Abstract
Studies of copy number variants (CNVs) in miscarriages are rare in comparison to post-natal cases with developmental abnormalities. The overall characteristics of miscarriage CNVs (size, gene content and function) are therefore largely unexplored. Our goal was to assess and compare the characteristics of CNVs identified in 101 euploid miscarriages from four high-resolution array studies that documented both common miscarriage CNVs (i.e. CNVs found in controls from the Database of Genomic Variants, DGV) and rare miscarriage CNVs (not reported in DGV). Our miscarriage analysis included 24 rare CNVs with 93 genes, and 372 common CNVs (merged into 119 common CNV regions; CNVRs) with 354 genes. The rare and common CNVs were comparable in size (median size of ∼0.16 and 0.14 Mb, respectively); however, rare CNVs showed a significantly higher gene density, with 56 genes/Mb in rare and 24 genes/Mb in common CNVs (P = 0.03). Rare CNVs also had two times more genes with mouse knock-out models which were reported for 42% of rare and 19% of common CNV genes. No specific pathway enrichment was noted for 24 rare CNV genes, but common CNV genes showed significant enrichment in genes from immune-response related pathways and pregnancy/reproduction-related biological processes. Our analysis of CNVs from euploid miscarriages suggests that both rare and common CNVs could have a role in miscarriage by impacting pregnancy-related genes or pathways. Cataloguing of all CNVs and detailed description of their characteristics (e.g. gene content, genomic breakpoints) is desirable in the future for better understanding of their relevance to pregnancy loss.
Keywords: miscarriage, CNV, bioinformatics, genomics, microarray
Introduction
Epidemiological evidence suggests that genetic factors play a significant role in pathogenesis of miscarriage (Yamada et al., 2005), and that both the fetal/placental and the parental genotypes are involved (Christiansen et al., 2008). Chromosome errors, such as trisomy, monosomy and polyploidy, are usually detected by conventional cytogenetic analysis, and account for 50–70% of miscarriages of <10 weeks of gestation (Warburton and Fraser, 1964). Traditional chromosome analysis is hampered by low chromosome resolution, long-term culture, tissue culture failure or artifacts, and no diagnosis in ∼30–40% of miscarriages that have a normal chromosome finding (euploid).
The advent of chromosome microarray analysis allows an unbiased search for new genetic changes, gains and losses of minute, submicroscopic amounts of DNA (also called copy number variants or CNVs), across the whole genome (Cheung et al., 2005; Rajcan-Separovic, 2012). This technology complements cytogenetic analysis when no chromosome errors are found using conventional chromosome analysis. In many clinical genetic centres worldwide, genomic arrays replaced conventional cytogenetic analysis and became a standard of practice for patients with neurodevelopmental disorders (e.g. intellectual disability (ID) or autism) because such technology can identify causative, pathogenic CNVs in 15% of ID patients with euploid karyotypes. However, owing to limited data, chromosome microarrays are not yet recommended in clinical practice to determine the cause of first or second trimester miscarriage (American College of Obstetrics and Gynecology, ACOG, 2015) or recurrent pregnancy loss.
Knowing the CNV size, their gene content and function, association with established developmental phenotypes, presence in parents or databases of controls (e.g. Database of Genomic Variants, DGV) is essential for their interpretation (Kearney et al., 2011). Generally, CNVs that are larger (>1 Mb), de novo (occur in the affected subject as a new event), with clinically relevant gene content are considered pathogenic, while those that are common in healthy controls (e.g. catalogued in the DGV) are considered benign. Putatively pathogenic CNVs have unclear clinical significance and include those that are rare and can predispose to disease. This third group includes CNVs that are inherited from a normal parent, and could affect regions that are preferentially expressed from one parent (i.e. imprinted).
To our knowledge, there have been 16 published studies which used microarray technology in a total of ∼3000 (<20 weeks of gestation) miscarriages worldwide (Schaeffer et al., 2004; Benkhalifa et al., 2005; Ballif et al., 2006; Shimokawa et al., 2006; Menten et al., 2009; Robberecht et al., 2009, 2012; Warren et al., 2009; Zhang et al., 2009; Rajcan-Separovic et al., 2010a, b; Gao et al., 2012; Lathi et al., 2012; Viaggi et al., 2013; Bug et al., 2014; Levy et al., 2014). These studies demonstrated the benefit of chromosome microarray analysis (CMA) for detecting large chromosomal changes; however the significance of detected submicroscopic gains and losses remained largely unexplored. Early low-resolution CMA studies reported CNVs in 1–13% of early miscarriages (Schaeffer et al., 2004; Shimokawa et al., 2006; Menten et al., 2009; Robberecht et al., 2009) and in 6% of miscarriages that failed to grow in culture (Benkhalifa et al., 2005; Zhang et al., 2009); however, these studies did not determine the origin of the reported CNVs by parental analysis and did not identify the precise size/genomic breakpoints of the CNVs, nor their gene content.
More recent high-resolution CMA studies provided more details on miscarriage CNVs, which is particularly important for chromosomally normal miscarriages, in search for their cause (Rajcan-Separovic et al., 2010a, b; Robberecht et al., 2012; Viaggi et al., 2013; Bug et al., 2014; Levy et al., 2014). So far, clinically relevant CNVs (described as corresponding to post-natal microdeletion and microduplication syndromes, clinically significant phenotypes with reduced penetrance or greater than 5 Mb) were found in 1.6% euploid miscarriages, while rare CNVs of unknown clinical significance (VUS), were noted in ∼1–40% of sporadic and recurrent euploid miscarriages (Rajcan-Separovic et al., 2010a, b; Robberecht et al., 2012; Viaggi et al., 2013; Bug et al., 2014; Levy et al., 2014). The majority of studies, including our two previous publications (Rajcan-Separovic et al., 2010a, b) focused on rare miscarriage CNVs, not present in control databases, while common CNVs, reported in controls, were not listed. Considering the recently reported enrichment of a common CNV containing genes involved in placenta function in females with recurrent miscarriage (RM) (Nagirnaja et al., 2014), we were interested to determine and compare the genomic characteristics and potential contribution to miscarriage of both common and rare CNVs. To achieve this, we have analysed the size, gene content, in silico gene function from these two groups of CNVs from 101 euploid miscarriages from four recent high-resolution CMA studies that catalogued all CNVs, including our two previous studies (Rajcan-Separovic et al., 2010a, b; Robberecht et al., 2012; Viaggi et al., 2013) in order to understand their overall role in miscarriage.
Materials and Methods
Cohorts
Rajcan-Separovic et al. (2010a)—Twenty-three couples were recruited based on the following criteria: (1) a history of idiopathic RPL, based on a negative evaluation, as previously described (Stephenson, 1996) and (2) at least one miscarriage with a normal karyotype (46,XY or 46,XX, confirmed with microsatellite analysis). The obstetric history and details on miscarriages can be found in Rajcan-Separovic et al. (2010a).
Rajcan-Separovic et al. (2010b)—Seventeen euploid embryonic miscarriages, defined as a crown-rump length between 4 and 30 mm without cardiac activity on transvaginal ultrasonography, were included in the study. All of these miscarriages had abnormal morphology, based on embryoscopy findings. The obstetric history and details on miscarriages can be found in Rajcan-Separovic et al. (2010b).
Robberecht et al. (2012)—Thirty-two miscarriages with morphological findings had a low resolution array and 11/32 had a normal result. The 11 euploid miscarriages were further analysed using high resolution array. Common CNVs and rare CNVs (found in 8/11 and 6/11 miscarriages respectively) were followed up by qPCR in parents.
Viaggi et al. (2013)—Forty first-trimester miscarriages with a normal karyotype were investigated by array-CGH. No sample from the parents could be retrieved.
CNV data acquisition
Four recent high-resolution CMA studies of miscarriages (Rajcan-Separovic et al., 2010a, b; Robberecht et al., 2012; Viaggi et al., 2013) were used because they provided breakpoints for the CNVs, classified the CNVs as rare or common based on the Database of Genomic Variants (DGV, http://dgv.tcag.ca), and provided origin for some of the CNVs. Only CNVs >∼1 Kb were included.
Two studies were our previous publications (Rajcan-Separovic et al., 2010a, b) from which we used unpublished data on common CNVs in miscarriages. Miscarriage CNVs overlapping with CNVs from at least two control cohorts were considered common; the remaining CNVs were considered rare. All of the included CNVs were reviewed and reclassified, if required, using the latest version of DGV to determine their current gene content and presence in healthy controls catalogued in DGV. Overlapping common CNVs were merged into CNV regions (CNVRs).
CNV gene content
Gene names and chromosomal coordinates for CNVs were mined from the human reference sequence (human genome 19 assembly, hg19, University of California, Santa Cruz (UCSC) database). Gene content of a CNV was defined based on genes located within CNVs.
Functional gene enrichment analyses
Functional enrichment and pathway analysis for common and rare CNV groups was performed using the Database for Annotation, Visualisation and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003). Gene-enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms (biological process, cellular component and molecular function) was carried out for rare and common CNV groups separately using the ‘Functional annotation’ tool from the DAVID (Dennis et al., 2003). An adjusted P-value of <0.05 was considered statistically significant for pathway analysis and GO terms. Data were reported as significantly enriched pathways and GO terms.
Gene characteristics
We assessed the functional relevance of common and rare CNVs by determining (i) the expression pattern of their integral genes using databases TiGER (Tissue-specific Gene Expression and Regulation: http://bioinfo.wilmer.jhu.edu/tiger/) (Liu et al., 2008) and TiSGeD (Tissue-Specific Genes Database: http://bioinf.xmu.edu.cn:8080/databases/TiSGeD/search.php) (Xiao et al., 2010); and (ii) the consequences of murine knock-out studies, when available (e.g. embryonic lethality and/or pregnancy-related abnormalities) using Mouse Genome Informatics (MGI) (http://www.informatics.jax.org).
Statistical analysis
All statistical analyses were performed in R 2.12.0 for Windows (The R project for Statistical Computing: http://cran.r-project.org/bin/windows/base/old/2.12.0/). CNV size was described with medians and interquartile ranges (IQR), and CNV gene density with a mean. CNV size and gene density comparison was performed using Wilcoxon rank-sum test.
Results
A total of 101 euploid miscarriages were included in our analysis collected from four high-resolution array studies of miscarriages which recorded common and rare CNVs (Rajcan-Separovic et al., 2010a, b; Robberecht et al., 2012; Viaggi et al., 2013). This included 50 miscarriages from Rajcan-Separovic et al. (2010a, b), 11 from Robberecht et al. (2012), and 40 from Viaggi et al. (2013). The rare and common CNV/CNVR gene content is shown in Supplementary Tables SI–SIII.
Out of 44 CNVs initially reported as rare, 24 were rare after re-assessment using DGV: 14 were familial, 1 de novo, and 9 with unknown origin (Supplementary Table SI). The de novo CNV was a 1.5 Kb gain disrupting G Protein-Coupled Receptor 180 (GPR180) gene, known to be associated with response to vascular injury, but with no evidence of a role in pregnancy and reproduction (Rajcan-Separovic et al., 2010b). Three CNVs initially reported as rare and de novo were either excluded (one was ∼100 bp in size, detectable by custom array only and found to disrupt the WD Repeat Domain 37 (WDR37) gene, not known to be associated with miscarriage (Rajcan-Separovic et al., 2010b)) or were reclassified as common (Chr11:50414030–51372036 and Chr11:2904010–2906824, hg19) covering no genes and Cyclin-Dependent Kinase Inhibitor 1C (CDKN1C), respectively (reported in Robberecht et al., 2012).
Three hundred and seventy-two individual common CNVs in miscarriages were noted in the four studies. CNV from 15q11.2 (mapping to ∼20–22 Mb) was the most common and was present in ∼31/50 (62%) and 17/40 (42%) of all miscarriages studied by Rajcan-Separovic et al. (2010a, b) and Viaggi et al. (2013), respectively. The common CNVs were merged into 119 non-redundant common CNV regions (CNVRs) (Supplementary Tables SII and SIII). The paternal/maternal origin of these common miscarriage CNVs could not be conclusively determined, such as when the common CNV was present in both partners.
We compared the 24 rare and 372 common miscarriage CNVs in regard to size and gene content. Ninety-three and 354 genes were found in these CNVs, respectively (Table I). Overall, the rare and common CNVs did not differ significantly in their size (median of 0.16 versus 0.14 Mb, respectively) but the mean gene density for rare CNVs was significantly higher than common CNVs (56 versus 24 Genes/Mb) respectively (P = 0.03, Table I).
Table I.
Comparison of the CNV characteristics in 101 miscarriages from 4 studies.
| Rare | Common | |
|---|---|---|
| Number of CNVs/CNVRs | 24/24 | 372/119 |
| Median CNV size (Mb) | 0.16 (IQR 0.08–0.51) | 0.14 (IQR 0.08–0.55) |
| Number of genes | 93 | 354 |
| Gene density (No. of genes/Mb) | 56* | 24 |
| GO term enrichment | NS | Immune-response and reproductive-processes* |
| Pathway enrichment# | NS | Immune-response* |
| Genes with phenotypes in MGI§ | 39/93 (42%) | 67/354 (19%) |
| Genes with embryonic lethality phenotype§ | 11/39 (28%) | 15/67 (22%) |
| Genes with reproduction/pregnancy-related abnormalities§ | 8/39 (21%) | 15/67 (22%) |
| Genes expressed in placenta§§ | 3/93 (3%) | 3/354 (1%) |
NS, not significant; IQR, interquartile range.
*P<0.05.
#KEGG pathway-analysis using DAVID web-tool (http://david.abcc.ncifcrf.gov/).
§MGI web-tool (http://www.informatics.jax.org) was used to identify the number of genes with mouse-knock out phenotypes.
§§TiGER (http://bioinfo.wilmer.jhu.edu/tiger/) and TiSGeD (http://bioinf.xmu.edu.cn:8080/databases/TiSGeD/search.php) web-tools were used to identify the number of genes which have an exclusive or high expression in placenta.
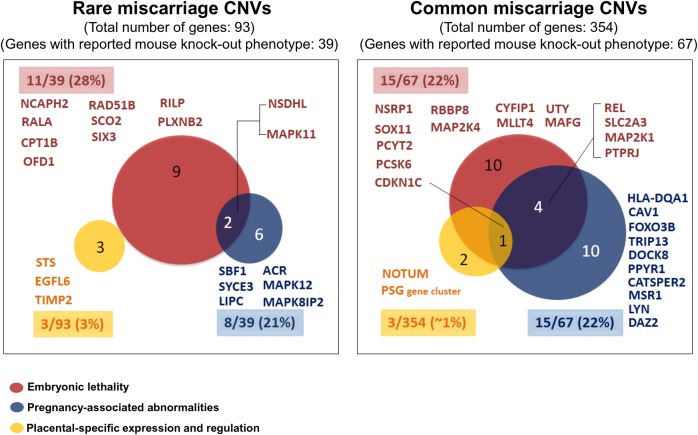
The 93 genes from rare CNV genes were not significantly enriched in any specific pathways, and did not result in any significant GO terms. On the other hand, the 354 common CNV genes were significantly enriched in immune-response related pathways (including graft-vs-host disease, allograft rejection, and antigen processing and presentation) (Tables I and II) and in immune-response and reproductive processes (Table I; Supplementary Table SIV). Forty-two per cent (39/93) of the genes in the rare CNVs and 19% (67/354) of all the genes in the common CNVs had reported mammalian phenotypes in MGI (Table I). The number of genes which resulted in murine embryonic lethality and in reproduction/pregnancy-associated abnormalities using a knock-out model was comparable in rare and common miscarriage CNVs (28 versus 22% and 21 versus 22% of genes with a knock-out model, respectively (Table I; Fig. 1; Supplementary Tables SV and SVI). Similarly, the number of genes with placental-specific or placental expression was not different in rare versus common miscarriage CNVs, 3 versus 1% (Table I; Fig. 1; Supplementary Table SVII). Strong or almost exclusive expression in the placenta was noted for genes EGF-Like–Domain, Multiple 6 (EGFL6), Steroid Sulfatase (Microsomal), Isozyme S (STS), Tissue Inhibitor of Metalloproteinase 2 (TIMP2) from rare CNVs and for Notum Pectinacetylesterase Homolog (Drosophila) (NOTUM), CDKN1C, and the pregnancy-specific glycoprotein (PSG) gene cluster, from common CNVs (Fig. 1; Supplementary Table SVII).
Table II.
Enriched functional pathways for genes from rare/common miscarriage CNVs using the DAVID.
| CNV | Significantly enriched pathways (P < 0.05) | CNV genes involved in pathways | CNV characteristics (type, origin and frequency) | Gene function | Mouse model (Pubmed reference) |
|---|---|---|---|---|---|
| Rare | No significant pathway enrichment | – | – | – | – |
| Common |
|
KLRC1, KLRC2 | 12p13.2 (loss)—unknown origin in miscarriage | Receptors for recognition of MHC class I HLA-E molecules by Natural Killer (NK) cells and several cytotoxic T-cells. | – |
| HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DQA1 | 6p21.32 (loss/gain) *—present in 18/101 (18%) miscarriages (unknown origin in miscarriage) | Key immune response regulators due to their facilitating role in antigen processing and presentation. | HLA-DQA1: Abnormal T-cell morphology and subtype cell number (PMID: 8398989) | ||
| RNASE3 | 14q11.2 (loss)—paternal, transmitted to miscarriage | Involved in immune response by its antibacterial, cytotoxic, and low ribonuclease activity. Allograft-rejection is among the super-pathways related to this gene | – | ||
| CAV1 | 7q31.2 (gain)—paternal transmitted to miscarriage | Facilitates cell cycle progression and stimulates the activation and proliferation of T-cells | Increased susceptibility to bacterial infection induced mortality (PMID: 16982844), reduced fertility (PMID: 18849439), and vasculature abnormalities (PMID: 12167625) | ||
| H2BFM, H2BFWT | Xq22.2 (gain)—unknown origin, present in 4/101 (4%) miscarriages | Members of the H2B histone. H2BFWT is specifically expressed in sperm nuclei and its 5’ UTR polymorphism has been linked to male infertility. | – |
*Variably deleted or duplicated: In some cases only two HLA subtypes were affected while in others all were affected.
Figure 1.
Analysis of the function of CNV genes. Rare and common miscarriage CNV genes associated with embryonic lethality, pregnancy-associated abnormalities, and/or placental-specific expression were identified. This was determined by assessing the 39/93 rare miscarriage CNV genes and 67/354 common miscarriage CNV genes that had reported mammalian phenotypes in mouse knock-out studies, catalogued on MGI, as well as assessing all 93 rare and 354 common genes against human placental-specific genes listed on TiGER, and TiSGeD. For details of the CNVs in which these genes are involved refer to Supplementary Tables SV–SVII. The number and percentage of genes (highlighted) associated with each category is indicated inside and outside of the corresponding colour-coded circles, respectively.
Discussion
Our study represents a unique analysis of rare and common CNVs and their integral genes detected in 101 euploid miscarriages from 4 studies in order to evaluate their overall genomic features and in silico analysis-based function. Collectively, the rare and common CNVs were small (median ∼0.14–0.16 Mb) in these four studies. Although the number of available rare CNVs was small in this analysis, we noted a significantly (P = 0.03) higher mean gene density and two times more rare CNV genes with an abnormal phenotype in mouse knock-out models (42 versus 19%) compared with common CNV genes, which could suggest their importance in pregnancy failure.
The in silico analysis of the miscarriage CNVs showed that rare CNV genes were not enriched in specific pathways associated with pregnancy, although this could be due to the limited number of rare CNVs and their genes. However, common CNV genes were significantly enriched in immunological pathways, such as graft-vs.-host, allograft rejection and antigen processing and presentation, and biological processes related to immune response and reproduction. Significant enrichment of loci related to innate immunity and immunoregulatory pathways essential for immune tolerance at fetomaternal interface was also reported in CNVs from females with RM (Nagirnaja et al., 2014). The most frequent common CNV in the four studies maps to 15q11.2 (∼20–22 Mb) and was reported by Viaggi et al. (2013) to occur in miscarriages in significantly higher proportion than in controls from DGV. Two of the genes from this CNV, POTE Ankyrin Domain Family, Member B (POTEB) and POTEB2 could have a role in reproduction as they belong to a family of genes expressed in reproductive tissues (prostate, testis and placenta) (Bera et al., 2002, 2004). However, comparison to the frequency of the above CNV with the frequency in controls is challenging as the studies in DGV differ in the array platforms used, analysis algorithms and populations. Furthermore, the studies included in DGV change over time as their validity is tested in general use. For example, in the current version of the DGV, the frequency of the 15q11.2 CNV in the control studies varies from <1% (Cooper et al., 2011) to almost 80% (Pinto et al., 2007). Control fertile populations and/or control pregnancies studied using the same methods as the ones used for the subjects would allow for a more ideal comparison. The most frequent common CNV from 5p13.3 found in women with RM, as reported by Nagirnaja et al. (2014), was not found in the miscarriages we analysed.
Our re-analysis of CNVs from euploid miscarriages from four studies did not identify any single miscarriage CNV that was clearly pathogenic, which is similar to the study by Bug et al. (2014) but in contrast to Levy et al. (2014) which was based on a SNP-array study of 2392 miscarriages, reported that 1.6% of euploid miscarriages have a CNV that could be clinically significant, and ∼3% had variants of unknown clinical relevance. Overall, the frequency of clinically significant miscarriage CNVs in euploid miscarriages appears to be smaller than in cohorts of post-natal cases with developmental delay, where 10–15% of subjects have a clinically relevant CNV (Vulto-van Silfhout et al., 2013). Similarly, de novo rare miscarriage CNVs appear to be infrequent in euploid miscarriages as the vast majority of rare CNVs with a known origin were familial (14/15 in 4 studies we re-analysed and 10/12 CNVs in the study by Levy et al. (2014)). CNVs carried by parents are typically considered benign but could impair normal pregnancy development if they contain genes which are relevant for parental reproduction, are imprinted in pregnancy tissues, show variable expression or carry mutations on the second allele in miscarriage.
The contribution of CNVs to miscarriages seems to be complex, with common, rare and parental CNVs potentially playing a role. Future array studies of additional miscarriages and couples should therefore enlist all CNVs with their characteristics (gene content, size and origin). The interpretation of CNVs detected in parents could be facilitated with a database of CNVs in controls with a known reproductive history, as very little is known about the DGV controls other than they are healthy. One recent example of such effort was provided by Migita et al. (2014) who recorded CNVs in 411 Japanese women presumed fertile based on one or more live born children. Finally, the in silico analysis of the functional impact of the CNV should be complemented by in vivo analysis of the function of genes integral to CNVs (e.g. RNA and protein expression) in miscarriage tissues as recently reported (Wen et al., 2015) but also in reproductive tissues of carrier parents.
In conclusion, this study showed that both rare and common miscarriage CNVs could have a role in miscarriage. Rare CNVs significantly have higher gene density and contain more genes studied in mouse knock-out models when compared with common miscarriage CNVs, despite having a comparable size. But, common miscarriage CNVs were found to be significantly enriched in genes involved in pathways and biological processes relevant to pregnancy. No CNVs of clearly pathogenic role were identified. Future studies of euploid miscarriages and couples should record a complete CNV burden (rare and common CNVs) and characteristics (size, gene content, origin) for a more comprehensive assessment of their role in miscarriage.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors' roles
H.B. performed data acquisition and analysis as well as drafting of manuscript; E.M. performed statistical and bioinformatics analyses; Y.Q. participated in data collection and preparation; M.D.S. recruited subjects and critically revised the manuscript; E.R.-S. designed and supervised the study, assisted with data interpretation, and critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the Canadian Institutes of Health Research (CIHR) (MOP 20R63785; PI: E.R.-S.). E.R.-S. appreciates the Michael Smith Foundation for Health Research Career Scholar award.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
The authors appreciate the collaboration and support of the participating subjects and their families. This study makes use of data generated by four published studies which are included in the list of references.
References
- ACOG. American College of Obstetricians and Gynecologists Practice Bulletin 2015. 150 Early Pregnancy Loss.
- Ballif BC, Kashork CD, Saleki R, Rorem E, Sundin K, Bejjani BA, Shaffer LG. Detecting sex chromosome anomalies and common triploidies in products of conception by array-based comparative genomic hybridization. Prenat Diagn 2006;26:333–339. [DOI] [PubMed] [Google Scholar]
- Benkhalifa M, Kasakyan S, Clement P, Baldi M, Tachdjian G, Demirol A, Gurgan T, Fiorentino F, Mohammed M, Qumsiyeh MB. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat Diagn 2005;25:894–900. [DOI] [PubMed] [Google Scholar]
- Bera TK, Zimonjic DB, Popescu NC, Sathyanarayana BK, Kumar V, Lee B, Pastan I. POTE, a highly homologous gene family located on numerous chromosomes and expressed in prostate, ovary, testis, placenta, and prostate cancer. Proc Natl Acad Sci USA 2002;99:16975–16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera TK, Huynh N, Maeda H, Sathyanarayana BK, Lee B, Pastan I. Five POTE paralogs and their splice variants are expressed in human prostate and encode proteins of different lengths. Gene 2004;337:45–53. [DOI] [PubMed] [Google Scholar]
- Bug S, Solfrank B, Schmitz F, Pricelius J, Stecher M, Craig A, Botcherby M, Nevinny-Stickel-Hinzpeter C. Diagnostic utility of novel combined arrays for genome-wide simultaneous detection of aneuploidy and uniparental isodisomy in losses of pregnancy. Mol Cytogenet 2014;7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med 2005;7:422–432. [DOI] [PubMed] [Google Scholar]
- Christiansen OB, Steffensen R, Nielsen HS, Varming K. Multifactorial etiology of recurrent miscarriage and its scientific and clinical implications. Gynecol Obstet Invest 2008;66:257–267. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011;43:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:P3. [PubMed] [Google Scholar]
- Gao J, Liu C, Yao F, Hao N, Zhou J, Zhou Q, Zhang L, Liu X, Bian X, Liu J. Array-based comparative genomic hybridization is more informative than conventional karyotyping and fluorescence in situ hybridization in the analysis of first-trimester spontaneous abortion. Mol Cytogenet 2012;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011;13:680–685. [DOI] [PubMed] [Google Scholar]
- Lathi RB, Massie JA, Loring M, Demko ZP, Johnson D, Sigurjonsson S, Gemelos G, Rabinowitz M. Informatics enhanced SNP microarray analysis of 30 miscarriage samples compared to routine cytogenetics. PLoS One 2012;7:e31282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B, Sigurjonsson S, Pettersen B, Maisenbacher MK, Hall MP, Demko Z, Lathi RB, Tao R, Aggarwal V, Rabinowitz M. Genomic imbalance in products of conception: single-nucleotide polymorphism chromosomal microarray analysis. Obstet Gynecol 2014;124:202–209. [DOI] [PubMed] [Google Scholar]
- Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 2008;9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B, Swerts K, Delle Chiaie B, Janssens S, Buysse K, Philippé J, Speleman F. Array comparative genomic hybridization and flow cytometry analysis of spontaneous abortions and mors in utero samples. BMC Med Genet 2009;10:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita O, Maehara K, Kamura H, Miyakoshi K, Tanaka M, Morokuma S, Fukushima K, Shimamoto T, Saito S, Sago H et al. Compilation of copy number variants identified in phenotypically normal and parous Japanese women. J Hum Genet 2014;59:326–331. [DOI] [PubMed] [Google Scholar]
- Nagirnaja L, Palta P, Kasak L, Rull K, Christiansen OB, Nielsen HS, Steffensen R, Esko T, Remm M, Laan M. Structural genomic variation as risk factor for idiopathic recurrent miscarriage. Hum Mutat 2014;35:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet 2007;16:R168–R173. [DOI] [PubMed] [Google Scholar]
- Rajcan-Separovic E. Chromosome microarrays in human reproduction. Hum Reprod Update 2012;18:555–567. [DOI] [PubMed] [Google Scholar]
- Rajcan-Separovic E, Diego-Alvarez D, Robinson WP, Tyson C, Qiao Y, Harvard C, Fawcett C, Kalousek D, Philipp T, Somerville MJ et al. Identification of copy number variants in miscarriages from couples with idiopathic recurrent pregnancy loss. Hum Reprod 2010a;25:2913–2922. [DOI] [PubMed] [Google Scholar]
- Rajcan-Separovic E, Qiao Y, Tyson C, Harvard C, Fawcett C, Kalousek D, Stephenson M, Philipp T. Genomic changes detected by array CGH in human embryos with developmental defects. Mol Hum Reprod 2010b;16:125–134. [DOI] [PubMed] [Google Scholar]
- Robberecht C, Schuddinck V, Fryns JP, Vermeesch JR. Diagnosis of miscarriages by molecular karyotyping: benefits and pitfalls. Genet Med 2009;11:646–654. [DOI] [PubMed] [Google Scholar]
- Robberecht C, Pexsters A, Deprest J, Fryns JP, D'Hooghe T, Vermeesch JR. Cytogenetic and morphological analysis of early products of conception following hystero-embryoscopy from couples with recurrent pregnancy loss. Prenat Diagn 2012;32:933–942. [DOI] [PubMed] [Google Scholar]
- Schaeffer AJ, Chung J, Heretis K, Wong A, Ledbetter DH, Lese Martin C. Comparative genomic hybridization–array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet 2004;74:1168–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa O, Harada N, Miyake N, Satoh K, Mizuguchi T, Niikawa N, Matsumoto N. Array comparative genomic hybridization analysis in first-trimester spontaneous abortions with ‘normal’ karyotypes. Am J Med Genet A 2006;140:1931–1935. [DOI] [PubMed] [Google Scholar]
- Stephenson MD. Frequency of factors associated with habitual abortion in 197 couples. Fertil Steril 1996;66:24–29. [PubMed] [Google Scholar]
- Viaggi CD, Cavani S, Malacarne M, Floriddia F, Zerega G, Baldo C, Mogni M, Castagnetta M, Piombo G, Coviello DA et al. First-trimester euploid miscarriages analysed by array-CGH. J Appl Genet 2013;54:353–359. [DOI] [PubMed] [Google Scholar]
- Vulto-van Silfhout AT, Hehir-Kwa JY, van Bon BW, Schuurs-Hoeijmakers JH, Meader S, Hellebrekers CJ, Thoonen IJ, de Brouwer AP, Brunner HG, Webber C et al. Clinical significance of de novo and inherited copy-number variation. Hum Mutat 2013;34:1679–1687. [DOI] [PubMed] [Google Scholar]
- Warburton D, Fraser FC. Spontaneous abortion risks in man: data from reproductive histories collected in a medical genetics unit. Am J Hum Genet 1964;16:1–25. [PMC free article] [PubMed] [Google Scholar]
- Warren JE, Turok DK, Maxwell TM, Brothman AR, Silver RM. Array comparative genomic hybridization for genetic evaluation of fetal loss between 10 and 20 weeks of gestation. Obstet Gynecol 2009;114:1093–1102. [DOI] [PubMed] [Google Scholar]
- Wen J, Hanna CW, Martell S, Leung PC, Lewis SM, Robinson WP, Stephenson MD, Rajcan-Separovic E. Functional consequences of copy number variants in miscarriage. Mol Cytogenet 2015;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SJ, Zhang C, Zou Q, Ji ZL. TiSGeD: a database for tissue-specific genes. Bioinformatics 2010;26:1273–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Sata F, Saijo Y, Kishi R, Minakami H. Genetic factors in fetal growth restriction and miscarriage. Semin Thromb Hemost 2005;31:334–345. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Zhang YP, Gu Y, Guan FJ, Li SL, Xie JS, Shen Y, Wu BL, Ju W, Jenkins EC et al. Genetic analysis of first-trimester miscarriages with a combination of cytogenetic karyotyping, microsatellite genotyping and arrayCGH. Clin Genet 2009;75:133–140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.