Abstract
Degreening, caused by chlorophyll degradation, is the most obvious symptom of senescing leaves. Chlorophyll degradation can be triggered by endogenous and environmental cues, and ethylene is one of the major inducers. ETHYLENE INSENSITIVE3 (EIN3) is a key transcription factor in the ethylene signaling pathway. It was previously reported that EIN3, miR164, and a NAC (NAM, ATAF, and CUC) transcription factor ORE1/NAC2 constitute a regulatory network mediating leaf senescence. However, how this network regulates chlorophyll degradation at molecular level is not yet elucidated. Here we report a feed-forward regulation of chlorophyll degradation that involves EIN3, ORE1, and chlorophyll catabolic genes (CCGs). Gene expression analysis showed that the induction of three major CCGs, NYE1, NYC1 and PAO, by ethylene was largely repressed in ein3 eil1 double mutant. Dual-luciferase assay revealed that EIN3 significantly enhanced the promoter activity of NYE1, NYC1 and PAO in Arabidopsis protoplasts. Furthermore, Electrophoretic mobility shift assay (EMSA) indicated that EIN3 could directly bind to NYE1, NYC1 and PAO promoters. These results reveal that EIN3 functions as a positive regulator of CCG expression during ethylene-mediated chlorophyll degradation. Interestingly, ORE1, a senescence regulator which is a downstream target of EIN3, could also activate the expression of NYE1, NYC1 and PAO by directly binding to their promoters in EMSA and chromatin immunoprecipitation (ChIP) assays. In addition, EIN3 and ORE1 promoted NYE1 and NYC1 transcriptions in an additive manner. These results suggest that ORE1 is also involved in the direct regulation of CCG transcription. Moreover, ORE1 activated the expression of ACS2, a major ethylene biosynthesis gene, and subsequently promoted ethylene production. Collectively, our work reveals that EIN3, ORE1 and CCGs constitute a coherent feed-forward loop involving in the robust regulation of ethylene-mediated chlorophyll degradation during leaf senescence in Arabidopsis.
Author Summary
Yellowing, caused by chlorophyll degradation, is the most obvious symptom of senescent leaves. Chlorophyll degradation can be triggered by a broad range of endogenous and environmental cues, and ethylene is one of the major inducers. Yet, the molecular regulation of chlorophyll degradation remains largely unknown. Here, we report a feed-forward regulation of ethylene-mediated chlorophyll degradation that involves ETHYLENE INSENSITIVE3 (EIN3), ORE1/NAC2, and major chlorophyll catabolic genes. EIN3, a master positive regulator of ethylene signaling, could directly promote chlorophyll degradation by physically binding to the promoters of three major chlorophyll catabolic genes to activate their expressions. Meanwhile, ORE1, a direct target of EIN3, also activates the expression of the similar set of chlorophyll catabolic genes directly. Moreover, ORE1 activates the expression of a major ethylene biosynthesis gene ACS2 during senescence, and subsequently activates a positive feedback to ethylene synthesis. Our work reveals a feed-forward loop that promotes ethylene-mediated chlorophyll degradation during leaf senescence, advancing our understanding on the molecular mechanism of leaf yellowing.
Introduction
Leaf senescence occurs at the final stage of leaf development and involves a series of changes at the molecular, cellular and phenotypic levels. Senescence is initiated by characteristic degenerative processes, e.g. chlorophyll (chl) degradation and macromolecule breakdown, and particularly recycling of nutrients to actively growing tissues or storage organs [1]. Molecular and genetic studies of Arabidopsis thaliana have identified dozens of senescence-related mutants and hundreds of senescence-associated genes (SAGs) involved in light signaling, hormone signaling and chl catabolism [2–4]. The phenotypic change of senescing leaves is degreening due to the net loss of chl in chloroplasts.
A biochemical pathway of chl degradation was recently elucidated in Arabidopsis via the identification of chl catabolic genes (CCGs). As the initial step, chl b is converted into chl a through two reductive reactions that are catalyzed by chl b reductase (NYC1/NOL) and 7-hydroxymethyl chl a reductase (HCAR), respectively [5–7]. Then the Mg atom of chl a is removed by an enzyme not yet identified to form pheophytin a. The phytol tail of pheophytin a is subsequently removed by pheophytin pheophorbide hydrolase (PPH) to produce pheophorbide a [8,9]. The ring structure of this intermediate product is then oxygenolytically opened by pheophorbide a oxygenase (PAO) to generate red chlcatabolite (RCC), which is degraded further by RCC reductase (RCCR) [10]. The conversion of pheophorbide a to RCC leads to the loss of green color during chl catabolism. Recently, these major chl catabolic enzymes (CCEs) were found to physically interact with STAY-GREEN1 (SGR1, also known as NYE1), a general regulator of chl degradation [11]. SGR1/NYE1 is essential for recruiting CCEs onto thylakoid membranes in senescing chloroplasts to promote chl degradation [11,12].
Plant hormones have been extensively reported to regulate leaf senescence, with ethylene, abscisic acid, jasmonic acid, brassinosteroid and salicylic acid functioning as inducers and auxin, cytokinin, and gibberellic acid as inhibitors [1,13–16]. Ethylene has long been considered a key endogenous regulator of leaf senescence and fruit ripening. Exogenous ethylene treatment accelerates leaf senescence, and the ethylene biosynthetic genes 1-Aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ACC oxidase (ACO), were upregulated in senescing leaves, coupled with an increased ethylene level [15]. An octuple mutant of ACSs showed a stay-green phenotype [17]. In the absence of ethylene, ethylene receptors are in an activated form and activate a Raf-like kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), and CTR1 in turn suppresses the downstream ethylene response pathway. Ethylene binding results in the inactivation of the receptor function [18], leading to the deactivation of CTR1, which then releases the suppression of the downstream positive regulators ETHYLENE INSENSITIVE2 (EIN2) and EIN3 [19,20]. EIN2 is a central positive regulator of ethylene signaling that locates in the membrane of the endoplasmic reticulum [21], where it undergoes cleavage and nucleus translocation controlled by CTR1-directed phosphorylation [22–24]. EIN3 is the key transcription factor of ethylene signaling downstream of EIN2. The ethylene-insensitive mutant of the ethylene receptor gene ETR1, etr1-1, and loss-of-function mutants ein2 (also known as ore2) and ein3 were all isolated as delayed-senescence mutants [25–27], which suggests a key role of ethylene in the initiation and/or progression of leaf senescence.
The pivotal role of ethylene in leaf senescence was further confirmed by the revelation of a feed-forward loop, whereby EIN2 affects leaf senescence in part by regulating the expression of miR164 and one of miR164 target genes, ORE1/NAC2 [28]. ORE1/NAC2 is a member of NAC transcription factor family which has been shown to play an important role in leaf senescence. In particular, ORE1 is a positive regulator of leaf senescence, promoting the expression of senescence-associated genes BFN1, SAG29, and SINA1 by directly binding to their promoters [29,30]. EIN3 was later on found to be involved in this feed-forward regulation by directly suppressing the expression of miR164, which negatively regulates ORE1 at the post-transcriptional level [27]. Meanwhile, EIN3 also directly activates the expression of ORE1 by binding to its promoter to accelerate leaf senescence [31]. These findings improve our understanding of the transcriptional regulatory cascade of ethylene signaling during leaf senescence and suggest that EIN3 positively regulates leaf senescence by inducing the expression of ORE1 directly and indirectly. Although both mutants of EIN3 and ORE1 showed delayed senescence or stay-green phenotypes, the molecular mechanism of how these genes regulate chl degradation during senescence is still largely unknown.
In this study, we report that EIN3 promotes chl degradation via the direct up-regulation of major chl catabolic genes, NYE1, NYC1 and PAO, by binding to their promoters. Meanwhile, one of the EIN3 direct targets, ORE1/NAC2 [31], also directly activates the expression of NYE1, NYC1 and PAO as well as NOL. Moreover, EIN3 and ORE1 promote NYE1 and NYC1 transcriptions in an additive manner. Intriguingly, ORE1 can also promote the transcription of ACS2 for a positive feedback regulation of ethylene biosynthesis and signaling.
Results
EIN3 upregulates the transcription of ORE1
EIN3 directly activates the expression of ORE1/NAC2, a master senescence-associated NAC transcription factor with significantly increased expression in the senescing leaves [27,28,31]. Our qPCR analysis confirmed that ethylene-induced ORE1 expression was abolished in the leaves of 4-week-old ein3 eil1 plants (Fig 1A), and the 35S promoter-driven expression of EIN3 significantly increased the activity of ORE1 promoter in Arabidopsis mesophyll protoplasts (Fig 1B). These results are consistent with previously published data [31].
Fig 1. ORE1 is directly activated by EIN3 and repressed by miR164.
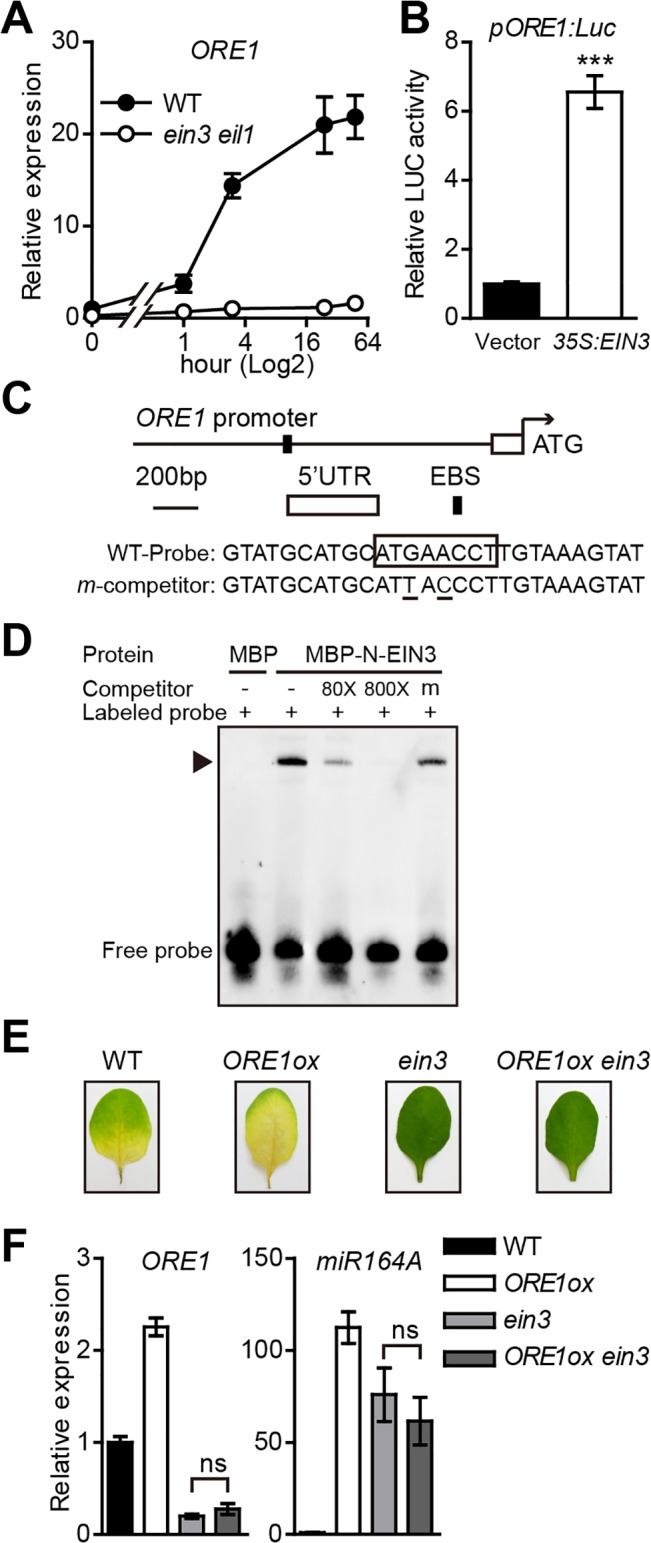
(A) Kinetic analysis of ORE1 expression in leaves of the wild-type (WT) and ein3 eil1 in response to ethylene treatment. Detached third and fourth rosette leaves from 4-week-old plants were treated with 100 μL/L ethylene for various periods of times. RT-qPCR was performed to quantify the ORE1 mRNA levels. ACT2 was used as an internal control to normalize different samples. The mRNA level of ORE1 in the WT at 0 hr was arbitrarily set to 1. The x axis is shown in log2 scale. Data are mean ± SEM of 3 biological replicates with technical duplicates for each. (B) Transient dual-luciferase transactivation of the ORE1 promoter by EIN3 in protoplasts from Arabidopsis leaves. Protoplasts were co-transformed with the pORE1:Luc reporter (1694 bp upstream from the translation start site of ORE1) and an effector overexpressing EIN3 (35S:EIN3). The 35S:REN was serving as an internal control. Relative reporter activity was normalized by the ratio of LUC/REN. An empty vector was used as a negative effector control, with LUC/REN ratio arbitrarily set to 1. Data are mean ± SEM of 3 biological replicates. *** p < 0.001 (t-test). (C) Schematic diagram of putative EIN3 binding site (EBS) in the ORE1 promoter. A 28-bp DNA fragment containing the EBS in ORE1 promoter was used as the probe for EMSA. The putative EBS in the WT probe sequence is boxed. The consensus nucleotides of EBS in the competitor sequence (underlined) were mutated. (D) EIN3 proteins physically interact with ORE1 promoter in EMSA. The N-terminus of EIN3 protein (aa 1–314 containing DNA binding domain) fused to maltose binding protein (MBP) was used to detect interaction (MBP-N-EIN3). MBP protein was used as a negative control. Biotin-labeled probes were added to each reaction mixture. WT competitor DNA was added in 80-fold and 800-fold molar excess. Mutated version of competitor DNA (m) was added in 800-fold molar excess. “‒” and “+” represent absence or presence, respectively. Triangle indicates the DNA-protein complex. (E) Representative leaves of WT, ORE1ox, ein3, and ORE1ox ein3 plants subjected to ethylene treatment for 3 days. (F) qRT-PCR analysis of relative gene expression of ORE1 and miR164A in leaves in (E). ACT2 was used as reference gene. Expression of each gene in the WT was set to 1. Data are mean ± SEM of 2 biological replicates. ns: not significant.
Although the association of EIN3 protein with the ORE1 promoter was first revealed in Y1H and ChIP assays [31], the evidence of direct binding was not clear. We scanned the ORE1 promoter and found a putative EIN3 binding site (EBS), ATGAACCT, located 1056~1064 bp upstream from the start codon (ATG) of the gene (Fig 1C). Electrophoretic mobility shift assay (EMSA) was used to determine the in vitro binding of EIN3 to its putative binding site with a recombinant truncated EIN3 that fused to MBP. This truncated version of EIN3 contains the first 314 amino acids of the N-terminus that covers the full-length DNA binding domain [32,33]. The MBP-N-EIN3 fusion protein could bind to the biotin-labeled wild-type (WT) probe (Fig 1D). Excessive amount of unlabeled competitor effectively competed with the binding, and the competition was dose-dependent: the binding signal was completely abolished with a sufficient amount of unlabeled competitor. Furthermore, once the putative EIN3 binding site was mutated, the unlabeled mutated probe could no longer compete for the binding. These results suggest that EIN3 positively regulates ORE1 expression by directly binding to the EBS on its promoter during ethylene-mediated leaf senescence in Arabidopsis.
We then tested whether EIN3 mutation affects the function of ORE1 protein. The ein3 single mutant displayed a stay-green phenotype with ethylene treatment partly because it may lack a functional ORE1 protein. As expected, overexpression of ORE1 in a WT background (ORE1ox) caused early leaf yellowing (Fig 1E). To check the effect of loss-of-function of EIN3 on the ORE1 protein function, we crossed the ORE1ox line with the ein3 mutant and examined the phenotype of ORE1ox ein3 with ethylene treatment. The ORE1ox ein3 line still exhibited a stay-green phenotype, which mimicked the phenotype of the ein3 mutant (Fig 1E). The ORE1 transcript was high in the ORE1ox line but very low in the ein3 and ORE1ox ein3 lines (Fig 1F). Considering the fact that these are the same transgenes in different genetic backgrounds, we reasoned that the ORE1 transcript in ORE1ox ein3 line was probably degraded. A previous study suggested that ORE1 mRNA is a target of miR164-mediated cleavage [28]. The miR164 family in Arabidopsis represses the expression of a group of NAC genes including ORE1. In addition, another recent paper reported that EIN3 negatively regulated miR164 transcription by directly binding to its promoter [27]. So the high level of miR164 in the ein3 background may have degraded the ORE1 transcript in ORE1ox ein3. Therefore, we checked the transcript level of miR164A, one of the 3 isoforms of the miR164 family, and found the pri-miR164A transcript level was low in the WT but high in ein3 or ORE1ox ein3 (Fig 1F). This result supports that EIN3 may also regulate ORE1 by repressing the miR164 function, as found previously [27]. We also found that the pri-miR164A transcript level was higher in ORE1ox line than that in WT (Fig 1F), suggesting a possible feedback upregulation of miR164 by highly accumulated ORE1. Taken together, EIN3 positively regulates ORE1 expression through 2 distinct ways during ethylene-mediated chl degradation: 1) directly binding to the ORE1 promoter and activating its expression, and 2) indirectly inhibiting the expression of miR164 and thereby activating ORE1.
EIN3 regulates chl degradation by directly activating major CCGs
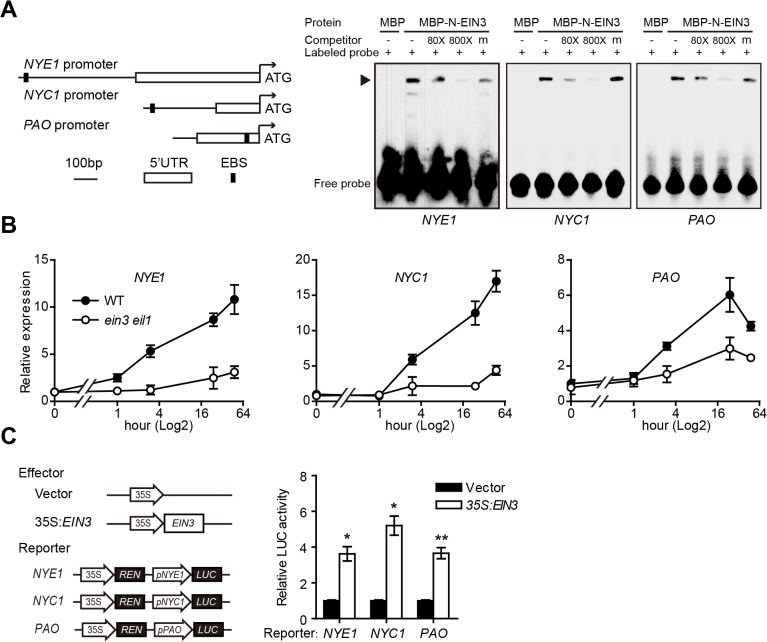
The above result indicated that ORE1 is a direct target of EIN3 during ethylene-mediated leaf senescence. Previously Li et al. found that the leaves of ein3 eil1 showed delayed senescence and a stay-green phenotype with ACC treatment [27]. Our results also confirmed the stay-green phenotypes in both the whole plant and detached leaves of ein3 eil1 with ethylene treatment (S1 Fig). Thus, EIN3 and EIL1 may play an indispensable role in chl degradation during ethylene-mediated senescence. The process of chl degradation is regulated by a number of CCGs, and some may represent direct targets of EIN3. EIN3/EIL1 transcription factors were reported to bind to a consensus DNA sequence of A[CT]G[AT]A[CT]CT [34,35]. We performed an in silico analysis to specifically scan for the consensus sequence in the non-coding regions of those CCGs and identified some putative EBS in the promoter or 5’-UTR regions of NYE1, NYC1 and PAO (Fig 2A). Therefore, we studied whether EIN3 could directly target these 3 CCGs to regulate their expression. EMSA results showed that the negative control MBP protein could not retard the mobility of biotin-labeled probes, whereas the MBP-N-EIN3 fusion protein caused a clear shift, which indicates the binding of the protein to probes (Fig 2A). Excessive amount of unlabeled probes successfully competed with the labeled probes in a dose-dependent manner, whereas excessive amount of mutated probes could not compete for binding (Fig 2A). Therefore, EIN3 physically bound to the promoters of NYE1, NYC1 and PAO in vitro.
Fig 2. EIN3 directly associates with and transactivates the promoters of NYE1, NYC1, and PAO.
(A) Left panel: Schematic diagrams of EIN3 binding site (EBS) in the promoter or 5’-UTR regions of NYE1, NYC1, and PAO. Right panel: EIN3 physically interacts with the promoters of NYE1, NYC1, and PAO in EMSA. About 30-bp DNA fragments containing the EBS in the promoters or 5’-UTR of NYE1, NYC1, and PAO were used as probes for EMSA. Mutated version of competitor DNA (m) was added in 800-fold molar excess. “‒” and “+” represent absence or presence, respectively. Triangle indicates the DNA-protein complex. (B) Kinetic analysis of NYE1, NYC1, and PAO expression in leaves of WT and ein3 eil1 in response to ethylene. Experiments were performed as in Fig 1A. The expression of each corresponding gene in the WT at 0 hr was set to 1. Data are mean ± SEM of 3 biological replicates with technical duplicates for each. (C) Left panel: Schematic diagrams of effector and reporter constructs used in the transient dual-luciferase assays. CaMV 35S promoter driving EIN3 (35S:EIN3) was used as effector, and the empty vector was used as a control. The dual-luciferase reporter constructs consist of 35S driving Renilla luciferase (REN) reporter gene for internal normalization, and the promoters of NYE1 (2012 bp), NYC1 (493 bp), PAO (365 bp) driving firefly luciferase (LUC) reporter gene. Right panel: transient dual-luciferase assay of EIN3 transactivating the promoters of NYE1, NYC1, and PAO in Arabidopsis protoplasts. The procedure was as in Fig 1B. Data are mean ± SEM of at least 3 biological replicates. * p < 0.05, ** p < 0.01 (t-test).
To examine the effect of EIN3/EIL1 on the expression of NYE1, NYC1 and PAO, we analyzed the kinetic expression of these CCGs in response to ethylene treatment and found that the transcript levels of NYE1, NYC1 and PAO were all greatly induced in WT plants; however, the ethylene induction of these CCGs was largely abolished in the ein3 eil1 double mutant (Fig 2B and S2 Fig). These results strongly suggest that EIN3 and EIL1 may positively regulate the transcription of these CCGs in ethylene-mediated chl degradation. To further investigate whether EIN3 indeed transcriptionally activated NYE1, NYC1 and PAO, transient dual-luciferase assays were carried out in Arabidopsis protoplasts. The NYE1, NYC1 and PAO promoters, which are 2012, 493 and 365 bp in length, respectively, were individually fused with the firefly luciferase (LUC) gene [36] and served as reporter constructs. Each reporter construct contained a separate expression cassette (35S:REN) with the Renilla luciferase (REN) gene under the control of a CaMV 35S promoter and functioned as an internal control to normalize the expression of each reporter gene (Fig 2C). A construct with 35S promoter driving the full-length EIN3 cDNA (35S:EIN3) was used as an effector, and the empty vector was included as a control (Fig 2C). The reporter and effector constructs were co-transformed into Arabidopsis protoplasts prepared from WT plants and luciferase activity (LUC/REN) was detected. Overexpression of EIN3 increased luciferase activity of each reporter as compared with the corresponding empty control (Fig 2C), which further demonstrates that EIN3 activated NYE1, NYC1 and PAO at transcriptional levels. Thus, EIN3 directly bound to promoters of the CCGs, NYE1, NYC1, and PAO and activated their transcription, which indicates that EIN3 play a crucial regulatory role during ethylene-mediated chl degradation.
ORE1 directly activates CCGs
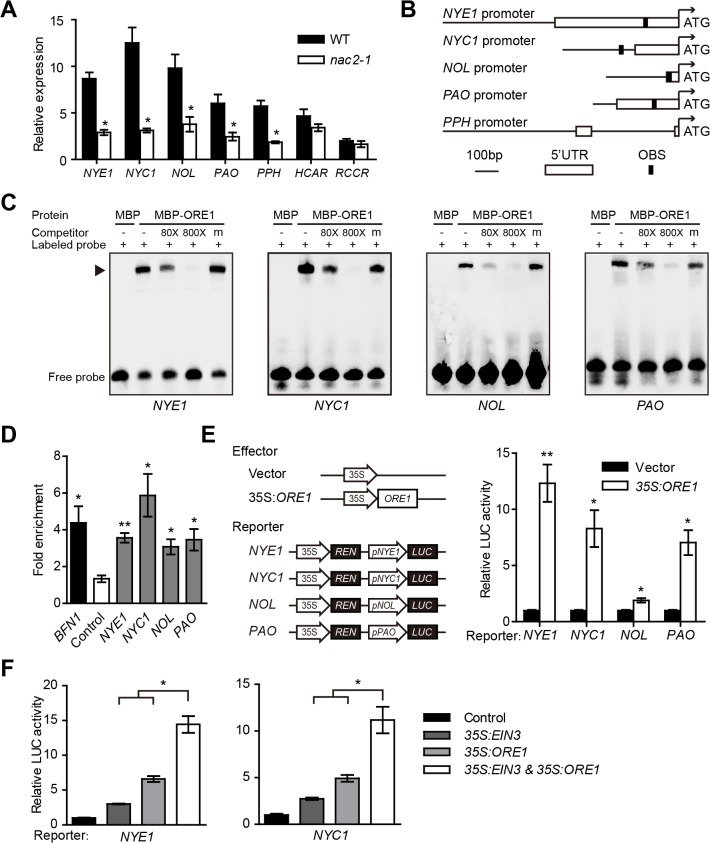
ORE1/NAC2 is a direct target of EIN3 in both age-dependent [31] and ethylene-mediated leaf senescence. The ORE1 mRNA level was rapidly induced in the WT but not the ein3 eil1 mutant with ethylene treatment (Fig 1A). Moreover, both the attached and detached leaves of nac2-1, a T-DNA insertion mutant of ORE1, showed a stay-green phenotype under ethylene treatment (S3 Fig), so ORE1 may also play a key role in ethylene-mediated chl degradation through CCGs. We checked the transcript levels of all CCGs in the nac2-1 mutant and found significantly repressed transcript levels of NYE1, NYC1, NOL, PAO and PPH with ethylene treatment (Fig 3A), so ORE1 is required for full induction of these CCGs. As a transcription factor, ORE1 was reported to bind to consensus DNA sequences of [ACG][CA]GT[AG]N{5,6}[CT]AC[AG] [29] or T[TAG][GA]CGT[GA][TCA][TAG] [37]. Among the promoters or 5’-UTR regions of NYE1, NYC1, NOL, PAO and PPH, all except PPH contain either of these two consensus putative ORE1 binding sites (OBS) (Fig 3B). Therefore, ORE1 may directly target these CCGs to regulate their expression. To test this possibility, we used EMSA with recombinant MBP-ORE1 proteins and DNA fragments of each promoter covering the putative OBS that were close to the ATG of each gene and found specific binding of MBP-ORE1 to the promoters of NYE1, NYC1, NOL and PAO (Fig 3C). As well, ChIP-qPCR assay revealed the association of ORE1 protein with these four promoters in 35S:ORE1-GFP transgenic Arabidopsis (Fig 3D).
Fig 3. ORE1 directly activates the expression of NYE1, NYC1, NOL and PAO.
(A) Relative expression of CCGs in leaves of WT and ein3 eil1with ethylene treatment for 24 hr. Gene expression was relative to that in the WT at 0 hr. Data are mean ± SEM of 3 biological replicates with technical duplicates for each. * p < 0.05 (t-test). (B) Schematic diagrams of ORE1 binding site (OBS) in the promoter or 5’-UTR regions of NYE1, NYC1, NOL and PAO. (C) ORE1 physically interacts with the promoters of NYE1, NYC1, NOL, and PAO in EMSA. About 30-bp DNA fragments containing the OBS in the promoter or 5’-UTR regions of NYE1, NYC1, NOL, and PAO were used as probes for EMSA, with purified MBP or MBP-ORE1 protein expressed in E. coli. “‒” and “+” represent in absence or presence, respectively. “m” represents mutated competitor. Triangle indicates the DNA-protein complex. (D) ORE1 associated with the promoters of NYE1, NYC1, NOL, and PAO in ChIP-qPCR assay. Chromatins isolated from 35S:ORE1-GFP transgenic line and WT control were immunoprecipitated with anti-GFP antibody followed by qPCR to amplify regions covering the putative ORE1 binding sites. Input sample was used to normalize the qPCR results in each ChIP sample. BFN1, reported as a direct target of ORE1, was used as a positive control. A retrotransposon (At4g03770) located within the heterochromatic region associated with di-methylated H3-K9 was used as a negative control. Fold enrichment was presented as a ratio of normalized results from 35S:ORE1-GFP plants and WT. Data are mean ± SEM of at least 3 technical replicates. * p < 0.05, ** p < 0.01 (t-test). The experiment was repeated twice with similar results. (E) Left panel: Schematic diagrams of effector and reporter constructs used in the transient dual-luciferase assays. CaMV 35S promoter driving ORE1 (35S:ORE1) was used as effector, and empty vector as a negative control. A 309-bp fragment upstream from ATG of NOL was used to make the pNOL:LUC reporter construct and all other reporters were as in Fig 2C. Right panel: Transient dual-luciferase assay of ORE1 transactivates the promoters of NYE1, NYC1, NOL, and PAO in Arabidopsis protoplasts. The procedure was as in Fig 2C. Data are mean ± SEM of at least 3 biological replicates. * p < 0.05, ** p < 0.01 (t-test). (F) EIN3 and ORE1 transactivate the promoters of NYE1 and NYC1 in Arabidopsis protoplasts in an additive manner. The transient expression procedure, and the constructs used for the assay were as in Figs 2C and 3E. The amount of each effector was half that used in Figs 2C and 3E. The same amount of corresponding empty vector was used if one effector was absent in a transformation so that the total amount of plasmids was the same among all assays. Data are mean ± SEM of at least 3 biological replicates. * p < 0.05 (t-test).
The direct transactivation effect of ORE1 on the expression of CCGs was further confirmed by transient dual luciferase assay in Arabidopsis protoplasts, with a construct overexpressing ORE1 (35S:ORE1) as an effector and promoters of NYE1, NYC1, NOL and PAO driving LUC as reporters (Fig 3E). It showed that ORE1 significantly transactivated the promoters of NYE1, NYC1, NOL and PAO (Fig 3E). Interestingly, EIN3 and ORE1 had additive effects in activating the promoters of both NYE1 and NYC1 (Fig 3F). Therefore, both EIN3 and ORE1 may actively regulate the transcription of NYE1 and NYC1 by directly binding to their promoters and contributing to chl degradation.
The EIN3- and ORE1-mediated chl degradation during ethylene-induced senescence is NYE1-dependent
The above data showed that both EIN3 and ORE1 directly promoted the transcription of three major CCGs, NYE1, NYC1, and PAO, during ethylene-mediated chl degradation. We then analyzed the relationship between CCGs and EIN3 or ORE1. Since NYE1 was reported to be essential for recruiting major CCEs into a possible multi-protein complex in senescing chloroplasts [11], and NYE1 responded earlier than did NYC1 and PAO to ethylene treatment and the responsiveness was significantly repressed in ein3 eil1 and nac2-1 mutants (Fig 2B and S4 Fig), and nye1 mutant showed stronger stay-green phenotype than that of nyc1 and pao mutants (Fig 4C and S7 Fig), NYE1 was then selected as a representative of CCGs. To investigate the relationship among NYE1, EIN3 and ORE1 in regulating ethylene-mediated chl degradation, we generated plant lines with different combination of these genotypes and measured their chl contents before (0 d) and after (4 d) ethylene treatment. The total chl contents of all tested lines were similar before any treatment, which were consistent with their leaf colors (Fig 4). An inducible NYE1 overexpression transgenic line (NYE1iox), which displayed an early-yellowing phenotype with ethylene treatment (Fig 4A and S5 Fig), was crossed with ein3 mutant. ein3 exhibited a stay-green phenotype with high chl content upon ethylene treatment (Fig 4A and 4B). Inducible expression of NYE1 efficiently reversed the stay-green phenotype of ein3 (Fig 4A and 4B). We also crossed an inducible EIN3 line (EIN3iox) to nye1 mutant. EIN3iox line displayed an early-yellowing phenotype with low chl content upon ethylene treatment (Fig 4C and 4D), which was opposite to that of nye1 mutant, whereas mutation of NYE1 suppressed EIN3-induced early-yellowing phenotype (Fig 4C and 4D). Similarly, inducible expression of NYE1 reversed the stay-green phenotype caused by ORE1/NAC2 mutation (Fig 4E and 4F), and NYE1 mutation repressed the early-yellowing phenotype caused by overexpression of ORE1 (Fig 4G and 4H). These results suggest that the ethylene-induced chl degradation through EIN3 and ORE1 depends on functional NYE1 gene.
Fig 4. Ethylene-induced chl degradation through EIN3 and ORE1 is NYE1-dependent.
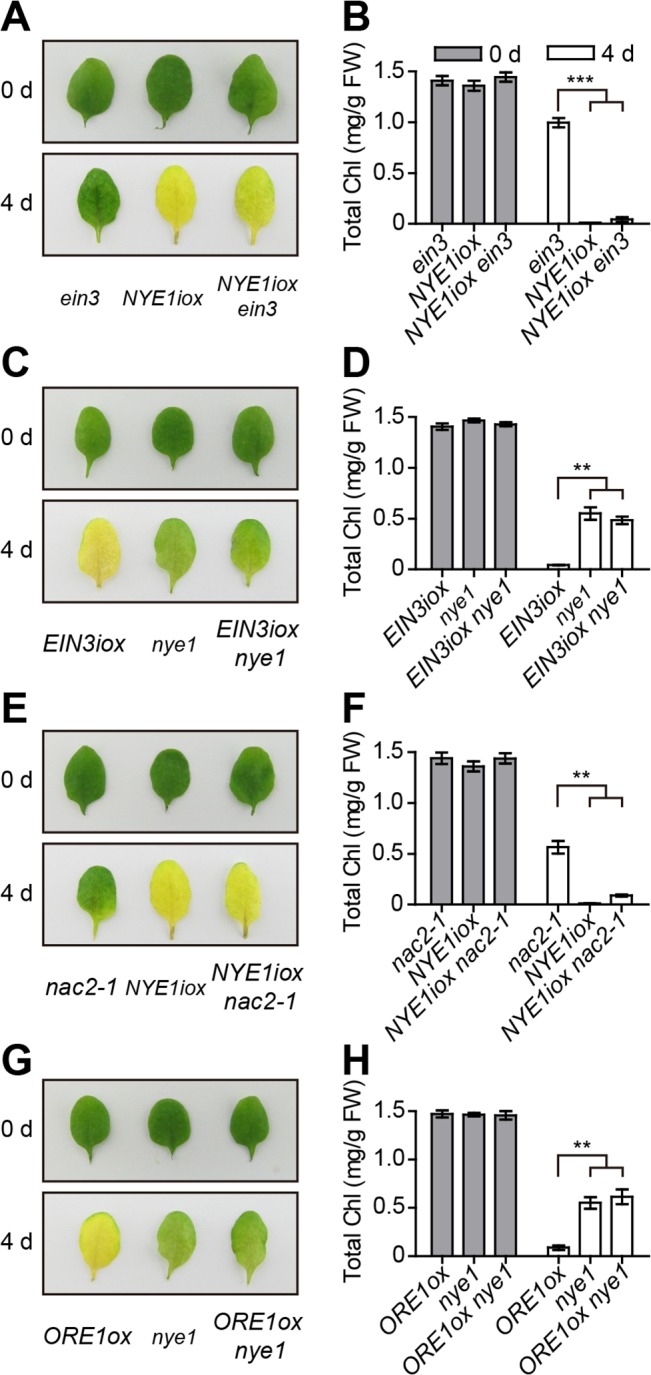
(A) Inducible overexpression of NYE1 reversed the stay-green phenotype of ein3 mutant under ethylene treatment. The dexamethasone (DEX)-inducible NYE1 line (NYE1iox) was crossed with ein3 to obtain NYE1iox ein3. The leaves of each indicated genotype were sprayed with 15 μmol/L DEX to induce NYE1 expression and underwent ethylene treatment for 4 d. (B) Quantification of total chl content in leaves of each indicated genotype and time point shown in (A). (C) The loss-of-function nye1 mutant repressed chl degradation of the inducible overexpression of EIN3 transgenic line during ethylene-induced senescence. The estradiol (EST)-inducible EIN3 line EIN3iox was crossed with nye1 to obtain EIN3iox nye1. Leaves were sprayed with 100 μmol/L EST to induce EIN3 expression and underwent ethylene treatments for 4 d. (D) Quantification of total chl content in leaves of each indicated genotype and time point shown in (C). (E) Inducible overexpression of NYE1 reversed the stay-green phenotype of nac2-1 mutant under ethylene treatment. The DEX-inducible NYE1 line (NYE1iox) was crossed with nac2-1 mutant to obtain NYE1iox nac2-1. The treatments were as in (A) except for different plant genotypes. (F) Quantitative analysis of total chl content in leaves of each indicated genotype and time point shown in (E). (G) The loss-of-function nye1 mutant repressed chl degradation caused by overexpression of ORE1 during ethylene-induced senescence. The leaves underwent ethylene treatment for 4 d. (H) Quantitative analysis of total chl content in leaves of each indicated genotype and time point shown in (G). For (A), (C), (E) and (G), the photographs were taken before (0 d) or after (4 d) induction and treatment. For (B), (D), (F) and (H), data are mean ± SEM (n > 4). ** p < 0.01, *** p < 0.001 (t-test). The total chl contents of all tested lines were similar before induction and treatment.
ORE1 directly activates the expression of ACS2
Previous data suggested a positive role of ORE1 in ethylene-mediated chl degradation. To identify possible ORE1 direct targets by scanning for putative ORE1 binding sites in the promoter regions of candidate genes induced by senescence, we identified some CCGs as putative direct targets but also located a putative ORE1 binding site [ACG][CA]GT[AG]N{5,6}[CT]AC[AG] [29] in the promoter region of ACS2, an ACC synthesis gene with the mRNA level increased with leaf aging (Fig 5A). We then examined the interaction of ORE1 protein with the promoter of ACS2. ChIP assay, with the 35S:ORE1-GFP transgenic line in Arabidopsis, showed an approximately four-fold enrichment of the ACS2 promoter region harboring the putative ORE1 binding site (Fig 5B). The direct binding of ORE1 protein to the ACS2 promoter was further confirmed by EMSA (Fig 5A). To examine the effect of ORE1 on the expression of ACS2 during ethylene-induced senescence, we analyzed the kinetic expression of ACS2 with ethylene treatment. The transcript level of ACS2 was greatly induced in the leaves of WT plants at a later stage of the treatment (Fig 5C); however, the ethylene induction of ACS2 was repressed in the loss-of-function nac2-1 mutant (Fig 5C), indicating that ORE1 may positively regulate the expression of ACS2. This finding was further confirmed by a dual luciferase assay in Arabidopsis protoplasts, with ACS2 promoter activity being monitored with a pACS2:LUC reporter construct. Overexpression of ORE1 significantly increased the activity of ACS2 promoter (Fig 5D). To further determine whether ORE1 mutation affects ethylene biosynthesis during senescence, we quantified ethylene production in the leaves of both WT and nac2-1 and found that nac2-1 leaves produced about 30% less ethylene within a period of 72 hr after detachment, in comparison to that produced by WT leaves (Fig 5E). These results support that ORE1 promotes ethylene synthesis via directly regulating the expression of ACS2 likely in a positive feedback manner.
Fig 5. ORE1 is associated with ACS2 promoter and transcriptionally activates its expression.
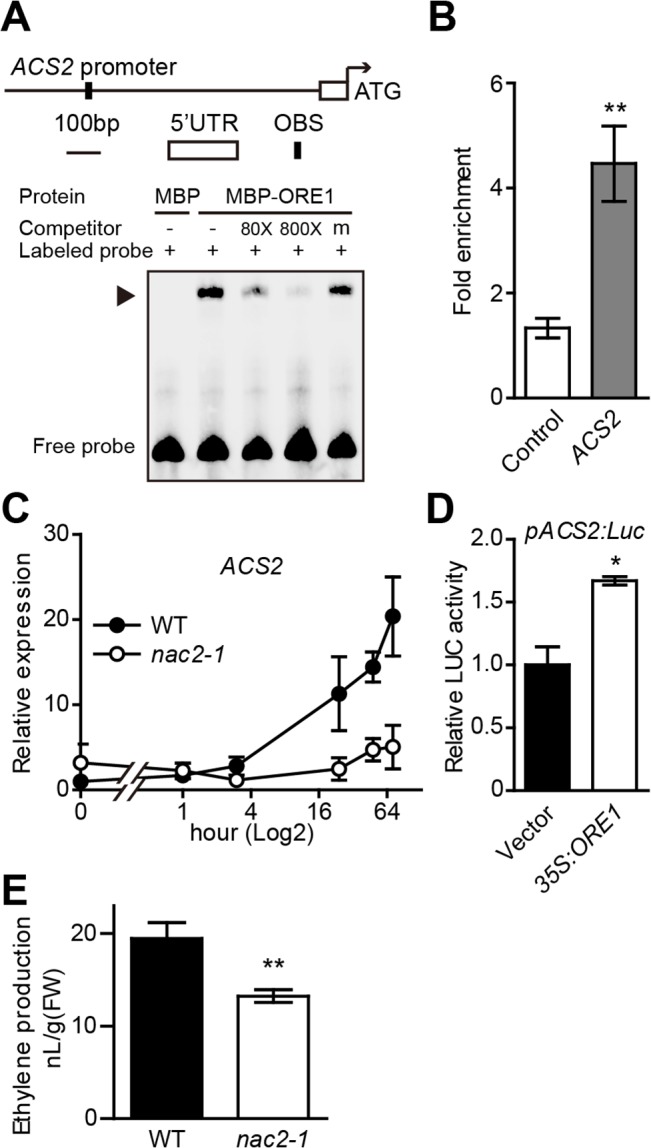
(A) EMSA detection of binding of ORE1 to ACS2 promoter in vitro. A 45-bp ACS2 promoter fragment containing the putative ORE1 binding site was biotin-labeled and used as a probe. Purified MBP-ORE1 protein expressed in E. coli was used in EMSA. MBP was included as a negative control. “‒” and “+” represent absence or presence, respectively. “m” represents mutated competitor. Triangle indicates the DNA-protein complex. (B) ChIP-qPCR analysis of ORE1 binding to ACS2 promoter in vivo. The ACS2 promoter region containing a putative ORE1 binding site was amplified to detect the enrichment. The ChIP procedure and qPCR data processing were as described in Fig 3D. Data are mean ± SEM of at least 3 technical replicates. ** p < 0.01 (t-test). The experiment was repeated twice with similar results. (C) Kinetic analysis of ACS2 expression in WT and nac2-1 with ethylene treatment for various periods of time (0, 1, 3, 24, 48, 72 hr). Expression in WT at 0 hr was set to 1. Data are mean ± SEM of 3 biological replicates with technical duplicates for each. (D) Transient dual-luciferase assay of transactivation of the ACS2 promoter by ORE1 in Arabidopsis protoplasts. A 1019-bp ACS2 promoter fragment covering the putative ORE1 binding site was used for making the pACS2:LUC reporter construct. The 35S:ORE1 effector construct was described in Fig 3E. Data are mean ± SEM of 3 biological replicates. * p < 0.05 (t-test). (E) Ethylene production in the leaves of WT and nac2-1 during senescence. Data are mean ± SEM (n = 9). ** p < 0.01 (t-test). The experiment was repeated twice with similar results.
Discussion
Our data reveal novel functions of existing transcription factors, EIN3 and ORE1, involved in activating the expression of several CCGs, which in turn accelerate ethylene-mediated chl degradation in Arabidopsis. We showed that EIN3 promotes ethylene-mediated chl degradation by activating the expression of some key CCGs, namely NYE1, NYC1 and PAO, via directly binding to their promoters. NYE1 was reported to be essential for recruiting CCEs into a possible multi-protein complex in the beginning of chl breakdown in senescing chloroplasts [11]. NYC1 is a chl b reductase that catalyzes the first step of chl degradation [6]. PAO is responsible for the ring opening reaction of the porphyrin macrocycle and is the key enzyme of the chl degradation pathway [10]. We found that EIN3 proteins bound to specific regions of the NYE1, NYC1 and PAO promoters (Fig 2A) and the relative expression of these genes was induced by ethylene in WT plants, but the expression considerably reduced in the ein3 eil1 double mutant even with ethylene treatment (Fig 2B). In addition, overexpression of EIN3 transactivated the expression of NYE1, NYC1 and PAO promoters (Fig 2C). Above all, we identified novel EIN3 direct targets, NYE1, NYC1 and PAO, which are the major CCGs in chl degradation pathway.
EIN3 was previously shown to promote leaf senescence by accumulating mRNA of ORE1 which has been reported to play a central role in leaf senescence and cell death [27–29,31,38]. We further found that ethylene-induced CCG expression was repressed in the loss-of-function nac2-1 mutant (Fig 3A and S4 Fig), so ORE1/NAC2 may play a positive role during chl degradation. Transient dual luciferase assay further confirmed that overexpression of ORE1 significantly transactivated the promoter activity of NYE1, NYC1, NOL and PAO (Fig 3E). Moreover, in vivo ChIP-qPCR and in vitro EMSA suggested that ORE1 directly bound to the promoters of these CCGs and activated their transcription (Fig 3C and 3D). Altogether, these results reveal four CCGs are novel direct targets of ORE1 in chl degradation during leaf senescence. We further found that EIN3 and ORE1 shared NYE1, NYC1 and PAO as common targets, and ORE1 targeted an additional CCG, NOL. In addition, EIN3 and ORE1 promoted the expression of NYE1 and NYC1 in an additive manner (Fig 3F). These results suggest that ORE1, a directly target of EIN3, strengthen and broaden the signal from EIN3.
Our data support that EIN3 promoted ethylene-mediated chl degradation by (1) directly activating the expression of CCGs or (2) indirectly activating the intermediate regulator ORE1, which in turn activates more CCGs and enhances their expressions. EIN3, ORE1, and CCGs constitute a coherent feed-forward loop in the regulation of ethylene-mediated chl degradation during leaf senescence (Fig 6). We noticed that ethylene-induced NYE1 mRNA accumulation increased earlier than did NYC1 and PAO levels (Fig 2B), indicating that different CCGs require different threshold values of EIN3 and/or ORE1 to initiate their transcription. Considering that NYE1 was reported to recruit CCEs such as NYC1 and PAO during chl degradation, the earlier induction of NYE1 by EIN3 and/or ORE1 might be necessary for its function. This fine-tuning mechanism may allow plants to robustly respond to ethylene-mediated chl degradation.
Fig 6. A working model of the EIN3-ORE1-CCGs coherent feed-forward loop in regulation of ethylene-mediated chl degradation.
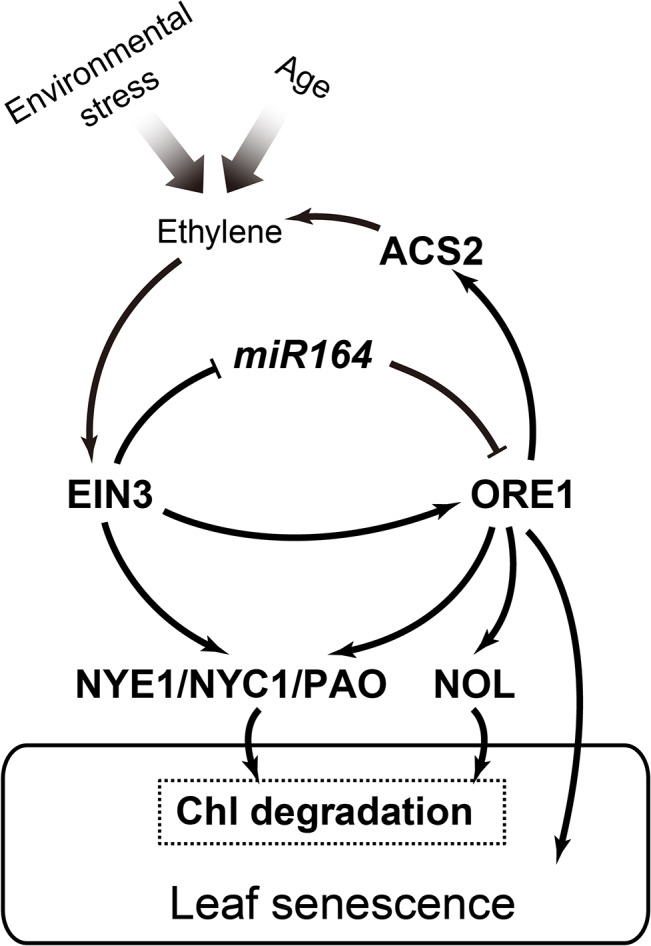
According to our study and previous reports [27,31], we propose a coherent feed-forward loop that involves EIN3 and ORE1 in regulating ethylene-mediated chl degradation. EIN3 directly represses the transcription of miR164, which negatively regulates ORE1 at the post-transcriptional level. Meanwhile, EIN3 can directly bind to the ORE1 promoter and induce ORE1 transcription. Three CCGs, NYE1, NYC1, and PAO, are the direct targets of EIN3. As a transcription factor downstream of EIN3, ORE1 shares these 3 common direct targets with EIN3. However, ORE1 also has its own distinct target, NOL, during the regulation of chl degradation. The broad range of expression of CCGs leads to chl degradation, the early step of leaf senescence. In addition, ORE1 directly activates the expression of ACS2, which presumably triggers a positive feedback regulation of ethylene synthesis. Arrows and bars represent positive and negative regulations, respectively.
In addition to EIN3 and ORE1 being able to bind to the promoters of NYE1, NYC1 and PAO to modulate ethylene-mediated chl catabolism, ABSCISIC ACID INSENSITIVE3 (ABI3), a B3 domain transcription factor that confers desiccation tolerance during seed maturation, regulates NYE1 expression during seed degreening [39]. ABI5 and ENHANCED EM LEVEL (EEL), two Group A bZIP transcription factors in the ABA signaling pathway, can bind to the promoters of NYE1 and NYC1 to directly accelerate chl degradation [4]. A more recent paper showed that the Phytochrome-interacting factor4 (PIF4), a bHLH transcriptional factors of light signal transduction pathway, binds to the promoter of NYE1 to activate chl catabolism [40]. Therefore, EIN3 and ORE1, together with other type of transcription factors from other signaling pathways, may promote key chl catabolic genes, such as NYE1, during chl breakdown. This mechanism allows plants to effectively and coordinately respond to environmental changes or stress conditions by hormone-mediated chl degradation.
We detected a lower level of ACS2 transcript level in the loss-of-function nac2-1 mutant than that in WT plants (Fig 5C) but a higher level in the ORE1 overexpression line (S6 Fig), suggesting that ORE1 might be a positive regulator of ACS2 expression. Transient dual luciferase assay indeed revealed that overexpression of ORE1 transactivated the expression of ACS2 (Fig 5D). Furthermore, a putative ORE1 binding site was found in the promoter of ACS2, and EMSA and ChIP-qPCR results suggested that ORE1 directly bound to the putative ORE1 binding site in the ACS2 promoter (Fig 5A and 5B). Moreover, we quantified ethylene production in detached leaves of WT and the nac2-1 mutant plants and found that nac2-1 indeed produced less ethylene than did WT (Fig 5E). Our data revealed that in addition to directly accelerating chl degradation and leaf senescence, ORE1 promotes ethylene synthesis by activating the expression of ACS2. In turn, ethylene further accelerates chl degradation and leaf senescence (Fig 6). The complicated network of interactions involving both positive feed-forward loop (EIN3-ORE1-CCGs) and positive feed-back regulation of ethylene synthesis (ORE1-ACS2) would likely lead to an irreversible process. This may facilitate plants to activate all available approaches to quickly trigger senescence processes during their final developmental stages.
In conclusion, our results reveal that EIN3 protein promotes expression of the CCGs, NYE1, NYC1 and PAO by directly binding to their promoters to advance ethylene-mediated chl degradation. Meanwhile, one of the EIN3 target genes, ORE1/NAC2 [31], can also directly activate the expression of NYE1, NYC1 and PAO as well as other CCGs such as NOL. EIN3 and ORE1 additively activate the expression of NYE1 and NYC1. In addition, ORE1 activates the expression of ACS2, presumably triggering a positive feedback regulation of ethylene production. Collectively, our work reveals a coherent feed-forward loop, involving EIN3, ORE1 and CCGs, which efficiently regulates ethylene-mediated chl degradation during leaf senescence in Arabidopsis.
Materials and Methods
Plant materials and growth conditions
All plants, including the WT, mutants and transgenic lines, were in Arabidopsis thaliana ecotype Columbia-0 (Col-0) background. The mutants ein3 eil1 [41], nye1 [12] and the EIN3iox transgenic line [42] were described previously. To generate NYE1iox transgenic line, the full-length NYE1 coding sequence (CDS) was PCR amplified, using WT cDNA as template, and cloned into the vector pTA7002 [43]. The final construct was transformed into nye1 mutant using the floral-dipping method [44]. The ORE1ox line [45] and 35S:ORE1-GFP line [29] were kindly provided by Feng Ming (Fudan University, China) and Bernd Mueller-Roeber (University of Potsdam, Germany), respectively. The mutant nac2-1 (SALK_090154) was obtained from the ABRC and the homozygosity of this line was confirmed by PCR-based genotyping. NYE1iox ein3, EIN3iox nye1, NYE1iox nac2-1 and ORE1ox nye1 were generated by genetic crossing. Homozygous lines were genotyped by PCR. Primers for genotyping are listed in S1 Table.
After surface sterilization, imbibed seeds were stratified for 3 days at 4°C to synchronize germination. Plants were grown in an environmentally controlled chamber at 22°C-24°C, at light intensity of approximately 120 μmol m-2 s-1 under a 16-hr light/8-hr dark photoperiod.
Ethylene-induced senescence
The third and fourth rosette leaves from 4-week-old plants were used for ethylene-induced senescence assay. Leaves were detached and placed on two layers of moist filter papers in Petri dishes to maintain moisture and were placed in a sealed glass desiccator. Ethylene was released by adding 1M ethephon stock solution into 5 mM Na2HPO3 buffer at the bottom of the desiccator [46]. The final concentration of ethylene was 100 μL/L. For kinetic analysis of gene expression, Petri dishes were placed in a sealed container that was injected with ethylene gas at a final concentration of 100 μL/L. Ethylene treatment was performed at 22°C to 24°C under a normal 16-hr/8-hr photoperiod for 0, 1, 3, 24, and 48 hr, unless otherwise stated.
Chl content measurement
Chl content was measured as previously described [47]. Briefly, leaves were incubated in DMSO at 65°C for 30 minutes, and absorbance was measured at 663 and 645nm. The concentration of total chl was calculated as follows: total chl (mg/L) = 20.2×D645 + 8.02×D663. The chl content was converted to microgram per gram fresh weight of leaf tissue (mg/g FW).
qRT-PCR
Total RNA was extracted by use of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized with the PrimeScript RT Master Mix (TaKaRa, China) and then used as templates for quantitative RT-PCR (qRT-PCR) with SYBR Premix Ex Taq II (Perfect Real-Time; TaKaRa, China) and the MyiQ2 Real Time PCR Detection System (Bio-Rad, Hercules, CA). ACT2 was an internal control for normalization. Primers used for qRT-PCR are listed in S1 Table.
Protein expression and electrophoretic mobility shift assay (EMSA)
For protein expression and purification, 1 to 942 bp of EIN3 CDS containing the DNA-binding domain and full-length CDS of ORE1/ANAC092 were cloned into pMAL-c5g (New England Biolabs). The empty vector pMAL-c5g was used for MBP expression alone as a negative control in EMSA. Plasmids were transformed into Escherichia coli strain Rosetta (DE3) pLysS (Merck). The expression of proteins was induced by 1mM isopropyl thio-β-D-galactoside (IPTG) at 20°C for 10 hr in 200 mL LB medium. Cells were then collected and sonicated. Protein was purified with Amylose resin (New England Biolabs) following the manufacturer’s instructions.
EMSA was carried out using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). Briefly, 400 ng purified protein was incubated with 12.5 fmol 5’-biotin-labeled probe DNA and 1 μg poly (dI-dC) in binding buffer for 15 min. Binding reactions were resolved on a polyacrylamide gel and electrophoretically transferred to nylon membrane. The transferred DNA was cross-linked to membrane by use of CL-1000 Ultraviolet Crosslinker (UVP, Upland, CA, USA). Biotin-labeled DNA was detected by chemiluminescence and exposed with a ChemiScope 3500 Mini Imaging System (Clinx Science Instruments, China).
Dual-luciferase transient expression assay in Arabidopsis protoplasts
To generate luciferase reporter constructs, the promoters of ORE1 (1694 bp), NYE1 (2012 bp), NYC1 (493 bp), NOL (309 bp), PAO (365 bp), and ACS2 (1019 bp) were amplified from Col-0 genomic DNA and cloned into the transient expression vector pGreenII 0800-Luc, with the expression of two cassettes—the target promoters driving a firefly luciferase (LUC) reporter gene and a CaMV 35S promoter driving an Renilla luciferase (REN) gene as an internal control [36]. To generate 35S:EIN3 effector construct, the full-length EIN3 coding sequence (CDS) was PCR amplified, using WT cDNA as template, and cloned into pDONR221 (Invitrogen, Carlsbad, CA), then into the destination vector pEarleyGate203 [48] to get pEarley203-EIN3 by recombination. The 35S:ORE1 effector construct pHB-ANAC092 was described previously [45]. Empty vectors pEARLY203 and pHBwere controls, respectively. Primers for all constructs are listed in S1 Table.
Arabidopsis mesophyll cell protoplast isolation and transformation were as described [49]. Rosette leaves of 4-week-old plants were cut into leaf strips, then digested in an enzyme solution containing 1.5% (w/v) cellulase R10 and 0.4% (w/v) macerozyme R10 (Yakult Honsha, Tokyo). Plasmids were introduced into protoplasts by PEG-mediated transformation. Transformed protoplasts were incubated overnight, and firefly and Renilla luciferase activity was quantified using Dual-Luciferase Reporter Assay System (Promega, USA) and detected with a Synergy 2 multi-mode microplate (Bio-Tek) according to the manufacturer’s instructions.
ChIP-qPCR
The third and fourth leaves of 5-week-old WT and 35S:ORE1-GFP plants were harvested and cross-linked with 1% formaldehyde. ChIP was carried out using the EpiQuik Plant ChIP Kit (Epigentek, Brooklyn, NY, USA) with the antibody against GFP (ab290; Abcam). Input samples and immunoprecipitated samples were analyzed by qPCR. Primers flanking the ORE1 binding sites in NYE1, NYC1, NOL, PAO and ACS2 promoters were used to detect ORE1 enrichment. Primers amplifying a fragment in the heterochromatic region (At4g03770) were used for a negative control. The primer sequences are listed in S1 Table. Anti-dimethyl H3-K9 antibody supplied by the ChIP kit was used as a negative experimental control, and no enrichment was detected in either WT or 35S:ORE1-GFP plants among all tested regions compared with using IgG (no antibody was conjugated). ChIP-qPCR results were first normalized with input sample as follows: cycle threshold (Ct) = CtChIP − CtInput. Relative enrichment was then calculated by the ratio of normalized results from 35S:ORE1-GFP plants and WT control.
Quantification of ethylene production
The third and fourth rosette leaves of 4-week-old plants of WT and nac2-1 were used for quantification of ethylene production. The detached leaves were weighed and incubated in 2 mL Agilent vial (clear glass) at 22°C to 24°C under a normal 16-hr/8-hr photoperiod. Ethylene produced within 72 hr period after detachment was measured with a sensitive laser-based ethylene detector (ETD-300, Sensor Sense BV, Nijmegen, the Netherlands) following the manufacturer’s instructions.
Statistical analysis
Data are given as mean ± SEM and were analyzed by two-tailed Student’s t-test or one-way ANOVA. p < 0.05 was considered statistically significant.
Accession numbers
Genes and their associated accession numbers in the Arabidopsis Genome Initiative or GenBank/EMBL are as follows: EIN3 (AT3G20770), EIL1 (AT2G27050), ORE1/NAC2/ANAC092 (AT5G39610), NAP (AT1G69490), miR164A (AT2G47585), NYE1/SGR (AT4G22920), NYC1 (AT4G13250), NOL (AT5G04900), PAO (AT3G44880), PPH (AT5G13800), HCAR (AT1G04620), RCCR (AT4G37000), ACS2 (AT1G01480), ACT2 (AT3G18780).
Supporting Information
(A) Whole plants of 4-week-old ein3 eil1 mutant showed a stay-green phenotype compared to the wild type (WT) with 100 μL/L ethylene treatment for 4 d. (B) Detached third and fourth rosette leaves from 4-week-old WT and ein3 eil1 plants treated with 100 μL/L ethylene for 4 d. (C) Quantitative analysis of total chl content in leaves of each genotype shown in (B). Data are mean ± SEM (n>4). *** p < 0.001 (t-test).
(TIF)
The expression level of each corresponding gene in the WT at 0 hr was set to 1. The scales of y-axes are consistent with that in Fig 2B. Data are mean ± SEM of 3 biological replicates.
(TIF)
(A) Whole plants of 4-week-old nac2-1 mutant showed a stay-green phenotype compared to the WT with 100 μL/L ethylene treatment for 4 d. (B) Detached third and fourth rosette leaves from 4-week-old WT and nac2-1 plants treated with 100 μL/L ethylene for 4 d. (C) Quantitative analysis of total chl content in leaves of each genotype shown in (B). Data are mean ± SEM (n>4). *** p < 0.001 (t-test).
(TIF)
Detached third and fourth Leaves from 4-week-old plants were treated with 100 μL/L ethylene for various times. RT-qPCR was performed to quantify the mRNA levels of each gene. ACT2 was used as an internal control to normalize different samples. The mRNA levels of each corresponding gene in WT at 0 hr were arbitrarily set to 1. Data are mean ± SEM from 3 biological replicates with technical duplicates for each.
(TIF)
Four-week-old NYE1iox transgenic line was sprayed with 15 μM DEX or 0.05% Methanol (mock) and incubated for two days. The transcript level of NYE1 in the third and fourth leaves was examined by RT-qPCR. ACT2 was used as an internal control for normalization. The transcript level of NYE1 with mock treatment was arbitrarily set to 1. Data are mean ± SEM of 3 biological replicates. ** p < 0.01 (t-test).
(TIF)
qRT-PCR analysis of the ACS2 transcript levels in third and fourth leaves of 4-week-old WT, nac2-1, and ORE1ox with 100 μL/L ethylene treatment for 4 d. The ACS2 transcript level in WT was arbitrarily set to 1. Data are mean ± SEM from 2 biological replicates (one-way ANOVA). Levels not connected by same letter are significantly different. p < 0.01.
(TIF)
(A) Detached third and fourth rosette leaves from 4-week-old WT, nyc1, and pao plants treated with 100 μL/L ethylene for 3 d. (B) Quantitative analysis of total chl content in leaves of each genotype shown in (A). Data are mean ± SEM (n = 3). * p < 0.05 (t-test).
(TIF)
(XLSX)
Acknowledgments
We thank Prof. Feng Ming (Fudan University, China) for kindly providing the pHB and pHB-ORE1 vectors and ORE1ox transgenic line. We thank Prof. Bernd Mueller-Roeber (University of Potsdam, Germany) for kindly providing the 35S:ORE1-GFP transgenic line. We are grateful to Prof Jianmin Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing the EIN3iox transgenic line. We thank Dr. Wei Zhang (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for assistance with ethylene quantification. We also thank Prof. Hongwei Guo and Dr. Zhonghai Li for helpful advice (Peking University, China), and Prof. Chi-Kuang Wen (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for helpful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by State Key Laboratory of Genetic Engineering and Fudan University to XZho and BK, and by Science and Technology Commission of Shanghai Municipality (13JC400900, http://www.stcsm.gov.cn/) and National Natural Science Foundation of China (3117021, http://www.nsfc.gov.cn/publish/portal0/default.htm). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gan S, Amasino RM. Making Sense of Senescence (Molecular Genetic Regulation and Manipulation of Leaf Senescence). Plant Physiology. 1997;113: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58: 115–136. [DOI] [PubMed] [Google Scholar]
- 3. Guo Y, Gan SS. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012;35: 644–655. 10.1111/j.1365-3040.2011.02442.x [DOI] [PubMed] [Google Scholar]
- 4. Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014;5: 4636 10.1038/ncomms5636 [DOI] [PubMed] [Google Scholar]
- 5. Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009;57: 120–131. 10.1111/j.1365-313X.2008.03670.x [DOI] [PubMed] [Google Scholar]
- 6. Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem. 2009;284: 17449–17456. 10.1074/jbc.M109.008912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell. 2011;23: 3442–3453. 10.1105/tpc.111.089714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, et al. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell. 2009;21: 767–785. 10.1105/tpc.108.064089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren G, Zhou Q, Wu S, Zhang Y, Zhang L, Huang J, et al. Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. J Integr Plant Biol. 2010;52: 496–504. 10.1111/j.1744-7909.2010.00945.x [DOI] [PubMed] [Google Scholar]
- 10. Pruzinska A, Tanner G, Anders I, Roca M, Hortensteiner S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci U S A. 2003;100: 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andres CB, et al. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell. 2012;24: 507–518. 10.1105/tpc.111.089474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, et al. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiology. 2007;144: 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kusaba M, Tanaka A, Tanaka R. Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth Res. 2013;117: 221–234. 10.1007/s11120-013-9862-x [DOI] [PubMed] [Google Scholar]
- 14. Jibran R, Hunter DA, Dijkwel PP. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol. 2013;82: 547–561. 10.1007/s11103-013-0043-2 [DOI] [PubMed] [Google Scholar]
- 15. van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R. Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology. 2006;141: 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jing HC, Schippers JH, Hille J, Dijkwel PP. Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J Exp Bot. 2005;56: 2915–2923. [DOI] [PubMed] [Google Scholar]
- 17. Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, et al. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics. 2009;183: 979–1003. 10.1534/genetics.109.107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94: 261–271. [DOI] [PubMed] [Google Scholar]
- 19. Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72: 427–441. [DOI] [PubMed] [Google Scholar]
- 20. Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- 21. Bisson MM, Bleckmann A, Allekotte S, Groth G. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J. 2009;424: 1–6. 10.1042/BJ20091102 [DOI] [PubMed] [Google Scholar]
- 22. Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109: 19486–19491. 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, et al. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22: 1613–1616. 10.1038/cr.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338: 390–393. 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grbic V, Bleecker AB. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995;8: 595–602. [Google Scholar]
- 26. Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12: 527–535. [DOI] [PubMed] [Google Scholar]
- 27. Li Z, Peng J, Wen X, Guo H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013;25: 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323: 1053–1057. 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- 29. Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue GP, Droge-Laser W, et al. NAC Transcription Factor ORE1 and Senescence-Induced BIFUNCTIONAL NUCLEASE1 (BFN1) Constitute a Regulatory Cascade in Arabidopsis. Mol Plant. 2013;6: 1432–1452. [DOI] [PubMed] [Google Scholar]
- 30. Farage-Barhom S, Burd S, Sonego L, Mett A, Belausov E, Gidoni D, et al. Localization of the Arabidopsis senescence- and cell death-associated BFN1 nuclease: from the ER to fragmented nuclei. Mol Plant. 2011;4: 1062–1073. 10.1093/mp/ssr045 [DOI] [PubMed] [Google Scholar]
- 31. Kim HJ, Hong SH, Kim YW, Lee IH, Jun JH, Phee BK, et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot. 2014;65: 4023–4036. 10.1093/jxb/eru112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, et al. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A. 2009;106: 21431–21436. 10.1073/pnas.0907670106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55: 821–831. 10.1111/j.1365-313X.2008.03551.x [DOI] [PubMed] [Google Scholar]
- 34. Kosugi S, Ohashi Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamasaki K, Kigawa T, Inoue M, Yamasaki T, Yabuki T, Aoki M, et al. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J Mol Biol. 2005;348: 253–264. [DOI] [PubMed] [Google Scholar]
- 36. Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olsen AN, Ernst HA, Leggio LL, Skriver K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Science. 2005;169: 785–797. [DOI] [PubMed] [Google Scholar]
- 38. Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010;62: 250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- 39. Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, et al. ABI3 controls embryo degreening through Mendel's I locus. Proc Natl Acad Sci U S A. 2013;110: E3888–3894. 10.1073/pnas.1308114110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B. Age-Triggered and Dark-Induced Leaf Senescence Require the bHLH Transcription Factors PIF3, 4, and 5. Mol Plant. 2014;7: 1776–1787. 10.1093/mp/ssu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301: 653–657. [DOI] [PubMed] [Google Scholar]
- 42. Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21: 2527–2540. 10.1105/tpc.108.065193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11: 605–612. [DOI] [PubMed] [Google Scholar]
- 44. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16: 735–743. [DOI] [PubMed] [Google Scholar]
- 45. Li J, Chen X, Luo LQ, Yu J, Ming F. [Functions of ANAC092 involved in regulation of anther development in Arabidopsis thaliana]. Yi Chuan. 2013;35: 913–922. [DOI] [PubMed] [Google Scholar]
- 46. Zhang W, Wen CK. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol Biochem. 2010;48: 45–53. 10.1016/j.plaphy.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 47. Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11: 587–596. 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45: 616–629. [DOI] [PubMed] [Google Scholar]
- 49. Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2: 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Whole plants of 4-week-old ein3 eil1 mutant showed a stay-green phenotype compared to the wild type (WT) with 100 μL/L ethylene treatment for 4 d. (B) Detached third and fourth rosette leaves from 4-week-old WT and ein3 eil1 plants treated with 100 μL/L ethylene for 4 d. (C) Quantitative analysis of total chl content in leaves of each genotype shown in (B). Data are mean ± SEM (n>4). *** p < 0.001 (t-test).
(TIF)
The expression level of each corresponding gene in the WT at 0 hr was set to 1. The scales of y-axes are consistent with that in Fig 2B. Data are mean ± SEM of 3 biological replicates.
(TIF)
(A) Whole plants of 4-week-old nac2-1 mutant showed a stay-green phenotype compared to the WT with 100 μL/L ethylene treatment for 4 d. (B) Detached third and fourth rosette leaves from 4-week-old WT and nac2-1 plants treated with 100 μL/L ethylene for 4 d. (C) Quantitative analysis of total chl content in leaves of each genotype shown in (B). Data are mean ± SEM (n>4). *** p < 0.001 (t-test).
(TIF)
Detached third and fourth Leaves from 4-week-old plants were treated with 100 μL/L ethylene for various times. RT-qPCR was performed to quantify the mRNA levels of each gene. ACT2 was used as an internal control to normalize different samples. The mRNA levels of each corresponding gene in WT at 0 hr were arbitrarily set to 1. Data are mean ± SEM from 3 biological replicates with technical duplicates for each.
(TIF)
Four-week-old NYE1iox transgenic line was sprayed with 15 μM DEX or 0.05% Methanol (mock) and incubated for two days. The transcript level of NYE1 in the third and fourth leaves was examined by RT-qPCR. ACT2 was used as an internal control for normalization. The transcript level of NYE1 with mock treatment was arbitrarily set to 1. Data are mean ± SEM of 3 biological replicates. ** p < 0.01 (t-test).
(TIF)
qRT-PCR analysis of the ACS2 transcript levels in third and fourth leaves of 4-week-old WT, nac2-1, and ORE1ox with 100 μL/L ethylene treatment for 4 d. The ACS2 transcript level in WT was arbitrarily set to 1. Data are mean ± SEM from 2 biological replicates (one-way ANOVA). Levels not connected by same letter are significantly different. p < 0.01.
(TIF)
(A) Detached third and fourth rosette leaves from 4-week-old WT, nyc1, and pao plants treated with 100 μL/L ethylene for 3 d. (B) Quantitative analysis of total chl content in leaves of each genotype shown in (A). Data are mean ± SEM (n = 3). * p < 0.05 (t-test).
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.