Abstract
The mammalian cochlea exhibit minimal spontaneous regeneration, and loss of sensory hair cells (HCs) results in permanent hearing loss. In nonmammalian vertebrates, spontaneous HC regeneration occurs through both proliferation and differentiation of surrounding supporting cells (SCs). HC regeneration in postnatal mammalian cochleae in vivo remains limited by the small HC number and subsequent death of regenerated HCs. Here, we describe in vivo generation of 10-fold more new HCs in the mouse cochlea than previously reported, most of which survive to adulthood. We achieved this by combining the expression of a constitutively active form of β-catenin (a canonical Wnt activator) with ectopic expression of Atoh1 (a HC fate determination factor) in neonatal Lgr5+ cells (the presumed SC and HC progenitors of the postnatal mouse cochlea), and discovered synergistic increases in proliferation and differentiation. The new HCs were predominantly located near the endogenous inner HCs, expressed early HC differentiation markers, and were innervated despite incomplete alignment of presynaptic and postsynaptic markers. Surprisingly, genetic tracing revealed that only a subset of Lgr5+ cells that lie medial to the inner HCs respond to this combination, highlighting a previously unknown heterogeneity that exists among Lgr5+ cells. Together, our data indicate that β-catenin and Atoh1 mediate synergistic effects on both proliferation and differentiation of a subset of neonatal cochlear Lgr5+ cells, thus overcoming major limitations of HC regeneration in postnatal mouse cochleae in vivo. These results provide a basis for combinatorial therapeutics for hearing restoration.
SIGNIFICANCE STATEMENT Hearing loss in humans from aging, noise exposure, or ototoxic drugs (i.e., cisplatin or some antibiotics) is permanent and affects every segments of the population, worldwide. However, birds, frog, and fish have the ability to recover hearing, and recent studies have focused on understanding and applying what we have learned from them for restoring hearing in humans. However, studies have been hampered by low efficiency, limited cell numbers, and subsequent death of these newly generated auditory cells. Here, we describe a combinatorial approach, which results in the generation of auditory cells in greater numbers than previously reported, with most of them surviving to adult ages in vivo. These results provide a basis for combinatorial therapeutics for hearing restoration efforts.
Keywords: canonical Wnt, combinatory therapy, direct conversion, Lgr5 stem cell, mitotic generation, Notch
Introduction
The mammalian hearing organ, the organ of Corti, is comprised of both mechanosensory hair cells (HCs) and nonsensory supporting cells (SCs), but the loss of HCs results in permanent hearing loss (Bohne et al., 1976; Hawkins et al., 1976; Kelley, 2007; Oesterle et al., 2008). In contrast, nonmammalian vertebrates are capable of HC regeneration by either direct SC to HC transdifferentiation or SC mitosis followed by SC to HC transdifferentiation (termed mitotic regeneration), and subsequently recover hearing function (Corwin and Cotanche, 1988; Ryals and Rubel, 1988; Stone and Cotanche, 2007; Rubel et al., 2013). Despite recent advances, mitotic regeneration in postnatal mammalian cochleae in vivo remains limited by both insufficient cell number and survival.
The canonical Wnt signaling pathway, mediated by β-catenin, is critical for proper development and maturation of the cochlea (Dabdoub et al., 2003; Ohyama et al., 2007; Jayasena et al., 2008; Munnamalai and Fekete, 2013; Shi et al., 2014). In the chicken basilar papilla and the zebrafish lateral line, Wnt activation plays a key role in promoting SC proliferation in response to ototoxic insult and the subsequent generation of new HCs (Head et al., 2013; Jacques et al., 2014). In the mouse, in vivo studies have shown that the expression of an active form of β-catenin only results in a transient proliferation response in Lgr5+ cells using the Lgr5CreER mouse line, whereas new HCs are generated from Lgr5+ cells using the Sox2CreER mouse line (Chai et al., 2012; Shi et al., 2013). However, it is unclear whether Sox2 haploinsufficiency contributes to the differential responses observed between the two aforementioned mice lines.
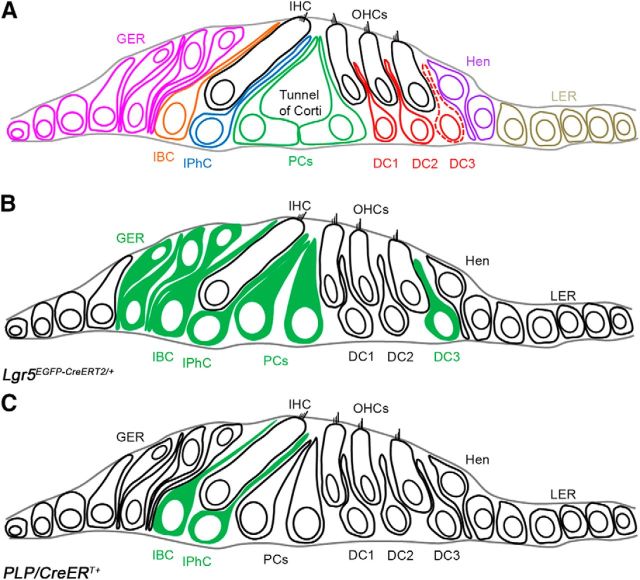
Lgr5 is characterized as a stem cell marker in the intestine and the hair follicle (Oshima et al., 2001; Barker et al., 2007). In the neonatal mammalian cochlea, Lgr5 is expressed in some nonsensory epithelial cells in the greater epithelial ridge (GER), the inner border cells (IBCs), the inner phalangeal cells (IPhCs), pillar cells, and the third row of Deiters' cells (DCs) (Fig. 1) (Chai et al., 2011; Shi et al., 2012). Isolated Lgr5+ cells from the neonatal organ of Corti are able to both proliferate and transdifferentiate into HCs in vitro, and recent reports have linked Lgr5+ cells to neonatal HC regeneration (Chai et al., 2012; Shi et al., 2012; Cox et al., 2014). These data suggest that Lgr5+ cells could represent progenitor-like cells in postnatal cochleae for regeneration.
Figure 1.
Diagram of the organ of the Corti and its SC subtypes. A, Schematic cross-section of organ of Corti of an adult mouse cochlea depicting the positions of SCs and the nonsensory epithelial cell subtypes relative to the IHC and OHCs. Red dotted line indicates the third row of Deiters' cells, where Lgr5 expression remains throughout adulthood. B, Schematic representation of Lgr5EGFP-CreERT2/+-targeted cells in green in neonatal cochlea. C, Schematic representation of PLP/CreERT+-targeted SCs in green in neonatal cochlea. OHCs, Outer hair cells; IHC, inner hair cell; GER, greater epithelial ridge; IBC, inner border cell; IPhC, inner phalangeal cell; PCs, pillar cells; DC1, first row of Deiters' cell; DC2, second row of Deiters' cell; DC3, third row of Deiters' cell; Hen, Hensen's cells; LER, lesser epithelial ridge.
The basic helix-loop-helix transcription factor, Atoh1, plays a key role in HC fate determination (Bermingham et al., 1999; Chen et al., 2002; Mulvaney and Dabdoub, 2012; Jarman and Groves, 2013; Jahan et al., 2015), and its ectopic expression in neonatal SCs induces SC to HC transdifferentiation. However, it is a slow and inefficient process, with most cells failing to transdifferentiate (<150 new HCs per cochlea) (Zheng and Gao, 2000; Kawamoto et al., 2003; Kelly et al., 2012; Liu et al., 2014).
To achieve both proliferation and transdifferentiation in postnatal cochleae, we ectopically coexpressed a constitutively active form of β-catenin and Atoh1 in neonatal cochlear Lgr5+ cells. We discovered that this combination acts synergistically on a subset of Lgr5+ cells by inducing both proliferation and differentiation, with the formation of 1338.48 ± 271.98 new HCs per cochlea, most of which survive to adulthood, thus illustrating an inherent heterogeneity that exists among the Lgr5+ cells. Together, our studies provide a foundation for future research using combinatorial therapeutics for mitotic regeneration in mammals for hearing restoration efforts.
Materials and Methods
Mice.
Lgr5EGFP-CreERT2/+ and PLP/CreERT+ mice were purchased from the The Jackson Laboratory (stock #008875 and #005975). Conditional Lgr5 deletion (Lgr5fl/+) mouse was obtained from Dr. Clevers (Hubrecht Institute, Utrecht, The Netherlands) and described previously (de Lau et al., 2011). Hes5nLacZ mouse was obtained from Dr. Kageyama and described previously (Imayoshi et al., 2010). Atoh1-HA mouse was described previously (Liu et al., 2012). Refer to the aforementioned references for details regarding the PCR genotyping. Tamoxifen (T5648-5G, Sigma; 3 mg/40 g body weight) was given by intraperitoneal injections at postnatal (P) day 0 (P0) and 1 (P1). Sample sizes were N = 3–7 mice of either sex for each group (control and experimental) at each time point for all experiments (except for P42-P44 where N = 2). All animal work conducted during the course of this study was approved by the Institutional Animal Care and Use Committee at St. Jude Children's Research Hospital and was performed according to National Institutes of Health guidelines.
Tissue preparation, immunofluorescence, and analysis.
Isolated cochleae were fixed in 2% (v/v) PFA (Electron Microscopy Services) in PBS (P3813, Sigma) for 3–4 h at room temperature or at 4°C overnight, and subsequently washed in PBS (3 × 5 min). Whole-mount preparations were performed as described previously (Liu et al., 2010). The following primary antibodies were used: anti-myosin-VII (rabbit, 1:200, 25-6790, Proteus Biosciences), anti-HA (rat,1:100, 11867423001, Roche), anti-Ctbp2 (mouse,1:500, 612044, BD Transduction Laboratories), anti-GFP (chicken, 1:1000, ab13970, Abcam), anti-GluR2 (mouse, 1:200, MAB397, Millipore), anti-prestin (goat, 1:200, sc-22692, Santa Cruz Biotechnology), anti-Sox2 (goat, 1:1000, sc-17320, Santa Cruz Biotechnology), vGlut3 (rabbit, 1:500, 135203, Synaptic Systems), and anti-Tuj1 (mouse, 1:1000, MMS-435P, Covance). All secondary antibodies were purchased from Invitrogen and used at 1:1000, except for AlexaFluor-405 goat anti-rabbit IgG and AlexaFluor-647 goat anti-rat IgG (1:500). BrdU detection was achieved using AlexaFluor-647-conjugated antibody (mouse, 1:20, B35133, Invitrogen). Confocal imaging was performed on a Zeiss LSM 700 or 710, and image processing was performed with Photoshop CS6 (Adobe Systems).
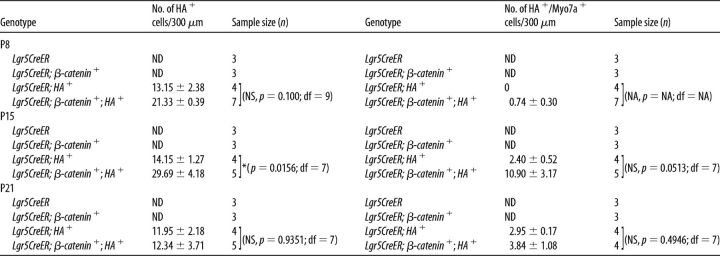
Quantification of ectopic HCs.
Cochleae were cut into two parts with the cut site close to the end of the first apical turn. The apical, middle, and base regions were first imaged <20× to identify regions of interest, and at least two Z-stacks from nonoverlapping regions were then obtained for each turn using the 40× objective (1 μm step). For each genotype and age, at least 3 cochleae from different mice were analyzed using Zeiss LSM image browser. We determined the number of ectopic HCs by scoring HA+ and HA+/Myosin7a+ cells along the z-axis and determined the average number of cells per 300 μm section based on an average measured cochlear length of 5.4 mm (from P8 and P21 whole mounts).
RNA isolation and real-time PCR.
A minimum of 3 littermates from at least 2 litters were assayed. Pups were killed by decapitation, and the ears were dissected in ice-cold HBSS. RNA was isolated using the RNAqueous-Micro RNA Isolation Kit (Ambion). cDNA was generated using Superscript First-Strand cDNA Synthesis system for RT-PCR (Invitrogen) with random primers. Relative expression levels were assayed using TaqMan Gene Expression Master Mix and TaqMan probes (Applied Biosystems) for Notch1, Sox2, Jagged1, Delta-like 1, and 18S. Reactions were run in triplicate in an Eppendorf Realplex2 Mastercycler System, which has a CT cutoff value of 40. In our analysis, we set the upper most limit of CT at 38 to be included in the analysis. Each sample was run in triplicate on each plate, and each sample was also run on multiple plates to ensure the validity of the results. The level of 18S was used as an internal control and was run as a multiplex reaction with each assayed gene. The difference in CT between the assayed gene and 18S for any given sample was defined as ΔCT(X). The difference in ΔCT(x) between two samples was defined as ΔΔCT(X), which represents a relative difference in expression of the assayed gene. The fold change of the assayed gene relative to 18S was defined as 2−ΔΔCT (Livak and Schmittgen, 2001). DataAssist software (Applied Biosystems) was used for statistical analysis and to confirm ΔCT(X) calculation.
Statistical analysis.
Statistical analysis was performed using the GraphPad Prism 5.0 (Graphpad Software). Nonparametric Student's t test with Bonferroni correction or two-way ANOVA with Bonferroni correction (for auditory brainstem response [ABR] measurements) was used to determine statistical significance.
Results
β-Catenin and Atoh1 synergistically increase the proliferation of Lgr5+ cells
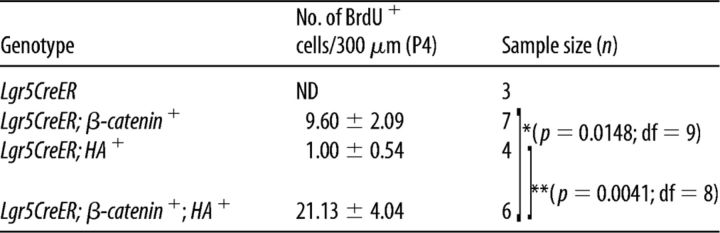
Our group and others have previously shown that in vivo expression of a constitutively active form of β-catenin in neonatal cochlear Lgr5+ cells results in a transient proliferative response with the formation of replication foci closely abutting the inner HCs (IHCs) (Chai et al., 2012; Shi et al., 2013). However, unlike cultured Lgr5+ cells (Chai et al., 2012), this induction paradigm did not generate new HCs in vivo. Atoh1 is critical for normal HC differentiation (Bermingham et al., 1999; Chen et al., 2002; Dabdoub et al., 2003; Mulvaney and Dabdoub, 2012; Jarman and Groves, 2013; Jahan et al., 2015); and because its ectopic expression induces the transdifferentiation of SCs in vivo, we generated Lgr5EGFP-CreERT2/+;Catnbflox(exon3)/+;CAG-loxP-Stop-loxP-Atoh1-HA mice (thereafter refer to as Lgr5CreER;β-catenin;HA+) to induce the transdifferentiation of β-catenin-induced proliferating Lgr5+ cells. The HA tag in the Atoh1-HA transgenic mice allowed for lineage tracing and provides a way to distinguish between ectopic and endogenous Atoh1 expression (Liu et al., 2012, 2014). We first assessed the effect of this combination on proliferation by performing a BrdU pulse-chase experiment. Tamoxifen induction was first performed at postnatal day 0 and 1 (P0-P1), followed by intraperitoneal injections of BrdU (4 injections 2 h apart) at P4. The resulting total of BrdU+ cells from P4 samples showed a significant increase (statistical significance was assessed by Student's t test with Bonferroni correction) in Lgr5CreER;β-catenin;HA+ mice compared with Lgr5CreER;β-catenin and Lgr5CreER;HA+ mice (Tables 1, 2). Similarly, the number of HA+/BrdU+ cells was significantly higher in Lgr5CreER;β-catenin;HA+ mice compared with Lgr5CreER;HA+ mice (Tables 1, 2). Of note, the number of HA−/BrdU+ cells is unexpectedly higher compared with the number of HA+/BrdU+ cells, which suggests the potential of a non–cell-autonomous affect via the recruitment of Cre-negative cells to the foci without subsequent proliferation. However, given that all cells in the foci were GFP+ (Cre+) in our previous study (Chai et al., 2012), this scenario is unlikely and we attribute this lower than expected number of HA+/BrdU+ cells to the low sensitivity of the HA antibody instead. Nonetheless, we conclude that β-catenin and Atoh1 act synergistically on the proliferation of Lgr5+ cells.
Table 1.
Synergistic effect of β-catenin and Atoh1 on Lgr5+ cells' proliferation in the IHC regiona
a Coexpression of β-catenin and Atoh1-HA in Lgr5+ cells results in a synergistic proliferation response. Number of BrdU+ cells per 300 μm section at P4 after 4 consecutive injections (2 h apart) of BrdU are shown with mean ± SEM. ND, Not detected. N = 3–7 for each time point and each genotype.
*p < 0.017 (unpaired Student's t test with Bonferroni correction).
**p < 0.003 (unpaired Student's t test with Bonferroni correction).
Table 2.
Synergistic effect of β-catenin and Atoh1 on Lgr5+ cells' proliferation in the IHC regiona
| Genotype | No. of HA+/BrdU+ cells/300 μm (P4) | Sample size (n) |
|---|---|---|
| Lgr5CreER | ND | 3 |
| Lgr5CreER; β-catenin+ | ND | 3 |
| Lgr5CreER; HA+ | 0 | 3 |
| Lgr5CreER; β-catenin+; HA+ | 3.40 ± 0.59 | 5 |
a Coexpression of β-catenin and Atoh1-HA in Lgr5+ cells results in a synergistic proliferation response. Number of HA+/BrdU+ cells per 300 μm section at P4 after 4 consecutive injections (2 h apart) of BrdU are shown with mean ± SEM. ND, Not detected. N = 3–7 for each time point and each genotype.
We next performed whole-mount analyses as described previously (Chai et al., 2012), with tamoxifen induction at P0-P1, and samples collected at P8, P15, and P21. The results show that replication foci (we use this term based on the results from the BrdU pulse-chase experiments) formed in Lgr5CreER;β-catenin;HA+ and Lgr5CreER;β-catenin mice, but not in Lgr5CreER;HA+ mice. The proliferation response was transient in both β-catenin ectopic expression models (the foci are no longer present at P21; Fig. 2F,L), but the replication foci were larger with poorly defined boundaries at P8 (Fig. 2D) and P15 (Fig. 2H) in Lgr5CreER;β-catenin;HA+ mice compared with Lgr5CreER;β-catenin mice (Chai et al., 2012) (Fig. 2B,F). Nonetheless, these results further support that the combination of β-catenin and Atoh1 synergistically increases the proliferation response in Lgr5CreER;β-catenin;HA+ mice, compared with Lgr5CreER;β-catenin mice.
Figure 2.
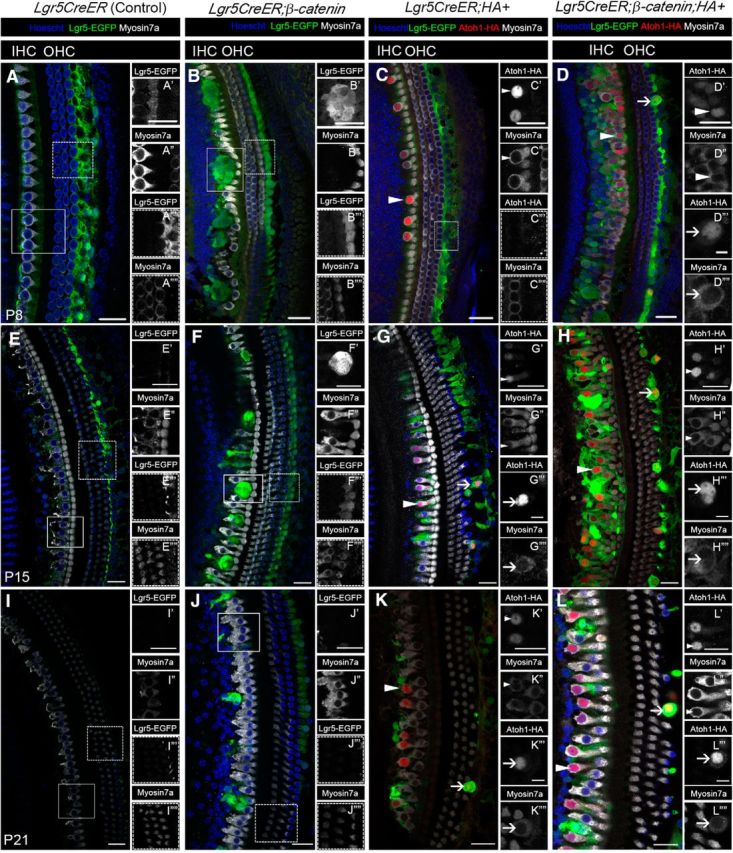
Lgr5+ cells differentiate into HCs in response to β-catenin and Atoh1. A–L, Confocal images of cochleae from Lgr5CreER (Control; similar phenotype observed in Lgr5CreER-negative;β-catenin;HA+) (A, E, I), Lgr5CreER;β-catenin (B, F, J), Lgr5CreER;HA+ (C, G, K), and Lgr5CreER;β-catenin;HA+ (D, H, L) mice that were induced with tamoxifen at P0-P1 and analyzed at P8 (A–D), P15 (E–H), and P21 (I–L). G, L, The optical sections do not show the entire third row of outer hair cells, although present. B, Lgr5+ cells only transiently proliferate and form foci in response to ectopic β-catenin at P8. C, Conversely, Lgr5+ cells differentiate into HCs (arrowheads) in response to Atoh1, but without apparent proliferation at P8. D, Replication foci from concurrent β-catenin and Atoh1 expression in Lgr5+ cells appear less defined (compared with β-catenin expression alone, B), suggesting an increased proliferation. In addition, Lgr5+ cells medial to the IHC and the third row of Deiters' cells lateral to the OHCs differentiate into HCs at P8 (D, arrowhead and arrow, respectively). The differentiation of the third row of Deiters' cells occurs at P8 only in the presence of β-catenin and Atoh1 (D, arrow). New HCs that formed medial to the IHC following Atoh1 alone or β-catenin and Atoh1 ectopic expression persist at P15 (G, H, arrowhead, respectively) and P21 (K, L, arrowhead, respectively). New HCs derived from the third row of Deiters' cells similarly persist at P15 (G, H, arrow, respectively) and P21 (K, L, arrows, respectively) following the same paradigm. L, At P21, the ectopic HCs adjacent to the endogenous IHCs have now formed an additional row of HCs. Scale bars, 25 μm. Scale bars: D″′, D″″, G″′, G″″, H″′, H″″, K″′, K″″, L″′, L″″, 10 μm.
Lgr5+ cells expressing β-catenin and Atoh1 give rise to new HCs
Because the proliferation response in Lgr5CreER;β-catenin;HA+ mice is increased, we next determined whether the coexpression of β-catenin and Atoh1 resulted in the transdifferentiation of proliferating Lgr5+ cells into HCs, using the Atoh1-HA-tag for lineage-tracing. At P8, new HCs expressing both the HA tag and Myosin7a (thereafter referred to as HA+/Myo7a+ HCs) were present in the IHC region along the cochleae of Lgr5CreER;HA+ mice (Fig. 2C, arrowhead), whereas new HCs in both the IHC and outer HC (OHC) regions along the cochleae (Fig. 2D, arrowheads and arrow, respectively) were present in Lgr5CreER;β-catenin;HA+ mice. The HA+/Myo7a+ HCs were still present in the IHC and OHC regions at P15 (Fig. 2G,H) and P21 (Fig. 2K,L). There was no significant difference in new HC numbers between cochlear turns (Student's t test with Bonferroni correction).
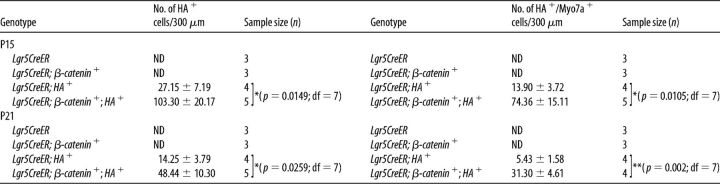
Because no new HCs formed in Lgr5CreER;β-catenin mice (Chai et al., 2012; Shi et al., 2013), we counted the numbers of HA+ and HA+/Myo7a+ cells in Lgr5CreER;HA+ and Lgr5CreER;β-catenin;HA+ mice (Tables 3, 4). In the IHC region, the numbers of HA+ and HA+/Myo7a+ cells per 300 μm section were significantly higher (statistical significance was assessed by Student's t test with Bonferroni correction) in Lgr5CreER;β-catenin;HA+ mice compared with Lgr5CreER;HA+ mice (Tables 3, 4). The total number of new HCs (HA+/Myo7a+) in the medial IHC region was as high as 1338.48 ± 271.98 cells per cochlea in Lgr5CreER;β-catenin;HA+ mice at P15 (e.g., 74.36 ± 15.11 cells/300 μm section; average measured cochlear length of 5.4 mm), and a significant number of the new HCs were also present at P21 (i.e., 563.4 ± 82.98 cells per cochlea). Given that no new HCs formed in the presence of ectopic β-catenin alone, our data suggest a potential synergistic effect between β-catenin and Atoh1-HA on the transdifferentiation of Lgr5+ cells in the IHC region. It is of interest to note that the total numbers of Atoh1-HA+ cells in Lgr5+ cells are more than those we reported previously in PLP/CreERT+; Atoh1-HA+ mice also induced at P0-P1 (Liu et al., 2014). This difference is most likely because Lgr5 is expressed in more nonsensory epithelial cells (GER, IBCs, and IPhCs) than in the PLP/CreERT+ model (IPhCs and IBCs only) (Fig. 1).
Table 3.
Synergistic effect of β-catenin and Atoh1 on Lgr5+ cells' differentiation in the IHC regiona
a β-Catenin and Atoh1-HA synergistically promote the differentiation of Lgr5+ cells located near the IHC region. Cell counts per 300 μm section for the number of HA+ cells and HA+/Myosin7a+ double-positive cells are shown with mean ± SEM. ND, Not detected. N = 3–7 for each time point and each genotype.
*p < 0.017 (unpaired Student's t test with Bonferroni correction).
**p < 0.003 (unpaired Student's t test with Bonferroni correction).
Table 4.
Synergistic effect of β-catenin and Atoh1 on Lgr5+ cells' differentiation in the IHC regiona
a β-Catenin and Atoh1-HA synergistically promote the differentiation of Lgr5+ cells located near the IHC region. Cell counts per 300 μm section for the number of HA+ cells and HA+/Myosin7a+ double-positive cells are shown with mean ± SEM. ND, Not detected. N = 3–7 for each time point and each genotype.
*p < 0.017 (unpaired Student's t test with Bonferroni correction).
**p < 0.003 (unpaired Student's t test with Bonferroni correction).
Because Lgr5CreER;β-catenin;HA+ mice have severely reduced body size and a high mortality rate (∼80%), which is likely due to detrimental effects on the intestinal lining, this greatly limited our long-term studies. Nonetheless, we managed to analyze two mice that survived to P42-P44 and observed an average of 28.60 ± 1.00 HA+/Myo7a+ new HCs per 300 μm cochlear section or 514.80 ± 18.00 new HCs per cochlea at P42-P44 (Fig. 3J,K), indicating that the new HCs that were observed at P21 survived up to those ages. Furthermore, HA+/Myo7a+/BrdU+ cells from animals that received BrdU at P4 (immunoprecipitation injections, 4 injections 2 h apart) were also present at P21 (18 ± 10 cells per 300 μm section) (Fig. 3L, arrowhead), suggesting that they were derived from proliferating and transdifferentiating Lgr5+ cells that coexpress β-catenin and Atoh1-HA.
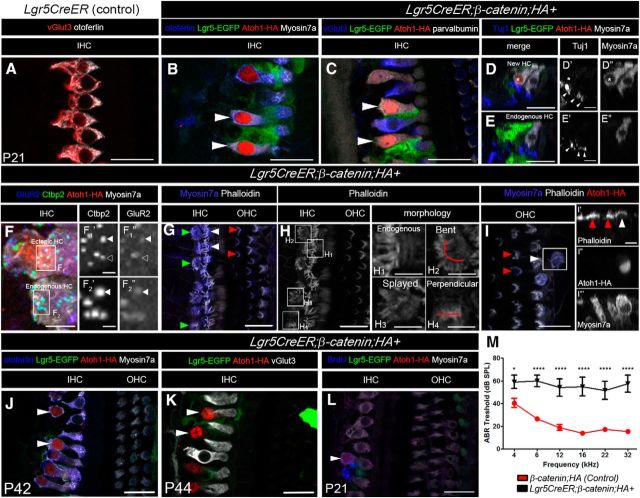
Figure 3.
Lgr5+ cells derived new HCs remain immature. A–L, Confocal images of immunostaining with multiple markers in cochleae from Lgr5CreER (Control) (A) and Lgr5CreER;β-catenin;HA+ (B–I) mice that were induced with tamoxifen at P0-P1 and analyzed at P21. A, IHCs express otoferlin and vGlut3 in control mice. New HCs (HA+) adjacent to the IHCs express Myosin7a (arrowheads in B and asterisk in D″), otoferlin (B, arrowheads), parvalbumin (C, arrowheads), but not the terminal IHC marker vGlut3 (C, arrowheads). In the new HCs, Tuj1 projections were present on the basolateral side of the ectopic HCs (D and arrowheads in D′). D–D″, Asterisk indicates the ectopic HC. In the endogenous IHCs, the synaptic markers Ctbp2 and GluR2 are both expressed in a matching pattern (arrowheads in F2′ and F2″, respectively). In the new HCs, some GluR2 puncta overlap with Ctbp2 puncta (arrowheads in F1′ and F1″, respectively), whereas others do not (open arrowhead in F1′ and F1″, respectively). G, The hair bundles form in the new HCs abutting the IHCs (green arrowheads in G), but their morphology differs from that of the endogenous IHCs (white arrowheads in G) and OHCs (red arrowheads in G). H, Phalloidin staining of the IHC region from G. Relative to the endogenous HCs (H1), the medially located new HCs display bent (H2), splayed (H3), or perpendicular morphology (H4). I, The ectopic HCs located in the OHC region (white arrowhead in I and I′) also display a peculiar hair cell bundle morphology, which does not resemble that of the traditional “V” shape found in normal endogenous OHCs (red arrowheads in I). I′–I″′, Cross-sectional view in which there is accumulation of actin at the cuticular plate in the endogenous (red arrowheads in I′) and ectopic HCs (white arrowhead in I′). J, K, New HCs are still present at P42-P44 (514.8 ± 18 per cochlea). Immunostaining of otoferlin (blue), Lgr5-EGFP (green), Atoh1-HA (red), Myosin7a (white in J), and vGlut3 (white in K) in the cochlea of a Lgr5CreER;β-catenin;HA+ mouse at P42-P44. L, New HCs generated from β-catenin and Atoh1 with BrdU pulse at P4 is still present at P21. M, ABR thresholds (± SEM) from P21 animals. Scale bars, 25 μm. Scale bars: D′, D″, E′, E″, H1–H4, I′–I″′, 10 μm; F, 5 μm; F1′, F2″, 2 μm. *p < 0.05 (two-way ANOVA with Bonferroni correction). ***p < 0.0001 (two-way ANOVA with Bonferroni correction). Refer to text for actual p values. N = 9 (control: β-catenin;HA+) and N = 6 (Lgr5CreER; β-catenin;HA+).
We analyzed the effect of β-catenin and Atoh1-HA on the transdifferentiation of the third row of Deiters' cells (Fig. 1A, dotted lined, B, SC-labeled DC3). We observed the formation of new HCs by P8 (Fig. 2D, arrow), which is in contrast to the Prox1CreERT2+;Atoh1-HA mouse line reported previously, where the transdifferentiation of similar SCs did not occur until P22 (Liu et al., 2012). HA+ and HA+/Myo7a cell counts (Table 5) showed no significant differences (statistical significance was assessed by Student's t test with Bonferroni correction) between Lgr5CreER;β-catenin;HA+ and Lgr5CreER;HA+ mice. We conclude that, although the ectopic expression of β-catenin and Atoh1-HA accelerated the transdifferentiation of the third Deiters's cells, it had little to no effect on proliferation or conversion efficiency of these cells.
Table 5.
Limited synergistic effect from β-catenin and Atoh1 on the third row of Deiters' Lgr5+ cellsa
a Limited synergistic effect was observed on the number of HA+ cells and the number of HA+/myo7a+ cells, except for the number of HA+/myo7a+ at P15. ND, Not detected; NS, not significant. N = 3–7 for each time point and each genotype. Data are mean ± SEM.
*p < 0.017 (unpaired t test with Bonferroni correction).
Lgr5-derived HCs from β-catenin and Atoh1-HA ectopic expression remain immature
To address the level of maturation of the new HCs, we analyzed the expression of various HC markers and synaptic components at P21 in Lgr5CreER;β-catenin;HA+ mice. The new HCs express the following HC markers: otoferlin (Fig. 3B, arrowheads), Myosin7a (Fig. 3B, arrowheads, D″, asterisk) and parvalbumin (Fig. 3C, arrowheads), but do not express the mature/terminal IHC marker, vGlut3 (Fig. 3C, arrowheads) or the mature/terminal OHC marker, prestin, in either the IHC or OHC regions, while retaining a 2.09 ± 0.38-fold increase in Sox2 expression compared with control littermates (p = 3.04 E-09; df = 46;n = 4 using a Student's t test with Bonferroni correction) (see Fig. 5H). The persistence of Sox2 is consistent with a previous study using the Atoh1-HA mouse line (Liu et al., 2012, 2014). The new HCs remained immature even at P44 (Fig. 3K).
Figure 5.
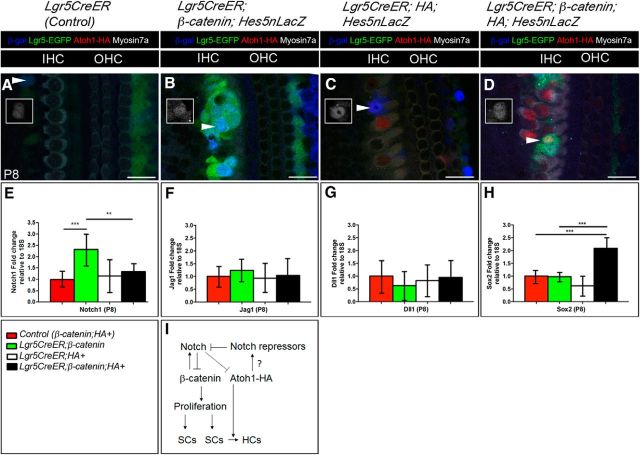
β-Catenin upregulates Notch, but its expression is abrogated by β-catenin and Atoh1-HA in Lgr5+ cells. A–D, Confocal images of immunostained cochleae from Lgr5CreER (Control) (A), Lgr5CreER;β-catenin;Hes5nLacZ (B), Lgr5CreER;HA+;Hes5nLacZ (C), and Lgr5CreER;β-catenin;HA+;Hes5nLacZ (D) mice that were induced with tamoxifen at P0-P1 and analyzed at P8. Hes5 upregulation in Lgr5+ foci from Lgr5CreER;β-catenin;Hes5nLacZ mouse (arrowhead and inset) suggests active Notch signaling mediated by β-catenin. Very few Hes5nLacZ-positive cells were observed in Lgr5CreER;HA+ (C, arrowhead and inset) and Lgr5CreER;β-catenin;HA+;Hes5nLacZ (D, arrowhead and inset). E, qPCR analysis of Notch1 from isolated P8 cochlea shows an increase and decrease in Lgr5CreER;β-catenin mice (CT value = 29.81 ± 0.98; 18S CT value = 20.59 ± 0.61) and Lgr5CreER;β-catenin;HA+ mice (CT value = 30.68 ± 0.96; 18S CT value = 20.68 ± 0.55) relative to control littermates (CT value = 31.13 ± 0.87; 18S CT value = 20.69 ± 0.47), respectively. For Lgr5CreER;HA+ mice: CT value = 30.56 ± 1.15; 18S CT value = 20.34 ± 0.49. F, G, qPCR analysis of Jagged1 (Jag1) and Delta-like 1 (Dll1) ligands from isolated P8 cochlea shows no change in expression between all genotypes. In the case of Jagged1: for control littermates: CT value = 31.11 ± 0.50; 18S CT value = 20.61 ± 0.75, for Lgr5CreER;HA+: CT value = 31.01 ± 0.84; 18S CT value = 20.42 ± 0.60, for Lgr5CreER;β-catenin+: CT value = 30.26 ± 0.79; 18S CT value = 20.08 ± 0.53, and for Lgr5CreER;β-catenin;HA+: CT value = 30.94 ± 1.10; 18S CT value = 20.51 ± 0.70. In the case of Dll1: for control littermates: CT value = 35.58 ± 1.22; 18S CT value = 20.76 ± 0.58, for Lgr5CreER;HA+: CT value = 35.28 ± 0.90; 18S CT value = 20.19 ± 0.35, for Lgr5CreER;β-catenin+: CT value = 35.68 ± 1.14; 18S CT value = 20.19 ± 0.45, and for Lgr5CreER;β-catenin;HA+: CT value = 35.56 ± 1.29; 18S CT value = 20.67 ± 0.63. H, qPCR analysis of Sox2 from isolated P8 cochlea shows a 2.09 ± 0.38 increase in Lgr5CreER;β-catenin;HA+ mice. The statistical significance of this increase was assessed by Student's t test with Bonferroni correction where the p = 3.04 E-09; df = 46; n = 4 (Control vs Lgr5CreER;β-catenin;HA+) and p = 7.52 E-04; df = 46; n = 3 (Lgr5CreER:HA+ vs Lgr5CreER;β-catenin;HA+). In this case, control littermates: CT value = 34.45 ± 0.41; 18S CT value = 21.50 ± 0.38, for Lgr5CreER;HA+: CT value = 35.04 ± 0.61; 18S CT value = 21.41 ± 0.37, for Lgr5CreER;β-catenin+: CT value = 34.37 ± 0.28; 18S CT value = 21.39 ± 0.30, and for Lgr5CreER;β-catenin;HA+: CT value = 33.36 ± 0.69; 18S CT value = 21.47 ± 0.29. I, Proposed model for the synergism of β-catenin and Atoh1 involving Notch upregulation (by β-catenin) and its downregulation by Atoh1 in Lgr5CreER;β-catenin;HA+ mouse. Dashed line of Notch to Atoh1-HA indicates the presumed low repression of Notch on the overexpressed Atoh1-HA. Question mark on the arrow between Atoh1-HA and Notch repressors indicates our current lack of targets. **p < 0.003 (unpaired Student's t test). ***p < 0.0003 (unpaired Student's t test). Refer to text for actual p values. N = 3 or 4 for each time point and each genotype. Data are mean ± SEM. Scale bars, 25 μm.
The new HCs closely abutting the endogenous IHCs were innervated as shown by the presence of Tuj1+ projection on their basolateral side (Fig. 3D,D′, arrowheads, where the asterisk denotes the new HC), and also expressed the presynaptic ribbon component, C-terminal binding protein 2 (Ctbp2) (Fig. 3F1′, arrowheads), as well as the postsynaptic marker glutamate receptor type 2 (GluR2) (Fig. 3F1″, arrowheads). However, there was some incomplete overlap/match of Ctbp2 and GluR2 puncta (Fig. 3F1′,F1″, open arrowheads, for Ctbp2 and GluR2, respectively).
Stereocilia were present at the apical surface of the new HCs located in the IHC region (Fig. 3G, green arrowheads), but their morphology differed from that of endogenous IHCs and OHCs (Fig. 3G, white and red arrowheads, respectively). We observed 3 types of stereocilia morphology relative to the endogenous HCs (Fig. 3H1): bent (Fig. 3H2), splayed (Fig. 3H3), and perpendicular (Fig. 3H4). Finally, the new HCs derived from the third row of Deiters' cells formed stereocillia as reflected by the accumulation of actin at the cuticular plate (Fig. 3I and white arrowhead for the new HCs vs red arrowheads for the endogenous HCs in Fig. 3I,I′). Auditory brainstem responses (ABR) measurements at P21 from Lgr5CreER;β-catenin;HA+ mice show significant higher hearing thresholds than littermate control mice (two-way ANOVA: 4 kHz: p = 0.293,df = 78; 6, 12, 16, 22, 32 kHz: p < 0.0001,df = 78) (Fig. 3M). This is consistent with our previous studies where the addition of immature new HCs resulted in elevated hearing thresholds (Liu et al., 2014). Together, these data indicate that the new HCs express some HC markers and synaptic components, are innervated, and contain stereocilia bundles. However, these cells remain immature and their presence results in elevated ABR thresholds.
Only a subset of Lgr5+ cells proliferate and differentiate in response to β-catenin and Atoh1 expression
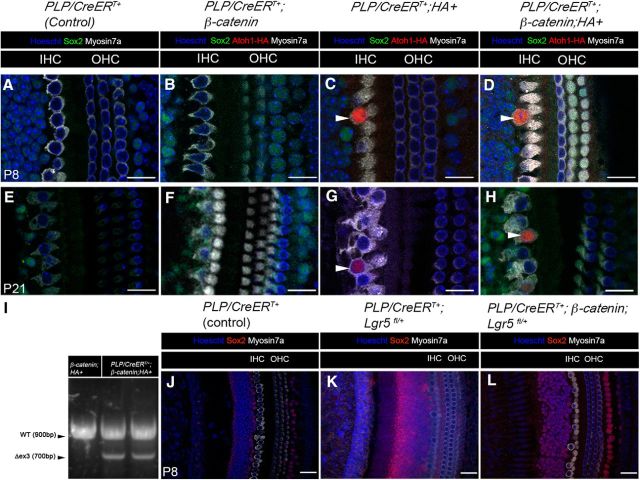
To assess the potential heterogeneity among Lgr5+ cell populations, we used another independent inducible Cre line, PLP/CreERT+ at P0-P1 (Gómez-Casati et al., 2010; Liu et al., 2014). PLP/CreERT+ targets the two SC subtypes directly abutting the IHCs, the IPhCs and IBCs (Fig. 1C), whereas Lgr5CreER targets a broader range of nonsensory epithelial cell subtypes (Fig. 1B) but includes the IPhCs and IBCs (Fig. 1B). Unlike the Lgr5CreER;β-catenin mice, we did not observe the formation of replication foci at P8 or P21 in any of the 3 genotypes generated: PLP/CreERT/+;β-catenin (Fig. 4B,F), PLP/CreERT+;CAG-loxP-Stop-loxP-Atoh1-HA (thereafter referred to as PLP/CreERT/+;HA+; Fig. 4C,G), and PLP/CreERT+;Catnbflox(exon3)/+;CAG-loxP-Stop-loxP-Atoh1-HA (thereafter referred to as PLP/CreERT/+;β-catenin;HA+; Fig. 4D,H). As expected, and based on previously published results (Liu et al., 2014), new HCs (HA+/Myo7a+) formed in the PLP/CreERT/+;HA+ (Fig. 4C,G, arrowhead, at P8 and P21, respectively) and in the PLP/CreERT/+;β-catenin;HA+ mice at P8 and P21 (Fig. 4D,H, arrowhead, at P8 and P21, respectively). HA+ and HA+/Myo7a+ cell counts showed no significant differences (statistical significance was assessed by Student's t test with Bonferroni correction) between PLP/CreERT/+;β-catenin;HA+ and PLP/CreERT/+;HA+ mice (Table 6). Because this line activates Atoh1-HA equally well in PLP/CreERT/+;β-catenin;HA+ and PLP/CreERT/+;HA+ mice, as reflected by the formation of new HCs, it is unlikely that low Cre efficiency underlies the lack of proliferation. Additionally, we were able to confirm the PLP/CreERT/+-mediated deletion of exon 3 in the β-catenin allele with the successful detection of the WT and the exon3-deleted β-catenin alleles in cochleae-extracted DNA by PCR from PLP/CreERT/+;β-catenin mice (Fig. 4I, WT and Δex3) as described by Harada et al. (1999). It may be of interest to note that we did observe differences in the number of ectopic HA+/Myo7a+ cells in PLP/CreERT/+;HA+ compared with a previous study (Liu et al., 2014), and these differences are likely due to variation between the mixed background strains used for the different studies or differences in sample processing for fixation and HA antibody labeling.
Figure 4.
Lack of synergistic effects on proliferation and differentiation on the IPhCs/IBCs. A–H, Confocal images of immunostained cochleae from PLP/CreERT+ (control) (A, E), PLP/CreERT+;β-catenin (B, F), PLP/CreERT+;HA+ (C, G), and PLP/CreERT+;β-catenin;HA+ (D, H) mice induced with tamoxifen at P0-P1 and analyzed at P8 (A–D) and P21 (E–H). There is no proliferative response at P8 (B) or P21 (F) in PLP/CreERT+;β-catenin mice. However, IPhCs/IBCs still transdifferentiate into HCs in the presence of Atoh1-HA at P8 (C, arrowhead) and persist at least until P21 (G, arrowhead). New HCs (HA+/Myo+) form at P8 (D, arrowhead) and persist at least until P21 (H, arrowhead) in PLP/CreERT+;β-catenin;HA+ mice. I, Both wild-type (WT; 900 bp) and mutant β-catenin alleles (Δex3; 700bp) were detected by PCR from the cochleae of PLP/CreERT/+;β-catenin; HA+ mice, whereas only the wild-type (WT) allele is detected in β-catenin; HA+ mice at P4. To address whether Lgr5 haploinsufficiency plays a role in the lack of proliferative response of the IBCs and IPhCs, we analyzed PLP/CreERT+;β-catenin;Lgr5fl/+ mice bearing one conditionally deleted Lgr5 allele to replicate the Lgr5EGFP-CreERT2/+ knock-in mice. Following tamoxifen induction at P0-P1, no replication foci were observed, and the IHC layer shows no endogenous HC displacement at P8 in the PLP/CreERT+ (J; control), PLP/CreERT+;Lgr5fl/+ (K), or the PLP/CreERT+;β-catenin;Lgr5fl/+ (L) mice. Scale bars, 25 μm.
Table 6.
Lack of synergistic effects on proliferation and differentiation of IPhCs/IBCsa
| Genotype | No. of HA+ cells/300 μm | Sample size (n) | Genotype | No. of HA+/Myo7a+ cells/300 μm | Sample size (n) |
|---|---|---|---|---|---|
| P8 | |||||
| PLP/CreERT/+; | ND | 3 | PLP/CreERT/+; | ND | 3 |
| PLP/CreERT/+; β-catenin | ND | 3 | PLP/CreERT/+; β-catenin | ND | 3 |
| PLP/CreERT/+; HA+ | 35.23 ± 10.05 | 3 (NS, p = 0.7223; df = 5) | PLP/CreERT/+; HA+ | 4.43 ± 2.15 | 3 (NS, p = 0.7013; df = 5) |
| PLP/CreERT/+; β-catenin; HA+ | 30.63 ± 7.54 | 4 | PLP/CreERT/+; β-catenin; HA+ | 5.98 ± 2.82 | 4 |
| P21 | |||||
| PLP/CreERT/+ | ND | 3 | PLP/CreERT/+ | ND | 3 |
| PLP/CreERT/+; β-catenin | ND | 3 | PLP/CreERT/+; β-catenin | ND | 3 |
| PLP/CreERT/+; HA+ | 36.62 ± 9.02 | 5 (NS, p = 0.9439; df = 9) | PLP/CreERT/+; HA+ | 23.40 ± 7.20 | 5 (NS, p = 0.4691; df = 9) |
| PLP/CreERT/+; β-catenin; HA+ | 35.70 ± 8.84 | 6 | PLP/CreERT/+; β-catenin; HA+ | 17.17 ± 4.59 | 6 |
a There is no synergistic effect on the proliferation or differentiation of the IPhCs/IBCs SCs at P8 or P21 in the presence of β-catenin and Atoh1. Statistical analysis of HA+ and HA+/Myo7a+ cell counts per 300 μm sections did not show any significant differences between PLP/CreERT+;Atoh1-HA+ and PLP/CreERT+;β-catenin; HA+ mice at P8 and P21. ND, Not detected; NS, not significant using unpaired Student's t test with Bonferroni correction. N = 3–6 for each time point and each genotype. Data are mean ± SEM.
We next asked whether Lgr5 haploinsufficiency could contribute to the absence of proliferation in the PLP/CreERT/+;β-catenin and PLP/CreERT/+;β-catenin;HA+ mice. To address this, we bred the Lgr5fl/+ mouse (de Lau et al., 2011) for the conditional deletion of one Lgr5 allele in PLP/CreERT/+; β-catenin; Lgr5 fl/+ mice. In these mice, we were unable to detect a proliferative response as reflected by the lack of IHC displacement at P8 (Fig. 4K,L), which we typically observed in the Lgr5CreER;β-catenin mice (Fig. 2). Therefore, this indicates that Lgr5 haploinsufficiency does not have a significant role in preventing Wnt mediated mitogenic effect on the IBCs/IPhCs, but rather that the IBCs/IPhCs are unresponsive to ectopic Wnt signaling. The lack of a GER-specific CreER mouse line further prevented us from testing the impact of Lgr5 haploinsufficiency on the proliferative response in these cells. Together, these data indicate that the Lgr5+, but PLP/CreER-negative nonsensory epithelial cells located medially to the IBCs or IPhCs in the GER region, are responsible for the synergistic proliferation and differentiation responses mediated by ectopic β-catenin and Atoh1 expression.
Notch upregulation by β-catenin is abrogated by β-catenin and Atoh1 in Lgr5+ cells
To address the potential mechanism underlying the synergistic effect of β-catenin and Atoh1-HA on the proliferation and differentiation of Lgr5+ cells, we focused on the Notch signaling pathway because it has been shown to regulate both β-catenin and Atoh1 (Lanford et al., 2000; Zheng et al., 2000; Zine and de Ribaupierre, 2002; Takebayashi et al., 2007; Sikandar et al., 2010; Kwon et al., 2011; VanDussen et al., 2012; Petrovic et al., 2014). Using Hes5nLacZ reporter mice (Imayoshi et al., 2010), we analyzed the expression of Hes5, one of the Notch downstream effectors (Jarriault et al., 1995; Kageyama and Ohtsuka, 1999; Ohtsuka et al., 1999) involved in HC differentiation (Zine et al., 2001; Zine and de Ribaupierre, 2002; Tateya et al., 2011). We observed the presence of Hes5+ cells in Lgr5CreER;β-catenin;Hes5nLacZ mice (Fig. 5B, arrowhead) compared with control mice (Fig. 5A). This suggests that Notch signaling is indeed activated by the canonical Wnt signaling effector, β-catenin, in the organ of Corti, when it is normally downregulated during the first week of postnatal development (Hartman et al., 2009; Maass et al., 2015). In contrast, there were very few Hes5+ cells in Lgr5CreER;HA+;Hes5nLacZ and Lgr5CreER;β-catenin; HA+;Hes5nLacZ mice (Fig. 5C,D).
In support of the observed β-catenin-mediated Notch activation (see above), we analyzed the expression of Notch1 since it has been implicated in cochlear development (Lanford et al., 1999; Zine et al., 2000). We analyzed Notch1 expression by qPCR analysis (CT values included in Fig. 5 legend) of P8 microdissected organ of Corti and found a significant increase in Notch1 expression in Lgr5CreER;β-catenin+ mice compared with littermate controls (2.33 ± 0.67 (n = 3); p = 0.001; df = 36 using a Student's t test with Bonferroni correction) (Fig. 5E), further supporting the notion that Wnt activation results in Notch activation. However, Notch1 expression is significantly decreased in Lgr5CreER;β-catenin; HA+ mice compared with Lgr5CreER;β-catenin (p = 0.0068; df = 39 using a Student's t test with Bonferroni correction) (Fig. 5E). We also analyzed the expression of Notch1 ligands, Jagged 1 (Fig. 5F) and Delta-like 1 (Fig. 5G) and found no significant differences between all genotypes (statistical significance was assessed by Student's t test with Bonferroni correction). Nonetheless, these data combined with the Hes5nlacZ results (Fig. 5A–D) suggest that Notch activation mediated by β-catenin is inhibited by the ectopic expression of Atoh1 in Lgr5+ cells, which may play a role in the synergistic effects that we described earlier. Understanding these opposing roles of Notch and Atoh1 in the inner ear warrants future investigation.
Discussion
In vivo HC regeneration in postnatal mammalian cochleae has been hampered by limited differentiation efficiency and spontaneous mitotic regeneration, and subsequent cell death of regenerated HCs (ranging from 12 to 150 new HCs generated per cochlea) (Kelly et al., 2012; Liu et al., 2012; Mizutari et al., 2013; Shi et al., 2013; Cox et al., 2014; Liu et al., 2014). Here we have shown that β-catenin and Atoh1 have synergistic effects on proliferation and transdifferentiation of neonatal Lgr5+ cells in vivo. Compared with the aforementioned studies, our results show new IHC numbers reaching 1338.48 ± 271.98 per cochlea, twice as many new HCs as endogenous IHCs (Ou et al., 2000). Moreover, most of these new HCs survive to adulthood with 514.80 ± 18.00 new HCs per cochlea remaining at P42-P44. Our study is the first to achieve substantial proliferation and transdifferentiation in a mammalian cochleae in vivo, which parallels the efficient mitotic HC regeneration found in nonmammalian vertebrates and therefore represents a significant advancement for in vivo generation of new auditory HCs in a mammalian system.
β-Catenin has been shown to positively regulate the expression of Atoh1 (Shi et al., 2010, 2013), so the absence of no new HCs generated in Lgr5CreER;β-catenin+ mice was puzzling (Chai et al., 2012; Shi et al., 2013). Because β-catenin promotes Notch activation in Lgr5CreER;β-catenin+ mice, we therefore attributed the absence of new HCs to Notch repression on β-catenin and presumed β-catenin-mediated Atoh1 activation. Indeed, Notch has been reported to post-translationally repress β-catenin activity (Kwon et al., 2011; Kim et al., 2012), and this further suggests the existence of a negative feedback loop to limit β-catenin activity to possibly prevent uncontrolled growth as observed in various cancers (Reya and Clevers, 2005). In addition, there is evidence that Hes family members, including Hes5, repress HC differentiation via Atoh1 repression in both auditory sensory epithelia and other tissues (Lanford et al., 2000; Zheng et al., 2000; Zine and de Ribaupierre, 2002; Takebayashi et al., 2007; Sikandar et al., 2010; VanDussen et al., 2012; Petrovic et al., 2014). Because we observed a decrease in Notch signaling with the synergistic effects of β-catenin and Atoh1-HA on proliferation and transdifferentiation in Lgr5+ cells in Lgr5CreER;β-catenin;HA+ compared with Lgr5CreER;β-catenin, this suggests a repressive role for Atoh1 on Notch signaling (incidentally activated by β-catenin). Repression of Notch would presumably promote the β-catenin mitogenic effect and provide an indirect role on proliferation. This could possibly be through the modulation of Notch repressors (i.e., Numb, Fbxw7) or activators (i.e., Rbpj or Mastermind) (for review, see Ranganathan et al., 2011) (Fig. 5I). Nonetheless, we speculate that the ectopic expression of the Atoh1-HA transgene in Lgr5CreER;β-catenin;HA+ abrogates the β-catenin-mediated Notch repression of endogenous Atoh1 and β-catenin. Furthermore, the ectopic expression of Atoh1 likely increases the levels of endogenous Atoh1, closer to a threshold from which the nonsensory epithelial cells can undergo HC differentiation. Consequently, this leads to an increase in β-catenin-expressing SCs competent to respond to pro-HC differentiation cues regulated by ectopic Atoh1. Of note, a recent report described the importance of Notch repression in the context of Wnt-dependent proliferation and HC generation in Sox2+ and Lgr5+ cells, which is largely consistent with our results (Li et al., 2015). Although the aforementioned report described an increase in Wnt signaling that promotes mitotic HC generation after Notch deletion alone, we attribute such discrepancy to several factors, including the use of Sox2CreER line in vivo, explant cultures for Lgr5CreER;Notch1f/f, and/or the use of use of γ-secretase inhibitors, which could affect additional signaling pathways. Nonetheless, Notch inhibition, through γ-secretase inhibitors, has been described to promote HC formation and maturation in the damaged cochlea (Korrapati et al., 2013; Mizutari et al., 2013; Bramhall et al., 2014), thus highlighting the impact of this pathway on HC regeneration. Together, we conclude that the combination of β-catenin and Atoh1 provides a platform from which to promote proliferation and differentiation of SCs, although this paradigm still requires further manipulation(s) for terminal maturation.
Although we describe an indirect role for Atoh1 on proliferation, via Notch repression, targetome analyses provide an extensive list of Atoh1-associated cellular processes (e.g., transcription, proliferation, migration, cytoskeletal rearrangements) (Klisch et al., 2011; Yoon et al., 2011; Cai et al., 2015). This suggests that Atoh1 is involved in other cellular events, which may also influence the synergistic effect observed in our model, although these cellular processes are also likely affected by β-catenin, especially in the context of proliferation.
Cellular heterogeneity has been shown for the Lgr5+ cells in the intestine, whereby only 5% of isolated Lgr5+ cells give rise to organoid (Sato et al., 2009), and lineage-tracing experiments have shown that only high GFP-expressing Lgr5+ cells exhibit clonal expansion capability (Snippert et al., 2010). Our results are consistent with this notion since. For instance, there is an absence of synergic effect on the proliferation and transdifferentiation rate in the third row of Deiters' cells, and in a subset of Lgr5+ cells, the IPhCs and IBCs. For the former, we attribute the lack of synergy to the presumed high Lgr5 expression (reflected by EGFP expression) in these cells (Chai et al., 2011), which could prevent their proliferation and transdifferentiation. In support, the new HCs generated in this study and those derived from isolated Lgr5+ cells cultured in the presence of Wnt3a no longer express EGFP (Chai et al., 2012). For the latter, we conjecture that IPhCs and IBCs may respond differently to β-catenin and Atoh1 compared with other Lgr5+ nonsensory epithelial cells, possibly because the endogenous β-catenin promoter may already be downregulated at the time of induction. This is important because the activation of the β-catenin allele used in this study is dependent upon the endogenous β-catenin promoter (Harada et al., 1999). In this scenario, Lgr5-EGFP expression could merely reflect residual Wnt activity because Lgr5 expression is normally downregulated in IPhCs and IBCs during the first postnatal week. This inherent cellular heterogeneity within the Lgr5+ cells is also supported by other studies (Liu et al., 2012; Shi et al., 2013). Regardless, because the Lgr5+ cells are thought to be the stem/progenitor cells of the neonatal cochlea with higher proliferation and differentiation potential than the broader population of p27Kip1+ cells (White et al., 2006), these cells remain of high interest for hearing restoration efforts.
Similar to previous reports (Kelly et al., 2012; Liu et al., 2012, 2014), the Atoh1-induced new HCs described here underwent incomplete maturation because they do not express the terminal IHC (vGlut3) and OHC (prestin) markers and have some incomplete alignment of presynaptic and postsynaptic markers, Ctbp2 and GluR2. We propose the following possible explanations for the lack of terminal differentiation in the new HCs from this study. Ectopic Atoh1 and/or Wnt/β-catenin expressions are unnatural to HC terminal differentiation, as both genes are normally downregulated during development (Lanford et al., 2000; Lumpkin et al., 2003; Layman et al., 2013; Munnamalai and Fekete, 2013; Jansson et al., 2015). In our model, Atoh1-HA expression is driven by a constitutive promoter and is therefore unlikely to be downregulated after tamoxifen induction (Liu et al., 2012, 2014). In contrast, the expression of the Catnbflox(exon3)/+ allele is controlled by the β-catenin endogenous promoter, but because Lgr5 (and presumably Wnt) is downregulated after the first postnatal week, it is unlikely that de novo production of stable β-catenin is sustained past the first week of postnatal development in Lgr5+ cells. Nonetheless, transient ectopic expression of Atoh1 and β-catenin may be more effective in promoting HC maturation. Of note, Cre-mediated activation of β-catenin (using the Catnbflox(exon3)/+ mouse line), and the subsequent evaluation of the effects of Atoh1 and β-catenin in mature SCs will require different strategies because of the inactive β-catenin endogenous promoter in mature cochleae.
Traditionally, HC regeneration strategies have focused on manipulating individual pathways or genes, such as Wnt (Chai et al., 2012; Jacques et al., 2012; Shi et al., 2013; Jacques et al., 2014), Notch (Kiernan et al., 2005; Yamamoto et al., 2006; Takebayashi et al., 2007; Korrapati et al., 2013), Eya1-Six1 (Ahmed et al., 2012), or Atoh1 (Izumikawa et al., 2005; Kelly et al., 2012; Liu et al., 2012; Liu et al., 2014). However, multiple pathways work in concert to promote HC mitotic regeneration in nonmammalian vertebrates (Jiang et al., 2014; Ku et al., 2014; Steiner et al., 2014), further supporting the use of combinatorial therapies. The success of this type of combinatorial approach for the generation of new HCs has been successfully illustrated by Minoda et al. (2007), with the adenovirus-mediated expression of Skp2 and Atoh1; Masuda et al. (2012), with the combination of Atoh1 with TCF3 or GATA3; and Costa et al. (2015), with the combination of Atoh1, Gfi1, and Pou4f3. Moreover, successful regeneration in other systems (e.g., cardiomyocytes, β islet cells, and CNS neurons) also uses a combinatorial approach (Niu et al., 2013; Xin et al., 2013; Ziv et al., 2013). Therefore, the combination of β-catenin and Atoh1 is likely critical for successful HC regeneration and provides the basis for combinatorial therapeutics for hearing restoration in humans.
Footnotes
This work was supported by National Institutes of Health Grants 2R01DC006471 and 1R21DC013879 to J.Z., Grant P30CA21765 to St. Jude Children's Research Hospital, and Grant F32DC013232 to W.S.L., American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital, and Office of Naval Research Grants 1N000140911014, N000141210191, and N000141210775 to J.Z., J.Z. is a recipient of the Hartwell Individual Biomedical Research Award. We thank members of our laboratory for discussions and comments; A. Cheng (Stanford University School of Medicine), L. Zhu and X. Cao (St. Jude Children's Research Hospital), and B. Cox (Southern Illinois University School of Medicine) for critical reading and comments; and Mario A. Sauceda (St. Jude Children's Research Hospital) for mouse genotyping.
The authors declare no competing financial interests.
References
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bohne BA, Ward PH, Fernández C. Irreversible inner ear damage from rock music. Trans Sect Otolaryngol Am Acad Ophthalmol Otolaryngol. 1976;82:ORL50–ORL59. [PubMed] [Google Scholar]
- Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014;2:311–322. doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Jen HI, Kang H, Klisch TJ, Zoghbi HY, Groves AK. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J Neurosci. 2015;35:5870–5883. doi: 10.1523/JNEUROSCI.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Xia A, Wang T, Jan TA, Hayashi T, Bermingham-McDonogh O, Cheng AG. Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J Assoc Res Otolaryngol. 2011;12:455–469. doi: 10.1007/s10162-011-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, Liu Z, Taketo MM, Oghalai JS, Nusse R, Zuo J, Cheng AG. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109:8167–8172. doi: 10.1073/pnas.1202774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- Costa A, Sanchez-Guardado L, Juniat S, Gale JE, Daudet N, Henrique D. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development. 2015;142:1948–1959. doi: 10.1242/dev.119149. [DOI] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, Chalasani K, Steigelman KA, Fang J, Rubel EW, Cheng AG, Zuo J. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141:816–829. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A, Donohue MJ, Brennan A, Wolf V, Montcouquiol M, Sassoon DA, Hseih JC, Rubin JS, Salinas PC, Kelley MW. Wnt signaling mediates reorientation of outer hair cell stereociliary bundles in the mammalian cochlea. Development. 2003;130:2375–2384. doi: 10.1242/dev.00448. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Gómez-Casati ME, Murtie J, Taylor B, Corfas G. Cell-specific inducible gene recombination in postnatal inner ear supporting cells and glia. J Assoc Res Otolaryngol. 2010;11:19–26. doi: 10.1007/s10162-009-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Basak O, Nelson BR, Taylor V, Bermingham-McDonogh O, Reh TA. Hes5 expression in the postnatal and adult mouse inner ear and the drug-damaged cochlea. J Assoc Res Otolaryngol. 2009;10:321–340. doi: 10.1007/s10162-009-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JE, Jr, Johnsson LG, Stebbins WC, Moody DB, Coombs SL. Hearing loss and cochlear pathology in monkeys after noise exposure. Acta Otolaryngol. 1976;81:337–343. doi: 10.3109/00016487609119971. [DOI] [PubMed] [Google Scholar]
- Head JR, Gacioch L, Pennisi M, Meyers JR. Activation of canonical Wnt/beta-catenin signaling stimulates proliferation in neuromasts in the zebrafish posterior lateral line. Dev Dyn. 2013;242:832–846. doi: 10.1002/dvdy.23973. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Puligilla C, Weichert RM, Ferrer-Vaquer A, Hadjantonakis AK, Kelley MW, Dabdoub A. A dual function for canonical Wnt/beta-catenin signaling in the developing mammalian cochlea. Development. 2012;139:4395–4404. doi: 10.1242/dev.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques BE, Montgomery WH, 4th, Uribe PM, Yatteau A, Asuncion JD, Resendiz G, Matsui JI, Dabdoub A. The role of Wnt/beta-catenin signaling in proliferation and regeneration of the developing basilar papilla and lateral line. Dev Neurobiol. 2014;74:438–456. doi: 10.1002/dneu.22134. [DOI] [PubMed] [Google Scholar]
- Jahan I, Pan N, Fritzsch B. Opportunities and limits of the one gene approach: the ability of Atoh1 to differentiate and maintain hair cells depends on the molecular context. Front Cell Neurosci. 2015;9:26. doi: 10.3389/fncel.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson L, Kim GS, Cheng AG. Making sense of Wnt signaling-linking hair cell regeneration to development. Front Cell Neurosci. 2015;9:66. doi: 10.3389/fncel.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Groves AK. The role of Atonal transcription factors in the development of mechanosensitive cells. Semin Cell Dev Biol. 2013;24:438–447. doi: 10.1016/j.semcdb.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Romero-Carvajal A, Haug JS, Seidel CW, Piotrowski T. Gene-expression analysis of hair cell regeneration in the zebrafish lateral line. Proc Natl Acad Sci U S A. 2014;111:1383–1392. doi: 10.1073/pnas.1402898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Ishimoto S, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Cellular commitment and differentiation in the organ of Corti. Int J Dev Biol. 2007;51:571–583. doi: 10.1387/ijdb.072388mk. [DOI] [PubMed] [Google Scholar]
- Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- Kim HA, Koo BK, Cho JH, Kim YY, Seong J, Chang HJ, Oh YM, Stange DE, Park JG, Hwang D, Kong YY. Notch1 counteracts WNT/beta-catenin signaling through chromatin modification in colorectal cancer. J Clin Invest. 2012;122:3248–3259. doi: 10.1172/JCI61216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrapati S, Roux I, Glowatzki E, Doetzlhofer A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One. 2013;8:e73276. doi: 10.1371/journal.pone.0073276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku YC, Renaud NA, Veile RA, Helms C, Voelker CC, Warchol ME, Lovett M. The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci. 2014;34:3523–3535. doi: 10.1523/JNEUROSCI.2606-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Shailam R, Norton CR, Gridley T, Kelley MW. Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J Assoc Res Otolaryngol. 2000;1:161–171. doi: 10.1007/s101620010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, Sauceda MA, Zuo J. Epigenetic alterations by NuRD and PRC2 in the neonatal mouse cochlea. Hear Res. 2013;304:167–178. doi: 10.1016/j.heares.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, Li H. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A. 2015;112:166–171. doi: 10.1073/pnas.1415901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Owen T, Zhang L, Zuo J. Dynamic expression pattern of Sonic hedgehog in developing cochlear spiral ganglion neurons. Dev Dyn. 2010;239:1674–1683. doi: 10.1002/dvdy.22302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, Zindy F, Gan L, Roussel MF, Zuo J. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters' cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Fang J, Dearman J, Zhang L, Zuo J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic atoh1 expression. PLoS One. 2014;9:e89377. doi: 10.1371/journal.pone.0089377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/S1567-133X(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Maass JC, Gu R, Basch ML, Waldhaus J, Lopez EM, Xia A, Oghalai JS, Heller S, Groves AK. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front Cell Neurosci. 2015;9:110. doi: 10.3389/fncel.2015.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Pak K, Chavez E, Ryan AF. TFE2 and GATA3 enhance induction of POU4F3 and myosin VIIa positive cells in nonsensory cochlear epithelium by ATOH1. Dev Biol. 2012;372:68–80. doi: 10.1016/j.ydbio.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoda R, Izumikawa M, Kawamoto K, Zhang H, Raphael Y. Manipulating cell cycle regulation in the mature cochlea. Hear Res. 2007;232:44–51. doi: 10.1016/j.heares.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, Edge AS. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77:58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13:281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Fekete DM. Wnt signaling during cochlear development. Semin Cell Dev Biol. 2013;24:480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang CL. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol. 2008;9:65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/S0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Ou HC, Harding GW, Bohne BA. An anatomically based frequency-place map for the mouse cochlea. Hear Res. 2000;145:123–129. doi: 10.1016/S0378-5955(00)00082-4. [DOI] [PubMed] [Google Scholar]
- Petrovic J, Formosa-Jordan P, Luna-Escalante JC, Abelló G, Ibañes M, Neves J, Giraldez F. Ligand-dependent Notch signaling strength orchestrates lateral induction and lateral inhibition in the developing inner ear. Development. 2014;141:2313–2324. doi: 10.1242/dev.108100. [DOI] [PubMed] [Google Scholar]
- Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rubel EW, Furrer SA, Stone JS. A brief history of hair cell regeneration research and speculations on the future. Hear Res. 2013;297:42–51. doi: 10.1016/j.heares.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Shi F, Cheng YF, Wang XL, Edge AS. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32:9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110:13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Hu L, Jacques BE, Mulvaney JF, Dabdoub A, Edge AS. β-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci. 2014;34:6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, Lipkin SM. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Steiner AB, Kim T, Cabot V, Hudspeth AJ. Dynamic gene expression by putative hair-cell progenitors during regeneration in the zebrafish lateral line. Proc Natl Acad Sci U S A. 2014;111:1393–1401. doi: 10.1073/pnas.1318692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51:633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Yamamoto N, Yabe D, Fukuda H, Kojima K, Ito J, Honjo T. Multiple roles of Notch signaling in cochlear development. Dev Biol. 2007;307:165–178. doi: 10.1016/j.ydbio.2007.04.035. [DOI] [PubMed] [Google Scholar]
- Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol. 2011;352:329–340. doi: 10.1016/j.ydbio.2011.01.038. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud Å, Grosse AS, Gumucio DL, Ernst SA, Tsai YH, Dempsey PJ, Samuelson LC. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Tsuji M, Yabe D, Ito J, Honjo T. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med (Berl) 2006;84:37–45. doi: 10.1007/s00109-005-0706-9. [DOI] [PubMed] [Google Scholar]
- Yoon H, Lee DJ, Kim MH, Bok J. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol. 2011;44:69–78. doi: 10.5115/acb.2011.44.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- Zine A, de Ribaupierre F. Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear Res. 2002;170:22–31. doi: 10.1016/S0378-5955(02)00449-5. [DOI] [PubMed] [Google Scholar]
- Zine A, Van De Water TR, de Ribaupierre F. Notch signaling regulates the pattern of auditory hair cell differentiation in mammals. Development. 2000;127:3373–3383. doi: 10.1242/dev.127.15.3373. [DOI] [PubMed] [Google Scholar]
- Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, de Ribaupierre F. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Glaser B, Dor Y. The plastic pancreas. Dev Cell. 2013;26:3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]