Abstract
Hyperactivation of the mechanistic target of rapamycin (mTOR) kinase, as a result of loss-of-function mutations in tuberous sclerosis complex 1 (TSC1) or TSC2 genes, causes protein synthesis dysregulation, increased cell size, and aberrant neuronal connectivity. Dysregulated synthesis of synaptic proteins has been implicated in the pathophysiology of autism spectrum disorder (ASD) associated with TSC and fragile X syndrome. However, cell type-specific translational profiles in these disease models remain to be investigated. Here, we used high-fidelity and unbiased Translating Ribosome Affinity Purification (TRAP) methodology to purify ribosome-associated mRNAs and identified translational alterations in a rat neuronal culture model of TSC. We find that expression of many stress and/or activity-dependent proteins is highly induced while some synaptic proteins are repressed. Importantly, transcripts for the activating transcription factor-3 (Atf3) and mitochondrial uncoupling protein-2 (Ucp2) are highly induced in Tsc2-deficient neurons, as well as in a neuron-specific Tsc1 conditional knock-out mouse model, and show differential responses to the mTOR inhibitor rapamycin. Gelsolin, a known target of Atf3 transcriptional activity, is also upregulated. shRNA-mediated block of Atf3 induction suppresses expression of gelsolin, an actin-severing protein, and rescues spine deficits found in Tsc2-deficient neurons. Together, our data demonstrate that a cell-autonomous program consisting of a stress-induced Atf3-gelsolin cascade affects the change in dendritic spine morphology following mTOR hyperactivation. This previously unidentified molecular cascade could be a therapeutic target for treating mTORopathies.
SIGNIFICANCE STATEMENT Tuberous sclerosis complex (TSC) is a genetic disease associated with epilepsy and autism. Dysregulated protein synthesis has been implicated as a cause of this disease. However, cell type-specific translational profiles that are aberrant in this disease are unknown. Here we show that expression of many stress and/or activity-dependent proteins is highly induced while some synaptic proteins are repressed in neurons missing the Tsc2 gene expression. Identification of genes whose translation is abnormal in TSC may provide insights to previously unidentified therapeutic targets.
Keywords: autism, dendiritic spines, mTOR, protein synthesis, translation
Introduction
Dysregulation of the mechanistic target of rapamycin (mTOR) kinase is involved in many neurologic disorders, such as tuberous sclerosis complex (TSC), PTEN-hamartoma syndromes, neurofibromatosis 1 (NF-1), and epilepsy (Lipton and Sahin, 2014). As a serine/threonine kinase, mTOR integrates extracellular nutritional stimuli and intracellular energetic status to finely regulate protein synthesis and control cell growth rate. An increasing body of evidence suggests that dysregulated synthesis of certain synaptic proteins is one underlying cellular mechanism of autism spectrum disorder (ASD) (Auerbach et al., 2011; Tsai et al., 2012; Gkogkas et al., 2013; Santini et al., 2013; Ebrahimi-Fakhari and Sahin, 2015). Several specific proteins, such as neuroligin and Arc, have been implicated in the synaptic deficits associated with autism (Auerbach et al., 2011; Gkogkas et al., 2013; Ebrahimi-Fakhari and Sahin, 2015); however, systematic translational profiling in these mTOR pathology disease models is lacking.
TSC is an autosomal dominant multisystem disorder caused by loss-of-function mutations in either the TSC1 or TSC2 genes. TSC manifests as a tumor-hamartoma syndrome affecting multiple organs, including the brain, skin, eyes, kidneys, heart, and lungs. The pathognomonic brain lesion in TSC is the cortical tuber, and the most devastating neurologic and psychiatric morbidities in TSC patients include epilepsy, intellectual disability, and autism (Feliciano et al., 2013; DiMario et al., 2015). Approximately 30%–50% of patients with TSC are affected by ASD (Asato and Hardan, 2004; Jeste et al., 2008). As a monogenic model for both mTOR pathology and autism, TSC mouse and cellular models have offered valuable opportunities for understanding how mTOR-dependent protein synthesis is involved in the synaptic deficits characteristic of ASD (Tavazoie et al., 2005; Choi et al., 2008; Nie et al., 2010; Nie and Sahin, 2012; Tsai et al., 2012; Bateup et al., 2013). On the other hand, TSC results in epilepsy in ∼90% of patients (Chu-Shore et al., 2010); thus, studies of TSC mouse models may also help clarify the role of seizure activity in the etiology and/or progression of autism.
To determine what alterations in mRNA translation occur in response to TSC1/2 loss in neurons, we used Translating Ribosomal Affinity Purification (TRAP) methodology, which provides better temporal resolution and cell type specificity than conventional gene expression profiling (Heiman et al., 2008, 2014). In this study, we introduced the transgene L10a in a hippocampal neuronal culture system using lentiviral vectors, for temporally and spatially specific analysis of translation profiling. We analyzed the differential translational expression profile between Tsc2-deficient and control hippocampal neurons, generating a “TSC translatome.” Although reduced expression of synaptic proteins was anticipated, surprisingly we identified induction of a large number of stress and/or activity-induced proteins as well as a number of cytokines in the Tsc2-deficient neurons. Functionally, our data demonstrate that Atf3-gelsolin cascade downstream of TSC1/2 loss can negatively regulate dendritic spine morphology and impair neuronal connectivity. Our results suggest that hippocampal neurons, when challenged by mTOR hyperactivation, activate a stress-induced prosurvival program at the expense of neuronal connectivity.
Materials and Methods
Antibodies, constructs, and animals.
Antibodies to PSD-95 (Pierce), MAP2 (Sigma-Aldrich), GFP (Abcam, Roche, and Rockland), mCherry (Abcam, BioVision), puromycin (KeraFAST), ATF3 (Santa Cruz Biotechnology), Ucp2 (Abcam), p-cAMP response element binding (CREB) (Ser133) (Cell Signaling Technology), tubulin (Cell Signaling Technology), GAPDH (Santa Cruz Biotechnology), and FLAG (Agilent Technologies) were obtained commercially as indicated. Monoclonal anti-GFP antibodies (19C8 and 19F7) for the TRAP assay were purchased from the Memorial Sloan-Kettering Monoclonal Antibody Facility (New York). Gelsolin antibody was a generous gift from Prof. Walter Witke (University of Bonn, Bonn, Germany). For immunocytochemistry, fluorescently labeled secondary antibodies (AlexaFluor-488, -594, or -647, Invitrogen) were used. TTX and rapamycin (both Sigma-Aldrich) and MK801 (Tocris Bioscience) were obtained commercially.
shRNAs against firefly luciferase or rat Tsc2 gene have been described previously (Nie and Sahin, 2012). GFP was replaced with mCherry at cloning sites NheI/EcoR I by using the primer set: 5′-TAGCTAGCCATGCTGTGCTGTATGAGA-3′ and 5′-GCGAATTCTTACTTGTACAGCTCGTCCA-3′. To clone GFP versus GFP-L10a transgenes into the lentivector pHAGE downstream of the UBC promoter, primer pairs were used as below: 5′-GAAAAAGCGGCCGCATCGCCACCATGGTGAGCAA-3′ and 5′-AAGTAAAAGGATCCCGGGCCCGCGGTACCGTCGACTGCAGAA-3′ for GFP; 5′-GAAAAAGCGGCCGCATCGCCACCATGGTGAGCAA-3′ and 5′-AATGGATCCCTAATACAGACGCTGGGGCTTGCCCCATG-3′ for GFP-L10a. The cloning sites NotI/BamHI were used.
shRNA (CAGAATAAACACCTCTGCCATC) targeting Atf3 was obtained from Sigma-Aldrich, validated and then subcloned into CSCGW vector, which has eGFP inserted downstream of a PGK promoter. Lentivirus particles were packaged and produced by the MGH Viral Core Facility, with a virus titer of 7.596 × 10−7 IU/ml. The pRK-ATF3 plasmid was a gift from Yihong Ye (AddGene, plasmid # 26115) (Wang et al., 2009).
SynI-Cre+Tsc1ww and Syn1-Cre+Tsc1cc mice were described previously (Meikle et al., 2007). Hippocampal lysates were generated from brains harvested at postnatal day 21 from both male and female animals. All animal procedures were in accordance with the Guide for the Humane Use and Care of Laboratory Animals, and the study was approved by the Animal Care and Use Committee of Boston Children's Hospital.
Neuronal culture, drug treatment, and Western blotting.
Hippocampal neuronal cultures from rat embryonic day 18.5 or mouse embryonic day 16.5 were obtained as previously described (Nie and Sahin, 2012). Neuron cultures were maintained in Neurobasal medium supplemented with antibiotics, B27 and glutamate. For drug treatments, drugs or vehicle were added to conditioned medium for the time periods indicated. Neurons were then quickly washed with 1× PBS and lysed with 1× RIPA buffer or directly with 1× SDS sample buffer. Lysates were boiled at 95°C for 5 min before SDS-PAGE and Western blotting.
Puromycin incorporation assay.
Hippocampal neurons infected with lentiviral sh-Luc versus sh-Tsc2 were treated with anisomycin (20 μm for 1 h), rapamycin (20 nm, overnight), or vehicle before puromycin incubation (1 μm for 30 min). Cells were snap frozen on dry ice, lysed in RIPA buffer (with protease inhibitors), and homogenized. After centrifugation at 13,000 rpm, 4°C, for 10 min, supernatants were collected, mixed with half volumes of 3× sample buffer, and boiled immediately at 95°C for 5 min. Total protein concentrations were determined by Bradford protein assay kit. Equal amounts of proteins were loaded onto 4%–20% gradient SDS-PAGE gels (Bio-Rad). Anti-puromycin antibody (KeraFAST) at 1:5000 dilutions was used for detection of newly synthesized proteins by Western blotting, and postblotting membrane staining by EZBlue reagent (Sigma-Aldrich).
Neuronal transfection, lentivirus infection, and immunocytochemistry.
Plasmids and Lipofectamine 2000 were diluted in plain Neurobasal medium at a ratio of 1 μg:2 μl for each 13 mm coverslip in a 24-well plate. The plasmid medium was added to each matching Lipofectamine tube. The mixtures were incubated for 20 min at room temperature. Medium in wells to be transfected was replaced with 500 μl prewarmed plain Neurobasal and the original medium was saved in a 15 ml Falcon tube. The plasmid:Lipofactamine transfection mixture was subsequently added to the top of each well (100 μl per well) and incubated for 2 h. The transfection medium was then washed off and replaced with the saved conditioned medium. In general, transfection was performed on day 7 in vitro (7 DIV) or day 11 (11 DIV) for experiments shown in Figure 7.
Figure 7.
Blocking ATF3 induction reverses the spine density deficit in Tsc2-deficient neurons. A, Representative Western blots show the induction of ATF3 and gelsolin in Tsc2-deficient neurons. shRNA-mediated Atf3 knockdown suppressed protein levels of ATF3 as well as gelsolin. B, Representative images show expression of mCherry positive sh-Tsc2 (red) and/or GFP positive sh-Atf3 (green), and/or hATF3-FLAG (blue). Coexpression of sh-Atf3 increases spine density in Tsc2-deficient neurons. Overexpression of human FLAG-tagged ATF3 (resistant to Atf3 shRNA) under this condition reverses this phenotype by reducing spine density. Scale bars, 5 μm. C, Quantitation of spine density in sh-Tsc2 alone, sh-Atf3 alone, sh-Tsc2 and sh-Atf3, and sh-Tsc2 and sh-Atf3 plus ATF3-FLAG-expressing neurons at DIV 16. Spine densities per 100 μm in sh-Tsc2, sh-Atf3, and sh-Tsc2 and sh-Atf3, and sh-Tsc2 and sh-Atf3 plus ATF3-FLAG-expressing cultures are 8.64 ± 2.24, 35.17 ± 7.67, 44.15 ± 5.49, and 20.04 ± 1.84 (mean ± SEM). **p < 0.01 (one-way ANOVA with Tukey's multiple-comparison test). ***p < 0.001 (one-way ANOVA with Tukey's multiple-comparison test).
Lentiviral particles encoding hU6-driven shRNAs were produced in HEK293T cells as previously described (Nie et al., 2010; Nie and Sahin, 2012). These included GFP or mCherry expressing sh-Luc (against firefly luciferase) versus sh-Tsc2 (against rat Tsc2 gene). Neuronal cultures were infected for 4–6 h on 2 DIV and 9 DIV (for experiments shown in Fig. 7). After washing off the virus-containing medium, cultures were maintained as described above.
Transfected or infected neurons were fixed at 16–21 DIV with 4% PFA plus 4% sucrose. Images were captured using Zeiss LSM700 confocal microscope system and analyzed using National Institutes of Health ImageJ.
Modified TRAP assay.
We followed the TRAP protocol for cell cultures with minor modifications (Heiman et al., 2008, 2014). Before harvesting, neuronal cultures in 10 cm dishes were incubated with 100 μg/ml of CHX for 20 min. After washing twice with PBS containing 100 μg/ml of CHX, cells were lysed with 1 ml ice-cold lysis buffer (20 mm HEPES KOH, pH 7.4, 150 mm KCl, 5 mm MgCl2, 1% NP-40, EMD Biosciences; 0.5 mm dithiothreitol, 100 μg/ml CHX, protease inhibitors, and RNase inhibitors). The cell lysates were then homogenized on ice by passing through differently gauged needles. Homogenates were centrifuged for 10 min at 2000 × g at 4°C, to pellet large cell debris, and 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC; Avanti Polar Lipids) was added to the supernatant at a final concentration of 300 mm. After mixture and incubation on ice for 5 min, the lysate was further centrifuged for 10 min at 20,000 × g at 4°C, to pellet unsolubilized material. Monoclonal anti-GFP-coated protein G Dynal magnetic beads (Invitrogen) were added to the supernatant, and the mixture was incubated at 4°C with end-over-end rotation for 30 min. Beads were subsequently collected on a magnetic rack, washed three times with high-salt polysome wash buffer (20 mm HEPES-KOH, pH 7.4, 350 mm KCl, 5 mm MgCl2, 1% NP-40, 0.5 mm dithiothreitol, and 100 mg/ml CHX) and immediately subjected to RNA extraction using absolutely RNA nanoprep kit (Agilent Technologies) with in-column DNase digestion according to the manufacturer's instructions. The quality of purified RNAs was evaluated based on RNA integrity numbers read by an Agilent Technologies BioAnalyzer 2100.
Microarray analysis and bioinformatics.
Three biological replicates were performed for each experimental condition (sh-Luc, sh-Tsc2, sh-Tsc2 with rapamycin). Purified RNAs were amplified with the Ovation system (NuGEN) according to the manufacturer's instructions. Biotin-labeled cDNAs were hybridized onto the Affymetrix rat gene 1.0 ST array. Differential gene expression analysis was conducted between sh-Tsc2 and sh-Luc, and between sh-Tsc2 with rapamycin and sh-Luc, using edgeR with TMM normalization (Robinson and Oshlack, 2010; Robinson et al., 2010) using a corrected p value (Benjamini and Hochberg, 1995) and a cutoff of 1e-5.
Protein and RNA extraction from brain samples.
Syn1-Cre+Tsc1ww and Syn1-Cre+Tsc1cc mice were killed on postnatal day 21. Cortex and hippocampus were quickly dissected and cut into two halves. One half was snap frozen in liquid nitrogen for protein extraction, and the other half was further chopped and dipped into RNALater stabilization solution (Ambion) for total RNA extraction. Lysis for protein extraction was performed with lysis buffer (150 mm NaCl, 20 mm Tris-HCl, pH 8.8, 1% NP-40, 1 mm CaCl2, 10% glycerol, 1 mm sodium fluoride, 1 mm sodium orthovanadate, and protease inhibitors). Protein samples were centrifuged at 13,000 rpm for 10 min, and supernatants were collected. Protein concentration was determined by Bradford protein assay. RNA extraction was performed using the RNease miniprep kit (QIAGEN) following the manufacturer's instructions, and RNA quality was evaluated using NanoDrop spectrometry.
Quantitative real-time PCR.
Quantitative real-time PCR (qPCR) reactions were performed to validate array results with RNA extractions from independent TRAP experiments as well as total RNA isolations. Total RNA was harvested using RNeasy Mini Plus Kit (QIAGEN). RNA was then converted to cDNA using a High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems) according to the manufacturer's instructions for use with RNase inhibitor. qPCR was done using SYBR Green Master Mix (Applied Biosystems) and primer pairs listed in Table 1. Samples were prepared in triplicates in 96-well optical reaction plates and run on an ABI 7300 Real-time PCR system. For analysis, all triplicate samples were normalized to GAPDH and then standardized to the control condition (sh-Luc nontreated); 18S was used as an additional internal control for confirmation. Relative quantifications are graphed as mean ± SEM using GraphPad Prism. Two-way ANOVA tests were used to determine statistical significance.
Table 1.
The primers used for qPCR
| Rat gene | Accession no. | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|---|
| Tsc2 | NM_012680.2 | GAGCTGTGTGACCAGAACGA | TGTTGATGAGGAGCACGAAG |
| GAPDH | NM_017008 | AGACAGCCGCATCTTCTTGT | CTTGCCGTGGGTAGAGTCAT |
| 18S | NR_046237.1 | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| Atf3 | NM_012912.2 | GACAGACAGCCCGCCTCTA | GACCTGGCCTGGATGTTGAA |
| Ucp2 | NM_019354.2 | ACCATTGCACGAGAGGAAGG | TCTTGACCACATCAACGGGG |
| Snap91 | NM_031728 | ATAGACCTTTTTGGGGATGC | GCAGGTTATTCTGAGCTGGA |
| Prelp | NM_053385.1 | AACAAGCTGTCGGATCGAGG | ACATCATGAGGTCCAACGGG |
| Il1r1 | NM_013123.3 | TTTGTTCCCGCCAAGATGGA | AGGACGTGCGGCAAGTATAG |
| Emp3 | NM_030847.1 | ATCTGGCAGCCATGTCACTC | ACGTGCAGTCATACCACAGG |
| Hspb1 | NM_031970.3 | AGCACGAAGAAAGGCAGGAT | ATTGTGTGACTGCTTTGGGC |
Results
Global increase in de novo protein synthesis in Tsc2-deficient neurons
mTOR is a master regulator of mRNA translation and protein synthesis (Thoreen et al., 2012). We have previously established a neuronal culture model of TSC that allows us to monitor mTOR activation after knocking down the Tsc2 gene (Nie and Sahin, 2012). To investigate the net effect of Tsc2 knockdown on protein synthesis, we now performed metabolic puromycin incorporation assays (Fig. 1). De novo protein synthesis, as quantitated based on Western blotting with an antipuromycin antibody, was significantly increased in hippocampal neuronal cultures infected with a lentiviral vector carrying Tsc2-shRNA (sh-Tsc2) compared with control neurons infected with shRNA against firefly luciferase (sh-Luc) (1.159 ± 0.054 normalized to sh-Luc). Furthermore, rapamycin treatment (20 nm, overnight) suppressed puromycin incorporation and corrected the difference between Tsc2 knockdown and control cultures (0.797 ± 0.076 vs 0.711 ± 0.075). As a control, anisomycin, a known protein synthesis inhibitor, reduced de novo protein synthesis by nearly 90% (0.115 ± 0.024). These data indicate that mTOR activation, as a consequence of Tsc2 knockdown, results in increased de novo protein synthesis in hippocampal neuronal cultures.
Figure 1.
De novo protein synthesis is globally increased in Tsc2-deficient rat hippocampal neurons in an mTOR-dependent manner. A, Hippocampal neurons infected with lentivirus that knock down either firefly luciferase (sh-Luc, control) or rat Tsc2 gene (sh-Tsc2), or left uninfected as indicated. At days 12–16 d in vitro (DIV12-16), neurons were pretreated with anisomycin (20 μm for 1.5 h), DMSO or rapamycin (20 nm overnight), incubated with 5 μg/ml puromycin for 30 min immediately before harvesting. Incorporation of puromycin in newly synthesized protein was detected by Western blotting with an anti-puromycin antibody. Total protein loaded on the gel was visualized by staining the postblotting membranes with EZblue dye. B, Quantification of puromycin blotting signals normalized to that of sh-Luc-infected neurons with DMSO treatment. Bars represent means ± SEM (n = 5; 0.115 ± 0.024 for anisomycin-treated wild-type, 1.159 ± 0.054 for DMSO treated sh-Tsc2 knockdown, 0.797 ± 0.076 for rapamycin treated sh-Luc, and 0.711 ± 0.075 for rapamycin treated sh-Tsc2 knockdown). *p < 0.05 (Student's t tests). **p < 0.01 (Student's t tests). ns, Not significant.
Application of a modified TRAP approach for translational profiling
Loss of neuronal Tsc1 or Tsc2 causes abnormal axon specification, aberrant axon path finding, defects in dendritic morphology, reduced myelination, hyperexcitability, and seizure activity, as well as autistic behaviors in mouse models (Tavazoie et al., 2005; Meikle et al., 2007; Choi et al., 2008; Nie et al., 2010; Tsai et al., 2012; Bateup et al., 2013). Whereas extracellular and intracellular regulators of the TSC-mTOR pathway have been relatively well investigated, many downstream effectors that are regulated by mTOR remain to be explored in detail (Thoreen et al., 2012). We hypothesized that translation of a subset of mRNAs is directly regulated by mTOR in postmitotic neurons when the TSC complex is functionally disrupted. To test this, we used the TRAP assay by introducing a lentivector-based L10a transgene into neuronal cultures (Heiman et al., 2008). This technique analyzes mRNAs associated with the ribosome rather than steady-state mRNA abundance and thus represents a more direct assessment of ongoing mRNA translation. We found that the GFP-L10a signal expression was enriched in nucleoli and also distributed throughout the soma and within the neurites, even though the latter is much weaker (Fig. 2A). We did not observe a significant increase in cell death or apoptosis in neurons with L10a transgene expression as measured by cleaved caspase-3 staining (data not shown). To perform anti-GFP immunoprecipitation in the Tsc2-deficient culture, we infected hippocampal neurons with two different lentiviral vectors: one coexpresses mCherry and shRNA (sh-Luc vs sh-Tsc2) and the other expresses GFP-L10a (Fig. 2B). The efficiency of Tsc2 knockdown and L10a transgene expression was confirmed by Western blotting. Consistent with previous studies (Tavazoie et al., 2005; Choi et al., 2008; Di Nardo et al., 2009), Tsc2 knockdown in hippocampal neurons resulted in enlarged cell body and phosphorylation of the mTORC1 substrate S6 kinase 1 (S6K1) (Fig. 2C). Furthermore, we performed an immunoprecipitation assay with anti-GFP antibody in both GFP and GFP-L10a expressing neurons. The enrichment of GFP-L10a or GFP is highly specific without significant background staining on the Western blots (Fig. 2D). By using RNA preserving buffer and RNase inhibitors, we performed RNA purification from both immunoprecipitation experiments. With the anti-GFP antibody, we were able to pull down RNAs only from GFP-L10a-expressing neuronal cultures, but not from GFP-expressing cultures (Fig. 2D). Anti-GFP immunoprecipitation in GFP-L10a-expressing neurons allowed us to purify ribosome-associated RNAs and profile the “translatome” in Tsc2-deficient hippocampal neurons compared with wild-type neurons.
Figure 2.
Application of TRAP assay for neuronal translational profiling. A, Expression of GFP-L10a transgene is seen in nucleoli as well as throughout the cytoplasm and neurites of cultured hippocampal neurons with MAP2 counterstaining. No significant cell death was observed in eGFP-L10a expressing cultures. Scale bar, 20 μm. B, Representative images show coexpression of GFP-L10a and mCherry-positive sh-Luc or sh-Tsc2. Scale bar, 50 μm. C, Representative Western blots confirm the coexpression of two lentivectors as indicated. D, TRAP assay using anti-GFP immunoprecipitation (IP). Representative Western blots show purification of GFP-L10a versus GFP from hippocampal neuronal cultures. Total RNAs were recovered from IP-purified ribosomes and amplified before microarray hybridization. The BioAnalyzer 2000 image confirms purification of RNAs from GFP-L10a-transfected cultures, but not from GFP-transfected cultures.
The TRAP genes display differential sensitivity to rapamycin
We purified ribosome-associated RNAs from three conditions (each in triplicate): (1) sh-Luc (control); (2) sh-Tsc2; and (3) sh-Tsc2 with overnight rapamycin (20 nm) treatment. The amplified cDNAs were subjected to conventional microarray study on Affymetrix rat gene 1.0 chips. The raw data revealed 82 genes differentially expressed at the ribosomal level among these three conditions at false discovery rate (FDR) < 1e-5. (Fig. 3A). The correlation plots from two independent TRAP experiments for each condition confirmed the high reproducibility among replicates (Fig. 3B). Based on the hierarchical clustering, the candidate genes were further classified into three groups. Group 1 contains 17 genes that were downregulated in Tsc2-deficient neurons. Several synapse or synaptogenesis-associated genes, such as Snap91 and Cbln1, are in this group. Group 2 contains 27 genes that were upregulated by Tsc2 loss, but not rescued by overnight rapamycin treatment. This group includes actin organizing proteins, such as gelsolin (Gsn) and FAM107a. Finally, in Group 3, there were 38 genes that were upregulated in response to Tsc2 loss and were suppressed by overnight rapamycin treatment. Among these, many stress responsive genes, including c-Fos, Ucp2, Hspb1/Hsp27, Hmox1, and Atf3, as well as cytokine genes, such as Ccl2 and Cxcl10, are of particular interest.
Figure 3.
TRAP microarray reveals upregulation of stress and activity responsive genes and downregulation of synapse-associated genes in Tsc2-deficient hippocampal neurons. A, Heatmap of differentially translated genes at FDR < 1e-5 (59 annotated) by comparing either sh-Luc versus sh-Tsc2, or sh-Luc versus sh-Tsc2 after rapamycin treatment. The candidates fall into three categories: (1) 17 mRNAs downregulated in sh-Tsc2 cultures compared with control cultures; (2) 27 mRNAs upregulated in sh-Tsc2 cultures and not suppressed by rapamycin treatment; and (3) 38 mRNAs upregulated in sh-Tsc2 cultures and then significantly suppressed by rapamycin treatment. B, Correlation plots from two independent TRAP experiments within sh-Luc, sh-Tsc2, or sh-Tsc2 knockdown after overnight treatment with rapamycin (20 nm). C, Scatter plot of fold changes between sh-Luc versus sh-Tsc2 knockdown cultures, and sh-Luc versus sh-Tsc2 cultures treated with rapamycin. Colored dots indicate mRNAs with significant changes at FDR < 1e-5. Red represents mRNAs that are differentially expressed in the sh-Tsc2 cultures and corrected by rapamycin treatment (based on twofold change cutoff). Green represents mRNAs that remain significantly altered in the Tsc2-deficient neurons regardless of rapamycin treatment (also based on a twofold change cutoff). Blue represents mRNAs that have altered expression merely due to rapamycin treatment.
We next focused on the annotated genes and used scatter plot analysis to determine which ones are corrected by mTORC1 inhibition by using a twofold change cutoff. Sixteen genes, including Atf3, were differentially expressed in the sh-Tsc2 cultures and corrected by rapamycin treatment (red dots in Fig. 3C; Log2 Fold < 1 when comparing sh-Tsc2 with rapamycin to sh-Luc). In contrast, 14 genes, including Ucp2, remained significantly altered in the Tsc2-deficient neurons despite rapamycin treatment (green dots; Log2 Fold > 1 when comparing sh-Tsc2 with rapamycin to sh-Luc; FDR < 1e-5). Both the activating transcription factor-3 (Atf3) and mitochondrial uncoupling protein-2 (Ucp2) are components of cellular stress response machinery. Together, these findings suggest that Tsc2 loss in neurons induces a potent stress response.
Atf3 is highly upregulated in neuronal TSC models
To further tease out translational versus transcriptional regulation, we purified total transcripts versus ribosome-associated transcripts after the TRAP procedure. Quantitative real-time PCRs for the candidate genes demonstrated a concordance between transcriptional and translational regulation for the genes we examined, except for the Il1r1 gene, whose translation is enhanced with no significant change in total transcripts. For example, both the total transcripts and the ribosome-associated transcripts of Atf3 and Ucp2 were elevated in Tsc2-deficient neurons. Interestingly, the two genes displayed different responses to rapamycin (20 nm, overnight). Overnight rapamycin treatment reversed the induction of both total transcripts and ribosome-associated transcripts of Atf3. The ribosome-associated transcripts of Ucp2, however, remained high following overnight rapamycin treatment even though its total transcript levels were dramatically reduced (Fig. 4A,B).
Figure 4.
Translational versus transcriptional regulation of the TRAP candidate genes. A, Fold change at ribosome-associated mRNA (“translating transcripts”) level for indicated genes in sh-Tsc2 cultures, sh-Tsc2 cultures treated with rapamycin compared with sh-Luc hippocampal cultures. B, Fold change at transcriptional (“total transcripts”) level for the same genes. *p < 0.05 (two-way ANOVA). **p < 0.01 (two-way ANOVA). ***p < 0.001 (two-way ANOVA). ns, Not significant. C, Representative Western blot image showing elevated protein levels of ATF3 and UCP2 in sh-Tsc2 hippocampal cultures and differential suppression by rapamycin treatment for overnight or 7 d. D, Densitometry quantification of ATF3 fold change normalized to tubulin (vehicle n = 13, Rapa O/N n = 7, Rapa 7d n = 4). Error bars indicate mean ± SEM. **p < 0.01 (two-way ANOVA). E, Increased protein levels of ATF3 (1.49 ± 0.09 (SD) fold) in Tsc1-deficient mouse hippocampus versus wild-type controls (CC: Syn1-Cre+Tsc1cc; WW: Syn1-Cre+/Tsc1ww; postnatal day 21). F, Densitometry quantification of ATF3 fold change normalized to GAPDH. *p < 0.05 (Student's t test). n = 3 per condition.
We further investigated protein levels of Atf3 in TSC models, as well as the responses to acute versus chronic rapamycin treatment. By Western blotting, we found that hippocampal neurons deficient in Tsc2 had a strong induction of ATF3 (Fig. 4C). This induction of ATF3 was reversed by overnight or prolonged rapamycin treatment (Fig. 4D). Furthermore, we found elevated expression of ATF3 in hippocampal lysates from a neuron-specific Tsc1 knock-out mouse model (Tsc1; Synapsin1-Cre, postnatal day 21; Fig. 4E). By quantification, we found that ATF3 levels in homozygous mutants (CC; Synapsin1-Cre+/Tsc1cc) were ∼50% higher than that of control littermates (WW; Synapsin1-Cre+/Tsc1ww) (Fig. 4F). Together, these data indicate that ATF3 is induced both in vitro and in vivo in TSC-deficient neurons.
In wild-type neurons, overexcitability due to excessive NMDA-receptor signaling activates the calcium-CREB pathway to eventually enhance the expression of Atf3 (Hashimoto et al., 2002; Zhang et al., 2009, 2011). Atf3 expression is also induced in sensory and motor neuron following axotomy (Tsujino et al., 2000; Seijffers et al., 2007). We thus asked whether the increase in ATF3 protein was due to neuronal hyperexcitability in the Tsc2-deficient cultures. To test whether neuronal hyperexcitability contributed to Atf3 expression in Tsc2-deficient neurons, we treated neurons with TTX (1 μm), which blocks sodium channels and consequently neuronal excitability. As expected, TTX treatment for 24 h reduced activation of CREB, as shown by Western blotting for phospho-CREB (Fig. 5A). However, levels of ATF3 and gelsolin proteins were not significantly altered (Fig. 5A–C). These results indicate that activation of the ATF3/gelsolin pathway is independent of neuronal excitability but likely due to mTOR hyperactivity, given that ATF3 induction is corrected by rapamycin treatment (Fig. 4D).
Figure 5.
Increased levels of ATF3 and gelsolin are activity-independent in the Tsc2-deficient neurons. A, Representative Western blot of control and Tsc2-deficient neurons untreated or treated with 1 μm TTX for 24 h. S6 phosphorylation at Ser240/244 (p-S6) indicates mTORC1 activation. Decreased CREB phosphorylation at Ser133 confirms reduced synaptic activity in the TTX-treated neurons. B, C, Densitometry quantification of ATF3 (B) and gelsolin (C) shown as fold change. Values were normalized to total Akt level (n = 6). Error bars indicate mean ± SEM (two-way ANOVA). n.s., Statistically not significant.
Dendritic spine deficits in Tsc2-deficient neuronal culture
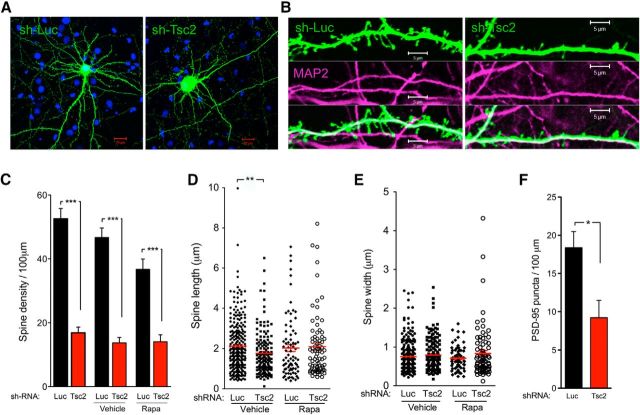
Loss of Tsc1 or Tsc2 in pyramidal neurons of hippocampal organotypic slice cultures results in enlarged soma size, increased spine length and head width, but decreased spine density (Tavazoie et al., 2005). In our dissociated cultures, we found similar morphologic deficits in the Tsc2-deficient hippocampal neurons. The soma size of neurons infected with lentivirus carrying sh-Tsc2 appeared larger than that of sh-Luc expressing neurons (Fig. 6A). The spine densities were significantly reduced in sh-Tsc2 compared with sh-Luc neuronal cultures (52.57 ± 3.204 and 16.85 ± 1.794 per 100 μm in sh-Luc vs sh-Tsc2 cultures at 16–21 DIV, respectively). We did not detect an increase of spine density in response to overnight rapamycin treatment. We also found a significant reduction of spine length in Tsc2-deficient neurons compared with control neurons (2.13 ± 0.079 μm in sh-Luc vs 1.77 ± 0.089 μm in sh-Tsc2 neuronal cultures). Rapamycin did rescue the deficit in spine length (2.03 ± 0.172 μm for sh-Luc vs 2.10 ± 0.175 μm for sh-Tsc2). By contrast, we did not observe differences in the spine width between Tsc2-deficient and control neurons that were treated with either rapamycin or vehicle (Fig. 6B–E).
Figure 6.
Acute rapamycin treatment rescues the reduction of dendritic spine length but not of spine density in Tsc2-deficient neurons. A, Hippocampal neurons were transfected with lentivector-based sh-Luc versus sh-Tsc2. Representative images show a dramatic decrease in the dendritic spine density as well as increased soma size in Tsc2-deficient neurons. Scale bars, 20 μm. B, High-magnification images showing the spine density and morphology in sh-Luc versus sh-Tsc2-transfected neurons. MAP2 stains dendrites. Scale bars, 5 μm. C, Quantification of spine densities in nontreated hippocampal neuronal cultures or cultures treated with vehicle versus rapamycin. Acute rapamycin treatment failed to rescue the deficits in spine density in Tsc2-deficient neurons compared with control cultures. ***p < 0.001 (Student's t test). D, Rapamycin rescued the spine length that decreases in Tsc2-deficient hippocampal neurons. **p < 0.01 (Student's t test). E, No significant differences in spine width between sh-Luc versus sh-Tsc2 cultures were detected, regardless of treatments with vehicle or rapamycin. F, The densities of PSD-95 puncta per 100 μm in sh-Luc versus sh-Tsc2 cultures are 18.38 ± 2.12 and 9.22 ± 2.27 (mean ± SEM). *p < 0.05 (Student's t test).
Given the reduction in spine density in Tsc2-deficient neurons, we asked whether molecular markers of excitatory synapses were affected. In neuronal cultures infected with sh-Tsc2 versus sh-Luc virus, we found a significant reduction in the density of PSD-95 puncta in mutant neurons (18.38 ± 2.120 per 100 μm in sh-Luc vs 9.22 ± 2.274 per 100 μm in sh-Tsc2 neurons; Fig. 6F), consistent with a loss of dendritic spines. Together, these data suggest structural and functional abnormalities of excitatory synapses in cultured Tsc2-deficient hippocampal neurons.
Blockade of the Atf3-gelsolin cascade rescues spine density deficit in Tsc2-deficient neurons
Actin-organizing proteins, such as gelsolin and FAM107a, were also found to be upregulated in our TRAP profiling study. Gelsolin, an actin-severing protein, regulates stability of the actin cytoskeleton and is a direct target of Atf3's transcriptional activity (Yuan et al., 2013). Therefore, we hypothesized that dysregulation of the Atf3-gelsolin cascade could contribute to the reduced spine density in Tsc2-deficient neurons. To test this hypothesis, we infected hippocampal neurons with mCherry expressing sh-Luc or sh-Tsc2, followed by second infection with lentivirus that bicistronically expresses GFP and sh-Atf3. Western blotting revealed that the gelsolin protein level was significantly increased in Tsc2-deficient neurons. shRNA-mediated Atf3 knockdown in Tsc2-deficient neurons prevented the elevation of gelsolin expression with no effects on mTORC1 activity as measured by phospho-S6 (S240/244) levels (Fig. 7A). The expression of mCherry and GFP by different shRNA lentivectors facilitates the investigation of dendritic morphology. We found a robust rescue of spine density in Tsc2-deficient neurons when Atf3 expression was blocked (Fig. 7B,C). Confirming that this increase in spine density is indeed due to reduced endogenous Atf3 expression, sequential exogenous overexpression of human FLAG-tagged ATF3 (resistant to Atf3 shRNA) reduced spine density significantly. The spine densities per 100 μm were 8.635 ± 2.242 (mean ± SE) in sh-Tsc2, 35.17 ± 7.667 in sh-Atf3, 44.15 ± 5.493 in sh-Tsc2 plus sh-Atf3, and 20.04 ± 1.838 in shTsc2 plus sh-Atf3 and ATF3-FLAG add-back cultures at DIV 16 (Figure 7B,C). A lower spine density obtained from this experiment compared with Figure 6C is likely due to the age difference of the neuronal cultures (DIV 16 vs DIV 16–21). These data demonstrate a previously unappreciated negative effect of the Atf3-gelsolin cascade on spine number and neuronal connectivity.
Discussion
In this study, we applied a TRAP profiling approach to identify mRNAs that are regulated by TSC/mTOR signaling in hippocampal neurons. Our data in Tsc2-deficient hippocampal neurons uncover several functional groups of mRNAs that encode synapse-related proteins, which are downregulated, and stress-inducible proteins, which are upregulated. Importantly, our data link the activation of the Atf3-gelsolin signaling cascade to dendritic spine deficits of Tsc2-deficient neurons, which has potential therapeutic implications for correcting impaired neuronal connectivity in TSC and related mTORopathies.
Ribosome purification has proven a powerful unbiased tool to elucidate cell-type specific translational profiles (Doyle et al., 2008; Heiman et al., 2008; Sanz et al., 2009; Knight et al., 2012). Expression of eGFP-L10a driven by cell type-specific regulatory elements (TRAP mouse lines) has demonstrated the high sensitivity and robustness of this methodology in various types of neural cell populations (Doyle et al., 2008; Heiman et al., 2008). One limitation of the TRAP methodology as originally published is the low yield of mRNA; however, we were able to overcome this by using lentivector-mediated eGFP-L10a expression for tagging polysomes in large-scale primary hippocampal neuronal cultures (Fig. 2A).
The serine/threonine kinase mTOR is a master regulator of cellular mRNA translation. We found that shRNA-mediated Tsc2 knockdown robustly activated mTOR signaling and increased the overall de novo protein synthesis by ∼16% (Fig. 1). To identify the mRNAs or proteins that might contribute to the seizure susceptibility and/or neurocognitive impairment seen in patients with TSC, we applied the comparative TRAP assays in a well-established TSC neuronal culture model (Nie and Sahin, 2012). Expression of the transgene eGFP-L10a does not only assist in cell visualization, but it also allows isolation of ribosomes from the entire cell with no need of dissociation that would otherwise disrupt neuronal processes and their network (Fig. 2A,B). Compared with conventional total RNA expression profiling, the TRAP assay allows identification of translated mRNA content that more closely matches the protein content in a given cell (Heiman et al., 2008, 2014). This advantage is corroborated by the fact that we were able to validate expression changes at the protein level for all tested candidate genes (Fig. 4C; and data not shown). Given the advent of new RNA amplification and sequencing technologies, the TRAP methodology will further enable the study of local translation within axons and dendrites.
Synaptogenesis-related proteins are downregulated in Tsc2-deficient neurons
We have identified 13 proteins that are significantly underexpressed in Tsc2-deficient neurons despite overall increased protein synthesis (Fig. 1). Of particular interest are presynaptic proteins, such as Snap91 and Cbln1 (Yao et al., 2002; Matsuda and Yuzaki, 2011; Ito-Ishida et al., 2012). Snap91/AP180 levels and distribution are highly sensitive to neuronal activity (Wu et al., 2010). Knockdown of Snap91/AP180 impairs axonal development, synaptic vesicle size, and density (Morris et al., 1993; Petralia et al., 2013). Cbln1, also known as precerebellin, induces synaptogenesis in various types of neurons, including hippocampal, cerebellar, and cortical neurons (Matsuda and Yuzaki, 2011). In the cerebellum, trans-synaptic interaction of postsynaptic GluRδ2 with presynaptic neurexins through Cbln1 mediates parallel fiber synapses onto Purkinje cells. Downregulation of presynaptic proteins may be simply a consequence of impaired neuronal connectivity in TSC, or alternatively, it may reflect a role of the TSC-mTOR pathway in presynaptic differentiation. This might possibly link TSC and another family of autism genes, the neurexins.
Stress-responsive and anti-inflammatory proteins in TSC and autism
The vast majority of genes in our profiling dataset are stress and/or activity-related genes, such as c-Fos, Ucp2, Hspb1, Hmox1, and Atf3. Accumulating evidence suggests the presence of oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism (Chauhan and Chauhan, 2006; Rossignol and Frye, 2014; Masi et al., 2015). Induced by cellular stress and kainic acid, the mitochondrial uncoupling protein-2 (Ucp2) modulates mitochondrial membrane potential to decrease ATP production, leading to neuroprotection against oxidative stress (Kim-Han and Dugan, 2005; Chuang et al., 2012). Decreased synthesis of ATP has been shown by 31P-magnetic resonance spectroscopy (Minshew et al., 1993), suggesting an involvement of mitochondrial dysfunction in autism. Our previous study has demonstrated an mTOR-dependent induction of Hmox1 (heme oxygenase-1) in Tsc2-deficient neurons, indicating an enhanced oxidative stress response (Di Nardo et al., 2009). Atf3 is a transcription factor in the CREB protein family that is induced by action potential bursting and nuclear calcium signaling (Zhang et al., 2009; Zhang et al., 2011; Hunt et al., 2012). Hmox1 also has anti-inflammatory effects. Other anti-inflammatory proteins we have identified include Ccl2 and Cxcl10. Overall, upregulation of these proteins represents prosurvival or antiapoptotic mechanisms. Given an incidence of ∼50% of autism in patients with TSC (Asato and Hardan, 2004; Jeste et al., 2008), the revealed molecular signatures could have diagnostic or prognostic value for the neurocognitive comorbidity in these patients.
The role of the Atf3-gelsolin cascade in TSC pathophysiology
Changes in dendritic and synaptic architecture have been described in Tsc1- or Tsc2-deficient neurons. Whereas a number of reports have documented a decrease in spine density following knockdown of Tsc1/2 (Tavazoie et al., 2005; Yasuda et al., 2014; Sugiura et al., 2015), none or only moderate changes were reported in others (Meikle et al., 2007; Bateup et al., 2011). In the present study, we observed morphologic deficits in Tsc2-deficient hippocampal neuronal cultures. These neurons developed enlarged soma size and decreased density of dendritic spines compared with controls. These findings are similar to what has been described previously (Tavazoie et al., 2005). Furthermore, we found that overnight rapamycin treatment did not rescue spine density, even though it did correct the deficit in spine length (Fig. 6B–E). We also detected a reduction in the number of PSD-95-immunoreactive puncta, which is consistent with more sparse dendritic spines in these neurons (Fig. 6F). These data strongly indicate an impairment of synaptic connectivity in Tsc2-deficient neurons.
Consistent with our previous in vivo studies (Meikle et al., 2007), we found that acute rapamycin treatment does not rescue spine deficits in cultured Tsc2-deficient rat hippocampal neurons (Fig. 6C), although it did reduce ATF3 to levels that are not different from control neurons (Fig. 4C,D). Different effects of acute rapamycin treatment versus Atf3 knockdown could potentially be explained by a number of factors in our experimental system. It is likely that different mTOR-dependent and mTOR-independent pathways regulate spine density in TSC. Consistent with our results, Yasuda et al. (2014) recently reported that Tsc2+/− neurons show impaired spine synapse formation, which is unaffected by mTORC1 inhibition. The same group went on to show that this is mediated through Rheb-syntenin signaling (Sugiura et al., 2015). Furthermore, rapamycin could affect spine formation, stability, and pruning in ways independent of Atf3. Detailed time-lapse experiments over a greater time period would help to clarify the roles of Atf3, rapamycin, and other players.
Dendritic spines are motile structures that contain high concentrations of filamentous actin. The importance of the actin skeleton for dendritic architecture is exemplified by findings by Zhang et al. (2014), which demonstrated an increased expression of actin-crosslinking protein filamin A (FLNA) responsible for abnormal dendritic morphogenesis in TSC-null mice. Although we did not detect increased FLNA expression in our TRAP screening assay, we found that gelsolin, an actin-severing protein, is one of the genes upregulated in the Tsc2-deficient neurons. Gelsolin was previously reported to be highly expressed in renal cystadenomas in a TSC mouse model (Onda et al., 1999). Gelsolin protein binds to and severs large actin filaments, by which it accelerates filopodia retraction. Interestingly, gelsolin is a direct target gene of Atf3 transcription activity (Yuan et al., 2013). Induction of Atf3 and gelsolin could be neuroprotective in certain contexts (Zhang et al., 2009; Harms et al., 2004; Seijffers et al., 2014). Loss of gelsolin in hippocampal neurons renders vulnerability to glutamate toxicity and exacerbates seizure-induced damage in the brain (Furukawa et al., 1997), suggesting a potent cell-autonomous program that protects neurons from dying of excitotoxicity or seizures.
Our data demonstrate that blocking Atf3 induction prevents gelsolin overexpression and subsequently corrects the loss of dendritic spines in Tsc2-deficient neurons. It is likely that activation of the Atf3-gelsolin cascade, regardless of initial prosurvival benefits, can reduce synaptic connectivity and contribute to long-term neurocognitive impairment seen in patients with TSC. Whether or not gelsolin is connected with mTOR complex-2 (mTORC2) or solely an indirect target of mTORC1 via Atf3 induction remains to be investigated. Given that the Atf3-gelsolin pathway is primarily prosurvival, temporal and local manipulation would be imperative for its use in a therapeutic context.
Footnotes
This work was supported by the John Merck Fund, Nancy Lurie Marks Family Foundation, National Institutes of Health Grant P30 HD018655, the Boston Children's Hospital Translational Research Program to M.S., and the Manton Center for Orphan Disease Research fellowship to D.N., D.E.-F. was supported by Ruprecht-Karls-University Heidelberg Faculty of Medicine Young Investigator Award Program, the Daimler and Benz Foundation (Project 32-05/13), and the Reinhard-Frank Foundation.
The authors declare no competing financial interests.
References
- Asato MR, Hardan AY. Neuropsychiatric problems in tuberous sclerosis complex. J Child Neurol. 2004;19:241–249. doi: 10.1177/088307380401900401. [DOI] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–522. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Di Nardo A, Kramvis I, Meikle L, Kwiatkowski DJ, Sahin M, He X. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 2008;22:2485–2495. doi: 10.1101/gad.1685008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang YC, Lin TK, Huang HY, Chang WN, Liou CW, Chen SD, Chang AY, Chan SH. Peroxisome proliferator-activated receptors gamma/mitochondrial uncoupling protein 2 signaling protects against seizure-induced neuronal cell death in the hippocampus following experimental status epilepticus. J Neuroinflammation. 2012;9:184. doi: 10.1186/1742-2094-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Shore CJ, Major P, Camposano S, Muzykewicz D, Thiele EA. The natural history of epilepsy in tuberous sclerosis complex. Epilepsia. 2010;51:1236–1241. doi: 10.1111/j.1528-1167.2009.02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario FJ, Jr, Sahin M, Ebrahimi-Fakhari D. Tuberous sclerosis complex. Pediatr Clin North Am. 2015;62:633–648. doi: 10.1016/j.pcl.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Kramvis I, Cho N, Sadowski A, Meikle L, Kwiatkowski DJ, Sahin M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J Neurosci. 2009;29:5926–5937. doi: 10.1523/JNEUROSCI.0778-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr Opin Neurol. 2015;28:91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- Feliciano DM, Lin TV, Hartman NW, Bartley CM, Kubera C, Hsieh L, Lafourcade C, O'Keefe RA, Bordey A. A circuitry and biochemical basis for tuberous sclerosis symptoms: from epilepsy to neurocognitive deficits. Int J Dev Neurosci. 2013;31:667–678. doi: 10.1016/j.ijdevneu.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fu W, Li Y, Witke W, Kwiatkowski DJ, Mattson MP. The actin-severing protein gelsolin modulates calcium channel and NMDA receptor activities and vulnerability to excitotoxicity in hippocampal neurons. J Neurosci. 1997;17:8178–8186. doi: 10.1523/JNEUROSCI.17-21-08178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille JC, Sonenberg N. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms C, Bösel J, Lautenschlager M, Harms U, Braun JS, Hörtnagl H, Dirnagl U, Kwiatkowski DJ, Fink K, Endres M. Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol Cell Neurosci. 2004;25:69–82. doi: 10.1016/j.mcn.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Zhang C, Kawauchi J, Imoto I, Adachi MT, Inazawa J, Amagasa T, Hai T, Kitajima S. An alternatively spliced isoform of transcriptional repressor ATF3 and its induction by stress stimuli. Nucleic Acids Res. 2002;30:2398–2406. doi: 10.1093/nar/30.11.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D, Raivich G, Anderson PN. Activating transcription factor 3 and the nervous system. Front Mol Neurosci. 2012;5:7. doi: 10.3389/fnmol.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Ishida A, Miyazaki T, Miura E, Matsuda K, Watanabe M, Yuzaki M, Okabe S. Presynaptically released Cbln1 induces dynamic axonal structural changes by interacting with GluD2 during cerebellar synapse formation. Neuron. 2012;76:549–564. doi: 10.1016/j.neuron.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Jeste SS, Sahin M, Bolton P, Ploubidis GB, Humphrey A. Characterization of autism in young children with tuberous sclerosis complex. J Child Neurol. 2008;23:520–525. doi: 10.1177/0883073807309788. [DOI] [PubMed] [Google Scholar]
- Kim-Han JS, Dugan LL. Mitochondrial uncoupling proteins in the central nervous system. Antioxid Redox Signal. 2005;7:1173–1181. doi: 10.1089/ars.2005.7.1173. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Tan K, Birsoy K, Schmidt S, Garrison JL, Wysocki RW, Emiliano A, Ekstrand MI, Friedman JM. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151:1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi A, Quintana DS, Glozier N, Lloyd AR, Hickie IB, Guastella AJ. Cytokine aberrations in autism spectrum disorder: a systematic review and meta-analysis. Mol Psychiatry. 2015;20:440–446. doi: 10.1038/mp.2014.59. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew NJ, Goldstein G, Dombrowski SM, Panchalingam K, Pettegrew JW. A preliminary 31P MRS study of autism: evidence for undersynthesis and increased degradation of brain membranes. Biol Psychiatry. 1993;33:762–773. doi: 10.1016/0006-3223(93)90017-8. [DOI] [PubMed] [Google Scholar]
- Morris SA, Schröder S, Plessmann U, Weber K, Ungewickell E. Clathrin assembly protein AP180: primary structure, domain organization and identification of a clathrin binding site. EMBO J. 1993;12:667–675. doi: 10.1002/j.1460-2075.1993.tb05700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie D, Sahin M. A genetic model to dissect the role of Tsc-mTORC1 in neuronal cultures. Methods Mol Biol. 2012;821:393–405. doi: 10.1007/978-1-61779-430-8_25. [DOI] [PubMed] [Google Scholar]
- Nie D, Di Nardo A, Han JM, Baharanyi H, Kramvis I, Huynh T, Dabora S, Codeluppi S, Pandolfi PP, Pasquale EB, Sahin M. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Indig FE, Bushlin I, Wu F, Mattson MP, Yao PJ. Reduction of AP180 and CALM produces defects in synaptic vesicle size and density. Neuromol Med. 2013;15:49–60. doi: 10.1007/s12017-012-8194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Evidence linking oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism. Front Physiol. 2014;5:150. doi: 10.3389/fphys.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Zhang J, Matthews JC, Chen A, Tamrazian E, Babaniyi O, Selig M, Hynynen M, Woolf CJ, Brown RH., Jr ATF3 expression improves motor function in the ALS mouse model by promoting motor neuron survival and retaining muscle innervation. Proc Natl Acad Sci U S A. 2014;111:1622–1627. doi: 10.1073/pnas.1314826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura H, Yasuda S, Katsurabayashi S, Kawano H, Endo K, Takasaki K, Iwasaki K, Ichikawa M, Kobayashi T, Hino O, Yamagata K. Rheb activation disrupts spine synapse formation through accumulation of syntenin in tuberous sclerosis complex. Nat Commun. 2015;6:6842. doi: 10.1038/ncomms7842. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mora-Jensen H, Weniger MA, Perez-Galan P, Wolford C, Hai T, Ron D, Chen W, Trenkle W, Wiestner A, Ye Y. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc Natl Acad Sci U S A. 2009;106:2200–2205. doi: 10.1073/pnas.0807611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Mattson MP, Yao PJ. Neuronal activity and the expression of clathrin-assembly protein AP180. Biochem Biophys Res Commun. 2010;402:297–300. doi: 10.1016/j.bbrc.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Coleman PD, Calkins DJ. High-resolution localization of clathrin assembly protein AP180 in the presynaptic terminals of mammalian neurons. J Comp Neurol. 2002;447:152–162. doi: 10.1002/cne.10217. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Sugiura H, Katsurabayashi S, Shimada T, Tanaka H, Takasaki K, Iwasaki K, Kobayashi T, Hino O, Yamagata K. Activation of Rheb, but not of mTORC1, impairs spine synapse morphogenesis in tuberous sclerosis complex. Sci Rep. 2014;4:5155. doi: 10.1038/srep05155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Yu L, Li J, Xie G, Rong T, Zhang L, Chen J, Meng Q, Irving AT, Wang D, Williams ED, Liu JP, Sadler AJ, Williams BR, Shen L, Xu D. ATF3 suppresses metastasis of bladder cancer by regulating gelsolin-mediated remodeling of the actin cytoskeleton. Cancer Res. 2013;73:3625–3637. doi: 10.1158/0008-5472.CAN-12-3879. [DOI] [PubMed] [Google Scholar]
- Zhang L, Bartley CM, Gong X, Hsieh LS, Lin TV, Feliciano DM, Bordey A. MEK-ERK1/2-dependent FLNA overexpression promotes abnormal dendritic patterning in tuberous sclerosis independent of mTOR. Neuron. 2014;84:78–91. doi: 10.1016/j.neuron.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Buchthal B, Lau D, Hayer S, Dick O, Schwaninger M, Veltkamp R, Zou M, Weiss U, Bading H. A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J Neurosci. 2011;31:4978–4990. doi: 10.1523/JNEUROSCI.2672-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]