Abstract
Xylella fastidiosa (X. fastidiosa) infects a wide range of plant hosts and causes economically serious diseases, including Pierce's Disease (PD) of grapevines. X. fastidiosa biocontrol strain EB92-1 was isolated from elderberry and is infectious and persistent in grapevines but causes only very slight symptoms under ideal conditions. The draft genome of EB92-1 revealed that it appeared to be missing genes encoding 10 potential PD pathogenicity effectors found in Temecula1. Subsequent PCR and sequencing analyses confirmed that EB92-1 was missing the following predicted effectors found in Temecula1: two type II secreted enzymes, including a lipase (LipA; PD1703) and a serine protease (PD0956); two identical genes encoding proteins similar to Zonula occludens toxins (Zot; PD0915 and PD0928), and at least one relatively short, hemagglutinin-like protein (PD0986). Leaves of tobacco and citrus inoculated with cell-free, crude protein extracts of E. coli BL21(DE3) overexpressing PD1703 exhibited a hypersensitive response (HR) in less than 24 hours. When cloned into shuttle vector pBBR1MCS-5, PD1703 conferred strong secreted lipase activity to Xanthomonas citri, E. coli and X. fastidiosa EB92-1 in plate assays. EB92-1/PD1703 transformants also showed significantly increased disease symptoms on grapevines, characteristic of PD. Genes predicted to encode PD0928 (Zot) and a PD0986 (hemagglutinin) were also cloned into pBBR1MCS-5 and moved into EB92-1; both transformants also showed significantly increased symptoms on V. vinifera vines, characteristic of PD. Together, these results reveal that PD effectors include at least a lipase, two Zot-like toxins and a possibly redundant hemagglutinin, none of which are necessary for parasitic survival of X. fastidiosa populations in grapevines or elderberry.
Introduction
Xylella fastidiosa (X. fastidiosa) is a fastidious, insect-vectored, plant xylem-limited bacterial pathogen that causes a variety of diseases in a wide range of plant species [1]. Among the most damaging X. fastidiosa diseases are Pierce’s disease (PD) of Vitis viniferea grapevines, leaf scorch of almonds, and citrus variegated chlorosis (CVC). In grapevines, X. fastidiosa cells multiply and spread widely from the site of infection to colonize the xylem of susceptible grapes. The symptoms of PD include leaf scorch and xylem occlusion, resulting in the eventual death of the vine [1]. Among the earliest symptoms caused by X. fastidiosa on grape is a characteristic leaf scorch that, although similar to symptoms of water stress, often occurs in bands, which is suggestive of either toxin or pathogenicity effector activity [2–4].
Genomic comparisons of various X. fastidiosa genomes to those of other bacterial pathogens have resulted in the identification of a limited number of potential virulence factors secreted through Type I, II and V secretion systems [5–7]. X. fastidiosa lacks genes encoding Type III secretion system machinery as well as Type III effectors. The Type I secretion system is involved in processes such as drug resistance via multidrug efflux as well as secretion of effectors [8]. X. fastidiosa tolC, which encodes the predicted outer membrane component of Type I secretion, was shown to be both functional and necessary for X. fastidiosa pathogenicity and in planta survival of X. fastidiosa in V. vinifera grapevines [9]. X. fastidiosa genomes include multiple genes encoding predicted type I effectors, including repeats in toxin (Rtx) family members, hemolysins and bacteriocins.
X. fastidiosa has also been shown to have all essential genes required for Type II secretion [5]. Proteins secreted by this system include proteases, cellulases, pectinases, phospholipases, lipases, and toxins. In general, extracellular enzymes are required for the hydrolysis of different components of the plant cell wall and contribute to cell damage and disease [10]. Lipases (EC 3.1.1.3) are lipolytic enzymes which constitute a special class of carboxylester hydrolases (EC 3.1.1) capable of releasing long-chain fatty acids from natural water-insoluble esters (lipids). Lipases typically carry a LIP domain and are secreted pathogenicity factors of both bacterial and fungal pathogens of animals and plants [11–14]. There are three putative lipases, PD1702, PD1703 and PD1211, found in the Temecula1 genome, but no reports of any potential role in pathogenicity of X. fastidiosa.
Type V secretion systems include the autotransporter pathway (type Va or AT-1), the two partner secretion pathway (type Vb), and the type Vc pathway (AT-2) [15]. X. fastidiosa proteins reported to be secreted through this secretion system include two comparatively large hemagglutinin-like proteins, PD2118 (3457 aa) and PD1792 (3377 aa), through type Vb pathway [16] and an AT-1 autotransporter (PD0528) through type Va pathway [7]. The two Type V secreted hemagglutinin-like proteins (PD1792 and PD2118) contributed to X. fastidiosa biofilm formation and colonization and were important for insect vector transmission [16–18]. Knockout mutations of either of these large hemagglutinins results in hypervirulence and greatly increased PD symptoms [16]. There are another four smaller predicted hemagglutinin-like proteins (PD0986, PD2108, PD2110 and PD2116; 376–438 aa) found in X. fastidiosa Temecula1 but at least one (PD0986) appeared missing in X. fastidiosa EB92-1 [19]. The potential role or possible functionality of these four, evidently redundant and smaller hemagglutinin proteins in X. fastidiosa pathogenicity and potential for secretion are unknown.
X. fastidiosa EB92-1 is a well characterized and effective PD biocontrol strain isolated from elderberry which can be inoculated in a single location in a grapevine and the entire plant is protected from PD for years [20]. EB92-1 causes only very slight symptoms on grapevines, even under ideal conditions, thus providing a natural platform strain to test hypotheses relating to pathogenicity effector function. The purpose of this study was to utilize this infectious but essentially nonpathogenic strain EB92-1 to attempt to identify pathogenicity effectors present in PD causing strain Temecula1 but missing from EB92-1. In this study, three of these missing genes, PD1703 (LipA), PD0928 (Zot) and PD0986 (hemagglutinin), were functionally tested and all three were found to enhance PD symptoms in EB92-1 on V. vinifera grapevines.
Materials and Methods
Bacterial strains, plasmids and media
All plasmids, X. fastidiosa strains, Xanthomonas citri B21.2 and E. coli strains used are listed in Table 1. X. fastidiosa cells were grown at 28°C in PD3 medium: tryptone (4 g/L), soytone peptone (2 g/L), trisodium citrate (1 g/L), disodium succinate (1 g/L), MgSO4·7H2O (1 g/L), K2HPO4 (1.5 g/L), KH2PO4 (1 g/L), and 0.1% (wt/vol) Hemin chloride (10 mL/L); pH was adjusted to 7.0, and soluble potato starch (2 g/L) and, optionally, agar (15 g/L) were added prior to sterilization by autoclave for 20 min [21]. Xanthomonas cells were routinely grown on nutrient Broth (Becton, Dickinson and Co., Sparks, MS, USA) at 28°C. E. coli strains were grown on Luria-Bertani medium at 37°C.
Table 1. Bacterial strains and plasmids used.
| Strains and plasmids | Relevant genotype or description a | Source or reference b |
|---|---|---|
| Xylella fastidiosa | ||
| Temecula1 | Wild type | [22] |
| EB92-1 | Biocontrol strain | [20] |
| Escherichia coli | ||
| Mach1-T1R | F- φ80(lacZ)ΔM15 ΔlacX74 hsdR(rk ‒mk +) ΔrecA1398 endA1 tonA | Invitrogen |
| BL21(DE3) | F–ompT hsdSB(rB–, mB–) gal dcm (DE3) | Invitrogen |
| Xanthomonas | ||
| B21.2 | X.citri 3213T (pthA::Tn5-gusA), SprKmrTcr, marker-exchanged mutant | [23] |
| Plasmids | ||
| pBBR1MCS-5 | Rep Bordetella, lacZ, Gmr | [24] |
| pGEM-T | Apr, pUC ori, lacZ | Promega Corp. |
| pET-27b(+) | Km, f1 and pBR322 ori, | EMD Millipore |
| pSZ24 | pGEM-T with 1982 bp PCR product of PD1703 including native promoter region, Apr | This study |
| pSZ25 | pGEM-T with 2286 bp PCR product of PD1702 including native promoter region, Apr | This study |
| pSZ26 | pBBR1MCS-5 with ApaI/ClaI fragment from pSZ24, Gmr | This study |
| pSZ28 | pBBR1MCS-5 with ApaI/ClaI fragment from pSZ25, Gmr | |
| pSZ40 | pGEM-T with 1446 bp PCR product of PD0928, including native promoter region, Apr | This study |
| pSZ41 | pBBR1MCS-5 with BamHI/SalI fragment from pSZ40, Gmr | This study |
| pPC2.5 | pGEM-T with 1194 bp PCR product of PD0986, including native promoter region, Apr | This study |
| pPC3.1 | pBBR1MCS-5 wtih EcoRI/ BamHI fragment from pSZ40, Gmr | This study |
| pSZ35 | pGEM-T with 1170 bp PCR product of revised PD1703 (lipA), without secretion signal. Apr | This study |
| pSZ37 | pET-27b(+) with MscI/NheI fragment from pSZ35. Kmr | This study |
a Gmr, Apr, and Kmr = gentamycin, ampicillin, and kanamycin resistant, respectively; PCR = polymerase chain reaction.
b Sources: Invitrogen Life Technologies, Carlsbad, CA, USA, Promega Corporation, Madison, WI, USA, EMD Millipore, Billerica, MA, USA,
Molecular and bioinformatics techniques
DNA primers used were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). Total DNA was extracted from Temecula1 and EB92-1 cells using the GenElute Bacterial Genomic DNA Kit (Sigma, St. Louis, MO, USA). Plasmid DNA was extracted from E. coli using the QIAGEN (Valencia, CA, USA) QIAprep Spin Miniprep kit. PCR purification and gel extraction were done using the QIAGEN QIAquick PCR purification and Gel Extraction kits. DNA fragment ligations were performed using pGEM-T Vector Systems (Promega Corp, Madison, WI, USA). Protein BLAST (Basic Local Alignment Search Tools; BLASTP) was used to search bacterial protein sequence databases (http://blast.ncbi.nlm.nih.gov). Secretion signals were predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP). Predictions of open reading frames (ORF) were conducted using GeneMark (http://exon.gatech.edu/GeneMark/). The phylogenetic tree was generated using AlignX, Vector NTI Advance 10 (Invitrogen, Carlsbad, CA, USA) with the default set of multiple alignment parameters.
Construction of shuttle and expression vectors
Primers used for PCR amplification of the target genes in this study are listed in Table 2. For Revised PD1703, a 1982 bp PCR fragment was amplified from Temecula1 genomic DNA using primers PDLPwp-F and PDLP03-R to amplify the region covering the PD1703 corrected putative ORF (including a predicted secretion signal leader sequence missing from the original annotation) and including its 691 bp native promoter region. This fragment was cloned into pGEM-T, resulting in pSZ24. This construct was verified by sequencing and the entire gene plus promoter was recloned using ApaI and ClaI into pBBR1MCS-5 (downstream from the lacZ promoter), to make pSZ26. Similarly, the constructs containing the native promoters and the ORF regions of revised PD1702, PD0928 and PD0986 were cloned into pGEM-T to form pSZ24, pSZ40 and pPC2.5, respectively (Table 1). The inserts from these three constructs were recloned into pBBR1MCS-5 using appropriate enzymes as listed in Table 1, resulting in pSZ28, pSZ41 and pPC3.1 respectively. For protein expression of revised PD1703 in E. coli BL21(DE3), the corrected putative ORF was amplified using primers PD1703-AF and PD1703-AR and cloned into pGEM-T without its predicted native secretion signal to form pSZ35. The sequence-verified insert was cloned into pET-27b (+) to create a translational fusion with the pelB leader to form pSZ37.
Table 2. Primers used in this study.
| Primer name | Sequence (5’-3’) | Gene target |
|---|---|---|
| PDLPwp-F | GGGCCCTGCCGCATTTGAGGCTGGC | PD1703 |
| PDLP03-R | ATCGATACGTTTGCAAACCCTATATCC | |
| PDLP02wp-F | GGGCCCGGATATAGGGTTTGCAAACGT | PD1702 |
| PDLP-R | ATCGATAGGGTACATTTACCAGACCG | |
| ZOT-F | GTCGACCTGGTTGGGCATTAATTGGG | PD0928 |
| ZOT-R | GGATCCTCATGGAAGCGACCCCGC | |
| PD0986-EcoRI-F2 | CGGAATTCAGGAAGCAGTGACGGGCTG | PD0986 |
| PD0986-BamHI-R | CGGGATCCTTAGTAGGGGGTGAGGGACT | |
| PD1703-AF | TGGCCAGCCGTGGGATGGTGATTAACAGA | PD1703 |
| PD1703-AR | GCTAGC CTAACGTTTCTTCTCTAGAAAC | |
| RST31 | GCGTTAATTTTCGAAGTGATTCGATTGC |
X. fastidiosa
marker gene [25] |
| RST33 | CACCATTCGTATCCCGGTG |
Preparation and Electroporation of X. fastidiosa and X citri cells
X. fastidiosa cells were grown on PD3 agar plates at 28°C. X. fastidiosa liquid cultures were grown in liquid PD3 with rotary shaking at 28°C at 120 rpm. To make X. fastidiosa cells competent for electroporation, 1 mL of 5 day old liquid X. fastidiosa culture was transferred into 35 mL of fresh PD3 broth and grown until the OD600 of the culture reached ~0.1. These liquid cultures (36 mL) were transferred to flasks containing 250 mL of PD3 broth and grown to OD600 = 0.1. The cell suspensions were chilled on ice for 30 min and cells recovered by centrifugation at 4°C for 15 min at 4,000 rpm. Cells were washed successively in 250 mL, then in 125 mL ice-cold sterile distilled water and then in 10 mL of ice-cold 10% glycerol; each wash was followed by centrifugation and resuspension by gentle hand shaking or tapping. Finally, the cells were resuspended in 1 mL of ice-cold 10% glycerol. Forty μL aliquots of concentrated cells were placed in 1.5 ml sterile tubes kept at -20°C before use and the aliquots were frozen in liquid nitrogen and either used immediately for electroporation or stored at -80°C for later use. Plasmids used for electroporation were treated with CpG Methyltransferase (New England Biolabs, Ipswich, MA, USA) [26]. One μL TypeOne Restriction Inhibitor (EPICENTRE, Madison, WI, USA) and less than 5 μL of 0.5 μg~1 μg treated plasmid DNA was added to competent cells thawed and kept on ice and mixed by pipetting. The X. fastidiosa cell-DNA mixture was transferred into 1mm electroporation cuvettes (pre-chilled on ice for 20 min before use) and electroporated at 1800 Volts (time constant usually 4.8~6.2 ms). One mL of PD3 broth without antibiotics was added into cuvette and mixed quickly by pipetting, then all cultures were transferred into 5 mL sterile tubes, sealed with parafilm and shaken at 120 rpm, 28°C for 24hrs. Cultured cells were spread (100 μL per plate) on solid PD3 medium with gentamycin at10 μg/mL. Plates were incubated at 28°C for 7–12 days and single colonies were picked for PCR analysis. X. citri B21.2 electrocompetent cells were prepared and electroporation conducted as described by Swarup et al [23].
Lipase indicator assays on plates
An in vitro lipase assay was conducted using Tween 20 as the substrate and 0.01% Victoria Blue B as indicator, essentially as described by Samad et al [27]. Agar plates containing the substrate and indicator were poured and wells were created by removal of agar with a sterile cork borer (0.7 cm diam.). For native secreted lipase assays, 50 μL supernatants from centrifuged E. coli, X. fastidiosa and X. citri cell cultures grown to late mid-log phase (OD600 = 0.7) were added to the wells and the plates were incubated overnight at 37°C for E. coli and 28°C for X. fastidiosa and X. citri supernatants. For recombinant, over-expressed lipase assays, crude protein was extracted from E. coli BL2 (DE3) carrying pET27b and expressing PD1703 using a QIAGEN (Valencia, CA, USA) Ni-NTA Spin Kit. Protein was resuspended in 200 μL of supplied buffer and quantified with a NanoDrop 2000 spectrophotometer (NanoDropTechnologies, Wilmington, DE, USA); some protein remained insoluble but was applied as a suspension and labeled "total protein". To recover soluble protein, total protein was centrifuged at 10,000 X g for 30 min at 4°C to pellet the insoluble protein, and the supernatant was used and labeled as "supernatant". Both 50 μL of total and 50 μL of supernatant protein were tested in the agar plate assays for recombinant, over-expressed lipase.
Hypersensitive reaction (HR) assays of crude protein extracts
Nicotiana tabacum cv. SR1 (also called Petite Havana) plants were maintained at 28°C with a 12-h photoperiod. At 4 to 5 weeks after germination from seeds, three nearly fully expanded upper leaves were pressure infiltrated through the stomata using the blunt end of a 1 ml tuberculin syringe gently pressed against the abaxial leaf surface. Approximately 10 μL of crude protein extracts from E. coli BL21 (DE3) carrying empty vector (pET-27b) or expressing revised PD1703 (pET-27b::lipA, pSZ37) were infiltrated into a zone of about 1.5 cm diameter. Similarly, three nearly fully expanded leaves of ca. 2' tall citrus trees in pots (Citrus sinensis L. Osbeck cv. Hamlin) were infiltrated with 10 μL of the same crude protein extracts. All the inoculation zones on inoculated tobacco or citrus plants were circled using a black marker to indicate the extent of infiltration.
X. fastidiosa pathogenicity assays on grapevines
For X. fastidiosa pathogenicity assays, grape (Vitis vinifera) cv. Carignane plants were inoculated by needle puncture as described [9]. In brief, 10μL droplets of X. fastidiosa bacterial suspensions (OD600 = 0.25) in SCP buffer (trisodium citrate, 1 g/L; disodium succinate, 1 g/L; MgSO4·7H2O, 1 g/L; K2HPO4, 1.5 g/L; and KH4PO4, 1 g/L; pH 7.0) were applied on opposite sides of each of 4–5 internodes of ca. 3 ft high grapevines in pots, starting with the second internode from the base. A sterile, tuberculin needle was used to puncture the stems to a depth of 1 to 3 mm through each of the droplets. This resulted in the droplets of suspension being drawn into the stem xylem. Plants were not watered for at least 36 hours prior to inoculation. Each experiment was conducted three times with 4–12 replications in each experiment. Inoculated plants were maintained in an air conditioned green house and carefully observed for the appearance of symptoms. Observations were recorded from the time the first visible symptoms appeared (ca. 4–6 weeks post inoculation) and continued for another 2 months. Disease severity was quantified and expressed as a % of diseased leaves (including bare petioles and bare nodes) on each inoculated plant by measuring the number of symptomatic leaves, the number of bare petioles and the number of bare nodes on each plant, and dividing by the total number of leaves (asymptomatic or symptomatic), bare petioles and bare nodes per plant.
Results
PCR and sequence evidence of 5 pathogenicity genes missing in EB92-1
Suspected pathogenicity genes found in Temecula1 that appeared to be missing from EB92-1 [19] were further investigated by designing PCR primers internal to and outside of the coding sequences (CDSs) of PD1703 (LipA), PD0956 (Serine protease), PD0915 (Zot), PD0928 (Zot) and PD0986 (hemagglutinin-like) (S1 Table). Two sets of primers were designed to attempt detect these genes in EB92-1; one set was designed internal to the CDS of each gene (92% of predicted EB92-1 proteins had more than 99% identity with Temecula1 proteins [19]) and at least one additional set was designed external to the CDS in Temecula1. PCR amplification results using DNA extracted from both Temecula1 and EB92-1 are shown (S1 Fig). In the cases of PD1703 and PD0956, the regions outside of the apparently missing genes were readily found based on gaps predicted by Mauve 2.3.1 within the existing EB92-1 contigs deposited in Genbank (Accession # AFDJ00000000.1). The regions of EB92-1 amplified by PCR primer sets PDLPwp-F / PDLP-R and XFEB114-F / XFEB114-R1 were sequenced to confirm that PD1703 and PD0956, respectively, were truly missing, at least from their locations in Temecula1 (additional sequence data not shown). Primers internal to the coding regions of PD1703 and PD0956 failed to amplify any products from EB92-1.
In the cases of PD0915 and PD0928, which are 100% identical in coding sequence and appear in nearly identical, 4.4 kb tandem direct repeats spaced ca. 4 kb apart in Temecula1, primer sets internal and external to both genes failed to amplify any products from EB92-1. Indeed, the entire 12.7 kb region containing genes PD0911–PD0929 appeared missing in EB92-1 (some data not shown). Similarly, in the case of PD0986, both the internal primer set and the external (2.0 kb) primer set failed to amplify; both flanking genes PD0985 and PD0987 appeared rearranged based on strong multiple BLASTn hits (3 contigs each) in EB92-1 (some data not shown).
PD1703 ORF analysis and re-annotation
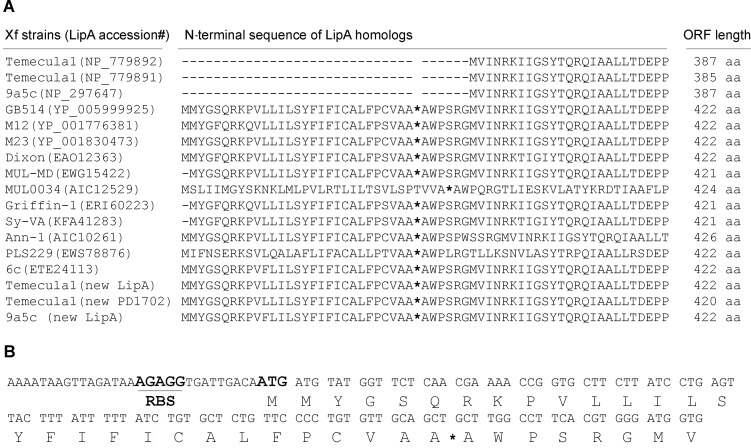
Protein BLAST (Basic Local Alignment Search Tools) revealed that the X. fastidiosa Temecula1 PD1703 product, which is missing from X. fastidiosa EB92-1, matched many proteins from various bacterial species annotated as secreted lipases and possessing a LIP domain (Pfam03583), including PD1702 of Temecula1 (Fig 1). The putative open reading frame (ORF) of PD1703 (GenBank accession no: NP_779892), located at nucleic acid positions 1,983,742 to 1,984,905 in the Temecula1 genome (GenBank accession no: NC_0045560) [5], encodes a predicted 387-amino-acid-long protein. The DNA sequence (nucleic acid positions 1,983,742 to 1,985,701) from X. fastidiosa Temecula1, which is annotated to contain the 3’ end of PD1704, was predicted using GeneMark to more likely belong to PD1703. This predicted LipA protein [labeled as “Temecula1 (new LipA)”, Fig 1A] is encoded by a putative ORF located in Temecula1 at nucleotides 1,983,742 to 1,985,010, with a putative ribosome biding site (RBS) “AGAGG” located 9 base pairs upstream of the ATG translational start codon. This revised LipA ORF encodes a 422-amino-acid-long protein and possesses a LIP domain (Pfam03583) between amino acid positions 132 and 203 (data not shown). Its Type II secretion signal was predicted using SignalP 3.0 with a probability of 0.993 with a likely cleavage site between positions 30th and 31st amino acids, entirely consistent with the other X. fastidiosa LipA homologs (Fig 1B). Similarly, Temecula1 PD1702 and XF0357 from X. fastidiosa 9a5c were found to likely be similarly mis-annotated. The N-terminal region of these revised predicted ORFs (labeled as "new PD1702" and "new LipA" are shown in Fig 1A). The revised predicted ORFs of PD1703 and PD1702 were used for further studies and functional verification in this work.
Fig 1. The N-terminal regions of LipA proteins from X. fastidiosa.
(A) Comparative sequences of 13 X. fastidiosa strains, with accession numbers, including citrus CVC strain 9a5c, PD strain GB514, almond leaf scorch strains M12, M23 and Dixon, mulberry leaf scorch strains MUL-MD and MUL0034, oak leaf scorch strain Griffin-1, sycamore leaf scorch strain Sy-VA, oleander leaf scorch strain Ann-l, pear leaf scorch strain PLS229 and coffee leaf scorch strain 6c, as well as three re-annotations (two from Temecula1 and one from CVC). The most likely signal peptide cleavage sites for each protein are indicated by asterisks. (B) Portion of the DNA sequence including 5’ untranslated region of revised PD1703. The putative RBS and the first start codon are indicated using bold and enlarged fonts.
Revised PD1703, but not PD1702, exhibited functional, secreted lipase activity in X. citri, E. coli and X. fastidiosa EB92-1
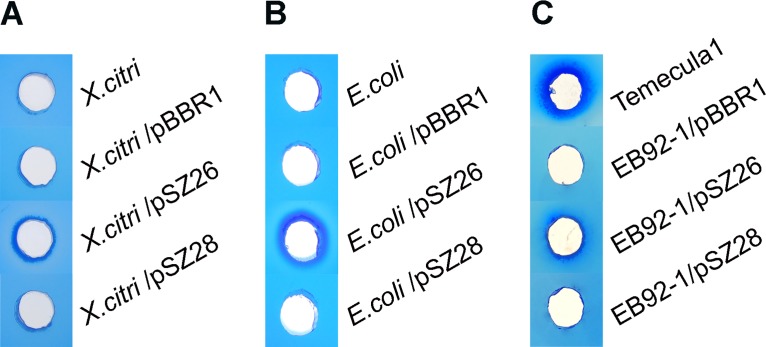
The revised putative PD1703 ORF with its native promoter region (691 bp) was cloned into pBBR1MCS-5 (and downstream from the lacZ promoter). Similarly, the revised putative PD1702 ORF with its native promoter region (1024 bp) was cloned into pBBR1MCS-5. Both constructs were used to transform X. citri B21.2, E. coli Mach1-T1R and X. fastidiosa EB92-1. The crude, cell free culture supernatants were assayed for lipase activity, without purification or concentration. As shown in Fig 2, only culture supernatants from cells carrying the revised PD1703, but not the revised PD1702, exhibited lipase activity, and in all three assayed strains (X. fastidiosa, E. coli and X. citri). Other than these, only culture supernatant from the positive control X. fastidiosa Temecula1 exhibited lipase activity (Fig 2C). Culture supernatants from the same strains or the same strains carrying the empty vector pBBR1MCS-5 (labeled as “pBBR1”) or PD1702 (labeled as "pSZ28") were all negative for lipase activity (Fig 2A, 2B and 2C).
Fig 2. Secreted lipase activity assays of culture supernatants from X. citri B21.2, E. coli Mach1-T1R and X. fastidiosa strains Temecula1 and EB92-1.
(A) Lipase assay of culture supernatants from untransformed X. citri B21.2 and transformants with empty vector pBBR1MCS-5 (pBBR1), revised PD1703 (pSZ26) and with revised PD1702 (pSZ28). (B) Lipase assay of culture supernatants from untransformed E. coli Mach1-T1R strains and transformants. (C) Lipase assay of culture supernatants from untransformed X. fastidiosa Temecula1 and EB92-1 transformants. Photos were taken at 24 hours after plating. The results shown are representative of data from three independent replicates.
Crude protein extracts of revised PD1703 overexpressed in E. coli exhibited lipase activity and induced HR in tobacco and citrus
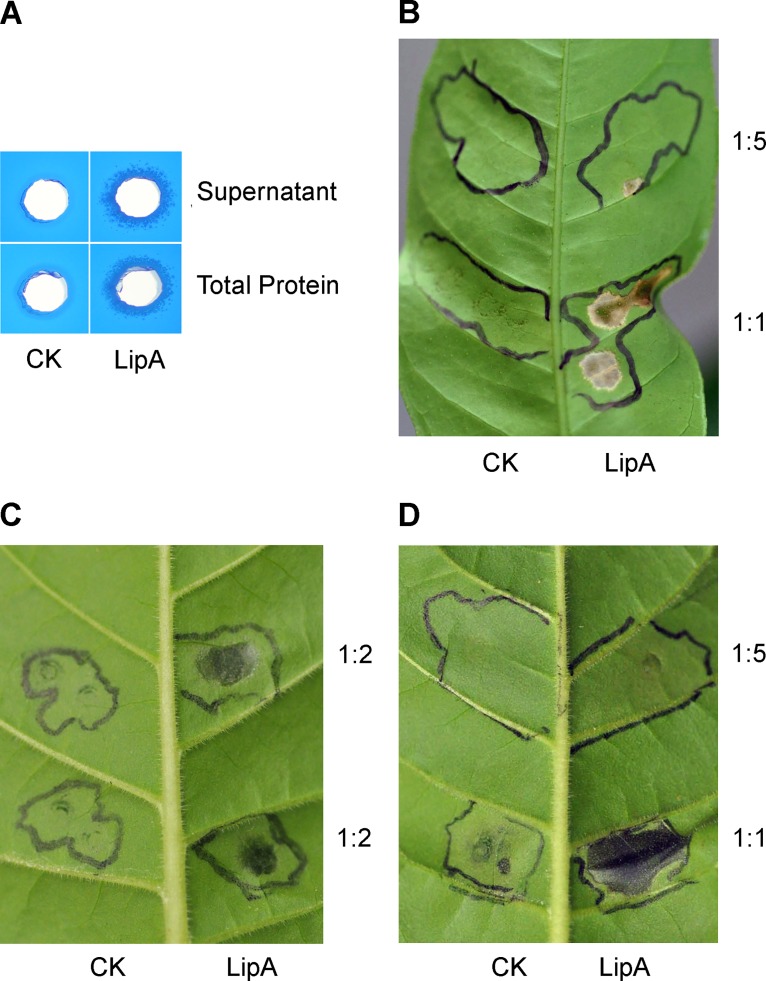
Crude protein was extracted from induced E. coli BL2 (DE3) cultures carrying pSZ37 (revised PD1703, LipA) and from empty pET27b vector control cells. SDS-PAGE gel analysis indicated that the LipA protein was induced, and much of it was insoluble (S2 Fig). Both "Total Protein" (30 mg/mL) extracts from E. coli/pSZ37 cultures expressing LipA as well as centrifuged, clarified supernatants ("Supernatants") from these extracts exhibited lipase activity in these assays, whereas slightly higher levels of "Total Protein" (36 mg/mL) extracted from the empty vector pET27b did not (Fig 3A).
Fig 3. Lipase activity and plant response assays using crude protein extracts from revised PD1703 (LipA) overexpressed in E. coli.
(A) Lipase activity assays of PD1703 recombinant crude protein extracts from E. coli BL21 (DE3) strains. Fifty μL of crude protein from E. coli BL21 (DE3) with empty vector pET-27b (labeled as “CK”, 36 mg/mL) or expressing pSZ37 (labeled as "Total Protein", 30 mg/mL) and applied as a suspension or clarified by centrifugation ("Supernatant") were applied in the agar assay wells. Photos were taken at 24 hours post plating. (B) Ten μL of undiluted crude protein (1:1) and 1:5 diluted extracts were inoculated into citrus leaves. Zones of inoculation were marked with black ink and the photo was taken 48 hours later (48 hpi). (C) Ten μL of 1:2 diluted crude protein extract (15–18 mg/mL total protein) was inoculated into tobacco leaves. Zones of inoculation were marked with black ink and the photo was taken 17 hours later (17 hpi). (D) Ten μL of undiluted crude protein (1:1) and 1:5 diluted extracts were inoculated into tobacco leaves. Zones of inoculation were marked with black ink and the photo was taken 48 hours later (48 hpi). Similar results were obtained in three independent experiments.
Ten μL of both "Total Protein" (30 mg/mL) and clarified supernatant extracts from E. coli pSZ37 ("LipA") cultures were injected into both citrus and tobacco leaves, along with total protein (36 mg/mL) and clarified supernatant extracts from E. coli empty vector ("CK") control cultures (refer Fig 3B, 3C and 3D). Both undiluted and 1:2 dilutions of the LipA extracts elicited a rapid HR that was visible at 14 hrs post inoculation (hpi) in tobacco (Fig 3C and 3D). Similarly, undiluted (1:1) LipA protein extracts inoculated into citrus (Fig 3B) also elicited a rapid HR that was visible at 14 hpi. A 1:5 dilution of crude LipA protein that was inoculated into both hosts caused a very slight necrotic response that was visible at 48 hpi.
Revised PD1703 (LipA), PD0928 (Zot), and PD0986 (hemagglutinin) all enhanced pathogenicity of EB92-1 on grapes
Revised PD1703 (LipA), PD0928 (Zot), and PD0986 (short hemagglutinin) were cloned with their native promoters in pBBR1MCS-5 (downstream from the lacZ promoter) and transformed into EB92-1 cells. EB92-1 transformants, including empty vector as a negative control, as well as Temecula1 as a positive control, were inoculated into V. vinifera Carignane grapevines. Visible symptoms of PD began to appear on plants inoculated with Temecula1 by 4–5 weeks post inoculation, and continued to develop up to 12 weeks. Most of the Temecula1 inoculated plants were killed by 12 weeks.
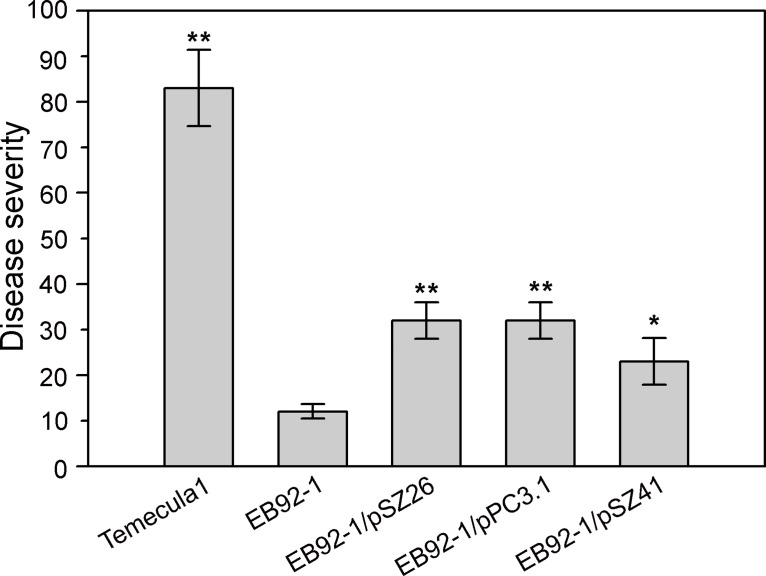
Plants inoculated with EB92-1/pSZ26 (LipA; refer Fig 4A) showed slightly delayed pathogenesis compared with Temecula1, and visible symptoms become evident by the end of six weeks. EB92-1/pSZ26 elicited typical leaf necrosis delimited with pinkish areas that were identical in appearance to plants inoculated with Temecula1. As the infection progressed the entire lamina underwent necrosis leading to defoliation (evident in Fig 4A with Temecula1). However, infection by EB92-1/pSZ26 progressed more slowly when compared to plants inoculated with Temecula1, the symptoms remained restricted to 9–10 internodes above the lowest point of inoculation, and disease severity (ca. 32%) was greatly reduced compared with Temecula1, where infections advanced to the tip of the vine and resulted in disease severity of 83% or more. Nevertheless, inoculations using EB92-1/pSZ26 resulted in disease symptoms affecting ca. 32% of the leaves, which was significantly higher (Student t-test, P<0.01) than the 12% levels observed with EB92-1 (EB92-1/pBBR1MCS-5 empty vector; Fig 5).
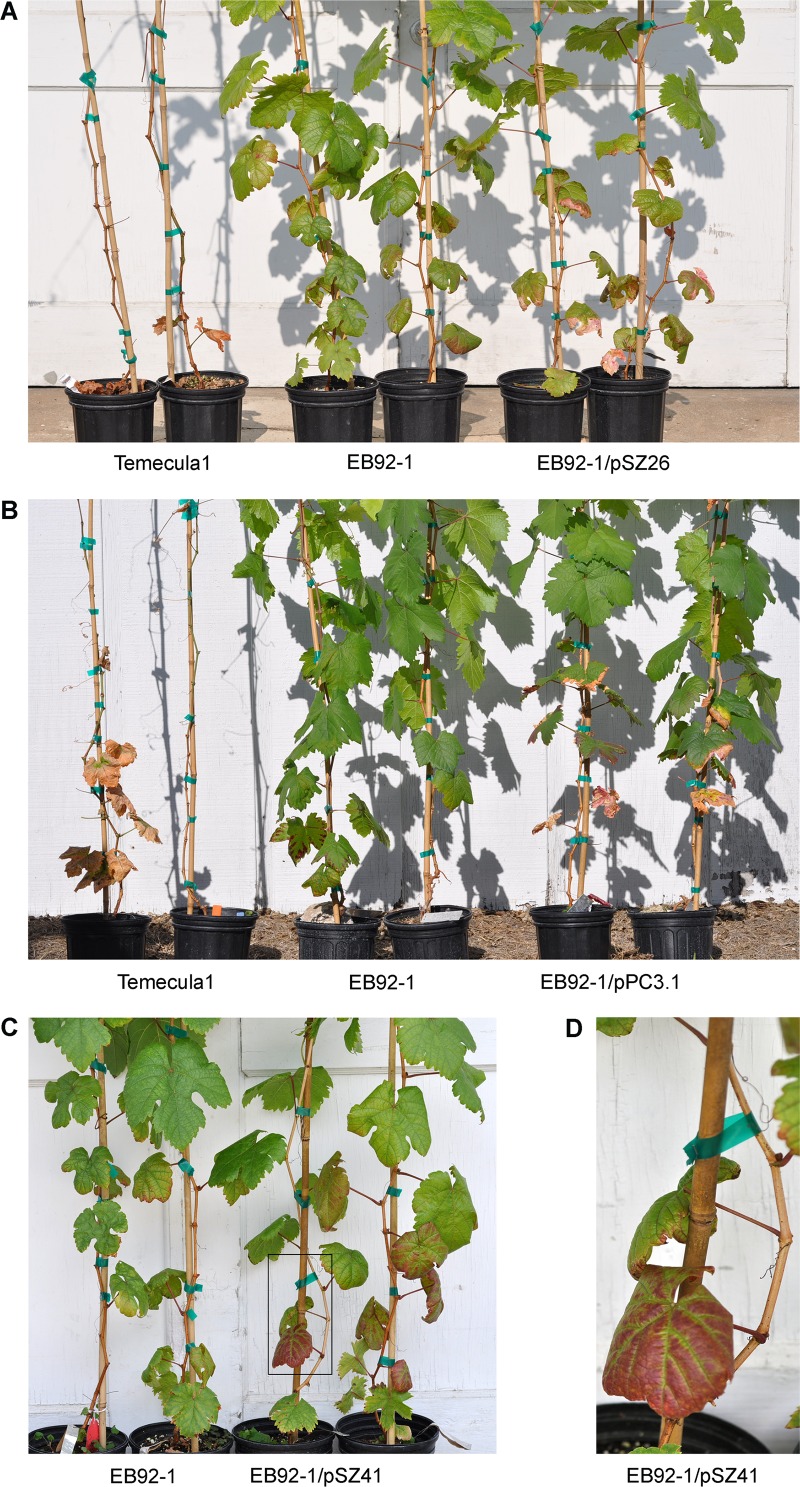
Fig 4. Pathogenic symptoms elicited by Temecula1, EB92-1 transformants and EB92-1 empty vector control on Carignane grapevines.
(A) Comparisons of plants inoculated with Temecula1, EB92-1 (EB92-1/pBBR1MCS-5 empty vector) and EB92-1/pSZ26 (revised PD1703; LipA). (B) Comparisons of plants inoculated with Temecula1, EB92-1 and EB92-1/pPC3.1 (PD0986; hemagglutinin). (C) Comparisons of plants inoculated with EB92-1 and EB92-1/pSZ41 (PD0928; Zot). (D) Enlarged region from boxed area in (C) illustrating unique symptoms; these lower leaves abscised 10 days later. All photos were taken 90 days post-inoculation.
Fig 5. Combined disease severity data of Temecula1, EB92-1 transformants and EB92-1 empty vector control on Carignane grapevines.
Temecula1, wild type PD strain; EB92-1, biocontrol strain EB92-1/pBBR1MCS-5 (empty vector); EB92-1/pSZ26, EB92-1/pSZ26 (revised PD1703, LipA); EB92-1/pPC3.1, EB92-1/pPC3.1 (PD0986, hemagglutinin), and EB92-1/pSZ41 EB92-1/pSZ41 (PD0928, Zot). Disease severity was calculated using combined data from 3 independent experiments with 4–12 replications of each inoculated strain in each experiment. Error bars indicate the standard deviations. Values marked with one or two asterisks are significantly different at the P < 0.05 and P<0.01 levels, respectively, using Student’s two-tailed t test.
Plants inoculated with EB92-1/pPC3.1 (hemagglutinin; refer Fig 4B) exhibited typical symptoms almost as rapidly as the wild type Temecula1 (by 5 weeks; slower than observed with EB92-1/pSZ26) but with a much slower rate of progression and affecting only ca. 32% of the leaves, as with EB92-1/pSZ26 (Fig 5). Again, these severity levels were significantly higher than the 12% levels observed with EB92-1 (P< 0.01).
Plants inoculated with EB92-1/pSZ41 (Zot; refer Fig 4C and 4D) exhibited unique, reduced visible symptoms that developed even more slowly than EB92-1/pSZ26 or EB92-1/pPC3.1, with visible symptoms becoming evident only by the end of 6 weeks. Instead of the initial necrotic symptoms typical of PD, the lower leaves (closest to the inoculation zones) of inoculated plants showed prominent anthocyanosis with green veinal areas (Fig 4C and 4D). Only as the infection advanced did the symptomatic leaves become necrotic, eventually resulting in limited defoliation and the disease did not progress very far past the inoculation zones. Nevertheless, the disease severity levels (ca 23%) were significantly higher (P< 0.05) than the 12% observed with EB92-1.
Discussion
EB92-1 is a biocontrol strain with a genome that is highly syntenic, and nearly identical in sequence, with pathogenic Temecula1 and is notably missing multiple potential pathogenicity factors found in Temecula1 [19]. Among the missing potential pathogenicity factors is PD1703, which was predicted to encode a lipase similar to LipA of X. oryzae pv. oryzae (X. oryzae.). X. oryzae LipA was reported to contribute to the virulence of X. oryzae on rice by synergistically enhancing rice cell wall xylan degradation in concert with X. oryzae xylanases [13–14, 28, 29]. Purified LipA induced innate defense responses in rice, including callose deposition and HR elicitation [14].
Bacterial lipases usually contain typical N-terminal signal peptides and are exported from the bacterial cell via a type II secretion system. Alignment of the predicted protein sequences of PD1703 and PD1702 from Temecula1 and > 90% identical protein sequences from several other sequenced X. fastidiosa strains revealed that PD1702, PD1703 and XF0357 from X. fastidiosa 9a5c were missing the first 35 amino acids from the annotation, including the Type II secretion signal sequence, predicted to be present in the other X. fastidiosa annotated LipA lipases (Fig 1). The revised X. fastidiosa Temecula1 PD1703 was cloned and demonstrably functional as a lipase when overexpressed in E. coli (Fig 3); revised PD1703 significantly enhanced both PD symptoms and disease severity elicited by transformed EB92-1 cells in inoculated grapevines (Figs 4A and 5).
X. fastidiosa is a rod-shaped bacterium with dimensions ranging from 250 to 500 X 1,000 to 4,000 nm [30] and requires cell wall degrading enzymes to enable intervessel migration, as the pore sizes of intact pit membranes of grape are too small to allow passive movement of the bacteria [31]. A polygalacturonase (pglA) was shown to be required for the colonization and pathogenicity of X. fastidiosa on grapevines [32] and later was proved to enlarge the pore size of intervessel pit membranes with endo-1, 4-b-glucanase in a synergistic manner [33]. Based on the observed hypersensitive response on nonhosts (Fig 3), and by analogy with LipA of Xoo, revised PD1703 likely contributes to disease symptoms by helping degrade cell walls and thereby generating endogenous elicitors, which elicit an HR on nonhosts (Fig 3).
PD1703 is negatively regulated in X. fastidiosa by gacA [34], encoding a transcriptional regulator of genes involved in quorum sensing, toxin production, motility, biofilm formation, and extracellular polysaccharide production in many plant bacterial pathogens [35–37]. Regulation of LipA, along with other degradative enzymes, may be required to constrain inter-vessel movement by X. fastidiosa strain Temecula1. Indeed, EB92-1 spreads more slowly and titers are 10–100 fold lower than Temecula1 in grapevine [38] and some unpublished data; we speculate that absence of LipA in EB92-1 may constrain movement as well as pathogenicity of the biocontrol strain.
In addition to PD1703, two hypothetical unknown proteins are present in both Temecula1 and EB92-1 with the potential to be lipases. PD1702 and PD1211 both have predicted LIP domains, including the canonical catalytic triad residues Ser-176, His-377, and Asp-336 of X. oryzae LipA [28] (S3 Fig). Although PD1702 is 91% identical to the protein sequence of PD1703, neither EB92-1 nor any transformants of E. coli or X. citri carrying PD1702 exhibited secreted lipase activity (using Tween-20 as substrate) from culture supernatants (S2 Fig). Since the native promoters of both PD1702 and PD1703 were used, both have predicted secretion signal peptides and both were sequenced prior to recloning into wide host range shuttle vector pBBR1MCS-5, these results indicate several possibilities: 1) PD1702 may be nonfunctional as a lipase capable of degrading Tween-20; 2) the PD1702 promoter is poorly functional in EB92-1, E. coli and X. citri; 3) PD1702 is poorly secreted from EB92-1, E. coli and X. citri, or 4) an unrecognized recloning artifact resulted in loss of function. Since the recloning methodology was standard restriction digestion followed by ligation and used for both PD1702 and 1703 and since PD1703 was well secreted from EB92-1, we favor the first possibility.
PD1211 is 61% identical to the protein sequence of PD1703, and EB92-1 carries a gene 100% identical to PD1211. Nevertheless, EB92-1 did not exhibit any detected lipase activity using Tween-20 as substrate in culture supernatants in these qualitative assays, unlike Temecula1 or EB92-1/ pSZ26 (PD1703), which clearly did (Fig 2C). Although Temecula1 appeared to exhibit somewhat more lipase activity than EB92-1/pSZ26 (PD1703) (Fig 2C), supernatant protein levels were not quantified, and substrates other than Tween-20 were not investigated.
Hemagglutinin-like proteins have been suggested to be involved in bacterial adherence, aggregation, motility, biofilm formation and virulence of plant pathogens [16–18, 39, 40]. Surprisingly, all six predicted hemagglutinin-like proteins appeared to be missing in EB92-1 [19], even though none of these genes are contiguous in Temecula1 (some are clustered). Two of these (PD1792 and PD2118) carry a type V, two-partner secretion (TPS) domain [41]; the remaining four have no TPS domain. Alignment of these six X. fastidiosa hemagglutinin like proteins indicated three related clades: one containing both large and small hemagglutinins, including PD0986 (394 aa) and PD1792 (3377 aa); another containing both large and small hemagglutinins, including PD2108 (376 aa) and PD2118 (3457 aa), and a third including PD2110 (436 aa) and PD2116 (438 aa; S4 Fig). In this study, PD0986 (394 aa) was confirmed missing in EB92-1.
Mutations of either large hemagglutinins, PD1792 or PD2118, in Temecula1 caused an increase in virulence and these two genes were identified as anti-virulence genes, causing loss of biofilm formation in vitro and in planta [16]. Surprisingly, transformation of EB92-1 with the small homolog PD0986 (394 aa), which is most closely related to PD1792 and missing in EB92-1, significantly enhanced pathogenicity of the transformed strain (Figs 4 and 5). The opposite effect might have been expected. The significance of the high degree of similarity of each pair of large and small hemagglutinins, if any, is unknown. It is also not clear how, or even if, the small ones are secreted. The two large hemagglutinins are secreted by TPS Type V secretion system, but the four small ones do not have TPS domains, nor do they appear to be autotransporters, nor do they carry a Type II signal sequence. It is possible that the highly similar ones are self-balancing anti-virulence/virulence pairs.
Zonular occludens toxin (Zot) family proteins are bacterial toxins represented by toxigenic strains of Vibrio cholera with the ability to reversibly alter intestinal epithelial tight junctions, allowing the passage of macromolecules through mucosal barriers [42–44]. Zot-like proteins are thought to be of viral origin and are likely to be laterally transferred among microorganisms through the action of filamentous phages [45]. Not surprisingly, the region encoding both Zot genes encodes various predicted phage genes, including PD0911 (replication initiation factor, PD0912 (phage coat protein B), PD0923 (replication initiation factor), PD0933 (phage related protein), and PD0936 (capsid protein). Interestingly, EB92-1 was missing both Zot genes, PD0915 and PD0928, which are identical in sequence, but retained the intervening genes from PD0916 to PD0927 (a total of 11 genes). The fact that both identical proteins in separate locations were missing from Temecula1 may indicate horizontal transfer followed by duplication in a progenitor strain similar to EB92-1, rather than independent deletion events in a Temecula1-like progenitor.
Based on annotation data alone, Zot-like proteins have been predicted and suggested as potential virulence factors in X. fastidiosa CVC strain 9a5c and other X. fastidiosa strains, as well as in X. campestris [45,46]. PD0915 and PD0928 possess a Zot domain (pfam05707) and a ATP-binding cassette (ABC) transporter nucleotide-binding domain (cl21455). ABC transporters are a large family of proteins involved in the Type I transport of a wide variety of different compounds, including sugars, ions, peptides, and more complex organic molecules [47]. pSZ41 (PD0928) conferred increased symptom production to EB92-1, and inoculated grapevines exhibited prominent anthocyanosis with green veinal areas in leaves adjacent to the inoculation zones (Fig 5C and 5D). To our knowledge there has been no previous functional evidence for a role in pathogenicity in X. fastidiosa, Xanthomonas, or any other plant pathogen. The duplication of an identical gene may indicate a selective advantage for X. fastidiosa strains carrying Zot.
Supporting Information
Lane 1, Temecula1; Lane 2, EB92-1; Lane 3, water; PCR primer sets (gene target and PCR product sizes) used were: (A) PD1702+3-F and PD1703-R (PD1703, 1.2 Kb); (B) PD0911-F and PD0916-R (PD0915/PD0928, 4.2 kb); (C) ZOT-F and ZOT-R (PD0915/PD0928, 1.4 kb); (D) PD0956-F and PD0956-R (PD0956, 1.0 kb); (E) XFEB114-F and XFEB114-R1 (PD0956, 1.9 kb); (F) PD0956-NF and PD0956-NR (PD0956, 1.5 kb); (G) PD0986-F and PD0986-R (PD0986, 1.2 kb); (H) PD0986-NF and PD0986NR (PD0986, 2.0 kb); (I) RST31 and RST33 (X. fastidiosa marker gene, 0.7 kb). Molecular weight markers used were Quick-Load 1 kb DNA Ladder from New England Biolabs Inc. (Beverly, MA).
(TIF)
Lane 1, protein marker (Precision Plus Protein All Blue Prestained Standards, #161–0373, Bio-Rad); Lane 2, total protein from empty vector pET27b cultures ("Total CK"); Lane 3, supernatant protein from empty vector pET27b ("Supernatant CK"); Lane 3, total protein from pSZ37 cultures ("Total LipA"); Lane 4, supernatant protein from pSZ37 cultures ("Supernatant LipA"). Ten μL protein marker and 150–180 ng (5 μL) protein were loaded for analysis. Protein was stained with Coomassie Blue R250. The arrow indicates the expected 42.4 kDa product of PD1703 (LipA).
(TIF)
(A). AlignX (Vector NTI Advance 10, Invitrogen, Carlsbad, CA) was used to generate the phylogenetic tree. (B). Residues marked in red are the canonical catalytic catalytic triad residues Ser176, His377, and Asp336 of X. oryzae LipA. Residues marked in green area are amino acids lining the tunnel with carbohydrate-anchoring pockets.
(TIF)
AlignX (Vector NTI Advance 10, Invitrogen, Carlsbad, CA) was used to generate the phylogenetic tree.
(TIF)
(DOCX)
Acknowledgments
We thank Dr. Zhilong Bao for his assistance in the statistical analysis of our data.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
University of California Pierce’s Disease Research Grants Program, project 2009-244. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Purcell AH, Hopkins DL. Fastidious xylem-limited bacterial plant pathogens. Annu Rev Phytopathol. 1996; 34: 131–151. [DOI] [PubMed] [Google Scholar]
- 2. Goodwin PH., DeVay JE, Meredith CP. Roles of water stress and phytotoxins in the development of Pierce’s disease of the grapevine. Physiol Mol Plant Pathol. 1988; 32: 1–15. [Google Scholar]
- 3. Stevenson J, Matthews M, Rost T. The developmental anatomy of Pierce's disease symptoms in grapevines: Green islands and matchsticks. Plant Dis. 2005; 89: 543–548. [DOI] [PubMed] [Google Scholar]
- 4. Thorne ET, Stevenson JF, Rost TL, Labavitch JM, Matthews MA. Pierce's disease symptoms: comparison with symptoms of water deficit and the impact of water deficits. Am J Enol Vitic. 2006; 57: 1–11. [Google Scholar]
- 5. Van Sluys MA, De Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, Camargo LEA, et al. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa . J Bacteriol, 2003; 185: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa . Annu Rev Phytopathol. 2008; 46: 243–271. 10.1146/annurev.phyto.45.062806.094342 [DOI] [PubMed] [Google Scholar]
- 7. Matsumoto A, Huston SL, Killiny N, Igo MM. XatA, an AT-1 autotransporter important for the virulence of Xylella fastidiosa Temecula1. MicrobiologyOpen. 2012; 1: 33–45. 10.1002/mbo3.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lubelsky J, Konings WN, Driessen AJM. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007; 71: 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reddy JD, Reddy SL, Hopkins DL, Gabriel DW. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol Plant Microbe Interact. 2007; 20: 403–410. [DOI] [PubMed] [Google Scholar]
- 10. Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001; 69: 3523–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stehr F, Felk A, Gácser A, Kretschmar M, M ähnss B, Neuber K. Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res. 2004; 4: 401–408. [DOI] [PubMed] [Google Scholar]
- 12. Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans . Mycoses. 2005; 48: 365–377. [DOI] [PubMed] [Google Scholar]
- 13. Jha G, Rajeshwari R, Sonti RV. Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol Plant Microbe Interact. 2005; 18: 891–898. [DOI] [PubMed] [Google Scholar]
- 14. Jha G, Rajeshwari R, Sonti RV. Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol Plant Microbe Interact. 2007; 20: 31–40. [DOI] [PubMed] [Google Scholar]
- 15. Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004; 68: 692–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guilhabert MR, Kirkpatrick BC. Identification of Xylella fastidiosa antivirulence genes: Hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005; 18: 856–868. [DOI] [PubMed] [Google Scholar]
- 17. Killiny N, Almeida RP. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl Environ Microbiol. 2009; 75: 521–528. 10.1128/AEM.01921-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voegel TM, Jeremy GW, Matsumoto A, Igo MM, Kirkpatrick BC. Localization and characterization of Xylella fastidiosa haemagglutinin adhesins. Microbiol. 2010; 156: 2172–2179. [DOI] [PubMed] [Google Scholar]
- 19. Zhang S, Flores-Cruz Z, Kumar D, Chakrabarty PK, Hopkins DL, Gabriel DW. The genome of Xylella fastidiosa biocontrol strain EB92-1 is very similar to Pierce’s Disease strains. J Bacteriol. 2011; 193: 5576–5577. 10.1128/JB.05430-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopkins DL. Biological control of Pierce’s disease in the vineyard with strains of Xylella fastidiosa benign to grapevine. Plant Dis. 2005; 89: 1348–1352. [DOI] [PubMed] [Google Scholar]
- 21. Davis MJ, Purcell AH, Thomson SV. Isolation media for the Pierce's disease bacterium. Phytopathology. 1980; 70: 425–429. [Google Scholar]
- 22. Guilhabert MR, Hoffman LM, Mills DA, Kirkpatrick BC. Transposon mutagenesis of Xylella fastidiosa by electroporation of Tn5 synaptic complexes. Mol Plant Microbe Interact. 2001; 6: 701–706. [DOI] [PubMed] [Google Scholar]
- 23. Swarup S, De Feyter R, Brlansky RH, Gabriel DW. A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit canker like lesions on citrus. Phytopathology. 1991; 81: 802–809. [Google Scholar]
- 24. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995; 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 25. Minsavage GV, Thompson CM, Hopkins DL, Leite RMVBC, Stall RE. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology. 1994; 84: 456–461. [Google Scholar]
- 26. Matsumoto A, Igo MM. Species-specific type II restriction-modification system of Xylella fastidiosa Temecula1. Appl Environ Microbiol. 2010; 76: 4092–4095. 10.1128/AEM.03034-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samad MYA, Razak CNA, Salleh AB, Yunus WMZW, Ampon K, Basri M. A plate assay for primary screening of lipase activity. J Microbiol Methods. 1989; 9: 51–56. [Google Scholar]
- 28. Aparna G, Chatterjee A, Sonti RV, Sankaranarayanan. A cell wall-degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. Plant Cell. 2009; 21: 1860–1873. 10.1105/tpc.109.066886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajeshwari R, Jha G, Sonti RV. Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol Plant Microbe Interact. 2005; 18: 830–837. [DOI] [PubMed] [Google Scholar]
- 30. Mollenhauer HH, Hopkins DL. Ultrastructural study of Pierce's disease bacterium in grape xylem tissue. J Bacteriol. 1974; 119: 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stevenson JF, Matthews MA, Greve LC, Labavitch JM, Rost TL. Grapevine susceptibility to Pierce’s disease II: progression of anatomical symptoms. Am J Enol Viticult. 2004; 55: 238–245. [Google Scholar]
- 32. Roper MC, Greve LC, Warren JG, Labavitch JM, Kirkpatrick BC. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol Plant Microbe Interact. 2007; 20: 411–419. [DOI] [PubMed] [Google Scholar]
- 33. Pérez-Donoso AG, Sun Q, Roper MC, Greve LC, Kirkpatrick BC, Labavitch JM. Cell wall-degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa-infected grapevines. Plant Physiol. 2010; 152: 1748–1759. 10.1104/pp.109.148791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi XY, Dumenyo CK, Hernandez-Martinez R, Azad H, Cooksey DA. Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by gacA . Appl Environ Microbiol. 2009; 75: 2275–2283. 10.1128/AEM.01964-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cui Y, Chatterjee A, Chatterjee AK. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol Plant Microbe Interact. 2001; 14: 516–526. [DOI] [PubMed] [Google Scholar]
- 36. Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001; 40: 1215–1226. [DOI] [PubMed] [Google Scholar]
- 37. Chatterjee A, Cui Y, Yang H, Collmer A, Alfano JR, Chatterjee AK. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol Plant Microbe Interact. 2003; 16: 1106–1117. [DOI] [PubMed] [Google Scholar]
- 38. Hopkins DL. Physiological and pathological characteristics of virulent and avirulent strains of the bacterium that causes Pierce’s disease of grapevine. Phytopathology. 1985; 75: 713–717. [Google Scholar]
- 39. Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci U S A. 2002; 20: 13142–13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE. 2009; 44: e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, Villeret V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci U S A. 2004; 101: 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uzzau S, Cappuccinelli P, Fasano A. Expression of a Vibrio cholerae zonula occludens toxin and analysis of its subcellular localization. Microb Pathog. 1999; 27: 377–385. [DOI] [PubMed] [Google Scholar]
- 43. Rossi M, Maurano F, Luongo D, Fasano A, Uzzau S, Auricchio S. Zonula occludens toxin (Zot) interferes with the induction of nasal tolerance to gliadin. Immunol let. 2002; 81: 217–221. [DOI] [PubMed] [Google Scholar]
- 44. Karyekar CS, Fasano A, Raje S, Lu R, Dowling TC, Eddington ND. Zonula occludens toxin increases the permeability of molecular weight markers and chemotherapeutic agents across the bovine brain microvessel endothelial cells. J Pharm Sci. 2003; 92: 414–423. [DOI] [PubMed] [Google Scholar]
- 45. Hagemann M, Hasse D, Berg G. Detection of a phage genome carrying a Zonula occludens like toxin gene (zot) in clinical strains of Stenotrophomonas maltophilia . Arch Microbiol. 2006; 185: 449–458. [DOI] [PubMed] [Google Scholar]
- 46. da Silva VS, Shida CS, Rodrigues FB, Ribeiro DC, de Souza AA, Coletta-Filho HD. Comparative genomic characterization of citrus-associated Xylella fastidiosa strains. BMC Genomics. 2007; 8: 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001; 42: 1007–1017. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lane 1, Temecula1; Lane 2, EB92-1; Lane 3, water; PCR primer sets (gene target and PCR product sizes) used were: (A) PD1702+3-F and PD1703-R (PD1703, 1.2 Kb); (B) PD0911-F and PD0916-R (PD0915/PD0928, 4.2 kb); (C) ZOT-F and ZOT-R (PD0915/PD0928, 1.4 kb); (D) PD0956-F and PD0956-R (PD0956, 1.0 kb); (E) XFEB114-F and XFEB114-R1 (PD0956, 1.9 kb); (F) PD0956-NF and PD0956-NR (PD0956, 1.5 kb); (G) PD0986-F and PD0986-R (PD0986, 1.2 kb); (H) PD0986-NF and PD0986NR (PD0986, 2.0 kb); (I) RST31 and RST33 (X. fastidiosa marker gene, 0.7 kb). Molecular weight markers used were Quick-Load 1 kb DNA Ladder from New England Biolabs Inc. (Beverly, MA).
(TIF)
Lane 1, protein marker (Precision Plus Protein All Blue Prestained Standards, #161–0373, Bio-Rad); Lane 2, total protein from empty vector pET27b cultures ("Total CK"); Lane 3, supernatant protein from empty vector pET27b ("Supernatant CK"); Lane 3, total protein from pSZ37 cultures ("Total LipA"); Lane 4, supernatant protein from pSZ37 cultures ("Supernatant LipA"). Ten μL protein marker and 150–180 ng (5 μL) protein were loaded for analysis. Protein was stained with Coomassie Blue R250. The arrow indicates the expected 42.4 kDa product of PD1703 (LipA).
(TIF)
(A). AlignX (Vector NTI Advance 10, Invitrogen, Carlsbad, CA) was used to generate the phylogenetic tree. (B). Residues marked in red are the canonical catalytic catalytic triad residues Ser176, His377, and Asp336 of X. oryzae LipA. Residues marked in green area are amino acids lining the tunnel with carbohydrate-anchoring pockets.
(TIF)
AlignX (Vector NTI Advance 10, Invitrogen, Carlsbad, CA) was used to generate the phylogenetic tree.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.