Abstract
CYP2C19 rs12769205 alters an intron 2 branch point adenine leading to an alternative mRNA in human liver with complete inclusion of intron 2 (exon 2B). rs12769205 changes the mRNA reading frame, introduces 87 amino acids, and leads to a premature stop codon. The 1000 Genomes project (http://browser.1000genomes.org/index.html) indicated rs12769205 is in linkage disequilibrium with rs4244285 on CYP2C19*2, but found alone on CYP2C19*35 in Blacks. Minigenes containing rs12769205 transfected into HepG2 cells demonstrated this single nucleotide polymorphism (SNP) alone leads to exon 2B and decreases CYP2C19 canonical mRNA. A residual amount of CYP2C19 protein was detectable by quantitative proteomics with tandem mass spectrometry in CYP2C19*2/*2 and *1/*35 liver microsomes with an exon 2 probe. However, an exon 4 probe, downstream from rs12769205, but upstream of rs4244285, failed to detect CYP2C19 protein in livers homozygous for rs12769205, demonstrating rs12769205 alone can lead to complete loss of CYP2C19 protein. CYP2C19 genotypes and mephenytoin phenotype were compared in 104 Ethiopians. Poor metabolism of mephenytoin was seen in persons homozygous for both rs12769205 and rs4244285 (CYP2C19*2/*2), but with little effect on mephenytoin disposition of CYP2C19*1/*2, CYP2C19*1/*3, or CYP2C19*1/*35 heterozygous alleles. Extended haplotype homozygosity tests of the HapMap Yorubans (YRI) showed both haplotypes carrying rs12769205 (CYP2C19*35 and CYP2C19*2) are under significant natural selection, with CYP2C19*35 having a higher relative extended haplotype homozygosity score. The phylogenetic tree of the YRI CYP2C19 haplotypes revealed rs12769205 arose first on CYP2C19*35 and that rs4244285 was added later, creating CYP2C19*2. In conclusion, rs12769205 is the ancestral polymorphism leading to aberrant splicing of CYP2C19*35 and CYP2C19*2 alleles in liver.
Introduction
CYP2C19 is an important drug-metabolizing enzyme that plays a critical role in the metabolism as well as drug-drug interactions of a variety of drugs, including proton pump inhibitors, antiepileptics, antiplatelet drugs, and antidepressants (Li-Wan-Po et al., 2010; Shah et al., 2012; Shirasaka et al., 2013). Similar to a variety of other cytochrome P450 (CYP) family members, CYP2C19 activity is polymorphic, with subpopulations of poor metabolizers (PMs) as well as intermediate, extensive, and ultra-rapid metabolizers (Wedlund, 2000; McGraw and Waller, 2012; Hicks et al., 2013). Multiple allelic variants of CYP2C19 have been described. CYP2C19*1 represents the wild-type allele. The frequent rs4244285 polymorphism, defining the CYP2C19*2 allele, creates an exon 5 aberrant splice site, altering the reading frame of the mRNA and leading to a premature stop codon and a nonfunctional protein (de Morais et al., 1994b).
There are numerous reports linking the CYP2C19*2 allele to altered substrate clearance (Hicks et al., 2013; Hirota et al., 2013; Owusu Obeng et al., 2014). Over 34 CYP2C19 variant alleles have been identified in a CYP database (http://www.cypalleles.ki.se/cyp2c19.htm). In addition, a recent study of 2203 African Americans (Gordon et al., 2014) found that CYP2C19 had the highest number of putative novel functional variants compared with 11 other drug metabolizing CYP genes. This result was interesting because it was recently reported that the CYP2C19*2 nonfunctional allele may have been positively selected in human evolution (Janha et al., 2014), and that inactivation of CYP2C19 might have afforded some survival advantage. Conversely, because CYP2C19 loss-of-function alleles confer increased risks for serious adverse cardiovascular events among clopidogrel-treated patients, Clinical Pharmacogenetic Implementation Consortium guidelines were issued for CYP2C19 genotype-directed drug therapy (Scott et al., 2013).
In an effort to identify additional function-disrupting CYP2C19 alleles, we sequenced CYP2C19 in human livers and identified a branch point single nucleotide polymorphism (SNP) (rs12769205; gene position 12662A>G) in intron 2 of CYP2C19 that leads to intron 2 retention. Interestingly, rs12769205 is found in combination with rs4244285 (the SNP that defines all CYP2C19*2 alleles) and 12662A>G is likely part of all CYP2C19*2 alleles. However, rs12769205 is also found without rs4244285 on CYP2C19*35 (allele designation assigned by the CYP allele nomenclature committee). Here, we have investigated the functional consequence of rs12769205, whether CYP2C19*35 is also under natural selection, and whether CYP2C19*35 is the ancestral CYP2C19 nonfunctional allele, arising before CYP2C19*2.
Materials and Methods
Human Liver Tissue.
A total of 335 human livers from 272 White and 63 Black donors were processed through the St. Jude Liver Resource at St. Jude Children’s Research Hospital and were provided by the Liver Tissue Procurement and Distribution System (National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Contract N01-DK92310) and by the Cooperative Human Tissue Network. The St. Jude Children’s Research Hospital Institutional Review Board approved the use of these human tissues for research purposes.
RNA Isolation and cDNA Preparation.
Total RNA was extracted from human livers with rs4244285 and rs12769205 genotypes using TRIzol reagent (Invitrogen, Carlsbad, CA). Then, 500 ng RNA was used to prepare cDNA using the Invitrogen Thermoscript real-time polymerase chain reaction (RT-PCR) system.
Genotyping of CYP2C19 Alleles in Human Livers.
Genomic DNA from human livers was isolated using a DNeasy tissue kit (Qiagen, Gaithersburg, MD). Genotyping of the rs4244285 and rs12769205 SNPs was performed by direct DNA sequencing. Primers used for PCR amplification and sequencing were forward primer (FP) 5′-CAACCAGAGCTTGGCATATTG-3′ and reverse primer (RP) 5′-TGATGCTTACTGGATATTCATGC-3′ for rs4244285 and FP 5′-AAAATATGAATCTAAGTCAGGCTTAGT-3′ and RP 5′-GGAGAGCAGTCCAGAAAGGTCAGTGATA-3′ for rs12769205. A general 25 µl PCR mixture consisted of 50 ng gDNA, 1 µM primers, and Platinum PCR supermix (Invitrogen). For quality control, minor allele frequencies for rs4244285 and rs12769205 were compared with existing population genotype data from the Exome Variant Server (http://evs.gs.washington.edu/EVS/) for Whites and Blacks. The observed and reported minor allele frequency values were in agreement.
Sequencing of CYP2C19 Exons from Genomic DNA.
CYP2C19 exons were PCR amplified in a 25 µl PCR mixture consisting of 50 ng gDNA, 1 µM primers, and Platinum PCR supermix (Invitrogen), and sequenced using CYP2C19-specific primers (Supplemental Table 1).
Genotyping of African Livers for CYP2C19*2 at the cDNA Level.
To confirm the CYP2C19*2 genotype assignments obtained by DNA sequencing we also genotyped the eight African livers used for subsequent analysis of the transcripts at the cDNA level using the primers and method reported by de Morais et al. (1994b).
CYP2C19 cDNA Amplification.
Various portions of CYP2C19 were PCR amplified from human liver cDNA using the following primer pairs and products, and were either directly sequenced or analyzed on 2% agarose gels. (1) Exon 4 (FP 5′-ATTGAATGAAAACATCAGGATTG-3′) and exon 6 (RP 5′-GTAAGTCAGCTGCAGTGATTA-3′) (de Morais et al., 1994b). CYP2C19*1 and CYP2C19*2 generate 284 and 244 base pair (bp) products, respectively. (2) Exon 2 (FP 5′-GAAGAGGCCATTTCCCACT-3′) and exon 4 (RP 5′-TTTCTGGAAAATAATGGAGCA-3′). Livers with and without rs12769205 generate 438 bp (retaining 169 bp intron 2) and 269 bp products, respectively. (3) Exon 2 (FP 5′-GAAGAGGCCATTTCCCACT-3′) and exon 6 (RP 5′-GTAAGTCAGCTGCAGTGATTA-3′) primers. CYP2C19*1 generates the canonical 597 bp product. CYP2C19*35 generates products of 766 bp (containing exon 2B) and 695 bp (exon 2B plus 70 bp deletion in exon 4). CYP2C19*2 generates products with the 40 bp deletion of exon 5 (557 bp) and additionally containing exon 2B with or without the 70 bp deletion in exon 4 (726 and 655 bp, respectively).
Sequencing the Full-Length CYP2C19*35 cDNA and Its Alternative Transcripts.
First, cDNA from the CYP2C19*1/*35 liver was used as a template and PCR amplified with exon 2 and 6 primers (as previously described). The PCR products were cloned into pCR 2.1 TOPO vector using the TOPO TA Cloning Kit (Invitrogen), and the products were transformed into One Shot Top 10 Chemically Competent cells (Invitrogen) and grown on Lysogeny Broth-ampicillin plates. Forty individual colonies were picked and colony PCR carried out using M13 primers FP 5′-GTAAAACGACGGCCAG-3′ and RP 5′-CAGGAAACAGCTATGAC-3′, and the PCR products were directly sequenced using the same primers.
Second, to determine whether the CYP2C19*35 allele carried other polymorphisms in the coding region, the full-length CYP2C19*35 cDNA from the CYP2C19*35 liver was PCR amplified using FP 5′-TTGTGGTCCTTGTGCTCTGTCTC-3′and RP 5′-GGAATGAAGCACAGCTGA-3′ and the PCR product was cloned into TOPO TA vector and sequenced using the M13 primers previously mentioned. The sequences obtained were then aligned using a software suite for sequence analysis (DNASTAR SeqMan Pro, version 9.0.4. 39, 418, Madison, WI).
CYP2C19 Genotypic Data Mining from the 1000 Genomes Server (Phase 3 Data).
rs4244285 and rs12769205 genotypes were downloaded for White descendants of Northern Europeans and all Africans and Asians from the 1000 Genomes browser (http://browser.1000genomes.org/index.html) to generate visual genotypes and study the linkage between these two alleles in different ethnic groups.
Linkage Disequilibrium (LD) Analysis.
Pairwise LD was calculated for 85 White descendants of Northern Europeans, 246 Africans, 88 Yorubans (YRI), and 97 Asians from the 1000 Genomes browser and displayed using Haploview 4.2 software (Broad Institute, Cambridge, MA) (http://haploview.software.informer.com/4.2/) (Barrett et al., 2005). The D′ value, along with its corresponding logarithm of odds score, and the r2 LD values were determined between rs4244285 and rs3758580, rs4417205, and rs12769205 SNPs.
Generation of CYP2C19 Minigenes.
The RHCglo minigene plasmid (Singh and Cooper 2006) was generously provided by Dr. Thomas A. Cooper (Baylor College of Medicine, Houston, TX). A 264 bp fragment consisting of the last 87 nucleotides (nt) of CYP2C19 intron 4 and full-length exon 5 was amplified from the DNA of a CYP2C19*2/*2 human liver using PCR primers FP 5′-ATATATGTCGACAGTTTTAAATTACAACCAGAGCTTGG-3′ [with a SalI site (underlined)] and RP 5′-ATATATCTCGAGCTTCTCCATTTTGATCAGGAAGC-3′ [with an XhoI site (underlined)], and used to replace the SalI/XhoI fragment of the RHCglo plasmid to generate the CYP2C19*2 minigene. The CYP2C19*2 minigene was used as the template to perform site-directed mutagenesis (SDM) in order to create the CYP2C19*1 wild-type minigene using the QuikChange II SDM kit (Agilent Technologies, Santa Clara, CA) and primers SDM-FP 5′-CCCACTATCATTGATTATTTCCCGGGAACCCATAACAAATTACTTAA-3′ and SDM-RP 5′-TTAAGTAATTTGTTATGGGTTCCCGGGAAATAATCAATGATAGTGGG-3′. The CYP2C19 exon 2/intron 2/exon 3 minigenes were generated by PCR amplifying this region from human liver DNAs with either CYP2C19*1 or CYP2C19*35 genotypes using primers FP 5′-ATATATGGATCCCTCTCAAAAATCTATGGCCCTG-3′ [with a BamHI site (underlined)] and RP 5′-ATATATCTCGAGCCTTGGTTTTTCTCAACTCC-3′ [with an XhoI site (underlined)]. The 482 bp amplified CYP2C19 fragments were used to replace the BamHI/XhoI fragment of the RHCglo plasmid to generate either CYP2C19*1 or rs12769205 minigenes. The sequences of all minigenes were confirmed by DNA sequencing.
Minigene Transfection Assays.
HepG2 human hepatoblastoma cells were cultured in minimal essential media supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin, and maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2. For transfection studies, 150,000 cells per well were seeded in 12-well culture dishes. Twenty-four hours later, cells were transfected with 1000 ng of different minigene plasmids using the LipoJet in vitro DNA and siRNA transfection kit, version II (SignaGen Laboratories, Gaithersburg, MD). Forty-eight hours later, cells were washed with phosphate-buffered saline and harvested with TRIzol reagent (Invitrogen). First-strand cDNA was prepared using 500 ng RNA and oligo (dT) primers (ThermoScript RT-PCR system, Invitrogen). PCR amplification was performed using FP RSV5U 5′-CATTCACCACATTGGTGTGC-3′ and RP TNIE4 5′-AGGTGCTGCCGCCGGGCGGTGGCTG-3′, which both anneal to the vector-expressed exons to selectively amplify only the plasmid-expressed mRNA transcripts (Singh and Cooper 2006). The amplified PCR fragments were electrophoresed on 1% agarose gel.
Quantitative PCR analysis.
Quantitative RT-PCR was used to quantitate the amount of canonical CYP2C19 transcript generated by the CYP2C19 wild-type and rs12769205 minigenes. The FP RSV5U 5′-CATTCACCACATTGGTGTGC-3 annealed in the vector and the RP 5′-CCATTGCTGAAAACGATTCCAA-3′ annealed across the CYP2C19 junction of exons 2 and 3 to specifically amplify only the canonical CYP2C19 transcript generated from the minigene. The glyceraldehyde-3-phosphate dehydrogenase primers were FP 5′-GGACCACCAGCCCCAGCAAGAG-3′ and RP 5′-GAGGAGGGGAGATTCAGTGTGGTG-3′. RT-PCR quantification was carried out using the SYBR GreenER quantitative PCR supermix (Invitrogen) and amplifications run on an ABI PRISM 7900HT Sequence Detection System (PE Applied Biosystems, Foster City, CA). The Ct values were analyzed by the comparative Ct method to obtain relative mRNA expression levels.
CYP2C19 Peptide Quantification.
CYP2C19 was quantified using three different surrogate peptides: (1) exon 2–specific peptide (IYGPVFTLYFGLER); (2) exon 8–specific peptide (GTTILTSLTSVLHDNK); and (3) exon 4–specific peptide (ASPCDPTFILGCAPCNVICSIIFQK). The surrogate peptides for liquid chromatography–tandem mass spectrometry (LC-MS/MS) quantification were selected based on previously reported criteria (Prasad et al., 2014). Trypsin digestion and sample preparation for LC-MS/MS analysis of the genotype-defined pooled human liver microsomal samples was performed using a previously reported protocol (Edson et al., 2013; Wang et al., 2015) with few modifications. Briefly, the pooled human liver microsomal sample (60 μl) was denatured and reduced with 40 μl of ammonium bicarbonate digestion buffer (100 mM, pH 7.8) and 10 μl of 100 mM dithiothreitol at 90°C (5 minutes). The sample was then alkylated by adding 20 μl iodoacetamide (200 mM) at room temperature for 20 minutes. The protein was then extracted using addition of ice-cold methanol (500 μl), chloroform (200 μl), and water (400 μl). The mixture was vortexed, centrifuged at 12,000g for 5 minutes, and the upper layer was removed. The protein pellet was washed with 500 μl ice-cold methanol followed by centrifugation at 12000g for 5 minutes. The final protein pellet was dissolved in ammonium bicarbonate (40 μl) and 3% sodium deoxycholate (10 μl) before digestion by trypsin (protein:trypsin ratio of 25:1) at 37°C for 16 hours. The reaction was quenched by addition of 20 μl of heavy peptide 2 (GTTILTSLTSVLHDNK[13C6,15N2]) internal standard solution (prepared in 70% acetonitrile in water containing 0.1% formic acid), and 10 μl of the neat solvent (70% acetonitrile in water containing 0.1% formic acid). The samples were centrifuged at 4000g for 5 minutes. All of the human liver microsomal samples were digested and processed in triplicates.
The CYP2C19 surrogate peptides were then quantified using triple-quadrupole LC-MS instruments [Xevo TQ-S coupled to ACQUITY UPLC (Waters, Milford, MA)] in Electrospray positive ionization mode. Approximately 10 µg of the trypsin digest (5 µl) was injected onto the column (ACQUITY UPLC HSS T3 1.8 µm, 2.1 × 100 mm, Waters) and eluted at 0.3 ml/min. A mobile phase consisting of water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B) was used. A flow rate of 0.3 ml/min was used with a gradient elution starting from 3% B and kept until 2.0 minutes, followed by a gradient program (B concentration) of 3%–15% (2.0–4.0 minutes), 15%–25% (4.0–10 minutes), and 25%–50% (10.0–14 minutes). This was followed by washing with 80% mobile phase B for 0.9 minutes and re-equilibration for 4.9 minutes. The peak retention times were confirmed by spiking either peptide standards (peptides 1 and 2) and/or trypsin-digested CYP2C19 protein standard (gratis sample from Dr. Nina Isoherranen, School of Pharmacy, University of Washington). The MS/MS analysis was performed by monitoring the surrogate peptides and the internal standard using instrument parameters provided (Supplemental Table 2). The LC-MS/MS data were processed using MassLynx 4.1 (Waters) by integrating the peak areas generated from the ion chromatograms for the surrogate peptides and normalized by the internal standard response. The peak response for two transitions from each peptide was averaged for quantification of the samples and the relative protein quantification was reported as the mean and S.D. of the peak area ratio values obtained in at least three experiments.
Ethiopian Cohort (n = 104).
Details of the healthy unrelated Ethiopian subjects of both sexes living in Ethiopia who participated in this study have been described previously (Persson et al., 1996; Aklillu et al., 2002). The study received ethics approval from the Human Ethics Committees at Huddinge University Hospital, Karolinska Institutet, Stockholm, Sweden, and the National Ethics Committees at the Ethiopian Science and Technology Commission, Addis Ababa, Ethiopia.
CYP2C19 Phenotyping of the Ethiopian Cohort.
Details on CYP2C19 phenotyping of the Ethiopian cohort have been previously published (Persson et al., 1996; Aklillu et al., 2002; Sim et al., 2006). Briefly, the subjects received 100 mg racemic mephenytoin (Mesantoin Sandoz Pharmaceuticals, Basel, Switzerland) after emptying their bladder just before bedtime. Total urine was collected for 0–8 hours after drug intake. Volume and pH were measured and 20 ml aliquots were stored at −20°C until analysis. The concentration ratio of S- and R-mephenytoin was measured by gas chromatography as described previously (Sanz et al., 1989). Urine samples with S/R ratio > 0.6 were reanalyzed after acid treatment (Tybring and Bertilsson, 1992). Subjects with an S/R ratio greater than 0.9 and not increasing above 1.4 after acid treatment were assigned as PMs.
CYP2C19 Genotyping of the Ethiopian Cohort.
A 10 ml venous blood sample was taken from each subject into an EDTA-containing vacutainer tube and DNA was isolated from peripheral leukocytes using a guanidinium-isothiocynate method. Genotyping for CYP2C19*2 and CYP2C19*3 was done using allele-specific PCR as described previously (De Morais et al., 1994a; de Morais et al., 1994b; Persson et al., 1996). Genotyping for CYP2C19*17 was performed as described previously (Sim et al., 2006). CYP2C19*35 (rs12769205) was genotyped using a custom Taqman SNP genotyping assay (Item No. 4331349) from Life Technologies. Taqman genotyping assay was done using a QuantStudio 12K Flex-RT-PCR system (Life Technologies, Carlsbad, CA). The final volume for each reaction was 10 µl, consisting of TaqMan fast advanced master mix (Applied Biosystems, Foster City, CA), TaqMan 1× drug metabolism genotyping assays mix (Applied Biosystems, Foster City, CA), and 20 ng genomic DNA. The PCR profile consisted of an initial step at 60°C for 30 seconds, hold stage at 95°C for 10 minutes, and a PCR stage for 40 cycles: step 1 at 95°C for 15 seconds, step 2 at 60°C for 1 minute, and postread stage at 60°C for 30 seconds.
Preparation of RNA-Sequencing (RNA-Seq) Libraries.
Liver RNA was isolated at St. Jude Children’s Research Hospital. Liver RNA was shipped to the Baylor College of Medicine Human Genome Sequencing Center where it was analyzed for integrity and used for RNA-Seq library preparation and sequencing as described in detail in a separate study (Chhibber et al., 2015). Raw reads were sent to the St. Jude Children’s Research Hospital Computational Biology Team for analysis.
RNA-Seq Analysis.
Fast all quality (FASTQ) sequences were mapped to the hg19 genome using an internal unpublished pipeline that employs STAR (http://code.google.com/p/rna-star/) (Dobin et al., 2013), TopHat2 (http://ccb.jhu.edu/software/tophat) (Kim et al., 2013), and other mappers and was developed for the Pediatric Cancer Genome Project (Downing et al., 2012). Mapped reads were counted with HTSeq (https://pypi.python.org/pypi/HTSeq) (Anders et al., 2015) coverage files, and gene-level fragments per kilobase of exon per million fragments mapped (FPKM) values were then computed and data were visualized using an integrative genomic viewer (IGV) (Robinson et al., 2011). Exon junction data were extracted through the RNApeg pipeline (Edmonson MN, Rush MC, Zhang J, manuscript in preparation).
Haplotypes, Relative Extended Haplotype Homozygosity (REHH), and Ancestral Analysis.
The Sweep program (http://www.broadinstitute.org/mpg/sweep/) was used to determine haplotypes and to generate REHH plots in order to look for positive selection in the YRI population and then in other African populations. All 216 chromosomes from the YRI in the 1000 Genomes phase 3 data (release #20130502) were used, compared with a recent study (Janha et al., 2014), which used 120 YRI chromosomes. The coordinates were lifted from hg19 to hg17 using the LiftOver program (https://genome.ucsc.edu/cgi-bin/hgLiftOver), and then data in chr10:96486608-96688055 (hg17 coordinates) were used for subsequent analysis, totaling 201,447 bp. SNP positions rs12769205, rs4244285, rs4417205, and rs3758580 defined the core haplotypes. REHH plots were generated from rs77989980 (downstream from CYP2C18) to rs9332103 (upstream of CYP2C9). For ancestral analysis, the Sweep program predicts the chimp allele to be the ancestral allele for all SNPs from HapMap Release 16 (http://hapmap.ncbi.nlm.nih.gov). Where ancestral information was available, the ancestral core haplotype was predicted and a phylogenetic tree of haplotypes was then created.
Results
A Polymorphic CYP2C19 Alternative mRNA Retaining Intron 2.
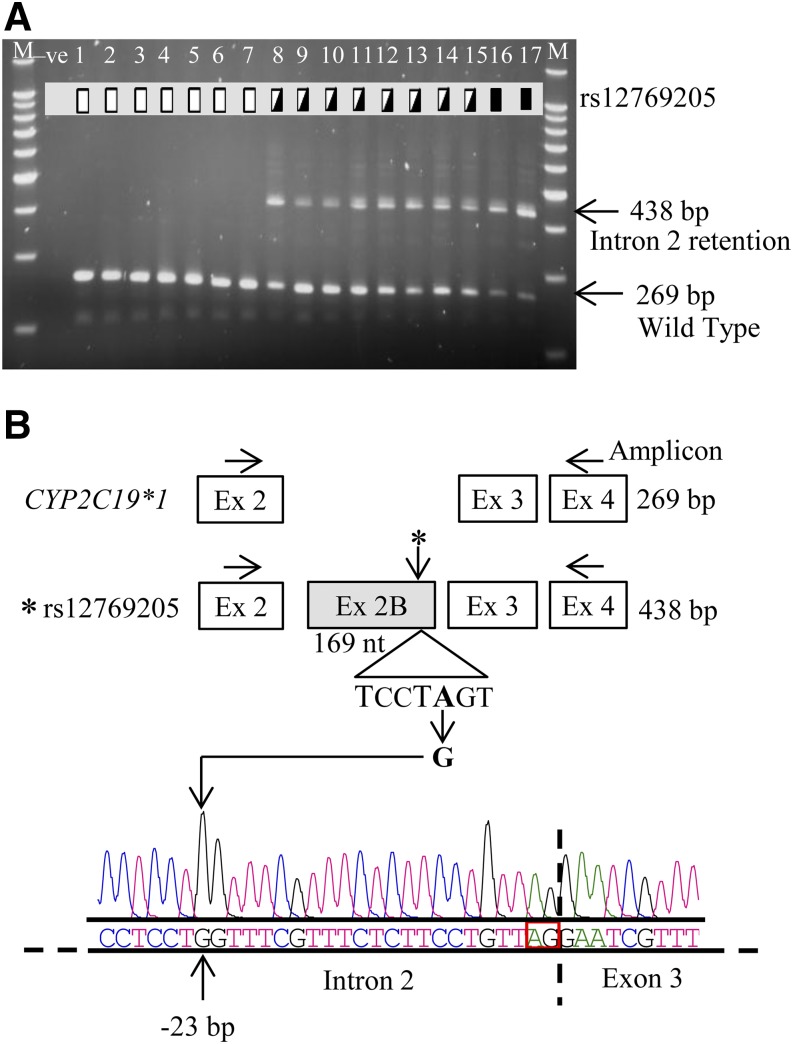
PCR amplification of the CYP2C19 cDNA from exons 2 to 4 revealed the expected 269 bp product in all livers, but with an additional 438 bp product polymorphically expressed in some (Fig. 1). Direct sequencing of the 438 bp fragment showed complete intron 2 retention generating exon 2B. All livers expressing the CYP2C19 alternative mRNA with exon 2B also carried a unique intron 2 SNP rs12769205; however, no other SNP in intron 2 or in exons 2 or 3 was found after sequencing exon 2 through exon 3 in genomic DNA. Sequencing of the entire CYP2C19 cDNA revealed that the rs12769205 was also found in those carrying the CYP2C19*2 allele.
Fig. 1.
CYP2C19*35 (rs12769205) leads to aberrant intron 2 retention (exon 2B). (A) CYP2C19 cDNA was amplified from human liver cDNAs by PCR using primers in exons 2 and 4 and the wild-type product (269 bp) and alternatively spliced product (438 bp) analyzed on agarose gel. Homozygous wild-type (lanes 1–7 are CYP2C19*1/*1), rs12769205 heterozygous (lanes 8–15), and homozygous (lanes 16 and 17) variant genotypes are indicated by the open, half-filled, and filled boxes, respectively. Lanes marked M and –ve represent the 100 bp DNA ladder and a negative control, respectively. (B) The 438 bp fragment was excised from the gel and directly sequenced and the resulting electropherogram and nucleotide sequence are shown. The cartoon illustrates the amplification strategy, the insertion of exon 2B in samples with rs12769205, and the location of rs12769205 at the branch point adenine −23 nt upstream of the intron 2 splice acceptor site.
rs12769205 Is in LD with rs4244285 on CYP2C19*2, but Occurs Independently on the CYP2C19*35 Allele in Some Blacks.
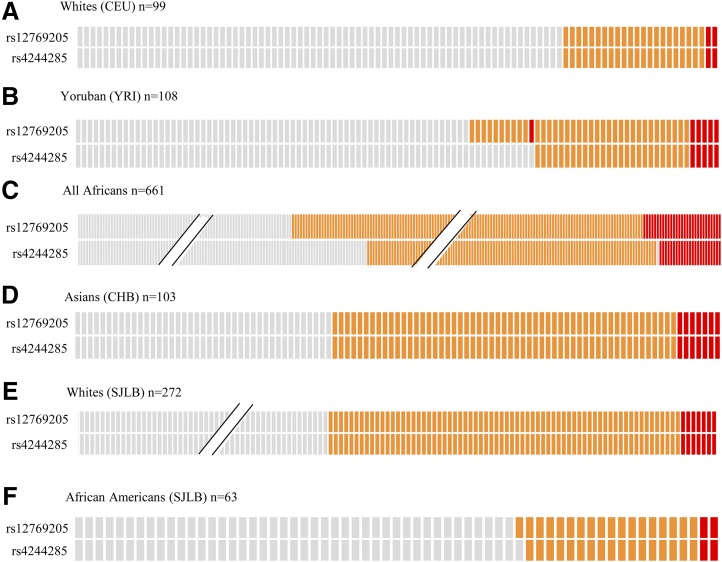
To observe the frequency of rs12769205 in different populations and determine whether rs12769205 ever occurred independent of CYP2C19*2, rs4244285 and rs12769205 genotypes were retrieved from different populations (Fig. 2) in the 1000 Genomes project database (phase 3) and plotted (Fig. 2 A–D). The rs4244285 and rs12769205 SNPs appeared to be linked in CYP2C19*2 in 99 White descendants of Northern Europeans; however, rs12769205 did occur independent of rs4244285 on a new allele (CYP2C19*35) in 11/108 Black descendants of YRI West Africans, and in 29/661 all Africans (which includes the YRI group). CYP2C19*2 and CYP2C19*35 appeared to be linked in 103 Asians. Next, we examined visual genotypes of rs4244285 and rs12769205 in each of the seven African populations representing the African super population in the 1000 Genomes project (Supplemental Fig. 1). The rs12769205 SNP had the highest frequency in YRI and the lowest frequency in the Esan in Nigeria. Interestingly, there were individuals in the Esan (in Nigeria), Gambian (in Western divisions in Gambia), Luhya (in Webuye), and Mende (in Sierra Leone) populations that were CYP2C19*35 homozygous, but CYP2C19*2 heterozygous, and even one YRI CYP2C19*35 homozygous, but CYP2C19*1 homozygous. The CYP2C19*2/*35 diplotypes as well as CYP2C19*2 and CYP2C19*35 allele frequency data in different populations is provided in Supplemental Table 3. To determine if the SNPs were in LD, the D′ value, along with its corresponding logarithm of odds score, and r2 LD values were determined between rs4244285 and rs12769205 SNPs in all populations. Haploview (Barrett et al., 2005) analysis showed that D′/logarithm of odds = 1.0 (Supplemental Fig. 2) for all pairwise comparisons of the two SNPs in all populations (note: the LD values in Supplemental Fig. 2 are scaled from 1.0 to 100 for visual clarity). The D′ value of 1.0 indicates that the CYP2C19*2 rs4244285 is co-inherited with rs12769205 (on the CYP2C19*2 haplotype) 100% of the time. The high correlation coefficients (r2 ≥ 0.6) between the two SNPs confirm they are significantly linked in each population. The r2 value is lower than the D′ value because the allele frequencies of rs4244285 and rs12769205 are not identical—rs12769205 is more frequent in YRI because rs12769205 can be found on CYP2C19*35 that lacks rs4244285. These results prompted us to genotype human livers from White and Black donors. rs12769205 and rs4244285 appeared to be in LD in Whites (Fig. 2E) and African Americans (Fig. 2F) with the CYP2C19*2 genotypes and there was a single African American who had rs12769205 (CYP2C19*35) without rs4244285. This liver was further analyzed to determine if exon 2B occurred when only rs12769205 was present.
Fig. 2.
Visual genotypes of rs12769205 and rs4244285 in different populations (1000 Genomes phase 3 data) and liver samples. Visual genotypes for rs12769205 and rs4244285 in the 1000 Genomes samples (genotypes downloaded from http://browser.1000genomes.org/index.html): (A) 99 White (CEU), (B) 108 YRI, (C) 661 all Africans, and (D) 103 Han Chinese in Beijing, China (CHB); and in the St. Jude Liver Bank (SJLB) samples from (E) 272 White and (F) 63 African Americans. Gray, orange, and red boxes indicate homozygous wild-type, heterozygous, and homozygous variant genotypes, respectively. The slanted lines (//) indicate not all subjects are shown for that particular diplotype and population.
CYP2C19*35 (rs12769205) Alone Leads to Exon 2B.
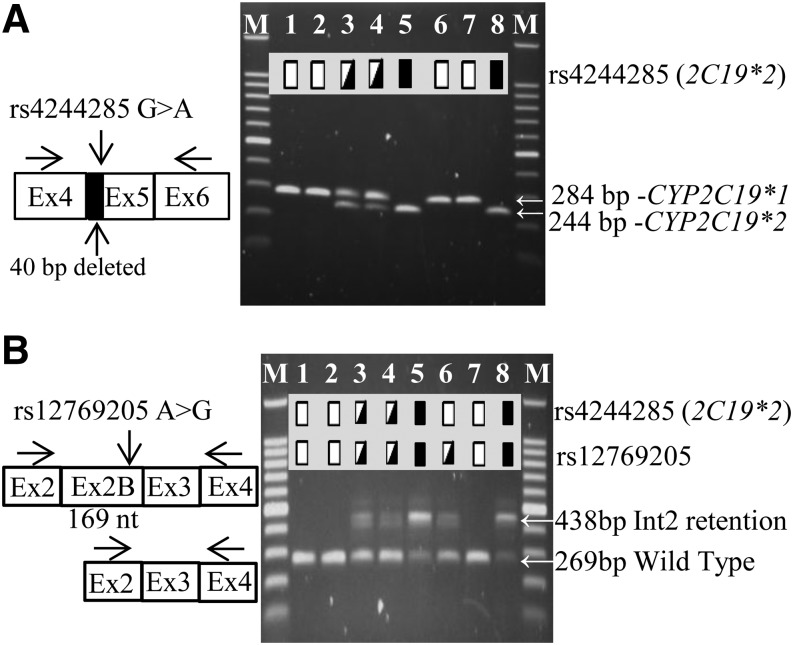
First, the CYP2C19*2 aberrant spliced mRNA (deletion of the first 40 bp of exon 5) was confirmed by PCR amplification of CYP2C19 exons 4–6 in all samples with the rs4244285 genotype (Fig. 3A). Next, CYP2C19 exons 2–4 (Fig. 3B) were amplified in the same samples, yielding both the canonical 269 bp product and a 438 bp amplicon retaining intron 2 (exon 2B). CYP2C19*2 exon 2B occurred in all livers that carried rs12769205 either alone (CYP2C19*35, lane 6), or in livers with rs4244285 (CYP2C19*2, lanes 3, 4, 5, and 8). Moreover, the relative abundance of canonical 269 bp product was always lower in amount in livers with the rs12769205 genotype. Lanes 3, 4, and 6 and lanes 5 and 8 indicate the smaller amount of residual wild-type mRNA amplified in samples heterozygous and homozygous for rs12769205, respectively, compared with samples 1, 2, and 7 homozygous for the CYP2C19*1 genotype.
Fig. 3.
Effect of rs4244285 and rs12769205 on CYP2C19 splicing. (A) Eight liver samples were analyzed by PCR for (A) the CYP2C19 exon 5–40 bp deletion, caused by rs4244285, using primers in exons 4 and 6, and (B) the CYP2C19 exon 2B insertion, caused by rs12769205, using primers in exons 2 and 4 and the products analyzed on agarose gels. Arrows indicate the migration of the canonical and splice variant bands. Homozygous wild-type, heterozygous, and homozygous variant genotypes are indicated by the open, half-filled, and filled boxes, respectively. In (B), the smaller residual amount of the 269 bp wild-type mRNA is seen in rs12769205 heterozygous (lanes 3, 4, and 6) or homozygous (lanes 5 and 8) samples compared with samples 1, 2, and 7 homozygous for the CYP2C19*1/*1 genotype.
Alternative mRNAs in Livers Carrying the rs12769205 Allele.
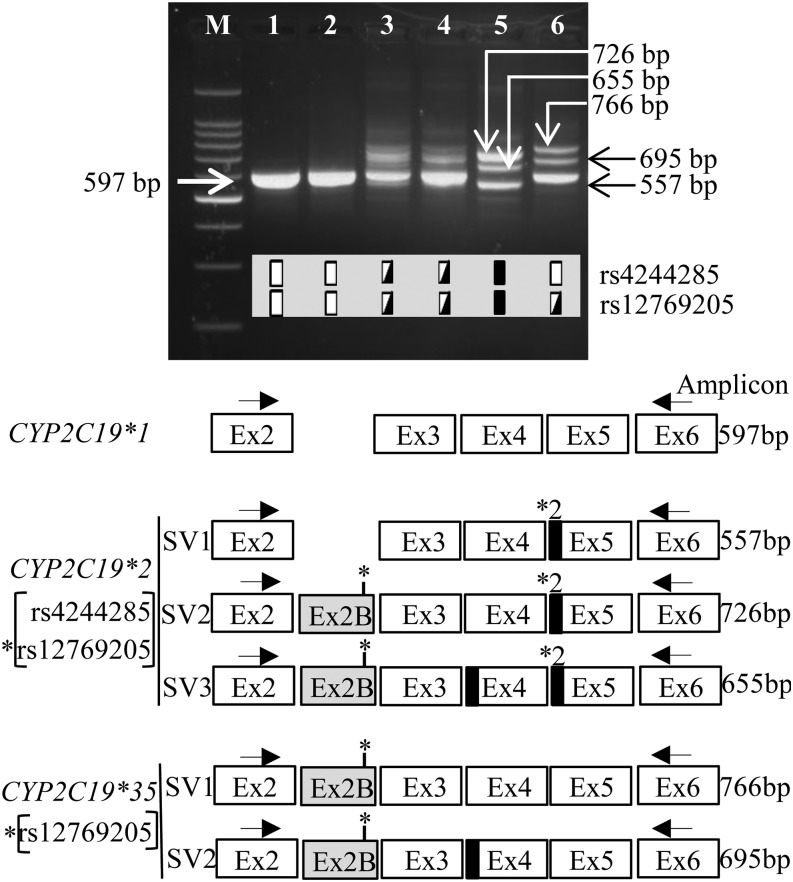
CYP2C19 was amplified from exons 2–6 and the PCR products sized on agarose gels to further determine the structure of all hepatic CYP2C19 alternative mRNAs in livers with the rs12769205 and rs4244285 polymorphisms. Samples homozygous for CYP2C19*1 yielded the single 597 bp wild-type product (Fig. 4). CYP2C19 PCR products from samples heterozygous for rs4244285 and rs12769205 yielded the 597 bp wild-type product; however, other bands were difficult to resolve. To unambiguously identify the alternative mRNAs arising from rs12769205, CYP2C19 exons 2–6 were amplified from the sample heterozygous for CYP2C19*35, the products were TOPO TA cloned, and individual clones sequenced. Only transcripts represented in 5% or more of clones are reported: 52/72 of them were wild type, 14/72 had aberrant exon 2B, and 4/14 lacked the first 71 bp of exon 4. These CYP2C19*35 mRNAs corresponded to the wild-type (597 bp), SV1 (766 bp), and SV2 (695 bp) bands, respectively (Fig. 4). The partial deletion of 71 bp of exon 4 (which alters the reading frame and truncates the protein after amino acid 165) appears to be a passenger splice variant because there were no additional SNPs associated with this spliced transcript. This strategy also allowed the unambiguous identity of the CYP2C19 mRNAs from samples homozygous for rs12769205 and rs4244285: the alternative mRNAs shared the 40 bp deletion of exon 5 (SV1-3); SV2 and SV3 had exon 2B and SV3 had an additional 71 bp deletion of exon 4. There was only a small amount of correctly spliced CYP2C19 wild-type transcript in livers homozygous for rs12769205 and rs4244285, and this can be seen in Fig. 1 (lanes 16 and 17) and Fig. 3.
Fig. 4.
Structure of alternative CYP2C19 mRNAs in livers with rs4244285 and rs12769205 genotypes. CYP2C19 was PCR amplified using exon 2 and 6 primers in liver samples with indicated genotypes and the products were analyzed on agarose gels. Arrows indicate the migration of the canonical and splice variant bands. Genotypes are illustrated as depicted in the Fig. 3 legend. The cartoon illustrates the structures of the canonical exons (open boxes), novel exon 2B (gray box), and partial deletions of 40 and 71 bp at the start of exons 5 and 4 (black boxes), respectively, in the splice variant (SV) transcripts.
Sequencing CYP2C19 from a CYP2C19*1/*35 Liver.
To determine whether the CYP2C19*35 allele carried other polymorphisms, the full-length CYP2C19*35 cDNA from the CYP2C19*35 liver was amplified, cloned into the TOPO TA vector and sequenced (Supplemental Table 4). The CYP2C19*35 mRNA contained exon 2B with the causative rs12769205, the common nonsynonymous Ile331Val, and the synonymous Pro33Pro. Additional variations were discovered on the other allele but are the focus of another study.
In Silico and In Vitro Analysis of the Effect of rs12769205 on CYP2C19 Splicing.
The online bioinformatics resource Human Splicing Finder, version 2.4.1 (http://www.umd.be/HSF/) (Desmet et al., 2009) was used to analyze intron 2 for splicing consensus sequences. The program identified a ctcctAg sequence 23 bp upstream of the end of intron 2 as the optimal branch point motif (recognized by the highest number of matrices) with a consensus value of 73.05 (on a 0–100 score range); however, rs12769205 will disrupt the branch point sequence and decrease the value to 43.42 (Gooding et al., 2006).
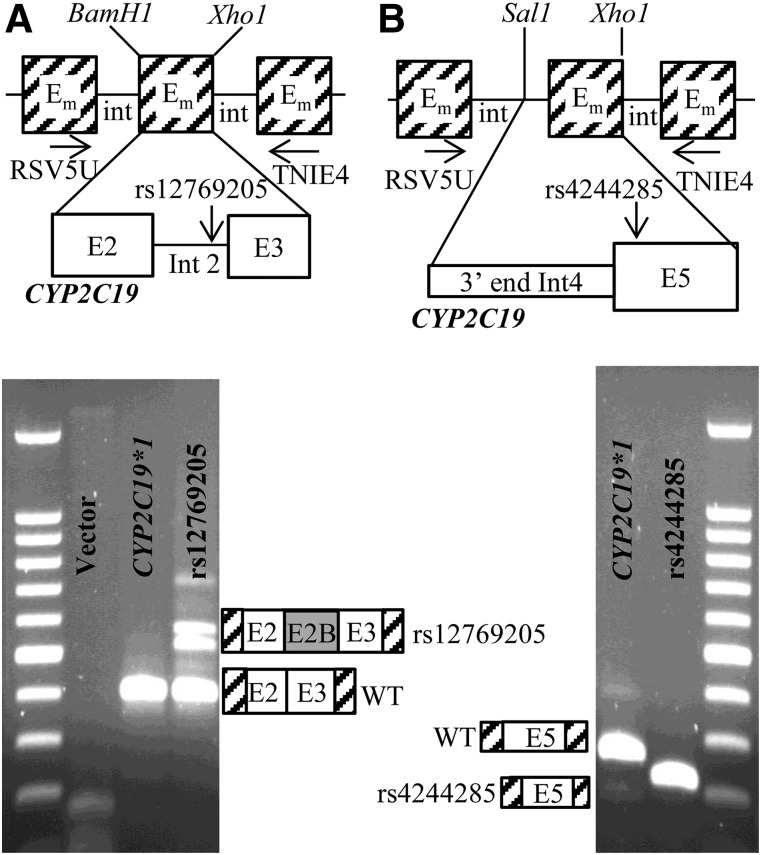
To test whether the rs12769205 branch point SNP alone leads to exon 2B, and to confirm that rs4244285 leads to a 40 bp deletion of exon 5, CYP2C19 minigenes were constructed using the RHCglo minigene vector (Singh and Cooper 2006). Two minigenes contained exons 2 and 3 and intron 2 and differed only by rs12769205 (Fig. 5A), and two minigenes contained exon 5 and the last 87 bp of intron 4 and differed only by rs4244285 (CYP2C19*2) (Fig. 5B). HepG2 cells were transfected with the four minigenes and RNA from the transfected cells was used for RT-PCR analysis using primers residing in the flanking minigene exons. The CYP2C19*1 intron 2 minigene generated the expected wild-type transcript, whereas the rs12769205 variant generated a larger transcript containing insertion of intron 2 and one other smaller alternatively spliced fragment (Fig. 5A). Quantitative PCR of the amount of residual wild-type transcript from the minigene revealed a 40% decrease in the amount of wild-type transcript from the rs12769205 versus CYP2C19*1 minigene. As expected, the CYP2C19*1 exon 5 minigene generated the correctly spliced exon 5 transcript, whereas the rs4244285 minigene demonstrated the 40 bp deleted fragment at the start of exon 5 (Fig. 5B).
Fig. 5.
RT-PCR analysis of minigene mRNA products. (A) Exon2-intron2-exon3 minigenes: two minigenes contained CYP2C19 exon 2 + intron 2 + exon 3 (differing only by rs12769205A>G) that replaced the BamH1/Xho1 fragment [minigene exon (Em)] of the RHCglo minigene. (B) Exon 5 minigenes: Two minigenes contained the last 87 nt of CYP2C19 intron 4 and all of exon 5 (differing only by rs4244285) that replaced the Sal1/Xho1 fragment of the RHCglo minigene. HepG2 cells were transfected with each of the four plasmids and minigene mRNA products were analyzed by RT-PCR using the RSV5U/TNIE4 primers and the products analyzed on agarose gels. The rs12769205 minigene generated a transcript with the exon 2B insertion (A), and the rs4244285 minigene generated a transcript with the 40 bp exon 5 deletion (B).
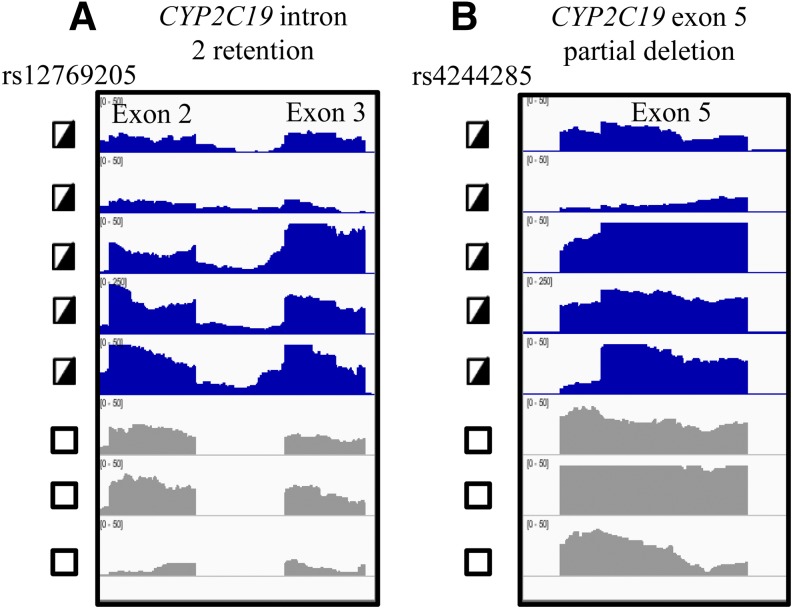
Can next-generation RNA sequencing analysis of CYP2C19 in human livers identify the 40 bp exon 5 deletion and the exon 2B splicing events? Twenty-four liver samples that had undergone next-generation RNA sequencing were analyzed for alternative CYP2C19 mRNAs and the results were visualized with the IGV. As is typical of RNA-Seq IGV results, there is nonuniformity in the exon peaks in part due to nonuniformity of the read coverage (even when the transcripts have very similar concentrations), sequence-specific read variability, and the transcriptional complexity for multigene family members, such as the CYPs. In general, the intron 2 retention (Fig. 6A) and the 40 bp deletion in exon 5 (Fig. 6B) was apparent by the IGV in mRNA samples heterozygous for rs12769205 and rs4244285, although it was much more apparent in some samples where CYP2C19 was more highly expressed. However, it would be difficult to accurately call either of the CYP2C19 alternative mRNAs in some samples (for example, the second sample from the top in Fig. 6) due to the low read coverage and nonuniformity of the exon architecture. Moreover, while visual inspection of each sample revealed there might be a small insertion of intron 2, when exon/intron junction analysis software was used to identify novel transcripts, the software did not call the novel exon 2B junction.
Fig. 6.
IGV visualization of human liver CYP2C19 mRNA. RNA-Seq results for human livers that were heterozygous (half-filled box) or homozygous wild type (open box). (A) rs12769205 were visualized across CYP2C19 exons 2-Exon3; and (B) rs4244285 were visualized across exon 5. Due to differences in CYP2C19 read coverage, IGV was scaled between 0 and 50 for six of the liver samples, and between 0 and 250 for one sample. Panel (A) shows that only samples heterozygous for rs12769205 had RNA sequences with intron 2 included; panel (B) shows that only samples heterozygous for rs4244285 had a decreased signal across the first 40 bp of exon 5, indicative of the heterozygous deletion of this region of the mRNA transcript.
Quantitation of CYP2C19 Protein in Pooled Human Liver Microsomes with Different CYP2C19 Genotypes.
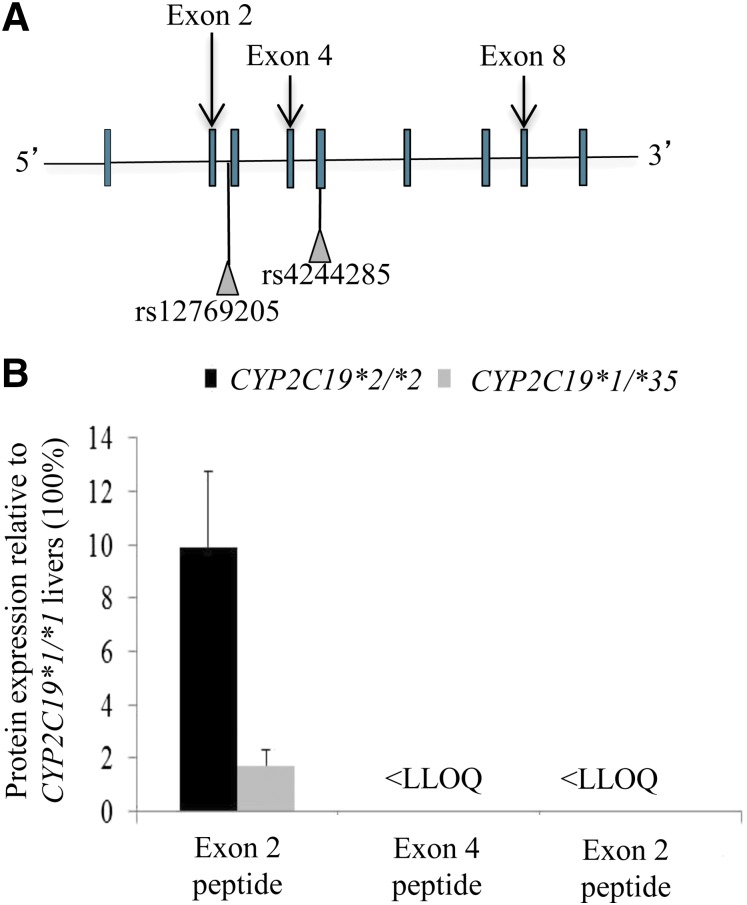
CYP2C19 was quantified in pooled human liver microsomes with different CYP2C19 genotypes by trypsin digestion and LC-MS/MS analysis using three different surrogate peptides specific for CYP2C19 exons 2, 4, and 8 (Fig. 7A). In theory, (1) the probes detect not just the amount of full-length CYP2C19, but also the truncated in-frame CYP2C19 translated from the splice variant mRNAs; and (2) because rs12769205 will frame shift the CYP2C19 protein after exon 2, and rs4244285 will frame shift the protein in the middle of exon 5, the exon 2 and 4 probes could distinguish between the functional effects of rs12769205 in intron 2 (downstream from the exon 2 probe, but upstream of the exon 4 probe) and rs4244285 in exon 5 (downstream from both the exon 2 and 4 probes) on the abundance of the residual CYP2C19 wild-type protein. As expected, CYP2C19 was detected with all exon probes in CYP2C19*1/*1 livers. The amounts of CYP2C19 protein detectable with the exon 2 probe in pooled livers homozygous for CYP2C19*2/*2 or heterozygous for CYP2C19*1/*35 were only 10% and 2%, respectively, of the amount of protein in CYP2C19*1/*1 pooled livers (Fig. 7B). The peptide quantities of CYP2C19 exons 4 and 8 were 95% and 81.5%, respectively, (relative to exon 2) in the CYP2C19*1/*1 samples. No wild-type CYP2C19 protein was detected in the CYP2C19*2/*2 or CYP2C19*1/*35 livers with either the exon 4 or 8 probe. The absence of detectable protein in the CYP2C19*1/*35 liver was surprising and suggested that the person carried an additional deleterious allele. That novel allele is the subject of a separate study. Nevertheless, the absence of detectable CYP2C19 protein in the CYP2C19*2/*2 livers with the exon 4 probe (but not the exon 2 probe) demonstrates that intron 2 rs12769205 has a functional effect on the CYP2C19 protein independent of the effect of rs4244285. In fact, it suggests that rs12769205, because it leads to insertion of exon 2B and creates 87 altered amino acids followed by a stop codon, confers the loss of CYP2C19 protein in CYP2C19*2/*2 livers because of its primacy (before rs4244285) in the RNA splicing event.
Fig. 7.
CYP2C19 peptides downstream from exon 2 failed to detect residual CYP2C19 wild-type protein in CYP2C19*2 and CYP2C19*35 liver microsomes. (A) The location of the peptide probes used relative to CYP2C19 exons and to rs12769205 and rs4244285. (B) Expression of CYP2C19 protein quantified in pooled human liver microsomes from CYP2C19*1/*1, CYP2C19*2/*2, and CYP2C19*1/*35 livers using exon 2, 4, and 8 specific peptide probes. Results are graphed relative to CYP2C19 protein in CYP2C19*1/*1 pooled liver microsomes (100%). Less than the lower limit of quantitation (<LLOQ).
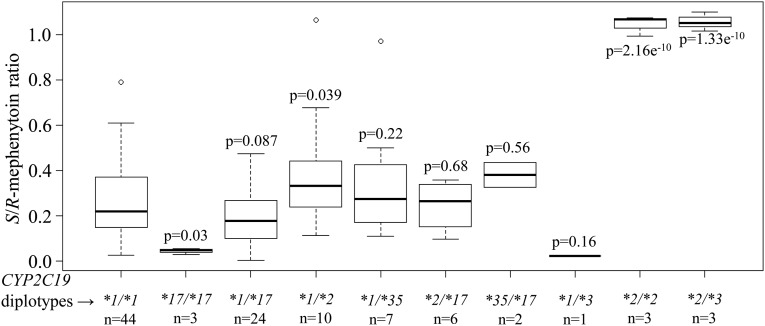
Relationship of CYP2C19 Genotype to Activity in Ethiopians.
Data on the S-mephenytoin hydroxylation phenotype and CYP2C19 genotypes for CYP2C19*2, CYP2C19*3, and CYP2C19*17 among 104 Ethiopians was already available from previous studies and detailed information about the cohort has been published (Persson et al., 1996; Aklillu et al., 2002; Sim et al., 2006). We genotyped this in vivo cohort for the new CYP2C19*35 allele in order to perform a phenotype/genotype association analysis (Fig. 8). Compared with CYP2C19*1/*1 individuals, those persons heterozygous for one nonfunctional CYP2C19 allele, either CYP2C19*1/*35 (P = 0.22) or heterozygous for CYP2C19*1/*3 (P = 0.16) did not show a significant increase in S/R-mephenytoin plasma concentrations, while those heterozygous for both rs12769205 and rs4244285 together (CYP2C19*1/*2) (P = 0.039) did. As expected, persons homozygous for rs12769205 and rs4244285 together were poor mephenytoin metabolizers; however, there were no persons homozygous for rs12769205 alone (CYP2C19*35/*35) to conclusively determine the independent functional effect of this SNP in vivo.
Fig. 8.
CYP2C19 activity in 104 Ethiopians with different CYP2C19 genotypes. S/R-mephenytoin ratio is plotted for each CYP2C19 diplotype group. Box plots indicate the 25th and 75th percentiles, the bold line within the box represents the median, and whiskers represent the range after excluding the outliers. Statistical analyses were performed using R version 3.1 (http://www.rproject.org). A general linear model was used to obtain P values for each group compared with the CYP2C19*1/*1 group. Number of subjects (n).
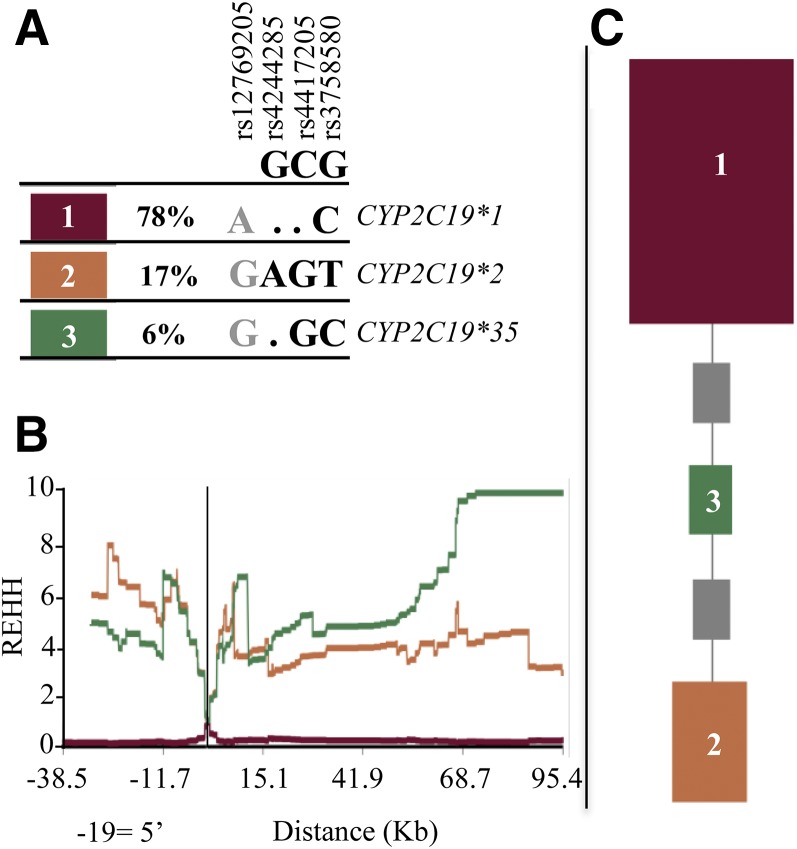
Extended Haplotype Homozygosity at the CYP2C19 Locus in YRI and Other African Populations.
It was recently reported that the CYP2C19*2 nonfunctional allele may be under positive selection with CYP2C19 inactivation, conferring an evolutionary advantage in Africa (Janha et al., 2014). To investigate if the signal for selection could be attributed to rs12769205, we used Sweep—a program that uses large-scale analysis of haplotype structure in the genome to detect evidence of natural selection—in order to reanalyze CYP2C19*2 in 108 YRI, and we included rs12769205. We first used Sweep to determine the CYP2C19 haplotype structure. Sweep detected haplotype blocks 1, 2, and 3 (defining CYP2C19*1, CYP2C19*2, and CYP2C19*35, respectively) (Fig. 9A). CYP2C19*2 rs4244285 is carried only on haplotype 2, while SNP rs12769205 is carried on haplotypes 2 (CYP2C19*2) and 3 (CYP2C19*35). Sweep then used the long-range haplotype test to analyze the haplotypes for long-range LD. CYP2C19 haplotypes 2 and 3 in YRI both displayed extended homozygosity as seen by the high REHH scores (Fig. 9B). The REHH plots for the three haplotypes, with the core for the haplotypes centered on the genomic position of the CYP2C19*2 variant, shows that both CYP2C19*2 (containing both rs4244285 and rs12769205) and CYP2C19*35 (containing only rs12769205) showed long-range LD, suggesting the haplotypes rose rapidly to a high frequency before recombination could break down associations with nearby markers (Sabeti et al., 2002).
Fig. 9.
CYP2C19 haplotype frequencies, extended haplotype homozygosity, and ancestral tree in 216 YRI chromosomes. (A) Sweep was used to determine CYP2C19 haplotypes (SNP positions rs12769205, rs4244285, rs4417205, and rs3758580) and their frequencies. The online dot (.) in the observed haplotypes represents nucleotides that match the ancestral allele. GCG below the SNP rsIDs are the Sweep predicted ancestral allele nucleotides based on HapMap Release 16. (B) REHH for each CYP2C19 haplotype with the core of the haplotypes centered on rs4244285. Both haplotypes containing SNP rs12769205 either alone [haplotype 3 (green, CYP2C19*35)], or with rs4244285 [haplotype 2 (orange, CYP2C19*2)] show extended haplotype homozygosity REHH. (C) Phylogenetic tree of the CYP2C19 haplotypes. Haplotypes closer to ancestral are at the top. The area of the squares is proportional to the frequency of the haplotype. The gray squares represent haplotypes not present in the data, but that are missing links in the phylogeny. The program determined the ancestral root of the tree was CYP2C19*1.
The region of longest extended homozygosity and highest REHH of 10 was seen 68–95 kb from the core with haplotype 3 (rs12769205 alone on CYP2C19*35), while across the same region for haplotype 2 with rs12769205 + rs4244285 (CYP2C19*2) the REHH was 5.5–2.5. Indeed, the significant REHH for both haplotypes 2 and 3 suggests rs12769205, both alone on CYP2C19*35 and together with rs4244285 on CYP2C19*2, confers an evolutionary advantage to these alleles.
The highest REHH scores were 3′ of CYP2C19. Although the most distal 3′ intergenic SNPs flanking CYP2C19 did not extend into CYP2C9, we determined whether either CYP2C9*2 or CYP2C9*3 were on these long-range CYP2C19-extended haplotypes. However, none of the 108 YRI from the HapMap project carried either the CYP2C9*2 (rs1799853) or CYP2C9*3 (rs1057910) nonfunctional SNPs, demonstrating that it is the two CYP2C19 haplotypes with rs12769205 that are under natural selection.
Sweep was next used to construct a phylogenetic tree of the CYP2C19 haplotypes (Fig. 9C). The haplotypes closer to the ancestral haplotype are at the top in Fig. 9C. CYP2C19*35 is calculated to be closer than CYP2C19*2 to the ancestral haplotype, and hence CYP2C19*35 is the ancestral haplotype, and rs12769205 arose before rs4244285.
Next, we performed the same analysis on the 396 individuals who represented other African populations still living on the African continent (Luhya in Webuye, Kenya; Gambian in Western divisions in Gambia; Mende in Sierra Leone; and Esan in Nigeria). The same three CYP2C19 haplotypes were present in the other African populations, and at a similar frequency to the YRI (Supplemental Fig. 3). Likewise, the combined other African populations displayed extended homozygosity for CYP2C19*35, as evidenced by the high REHH (7.6, at 95.4 kb from the core). Finally, Sweep ancestral tree analysis generated a CYP2C19 phylogenetic tree for the other African populations that was identical to that generated for the YRI, again showing the haplotype with rs12769205 alone in the ancestral haplotype, with rs4244285 added later to generate CYP2C19*2.
Discussion
We discovered that rs12769205 disrupts the branch site in CYP2C19 intron 2, creating a novel exon 2B. This alternative CYP2C19 mRNA will generate a nonfunctional protein since insertion of exon 2B creates an out-of-frame protein with 87 novel amino acid residues followed by a premature termination codon, resulting in a truncated 197 amino acid protein. Several lines of evidence showed that rs12769205 leads to intron 2 retention (exon 2B): (1) all livers with rs12769205 generated CYP2C19 exon 2B; (2) in silico splice-site strength analysis predicted rs12769205 perturbed the fidelity of intron 2 branch point splice-site recognition; and (3) minigenes with rs12769205 transfected into HepG2 cells showed intron 2 inclusion.
Interestingly, rs12769205, which alone defines CYP2C19*35, was found together with rs4244285 on the CYP2C19*2 allele. CYP2C19*2 was discovered in 1994 (de Morais et al., 1994b) and rs4244285, which clearly leads to altered splicing of exon 5, was the single variant thought to contribute to the CYP2C19*2 PM phenotype. Clearly, both rs12769205 and rs4244285 are functionally important because they can individually alter the CYP2C19 reading frame and produce a premature stop codon, resulting in a truncated nonfunctional protein. This leads to the obvious question of whether rs12769205 and rs4244285 contribute equally to the PM phenotype in livers with CYP2C19*2. Because we did not have any individuals homozygous for CYP2C19*35, we cannot at this time determine, using RNA, whether the residual pool of wild-type CYP2C19 transcript differed between those homozygous for the CYP2C19*2 versus CYP2C19*35 alleles. To address this question we took two approaches. First, we used peptide probes to quantify the amount of remaining CYP2C19 wild-type protein in persons carrying CYP2C19*2 and *35 alleles. While the exon 2 probe (upstream of both rs12769205 and rs4244285) detected residual CYP2C19 protein in livers homozygous for CYP2C19*2 or with CYP2C19*35 the exon 4 probe failed to detect CYP2C19 protein in the same livers. Because the exon 4 probe is downstream from intron 2 rs12769205 but upstream of exon 5 rs4244285, this result suggests the intron 2 SNP can lead to complete loss of CYP2C19 protein due to its primacy in the RNA splicing event. Second, we used extended haplotype homozygosity statistics and uncovered significant evidence that the haplotypes with rs12769205 alone (CYP2C19*35) and with rs4244285 on CYP2C19*2 have undergone positive selection, and that natural selection was not limited to YRI but was seen across African populations, having probably arisen in earlier human ancestors from which all other groups of Africans descended. Notably, the magnitude of evolutionary pressure for both haplotypes with rs12769205 was as great as that exerted on the human genome by infectious diseases (Janha et al., 2014), and implies that rs12769205 on both haplotypes confers an evolutionary advantage. Indeed, the signature of positive selection on the haplotype with rs12769205 alone (CYP2C19*35) reinforces that this polymorphism has a significant functional effect independent of rs4244285.
Ancestry analysis showed that, on an evolutionary timescale, rs12769205 is the original CYP2C19 deleterious polymorphism that arose first on CYP2C19*35, and then later added rs4244285 to create the new haplotype (CYP2C19*2). While it is possible that the gain of rs4244285 added to CYP2C19*2 nonfunction, evidence for natural selection on the haplotype with rs12769205 alone suggests it is sufficient to create the no function allele.
In vivo analysis was unable to conclusively demonstrate that rs12769205 alone contributes to the mephenytoin PM phenotype in Ethiopians because only persons heterozygous for CYP2C19*35 were available; there were no persons with CYP2C19*35 paired with another PM allele and there was only a modest effect of the heterozygous genotype (intermediate phenotype) on mephenytoin disposition. Poor metabolism of mephenytoin was seen in persons homozygous for both rs12769205 + rs4244285 (CYP2C19*2/*2). However, there was only a moderate effect of any of the heterozygous PM genotypes (CYP2C19*1/*2, CYP2C19*1/*35, or CYP2C19*1/*3) on mephenytoin disposition. Indeed, persons with CYP2C19 intermediate phenotypes (e.g., CYP2C19*1/*2) are the most challenging populations to address for proposing Clinical Pharmacogenetic Implementation Consortium guidelines (drug and dose recommendations) because of the wide interindividual variability in residual CYP2C19*1 activity (Scott et al., 2013). Consequently, the most informative subjects, individuals homozygous for CYP2C19*35, or heterozygous for CYP2C19*35 and another CYP2C19 PM allele, need to be CYP2C19 phenotyped before comparisons can be made with CYP2C19*2/*2 PMs and before any CYP2C19*35 genotype-directed recommendations can be made.
The discovery that rs12769205 leads to alternative CYP2C19 splicing adds to the growing list of hepatic CYPs in which we have identified SNPs leading to polymorphic splicing (CYP3A5*3, CYP3A5*6, and CYP2B6*6) (Kuehl et al., 2001; Lamba et al., 2003) and nonfunctional alleles. Sakabe and de Souza (2007) proposed that intron retention happens when introns and flanking exons are small. CYP2C19 exons 2 and 3 (163 and 150 nt, respectively), are not large, and while the average CYP2C19 intron size is 11,092 nt (range 169–38,498 nt), intron 2 is the smallest (169 nt) and is 6.8× smaller than the next-smallest intron (intron 4, 1161 nt). CYP2C19 rs12769205 is an interesting example of a branch point SNP leading to intron retention. There are numerous hereditary disease alleles where polymorphisms in branch point motifs lead to loss of splicing activity (Taggart et al., 2012). The branch point signal, located upstream of the polypyrimidine tract, is one of three obligatory signals required for appropriate pre-mRNA splicing. Approximately 96% of branch points fall between −15 and −55 nt relative to the 3′-splice site (with the peak at position −24 nt), and CYP2C19 intron 2 rs12769205 branch point A is located at −23 bp relative to the 3′-splice site. This SNP would disrupt the invariant branch point adenine, a nucleotide that is absolutely required to engage in a 2′-5′ phosphodiester bond with the 5′ end of the intron after the first catalytic step of the splicing reaction (Corvelo et al., 2010).
Since we have a large liver resource, it would be extremely useful to have a high-throughput RNA sequencing and analysis pipeline that could identify novel alternative splicing of ADME genes, such as CYP2C19, that might be caused by sequence variation. This resource is the ideal tissue to look for the consequence of any sequence variant that could lead to alternative splicing because (1) many ADME genes are highly expressed in liver; (2) liver is one of the tissues that generates the highest number of alternative mRNAs per genes; and (3) alternative splicing can show tissue specificity. Hence, any polymorphism with the potential to cause alternative splicing has the best chance of being detected in liver tissue. In theory, RNA-Seq analysis can discover new mRNA transcripts and it would be incredibly valuable if the analysis tools could unambiguously call novel transcripts, such as the inclusion of exon 2B, and report those livers that had these novel transcripts. The assembly of CYP2C19 mRNAs from short RNA-Seq reads is complicated by the high percent of similarity with other neighbor CYP2C family members. For example, exon 3 in CYP2C19 shows 99%, 95%, and 97% identity with 2C9, 2C8, and 2C18, respectively; and intron 2 in CYP2C19 shows 96% identity with CYP2C9 intron 2. Regardless, visual inspection of the IGV views of CYP2C19-assembled exons 2 and 3 revealed that there might be inserted nucleotides between these exons (Fig. 6). However, the IGV views still required visual analysis of the results, and we already had PCR results to guide this analysis. The exon junction program analysis (data not shown) correctly called the novel CYP2C19*2 exon 4/5 junction; however, that was because the CYP2C19*2 alternative mRNA was already in the reference database. The exon junction program did not observe a novel junction at exon 2 in rs12769205 livers; this would require that the transcript with exon 2B first be added to the reference database because the current programs do not have a reliable intron retention caller (D. Finkelstein, personal communication). Hence, because RNA-Seq requires a reference mRNA, and because small RNAs and long, noncoding RNAs can also inhabit introns, identification of polymorphic alternative mRNAs, particularly those with intron retention, may not yet be unequivocally identified by these high-throughput approaches. An additional complicating factor is that, while CYP2C19 is a highly expressed gene in human liver, the alternative CYP2C19 transcripts generated by rs12769205 ± rs4244285 ultimately lead to premature termination codons and these will trigger accelerated alternative mRNA degradation through nonsense-mediated decay, decreasing the amount of alternative mRNA. Indeed, it has been suggested that the rate of intron retention in mRNA transcripts is higher than reported because nonsense-mediated decay filters off some mRNA transcripts (Aten et al., 2013). Importantly, it also makes the identification of alternative mRNAs linked to premature termination codons and nonsense-mediated decay (those that are linked to functional consequences) harder to identify by an RNA-Seq approach.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical support of St. Jude Children’s Research Hospital (the Hartwell Center for DNA Sequencing and the Computational Biology and Bioinformatics Core); the Pharmacogenetics Research Network (http://www.nigms.nih.gov/Research/SpecificAreas/PGRN/Pages/default.aspx) [a network-wide sequencing project (Kathy Giacomini)]; and the Baylor College of Medicine Human Genome Sequencing Center, Houston, TX (Steve Scherer), for the RNA-seq data.
Abbreviations
- bp
base pair
- CYP
cytochrome P450
- FP
forward primer
- LD
linkage disequilibrium
- IGV
integrative genomic viewer
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- nt
nucleotide
- PM
poor metabolizer
- REHH
relative extended haplotype homozygosity
- RNA-Seq
RNA-sequencing
- RP
reverse primer
- RT-PCR
real-time polymerase chain reaction
- SDM
site-directed mutagenesis
- SNP
single nucleotide polymorphism
- YRI
Yorubans
Authorship Contributions
Participated in research design: Schuetz, Chaudhry, Thummel.
Conducted experiments: Chaudhry, Prasad, Aklillu, Sim, Shirasaka.
Contributed new reagents or analytic tools: Prasad.
Performed data analysis: Schuetz, Chaudhry, Fohner, Prasad, Finkelstein, Fan, Wu, Wang, Aklillu.
Wrote or contributed to the writing of the manuscript: Schuetz, Chaudhry, Prasad, Finkelstein, Wang, Aklillu, Thummel.
Footnotes
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM092666 (to E.G.S) and Grant GM32165 (to K.E.T)] and the Pharmacogenomics Research Network (PGRN) through the network-wide RNA Sequencing Project [GM61390] and [GM061388] at the Baylor College of Medicine Human Genome Sequencing Center; the National Institutes of Health National Cancer Institute [Cancer Center Support Grant P30 CA21765 (to E.G.S)]; and the American Lebanese Syrian Associated Charities (ALSAC) (E.G.S).
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aklillu E, Herrlin K, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M. (2002) Evidence for environmental influence on CYP2D6-catalysed debrisoquine hydroxylation as demonstrated by phenotyping and genotyping of Ethiopians living in Ethiopia or in Sweden. Pharmacogenetics 12:375–383. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten E, Sun Y, Almomani R, Santen GW, Messemaker T, Maas SM, Breuning MH, den Dunnen JT. (2013) Exome sequencing identifies a branch point variant in Aarskog-Scott syndrome. Hum Mutat 34:430–434. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Chhibber A, French CE, Yee SW, Gamazon ER, Theusch E, Qin X, Webb A, Papp AC, Wang A, Simmons CQ, et al. (2015) Transcriptomic variation of pharmacogenes in multiple human tissues and lymphoblastoid cell lines. Pharmacogenomics J. (in Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvelo A, Hallegger M, Smith CW, Eyras E. (2010) Genome-wide association between branch point properties and alternative splicing. PLOS Comput Biol 6:e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. (1994a) Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol 46:594–598. [PubMed] [Google Scholar]
- de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. (1994b) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269:15419–15422. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. (2012) The Pediatric Cancer Genome Project. Nat Genet 44:619–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson KZ, Prasad B, Unadkat JD, Suhara Y, Okano T, Guengerich FP, Rettie AE. (2013) Cytochrome P450-dependent catabolism of vitamin K: ω-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry 52:8276–8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Clark F, Wollerton MC, Grellscheid SN, Groom H, Smith CWJ. (2006) A class of human exons with predicted distant branch points revealed by analysis of AG dinucleotide exclusion zones. Genome Biol 7:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AS, Tabor HK, Johnson AD, Snively BM, Assimes TL, Auer PL, Ioannidis JP, Peters U, Robinson JG, Sucheston LE, et al. NHLBI GO Exome Sequencing Project (2014) Quantifying rare, deleterious variation in 12 human cytochrome P450 drug-metabolism genes in a large-scale exome dataset. Hum Mol Genet 23:1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, Skaar TC, Müller DJ, Gaedigk A, Stingl JC, Clinical Pharmacogenetics Implementation Consortium (2013) Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 93:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Eguchi S, Ieiri I. (2013) Impact of genetic polymorphisms in CYP2C9 and CYP2C19 on the pharmacokinetics of clinically used drugs. Drug Metab Pharmacokinet 28:28–37. [DOI] [PubMed] [Google Scholar]
- Janha RE, Worwui A, Linton KJ, Shaheen SO, Sisay-Joof F, Walton RT. (2014) Inactive alleles of cytochrome P450 2C19 may be positively selected in human evolution. BMC Evol Biol 14:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391. [DOI] [PubMed] [Google Scholar]
- Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, Fackenthal JD, Rogan PK, Ring B, Wrighton SA, et al. (2003) Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307:906–922. [DOI] [PubMed] [Google Scholar]
- Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. (2010) Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol 69:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw J, Waller D. (2012) Cytochrome P450 variations in different ethnic populations. Expert Opin Drug Metab Toxicol 8:371–382. [DOI] [PubMed] [Google Scholar]
- Owusu Obeng A, Egelund EF, Alsultan A, Peloquin CA, Johnson JA. (2014) CYP2C19 polymorphisms and therapeutic drug monitoring of voriconazole: are we ready for clinical implementation of pharmacogenomics? Pharmacotherapy 34:703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson I, Aklillu E, Rodrigues F, Bertilsson L, Ingelman-Sundberg M. (1996) S-mephenytoin hydroxylation phenotype and CYP2C19 genotype among Ethiopians. Pharmacogenetics 6:521–526. [DOI] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837. [DOI] [PubMed] [Google Scholar]
- Sakabe NJ, de Souza SJ. (2007) Sequence features responsible for intron retention in human. BMC Genomics 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz EJ, Villén T, Alm C, Bertilsson L. (1989) S-mephenytoin hydroxylation phenotypes in a Swedish population determined after coadministration with debrisoquin. Clin Pharmacol Ther 45:495–499. [DOI] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR, Clinical Pharmacogenetics Implementation Consortium (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah BS, Parmar SA, Mahajan S, Mehta AA. (2012) An insight into the interaction between clopidogrel and proton pump inhibitors. Curr Drug Metab 13:225–235. [DOI] [PubMed] [Google Scholar]
- Shirasaka Y, Sager JE, Lutz JD, Davis C, Isoherranen N. (2013) Inhibition of CYP2C19 and CYP3A4 by omeprazole metabolites and their contribution to drug-drug interactions. Drug Metab Dispos 41:1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. (2006) A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79:103–113. [DOI] [PubMed] [Google Scholar]
- Singh G, Cooper TA. (2006) Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques 41:177–181. [DOI] [PubMed] [Google Scholar]
- Taggart AJ, DeSimone AM, Shih JS, Filloux ME, Fairbrother WG. (2012) Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat Struct Mol Biol 19:719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybring G, Bertilsson L. (1992) A methodological investigation on the estimation of the S-mephenytoin hydroxylation phenotype using the urinary S/R ratio. Pharmacogenetics 2:241–243. [DOI] [PubMed] [Google Scholar]
- Wang L, Prasad B, Salphati L, Chu X, Gupta A, Hop CE, Evers R, Unadkat JD. (2015) Interspecies variability in expression of hepatobiliary transporters across human, dog, monkey, and rat as determined by quantitative proteomics. Drug Metab Dispos 43:367–374. [DOI] [PubMed] [Google Scholar]
- Wedlund PJ. (2000) The CYP2C19 enzyme polymorphism. Pharmacology 61:174–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.