistical model and numerical techniques, this study infers the strength and number of excitatory and inhibitory synaptic inputs onto electrotonically compact neurons given noisy current- or voltage-clamp recordings. [DOI] [PubMed] [Google Scholar]
- 47*.Pakman A, Huggins J, Smith C, Paninski L. Fast state-space methods for inferring dendritic synaptic connectivity. J Comput Neurosci. 2014;36:415–443. doi: 10.1007/s10827-013-0478-0. Pakman et al., building upon earlier work of Paninski et al. (2012), develop a method to infer the densities and strengths of synaptic inputs in large dendritic trees. Physiological features such as geometry and cable properties of neurons are incorporated in a statistical model that determines the locations of synapses from noisy measurements of the dendritic membrane potential. [DOI] [PubMed] [Google Scholar]
- 48.Vogelstein JT, Packer AM, Machado TA, Sippy T, Babadi B, Yuste R, Paninski L. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J Neurophysiol. 2010;104:3691–3704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Vidne M, Ahmadian Y, Shlens J, Pillow JW, Kulkarni J, Litke AM, Chichilnisky EJ, Simoncelli E, Paninski L. Modeling the impact of common noise inputs on the network activity of retinal ganglion cells. J Comput Neurosci. 2012;33:97–121. doi: 10.1007/s10827-011-0376-2. Even as the number of simultaneously recorded neurons grows it will remain challenging to record from all neurons in a large circuit such as the retina at once. Vidne et al. examine how to model a sample of neurons when either part of the sample is not known or an input layer cannot be measured. They employ a common input noise technique (with no direct coupling) to distinguish what recorded activity is attributable to intrinsic network connections/processing versus noise-related firing. Using a generalized linear model they report that the relative contributions of each input type can be known with good accuracy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran A, Katz DB. Sensory cortical population dynamics uniquely track behavior across learning and extinction. J Neurosci. 2014;34:1248–1257. doi: 10.1523/JNEUROSCI.3331-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Miller P, Katz DB. Stochastic transitions between neural states in taste processing and decision-making. J Neurosci. 2010;30:2559–2570. doi: 10.1523/JNEUROSCI.3047-09.2010. Miller and Katz analyze gustatory cortex data and use it to generate a hidden Markov model that can reliably distinguish between stimuli even in the presence of noise – a critical point because spike times are variable and unreliable. Indeed, the model predicts that noise can expedite the behavioral decision when presented with a given stimulus. This information is lost in across-trial averaging. This study demonstrates that Markov models can determine the nature of the presented stimulus with high reliability from recorded neural data alone without the need for trial averaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brookings T, Goeritz ML, Marder E. Automatic parameter estimation of multicompartmental neuron models via minimization of trace error with control adjustment. J Neurophysiol. 2014 doi: 10.1152/jn.00007.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossant C, Goodman DF, Platkiewicz J, Brette R. Automatic fitting of spiking neuron models to electrophysiological recordings. Front Neuroinform. 2010;4:2. doi: 10.3389/neuro.11.002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Mante V, Sussillo D, Shenoy KV, Newsome WT. Context-dependent computation by recurrent dynamics in prefrontal cortex. Nature. 2013;503:78–84. doi: 10.1038/nature12742. Mante et al. model the neural dynamics and decisions of monkeys in a motion and color discrimination task. In the relevant feature space (motion or color), the authors show how the correct decision serves as an attractor with limited variability in the trajectory, while in irrelevant feature space (e.g. motion, when color is cued) there is significant wandering in the trajectory. An abstract recurrent network model recapitulates broad features of the population activity when context data are fed into the network, providing clues to how context-dependent perceptual decision making works. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fee MS. Oculomotor learning revisited: a model of reinforcement learning in the basal ganglia incorporating an efference copy of motor actions. Front Neural Circuits. 2012;6:38. doi: 10.3389/fncir.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56**.Memmesheimer RM, Rubin R, Olveczky BP, Sompolinsky H. Learning precisely timed spikes. Neuron. 2014;82:925–938. doi: 10.1016/j.neuron.2014.03.026. The authors ask how single neuron plasticity mechanisms might allow learning of precise temporal sequences of spikes, and derive the sequence-encoding capacity of a set of synaptic weights. The work informs theories of how complex motor tasks – song learning in this case – can be learned robustly, such that small timing errors early in a learned sequence do not propagate and destroy the remaining sequence. Interestingly, applying the model to data permits reconstriction of an effective set of synaptic weights. [DOI] [PubMed] [Google Scholar]
- 57*.Kato S, Xu Y, Cho CE, Abbott LF, Bargmann CI. Temporal responses of C. elegans chemosensory neurons are preserved in behavioral dynamics. Neuron. 2014;81:616–628. doi: 10.1016/j.neuron.2013.11.020. The authors describe and model aspects of temporal coding in two sensory neurons of C elegans. Their models and experiments reveal that biochemical time-scales are often temporally similar to the behavior they control (e.g.: avoidance or attraction). The authors model fluorescence from simulated Ca2+ influx and find that a linear-non-linear combination of models best fits the observed data with respect to the relevant time scale. This enabled alternative behavioral models of chemotaxis to be evaluated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.O’Donnell C, Sejnowski TJ. Selective memory generalization by spatial patterning of protein synthesis. Neuron. 2014;82:398–412. doi: 10.1016/j.neuron.2014.02.028. O’Donnell and Sejnowski model the effects of spatially specific, activity dependent protein synthesis and diffusion on the persistence of new memories using a biologically plausible model of synaptic plasticity. The authors show how the model discriminates between strong and weak synaptic stimuli and their effects on plasticity-related proteins. Though rooted in low-level details, this biochemically plausible conceptual model offers a potential mechanism for a high-level phenomenon: memory generalization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Leary T, Williams AH, Caplan JS, Marder E. Correlations in ion channel expression emerge from homeostatic tuning rules. Proc Natl Acad Sci U S A. 2013;110:E2645–2654. doi: 10.1073/pnas.1309966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotaleski JH, Blackwell KT. Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nat Rev Neurosci. 2010;11:239–251. doi: 10.1038/nrn2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgess N, Barry C, O’Keefe J. An oscillatory interference model of grid cell firing. Hippocampus. 2007;17:801–812. doi: 10.1002/hipo.20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Domnisoru C, Kinkhabwala AA, Tank DW. Membrane potential dynamics of grid cells. Nature. 2013;495:199–204. doi: 10.1038/nature11973. Domnisoru et al. evaluate two proposed models of grid cell activity (the oscillatory interference and the continuous attractor models) by directly measuring intracellular neural activity during a virtual reality navigation task. They report that depolarization ramps associated with certain locations are the proximal cause of grid cell firing, but that the precise timing occurs at the peak of theta oscillations. However, in the absence of attractor-like depolarization theta oscillations are not sufficient to cause grid cells to fire. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samsonovich A, McNaughton BL. Path integration and cognitive mapping in a continuous attractor neural network model. J Neurosci. 1997;17:5900–5920. doi: 10.1523/JNEUROSCI.17-15-05900.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sreenivasan S, Fiete I. Grid cells generate an analog error-correcting code for singularly precise neural computation. Nat Neurosci. 2011;14:1330–1337. doi: 10.1038/nn.2901. [DOI] [PubMed] [Google Scholar]
- 65.Barnett WH, Cymbalyuk GS. A codimension-2 bifurcation controlling endogenous bursting activity and pulse-triggered responses of a neuron model. PLoS One. 2014;9:e85451. doi: 10.1371/journal.pone.0085451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sussillo D, Abbott LF. Generating coherent patterns of activity from chaotic neural networks. Neuron. 2009;63:544–557. doi: 10.1016/j.neuron.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


