Abstract
AgDNV is a powerful gene transduction tool and potential biological control agent for Anopheles mosquitoes. Using a GFP reporter virus system, we investigated AgDNV host range specificity in four arthropod cell lines (derived from An. gambiae, Aedes albopictus and Drosophila melanogaster) and six mosquito species from 3 genera (An. gambiae, An. arabiensis, An. stephensi, Ae. albopictus, Ae. aegypti and Culex tarsalis). In vitro, efficient viral invasion, replication and GFP expression was only observed in MOS55 An. gambiae cells. In vivo, high levels of GFP were observed in An. gambiae mosquitoes. Intermediate levels of GFP were observed in the closely related species An. arabiensis. Low levels of GFP were observed in An. stephensi, Ae. albopictus, Ae. aegypti and Cx. tarsalis. These results suggest that AgDNV is a specific gene transduction tool for members of the An. gambiae species complex, and could be potentially developed into a biocontrol agent with minimal off-target effects.
Densoviruses (DNVs) are non-enveloped single-stranded DNA viruses in the family Parvoviridae. DNVs are broadly distributed in invertebrates and are often pathogenic to their hosts1,2,3,4,5,6. Many DNVs have been isolated from various laboratory and field mosquitoes and cell lines1,7,8,9. The Anopheles gambiae densovirus (AgDNV) is highly infectious and capable of transducing exogenous genes in An. gambiae, the major human malaria vector in Sub-Saharan Africa9,10. Unlike most mosquito densoviruses, AgDNV exhibits negligible pathology in An. gambiae11. These features make AgDNV an attractive candidate for paratransgenesis, an approach that renders insects refractory to pathogens by using transgenic microbes12,13,14. The use of paratransgenesis in the field needs to be considered carefully and unwanted side effects such as off-target infections need to be investigated. Understanding basic aspects of viral ecology such as host range is crucial to evaluate feasibility of viral paratransgenesis in the field.
Densovirus host range has been studied with the Aedes aegypti densovirus (AeDNV) and Aedes albopictus densovirus (AalDNV). AeDNV and AalDNV are infectious to Aedes, Culex and Culiseta mosquitoes, but are not infectious to other insects or vertebrates1,6. Among mosquitoes, Ae. aegypti and Ae. albopictus show relatively high susceptibility to multiple mosquito densoviruses5,15,16,17. Ward et al. used a recombinant AeDNV expressing green fluorescent protein (GFP), and demonstrated that AeDNV can fully disseminate in Ae. aegypti but only infects the anal papillae or bristle cells and does not disseminate in An. gambiae18. Previous work by our lab led to the isolation of AgDNV and the development of GFP-expressing recombinant virus, which is capable of efficiently infecting and disseminating in An. gambiae9,10. In this study, we used this system to investigate the host range of AgDNV in multiple invertebrate cell lines and mosquito species.
Results
In vitro AgDNV host specificity
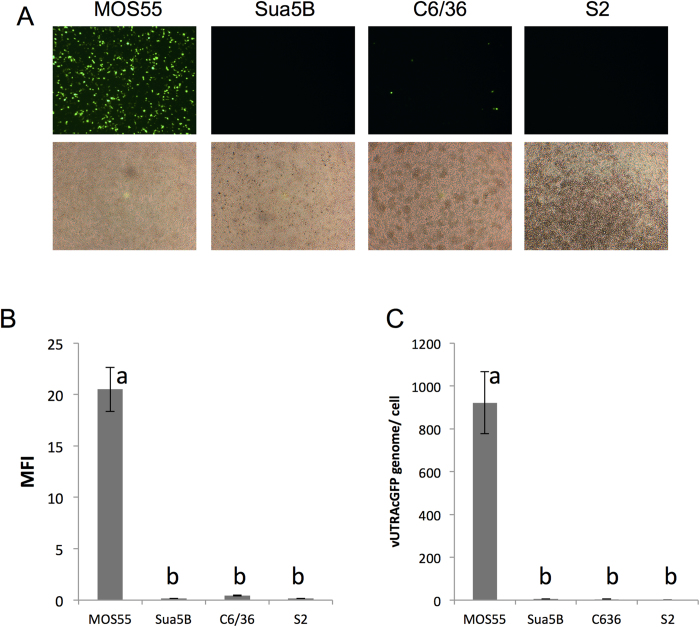
MOS55 (An. gambiae), Sua5B (An. gambiae), C6/36 (Ae. albopictus) and S2 (Drosophila melanogaster) cell lines were infected with 1 × 109 virions of recombinant GFP-expressing AgDNV (vUTRAcGFP)10. Three days post-infection, GFP expression levels were examined using fluorescence microscopy, flow cytometry and quantitative PCR (qPCR). MOS55 cells showed the highest GFP fluorescence (Fig. 1A,B). Low levels of GFP were observed in C6/36 cells and no fluorescence was observed in Sua5B or S2 cells (Fig. 1A,B). Using qPCR, levels of viral DNA copies of vUTRAcGFP matched results obtained by flow cytometry (Fig. 1C).
Figure 1. Infection of insect cell lines with vUTRAcGFP.
GFP expression was (A) visualized by fluorescent microscopy and (B) quantified by flow cytometry analysis in MOS55, Sua5B, C6/36 and S2 cells. MFI = mean fluorescence intensity. (C) vUTRAcGFP viral DNA copy number was quantified by qPCR analysis. Graphs show data mean and standard deviations. Data were analyzed by Analysis of Variance (ANOVA) with Bonferroni’s correction for multiple comparisons. Letters represent statistical significance (P < 0.05).
In vivo AgDNV host specificity
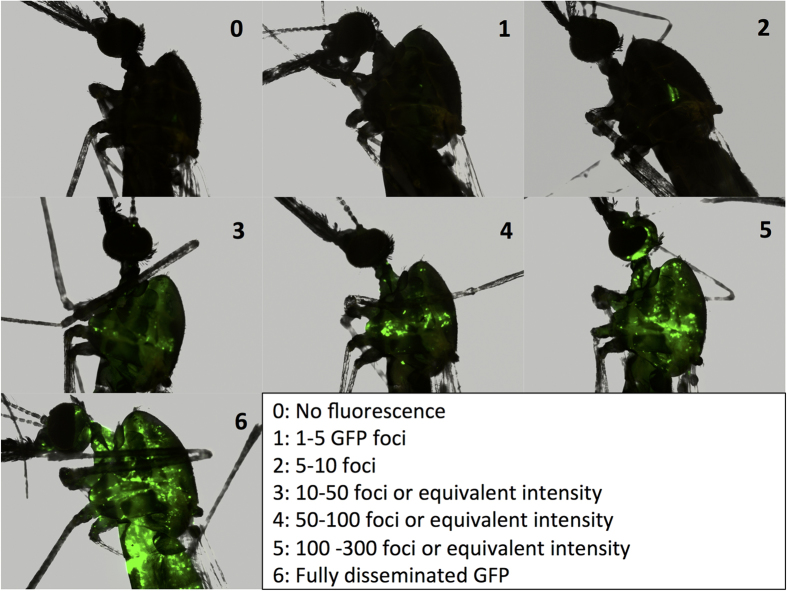
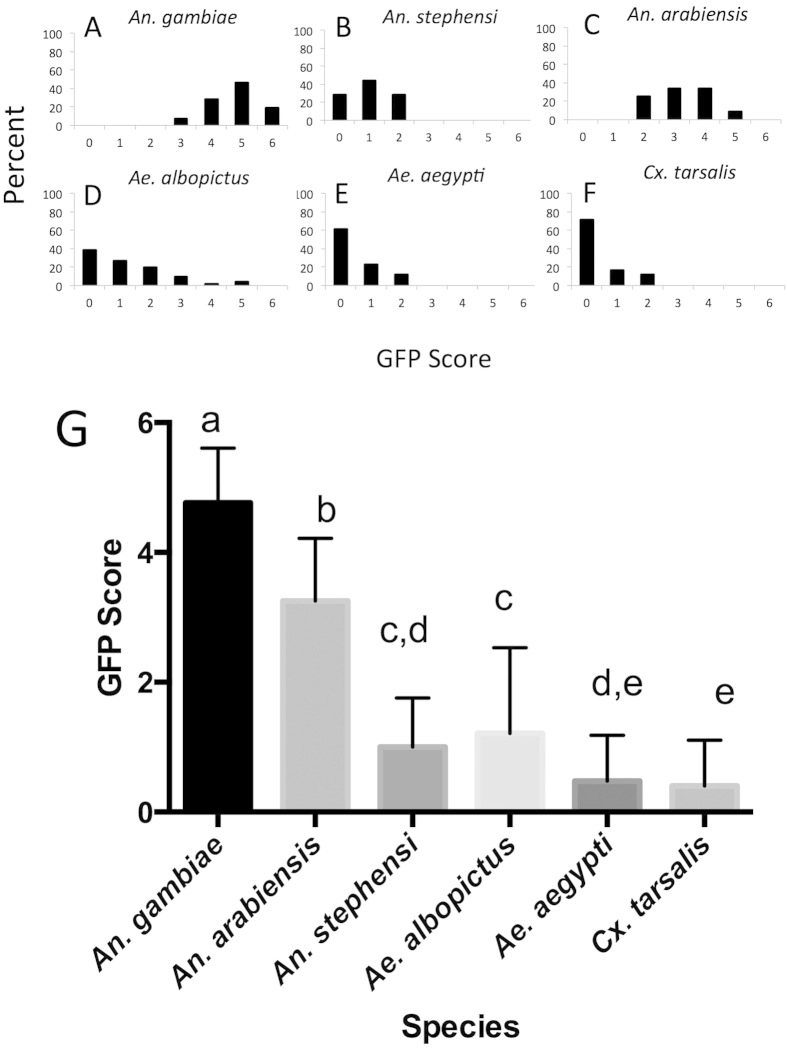
We next investigated viral host range among mosquito species in vivo using An. gambiae, An. stephensi, An. arabiensis, Ae. aegypti, Ae. albopictus and Culex tarsalis. 40–50 adult mosquitoes of each species were injected with 1 × 107 of vUTRAcGFP. At 7 days post injection, mosquitoes were visually examined for GFP expression using fluorescence microscopy. We defined a seven category scoring criteria (0–6) for the level of fluorescence expression in individual mosquitoes (Fig. 2). This scoring system allowed us to compare the viral infection levels semi-quantitatively and analyze the distribution of GFP expression level within each species. The known permissive mosquito, An. gambiae, exhibited scores ranging from 3 to 6 with an average score of 4.8 (Fig. 3A,G). The closely related species An. arabiensis exhibited scores ranging from 2 to 5 with an average score of 3.3 (Fig. 3C,G). In other mosquito species, the distributions were shifted and had statistically significantly lower ranges (Fig. 3). An. stephensi, the closest relative of An. gambiae and An. arabiensis examined in this study, had an average score of only 1.0 (Fig. 3B,G). Ae. aegypti, Ae. albopictus and Cx. tarsalis had average scores of 0.47, 1.2 and 0.40 respectively (Fig. 3D–G).
Figure 2. Representative images of vUTRAcGFP-infected mosquitoes for scoring GFP expression.
Fluorescence levels were categorized into 7 categories (0–6) based on the indicated criteria. An. stephensi (scores, 0–2) and An. gambiae (scores, 3–6) are shown as representative examples.
Figure 3. Comparison of GFP infection scores in six mosquito species.
(A) An. gambiae, (B) An. stephensi, (C) An. arabiensis, (D) Ae. albopictus, (E) Ae. aegypti and (F) Cx. tarsalis. (G) Mean infection score for each mosquito species. Data were analyzed by ANOVA with Bonferroni’s correction for multiple comparisons. Letters represent statistical significance (P < 0.05).
Discussion
Although AgDNV was originally isolated from An. gambiae Sua5B cells9, no fluorescence or viral DNA was detected after vUTRAcGFP infection of this cell line. However, Sua5B cells are permissive to viral replication of the naturally occurring virus present in the cell line or if transfected with a recombinant infectious clone plasmid9. These results suggest that Sua5B cells may have been originally infected with AgDNV from the original mosquito colony from which the cell line was established, however, during development of the cell line and/or over long-term serial passage the cells lost essential host factors (such as receptors) required for new infection. In contrast, the An. gambiae MOS55 cell line retains these factors and is permissive to infection. Comparison of these two cell lines may help identify the specific receptors required for AgDNV entry into cells.
The An. gambiae species complex consists of at least seven morphologically identical mosquito species, to which both An. gambiae and An. arabiensis belong19. We had initially hypothesized that AgDNV would in general infect Anopheline mosquitoes better than species belonging to other genera. However, this was not the case. While An. gambiae, and to a lesser extent An. arabiensis were susceptible to infection, the congeneric species An. stephensi was refractory to infection (Fig. 3B,G). An. stephensi is the major Asian vector of human malaria and is not part of the gambiae complex19. These observations suggest that AgDNV is specifically adapted to infect An. gambiae and closely related species.
We unexpectedly observed intermediate to high levels of GFP expression (scores of 3 to 5) in a low percentage of Ae. albopictus individuals (Fig. 3D), leading to a significantly higher mean infection score for Ae. albopictus compared to Ae. aegypti or Cx. tarsalis (P < 0.05) (Fig. 3D–G). These results complement our cell line data, where C6/36 cells (derived from Ae. albopictus) were also minimally permissive to viral infection. To date, there are no reports detailing the molecular mechanisms underlying host specificity of mosquito densoviruses. Clathrin-mediated endocytosis has been shown to be important for infection for mammalian and insect parvoviruses such as canine parvovirus (CPV) and Junonia coenia densovirus (JcDNV)20,21,22. The clathrin-mediated endocytosis pathway is likely used by mosquito densoviruses as well. Structural variation of the receptors and downstream molecules could determine the host specificity of AgDNV among mosquito species and remains to be investigated.
Unlike other densoviruses that are highly pathogenic to their hosts, AgDNV has a negligible impact on the life span of An. gambiae1,2,3,4,5,6,11. The highly infectious but non-pathogenic and specific nature of the interaction between An. gambiae and AgDNV suggests a history of co-evolution and host-specific adaptation in this system that is distinct from other studied mosquito densoviruses. Further experiments to elucidate the molecular mechanisms of this observed specificity will provide mechanistic insights into the evolution of host-specific pathogens, and will inform on the utility of using DNVs for targeted biocontrol of vector mosquitoes in the field.
Methods
Transducing virus production
Virions of recombinant GFP-expressing AgDNV (vUTRAcGFP) were produced by co-transfection of MOS55 cells with the recombinant virus plasmid pUTRAcGFP and the wild type AgDNV helper plasmid pBAgα as described9,10. Viral titer for these infection experiments was determined with qPCR using a standard curve as previously described10. Briefly, DNV samples were TURBO DNase (Ambion) treated to digest plasmid DNAs. Total DNA was extracted using DNEasy kits (Qiagen). qPCR was performed using the Quantitect SYBR Green Kit (Qiagen) on a Rotor-Gene Q (Qiagen) with EGFP primers: 5′ TCA-AGA-TCC-GCC-ACA-ACA-TC 3′, 5′ TTC-TCG-TTG-GGG-TCT-TTG-CT 3′. A standard curve was created using a dilution series of pUTRAcGFP ranging from 103 to 108 copies.
In vitro AgDNV infection quantitation
MOS55 (An. gambiae), Sua5B (An. gambiae), C6/36 (Ae. albopictus) and S2 (Drosophila melanogaster) cell lines were cultured in Schneider’s media with 10% fetal bovine serum. Cells were infected with 1 × 109 virions of recombinant GFP-expressing AgDNV (vUTRAcGFP)10. Viral DNA level in infected cells was determined by qPCR as described above. GFP mean fluorescence intensity (MFI) was determined using flow cytometry with FlowJo software. Statistical differences between treatments were determined using analysis of variance (ANOVA) with Bonferroni’s correction for multiple comparisons.
In vivo AgDNV infection quantitation
Mosquitoes of each species (An. gambiae [Keele strain], An. stephensi [Liston strain], An. arabiensis [Dongola strain], Ae. aegypti [Rock strain], Ae. albopictus [Houston strain] and Culex tarsalis [Yolo strain]) were held at 27 °C and 80% relative humidity and were maintained on expired human blood or commercially obtained bovine blood using a membrane feeding system, and were allowed access to 10% sucrose solution ad libitum through a cotton wick. For each species, 3–5 day old females were anesthetized by chilling and injected with 1 × 107 vUTRAcGFP using a glass capillary needle. Injected mosquitoes were held at 27 °C and 80% relative humidity with 10% sucrose. A 7 category scoring scale (Fig. 2) was used to visually quantify GFP expression using an Olympus BX40 epifluorescent microscope at 7 days post-injection. Statistical differences between treatments were determined using analysis of variance (ANOVA) with Bonferroni’s correction for multiple comparisons. An. gambiae injected with media were used as a negative control.
Additional Information
How to cite this article: Suzuki, Y. et al. In vitro and in vivo host range of Anopheles gambiae densovirus (AgDNV). Sci. Rep. 5, 12701; doi: 10.1038/srep12701 (2015).
Acknowledgments
We thank Mrs. Rhiannon Barry for insectary support. We appreciate the assistance of the Microscopy and Flow cytometry core facilities at the Huck Institutes of the Life Sciences at the Pennsylvania State University for assistance with FACS analysis. The following reagent was obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH: Anopheles arabiensis Dongola, MRA-856, deposited by M.Q. Benedict. This study was supported by NIH/NIAID grants R21AI111175, R21AI088311, R56AI116636 and R01AI067371 to JLR. T.K.B was in part supported by a Raman Postdoctoral Fellowship from the Indian Government University Grants Commission.
Footnotes
Author Contributions Y.S. designed and performed the experiments, analyzed the data and contributed to drafting the manuscript. T.K.B. performed the experiments and contributed to drafting the manuscript. R.M.J. provided technical support for experiments and contributed to drafting the manuscript. J.L.R. conceived the project, designed the experiments, assisted in data analysis and contributed to drafting the manuscript.
References
- Carlson J., Suchman E. & Buchatsky, L. Densoviruses for control and genetic manipulation of mosquitoes. Adv. Virus Res. 68, 368–2 (2006). [DOI] [PubMed] [Google Scholar]
- Hewson I. et al. Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. USA. 111, 17278–3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuel D. et al. Pathogenesis of Junonia coenia densovirus in Spodoptera frugiperda: a route of infection that leads to hypoxia. Virology 403, 137–4 (2010). [DOI] [PubMed] [Google Scholar]
- Lightner D. V., Redman R. M. & Bell T. A. Infectious hypodermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. J. Invertebr. Pathol. 42, 62–70 (1983). [DOI] [PubMed] [Google Scholar]
- Ledermann J. P., Suchman E. L., Black W. C. 4th & Carlson J. O. Infection and Pathogenicity of the Mosquito Densoviruses AeDNV, HeDNV, and APeDNV in Aedes aegypti Mosquitoes (Diptera: Culicidae). J. Econ. Entomol. 97, 1828–35 (2004). [DOI] [PubMed] [Google Scholar]
- Cornet M. A Parvo-like virus persistently infecting a C6/36 clone of Aedes albopictus mosquito cell line and pathogenic for Aedes aegypti larvae. Virus Res. 29, 99–114 (1993). [DOI] [PubMed] [Google Scholar]
- Ma M. et al. Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS One 6, e24758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y. G. et al. Isolation and characterization of the full coding sequence of a novel densovirus from the mosquito Culex pipiens pallens. J. Gen. Virol. 89, 195–9 (2008). [DOI] [PubMed] [Google Scholar]
- Ren X., Hoiczyk E. & Rasgon J. L. Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 4, e1000135 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Niu G., Hughes G. L. & Rasgon J. L. A viral over-expression system for the major malaria mosquito Anopheles gambiae. Sci. Rep. 4, 5127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Hughes G. L., Niu G., Suzuki Y. & Rasgon J. L. Anopheles gambiae densovirus (AgDNV) has negligible effects on adult survival and transcriptome of its mosquito host. PeerJ 2, e584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. & Rasgon J. L. Potential for the Anopheles gambiae densonucleosis virus (AgDNV) to act as an “evolution proof” biopesticide. J. Virol. 84, 7726–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W. et al. Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science 331, 1074–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. USA 109, 12734–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. L. et al. Insect densoviruses may be widespread in mosquito cell lines. J. Gen. Virol. 76, 2067–74 (1995). [DOI] [PubMed] [Google Scholar]
- Kittayapong P., Baisley K. & O’Neill S. A mosquito densovirus infecting Aedes aegypti and Aedes albopictus from Thailand. Am. J. Trop. Med. Hyg. 61, 612–7 (1999). [DOI] [PubMed] [Google Scholar]
- Afanasiev B. N., Ward T. W., Beaty B. J. & Carlson J. O. Transduction of Aedes aegypti mosquitoes with vectors derived from Aedes densovirus. Virology 257, 62–72 (1999). [DOI] [PubMed] [Google Scholar]
- Ward T. W. et al. Aedes aegypti transducing densovirus pathogenesis and expression in Aedes aegypti and Anopheles gambiae larvae. Insect Mol. Biol. 10, 397–405 (2001). [DOI] [PubMed] [Google Scholar]
- Neafsey D. E. et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A. et al. Densovirus infectious pathway requires clathrin-mediated endocytosis followed by trafficking to the nucleus. J. Virol. 83, 4678–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C. E., Chiorini J. A. & Parrish C. R. The parvovirus capsid odyssey: from the cell surface to the nucleus. Trends Microbiol. 16, 208–14 (2008). [DOI] [PubMed] [Google Scholar]
- Parker J. S. L. & Parrish C. R. Cellular uptake and infection by Canine Parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74, 1919–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]