Abstract
Transforming growth factor-β (TGF-β) and interleukin-6 (IL-6) are the pivotal cytokines to induce IL-17-producing CD4+ T helper cells (TH17); yet their signalling network remains largely unknown. Here we show that the highly homologous TGF-β receptor-regulated Smads (R-Smads): Smad2 and Smad3 oppositely modify STAT3-induced transcription of IL-17A and retinoic acid receptor-related orphan nuclear receptor, RORγt encoded by Rorc, by acting as a co-activator and co-repressor of STAT3, respectively. Smad2 linker phosphorylated by extracellular signal-regulated kinase (ERK) at the serine 255 residue interacts with STAT3 and p300 to transactivate, whereas carboxy-terminal unphosphorylated Smad3 interacts with STAT3 and protein inhibitor of activated STAT3 (PIAS3) to repress the Rorc and Il17a genes. Our work uncovers carboxy-terminal phosphorylation-independent noncanonical R-Smad–STAT3 signalling network in TH17 differentiation.
 TGF-ß and IL-6 are the essential cytokines for mediating the differentiation of IL-17-producing CD4+ T helper cells (TH17). Here, Yoon et al. provide more insights into this process and describe the opposing roles of TGFß-signalling intermediates Smad2 and Smad3 as STAT3 cofactors in Th17 differentiation.
TGF-ß and IL-6 are the essential cytokines for mediating the differentiation of IL-17-producing CD4+ T helper cells (TH17). Here, Yoon et al. provide more insights into this process and describe the opposing roles of TGFß-signalling intermediates Smad2 and Smad3 as STAT3 cofactors in Th17 differentiation.
Transforming growth factor-β (TGF-β) had been appreciated as the most potent immunosuppressive cytokine, suppressing the differentiation and functions of effector immune cells as inducing suppressor immune cells1,2. However, since identified as the requisite cytokine in combination with interleukin (IL)-6 for the differentiation of IL-17-producing CD4+ T helper cell (TH17) through inducing a master transcription factor, retinoic acid-related orphan receptor-γt (RORγt) and IL-17 (refs 3, 4), context-dependent multidirectional roles of TGF-β have been highlighted in immune regulation, similarly to its roles in carcinogenesis and cancer progression5. TH17 is a crucial effector CD4+ T-cell subset in inflammation, protective mechanisms against infections, tumour immunity and autoimmune responses6,7. Crucial pathogenic role of TH17 in autoimmune diseases such as rheumatoid arthritis (RA) has been well demonstrated by numerous studies including the pioneer work showing the attenuation of collagen-induced arthritis (CIA) in the mice deficient in IL-17A (ref. 8).
Intracellular signal transduction of TGF-β superfamily cytokines is initiated by two types of serine/threonine kinase transmembrane receptors9,10. TGF-β ligands bind to TGF-β type II receptor (TβRII), which transphosphorylates and activates TGF-β type I receptor (TβRI). TGF-β-specific receptor-regulated Smads (R-Smads), Smad2 and Smad3, are composed of N-terminal Mad homology-1 (MH1) domain, linker region and carboxy-terminal (C-terminal) MH2 domain that contains two serine residues phosphorylated by TβRI. Activated TβRI phosphorylates serine residues, SSXS in MH2 domains of R-Smads, which form the heterodimer complex with common-mediator Smad, Smad4, to regulate transcription of the target genes10,11. TβRI phosphorylates not only the C-termini of R-Smads but also activates various protein kinases including mitogen-activated protein kinases (MAPKs): extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK) and p38 MAPK (p38), which then phosphorylate the variable linker regions of R-Smads11,12,13. MAPKs are shared by T-cell receptor (TCR) and various cytokines including IL-6 as the crucial intracellular signalling mediators for effector T-cell differentiation14,15.
IL-6 and other cytokines such as IL-21 and IL-23 that induce and maintain TH17 activate STAT3, a critical transcription factor for TH17 differentiation and the pathogenesis of autoimmune diseases including RA16. In contrast to the established essential roles of STAT3-mediated IL-6 signalling in TH17 differentiation, molecular mechanisms by which R-Smads regulate TH17 differentiation still remain under debate. Despite their high amino-acid sequence homology, Smad2 and Smad3 exert both redundant and distinct functions in TGF-β signalling depending on the context17. Several reports have shown contradictory results regarding their roles in TH17 differentiation: Smad2 as the essential inducer18,19, Smad3 as the negative regulator20,21 or non-Smad signals as the crucial inducers with irrelevance of R-Smads22,23. Thus far, roles of canonical TGF-β signalling through C-terminal phosphorylation of R-Smads for TH17 differentiation have been examined. MAPK signalling pathways, which phosphorylate linker regions of R-Smads, play crucial roles in differentiation and functions of effector T cells1,2,24. However, whether diverse phosphorylation status of R-Smads, such as linker phosphorylation or unphosphorylation, affects TH17 differentiation remains largely undetermined.
In this study, we seek to determine the mechanisms whereby R-Smads regulate TH17 differentiation. We investigate the molecular mechanisms how Smad2 and Smad3 regulate the transcription of the essential genes for TH17 and examine the pathophysiological roles of R-Smads in TH17-related inflammatory disease by applying a CIA model to Smad2-deficient (Smad2−/−), Smad3-deficient (Smad3−/−) and control wild-type mice. We discover the opposing functions of Smad2 and Smad3 as transcription cofactors of STAT3 in TH17 differentiation independently of Smad4: the canonical partner of C-terminally phosphorylated R-Smads. Mechanistic studies show that the phosphorylation status of R-Smads distinctively modulates STAT3-induced transcription of the Rorc and Il17a genes. Linker-phosphorylated Smad2 (pSmad2L) at the residue Ser255 via ERK serves as a STAT3 co-activator in cooperation with p300, whereas C-terminally unphosphorylated Smad3 (unphosphorylated Smad3C) serves as a STAT3 co-repressor in cooperation with protein inhibitor of activated STAT3 (PIAS3), the negative regulator of STAT3 signalling.
Results
Opposing roles of Smad2 and Smad3 in CIA
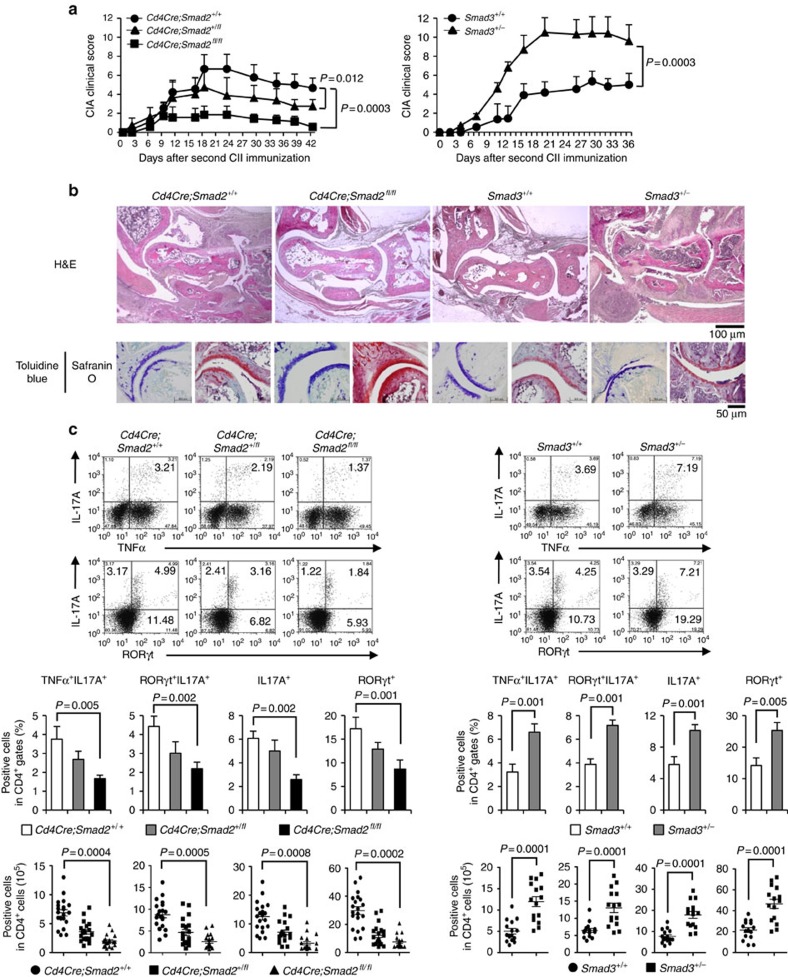
To examine the pathophysiological roles of R-Smads in TH17-mediated inflammatory disease, we applied a CIA model to T-cell-specific (Cd4Cre;Smad2+/+, +/fl, fl/fl), inducible systemic (Mx-1Cre;Smad2+/+, +/fl, fl/fl) Smad2 conditional knockout mice and Smad3 heterozygote (Smad3+/+, +/−) mice. They showed normal immune phenotypes with C57BL/6 background in a specific pathogen-free environment (Supplementary Fig. 1), indicating that R-Smads are dispensable for immune homeostasis. T-cell-specific and systemic deletion of Smad2 ameliorated, whereas Smad3 heterozygosity exacerbated CIA (Fig. 1a and Supplementary Fig. 2a). Because both systemic and T-cell-specific deletion of Smad2 showed the same phenotype (Supplementary Fig. 2a,b), we used Cd4Cre;Smad2+/+,+/fl. fl/fl mice for further study. Cd4Cre;Smad2fl/fl mice showed significant amelioration in joint lesions, whereas Smad3+/− mice showed proliferative detritic synovitis with mononuclear cell infiltration and joint destruction (Fig. 1b, upper). Evaluation of proteoglycan and mucopolysaccharide of cartilage by staining with toluidine blue and Safranin O showed the significant maintenance of cartilages in Cd4Cre;Smad2fl/fl mice and marked destruction of cartilages in Smad3+/− mice (Fig. 1b, lower). Accumulation of CD4+, RORγt+ and IL-17A+ cells in the joint lesions was ameliorated in Cd4Cre;Smad2fl/fl mice, whereas it was exacerbated in Smad3+/− mice (Supplementary Figs 3–5). Consistent with the joint lesions, IL-17A+, RORγt+, IL-17A+TNF-α+ and IL-17A+RORγt+ CD4+ T cells decreased in the draining lymph nodes of the arthritic joints of Cd4Cre;Smad2fl/fl mice and Mx-1Cre;Smad2fl/fl mice, whereas they increased significantly in those of Smad3+/− mice (Fig. 1c and Supplementary Fig. 2a). Smad genotypes did not affect other effector T-cell subsets, such as IL-6+CD4+, TNF-α+CD4+, TH1 (T-bet+IFN-γ+CD4+), natural and inducible Treg cells (CD103−Foxp3+CD4+, CD103+Foxp3+CD4+), naive and memory CD4+ (CD44lowCD62Lhigh, CD44highCD62Llow) and CD8+ (CD44low, CD44high) T cells in the draining lymph nodes of the arthritic joints (Supplementary Fig. 6). Thus, Smad2 and Smad3 have the opposing roles in TH17 differentiation in the pathogenesis of CIA.
Figure 1. Opposing effects of Smad2 and Smad3 on TH17 response in CIA.
Cd4Cre;Smad2+/+, +/fl, fl/fl and Smad3+/+, +/− mice were immunized with type II collagen emulsified in complete Freund adjuvant twice in 3 weeks interval to induce CIA. (a) CIA scoring courses of Cd4Cre;Smad2+/+, Cd4Cre;Smad2+/fl, Cd4Cre;Smad2fl/fl mice (left, n=11/Cd4Cre;Smad2 genotype) and Smad3+/+, Smad3+/− mice (right, n=13/Smad3 genotype) with P values (two-way analysis of variance (ANOVA) test). (b) Pathological analyses of the joint sections (haematoxylin and eosin, H&E, magnification, × 40, scale bar, 100 μm, toluidine blue and safranin O, magnification, × 200, scale bar, 50 μm). (c) Flow cytometry analyses of IL-17A+TNF-α+ CD4+ T cells and RORγt+IL-17A+ CD4+ T cells in the draining lymph nodes of Cd4Cre;Smad2+/+,+/fl, fl/fl (n=20/Cd4Cre;Smad2 genotype) and Smad3+/+,+/− mice (n=15/Smad3 genotype) on day 14 after second immunization. Graphs show the percentages and cell numbers of IL-17A+, RORγt+, IL-17A+TNF-α+ and IL-17A+RORγt+ in CD4+ gates in the draining lymph nodes. Data are from one experiment representative of seven (a,b), four (c, Cd4Cre;Smad2) or three (c, Smad3) independent experiments. Graphs show mean+s.d. with P values (unpaired Student's t-test).
Opposing functions of Smad2 and Smad3 as STAT3 cofactors
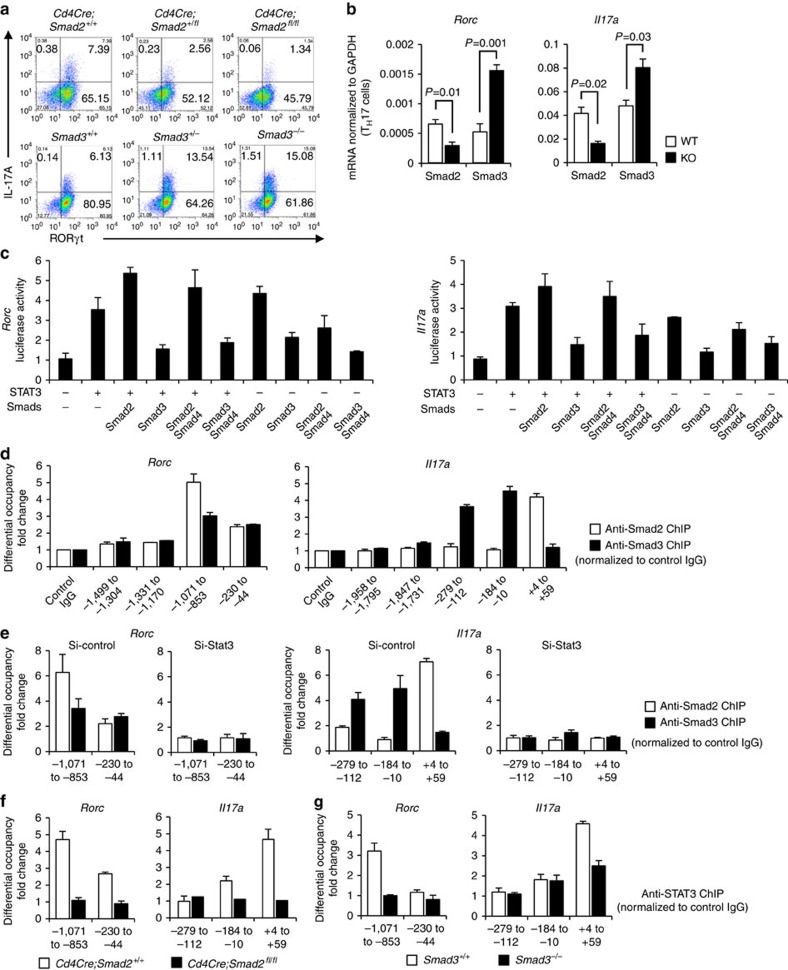
IL-6 is the main arthritogenic cytokine and TGF-β is produced and activated in the inflammatory lesions1,2,25. Because IL-6 and TGF-β are the pivotal cytokines to induce TH17 differentiation, we cultured Smad2−/− or Smad3−/− CD4+ T cells under TH17-polarizing condition with IL-6 and TGF-β (ref. 3) to examine the mechanisms whereby R-Smads regulate TH17 differentiation. Expression levels of protein and mRNA of RORγt and IL-17A decreased in Smad2−/− CD4+ T cells, whereas those increased in Smad3−/− CD4+ T cells (Fig. 2a,b). The mRNA levels of TH17-inducing genes (Batf, Il23r, Il6, Il6ra, Il21 and Il21r) and TH17-suppressing genes (Il2, Il2ra, Tbet and Eomesodermin) were unaffected in both Smad2−/− and Smad3−/− CD4+ T cells (Supplementary Fig. 7), suggesting that R-Smads regulate TH17 differentiation by specifically targeting the Rorc and Il17a genes. Because IL-6 or TGF-β alone has little effect on TH17 differentiation3 and STAT3-mediated IL-6 signalling is crucial for TH17 differentiation16, we examined whether R-Smads regulate STAT3-induced transcription of RORγt and IL-17A in CD4+ T cells cultured under TH17-polarizing condition by promoter assays with the luciferase reporters spanning 2 kb upstream of the first exons of the Rorc and Il17a genes (Fig. 2c). STAT3 or Smad2 alone induced their promoter activities, whereas Smad3 alone had no effect. Smad2 further enhanced, whereas Smad3 suppressed STAT3-induced reporter activation. Co-transfection of Smad4 with R-Smads and STAT3 did not show the additive effects. We next determined the binding of R-Smads to the proximal promoter regions of the Rorc and Il17a genes in TH17 cells by chromatin immunoprecipitation (ChIP) using the primers to detect the DNA-binding sequences of Smads and STAT3 (refs 10, 26, 27). Smad2 and Smad3 were bound to the same sites in the Rorc promoter, whereas they were bound to the distinct sites in the Il17a promoter (Fig. 2d). Active promoters are characterized by histone acetylation and trimethylation of H3K4, whereas repressed inactive chromatin is marked by methylation of H3K27 and H3K9 (ref. 28). Smad2-binding sites in the Il17a promoter showed higher acetylation of histone H3 and trimethylation of histone H3K4, which correlate with transcriptionally active chromatin (Supplementary Fig. 8a). By contrast, Smad3-binding sites in the Il17a promoter showed higher trimethylation of histone H3K27, which correlate with transcriptionally inactive chromatin (Supplementary Fig. 8b). These data suggest that Smad2 and Smad3 have the opposing roles in STAT3-induced transcription of the Rorc and Il17a genes.
Figure 2. Opposing roles of Smad2 and Smad3 in STAT3-induced TH17 differentiation.
Purified CD4+ T cells were activated under TH17-polarizing condition for 3 days. (a) Flow cytometry analyses of IL-17A and RORγt in Smad2+/+,+/−, −/− and Smad3+/+,+/−, −/− CD4+ T cells. (b) Quantitative RT–PCR analysis of the Il17a and Rorc mRNA in Smad2+/+, −/− and Smad3+/+, −/− TH17 cells (n=7). (c) Effects of Smads on STAT3-induced activation of the Rorc promoter and the Il17a promoter constructs transfected in TH17 cells were analysed using luciferase assay. (d) Binding of Smad2 and Smad3 to the proximal promoter regions of the Rorc gene and the Il17a gene in TH17 cells was determined using ChIP. (e) Requirement of STAT3 for the binding of Smad2 and Smad3 to the proximal promoter regions of the Rorc gene and the Il17a gene was determined with ChIP using STAT3 knockdown TH17 cells. Requirement of Smad2 and Smad3 for the binding of STAT3 to the proximal promoter regions of the Rorc gene and the Il17a gene was determined with ChIP using (f) Smad2−/− or (g) Smad3−/− TH17 cells. ChIP data are shown as differential occupancy fold changes. Data are from one experiment representative of seven (a,d), three (c), two (e) or five (f,g) independent experiments or pooled from seven experiments (b). Each experiment (a–g) was performed in triplicate (n=3). Data are mean+s.d. or mean+s.d. with P values (b, unpaired Student's t-test).
We next examined whether STAT3 was necessary for R-Smads to bind to these sites by STAT3 knockdown using short interfering RNA (siRNA) in TH17 cells (Supplementary Fig. 9). STAT3 knockdown completely abolished the binding of R-Smads to these sites (Fig. 2e). We then confirmed whether R-Smads are sufficient for STAT3 to bind to these sites using Smad2−/− and Smad3−/− TH17 cells. STAT3 bound to the Smad2/3-binding sites in the Rorc promoter or the Smad2-binding site in the Il17a promoter (Fig. 2f,g, white bars). Deficiency of Smad2 or Smad3 prevented STAT3 from binding to these sites (Fig. 2f,g, black bars). Thus, R-Smads and STAT3 are mutually required to bind to the proximal promoters of the Rorc and Il17a genes. Taken together, Smad2 functions as a transcription co-activator, whereas Smad3 functions as a transcription co-repressor of STAT3 in TH17 differentiation.
Linker-phosphorylated Smad2 induces TH17 differentiation
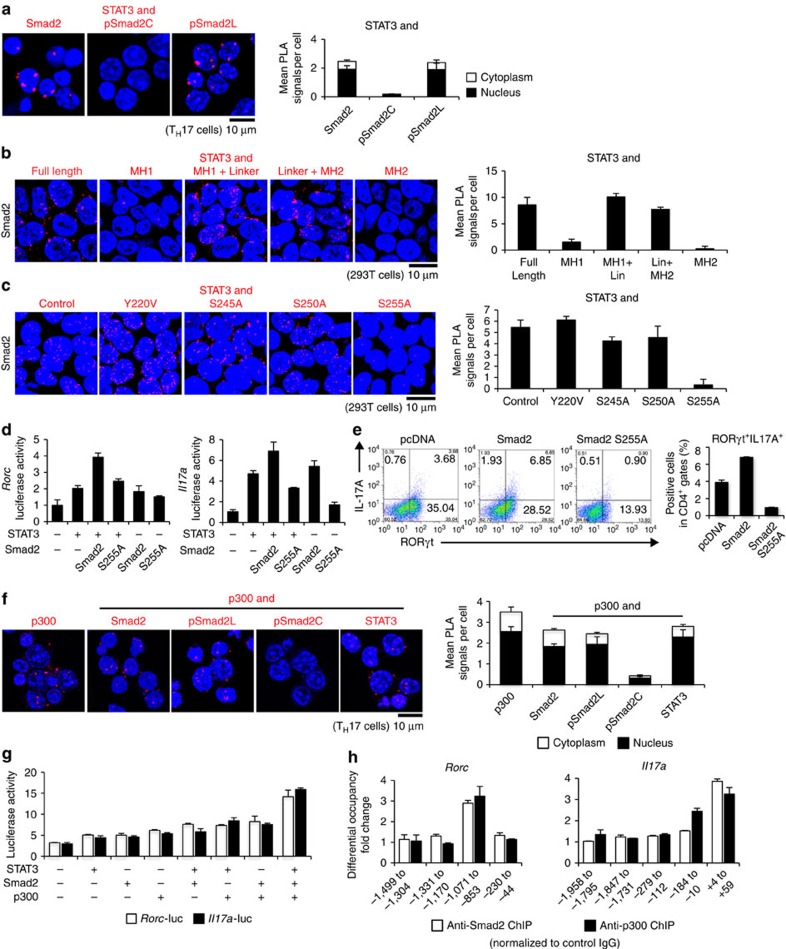
We investigated the mechanism how Smad2 functions as a transcription co-activator of STAT3. Proximity ligation assays (PLA) confirmed the endogenous close proximity between Smad2 and STAT3 in TH17 cells (Fig. 3a, left). We found that pSmad2L had close proximity with STAT3 in TH17 cells (Fig. 3a, right). By contrast, C-terminally phosphorylated Smad2 (pSmad2C) did not show close proximity with STAT3 (Fig. 3a, middle). PLA and immunoprecipitation of 293T cells transfected with the various deletion mutants of Smad2 showed that Smad2 linker deletion mutants (MH1 and MH2)29 failed to bind with STAT3 (Fig. 3b and Supplementary Fig. 10a). Transfection of the linker variants of Smad2 showed that the mutant of the linker serine residue 255 to alanine, Smad2 (S255A)30, failed to bind with STAT3 (Fig. 3c and Supplementary Fig. 10b). Luciferase reporter assays showed that Smad2 (S255A) failed to enhance STAT3-induced activation of the Rorc and Il17 a promoters (Fig. 3d). Overexpression of Smad2 (S255A) in CD4+ T cells cultured under TH17-polarizing condition impaired TH17 differentiation (Fig. 3e). Therefore, pSmad2L (Ser255) is essential for TH17 differentiation.
Figure 3. Linker-phosphorylated Smad2 (S255)–STAT3–p300 complex transactivates the Rorc and Il17a.
Interactions of endogenous proteins in TH17 cells and exogenous proteins in 293T cells were determined by PLA. PLA signals (a–c,f) were quantified using the BlobFinder software (scale bars, 10 μm; nucleus, black; cytoplasm, white, n=10 fields). (a) Endogenous interaction between Smad2 and STAT3 in TH17 cells. (b) Effects of truncated mutations in Smad2 on the interaction with STAT3 in 293T cells. (c) Effects of linker domain variations in Smad2 on the interaction with STAT3 in 293T cells. (d) Effects of Smad2 (S255A) on STAT3-induced activation of the Rorc promoter and the Il17a promoter constructs transfected in TH17 cells were analysed by luciferase assay. (e) Flow cytometry analyses of IL-17A+RORγt+CD4+ T cells transduced with the indicated DNA constructs using Nucleofector (n=2). (f) Endogenous interactions between p300 and Smad2 or STAT3 in TH17 cells were determined using PLA. (g) Effects of p300 on Smad2/STAT3-induced activation of the Rorc promoter (white) and the Il17a promoter (black) constructs transfected in 293T cells were analysed by luciferase assay. (h) Binding of Smad2 (white) and p300 (black) to the proximal promoter regions of the Rorc gene and the Il17a gene in TH17 cells was determined using ChIP. ChIP data are shown as differential occupancy fold changes. Data are from one experiment representative of six (a), three (b–d) or two (e–h) independent experiments. Each experiment (d,g,h) was performed in triplicate (n=3). Data are mean+s.d.
The histone acetyl-transferase p300 is a crucial transcription co-activator of Smads9,31. PLA showed that STAT3 and pSmad2L, but not pSmad2C, had the close proximity with p300 in TH17 cells (Fig. 3f). Luciferase reporter assays confirmed that p300 further enhanced Smad2/STAT3-induced activation of the Rorc and Il17 a promoters in 293T cells (Fig. 3g). Smad2, STAT3 and p300 bound to the same sites in the proximal promoters of the Rorc and Il17a genes in TH17 cells (Fig. 3h). Thus, pSmad2L (Ser255) forms complex with p300 and STAT3 to bind to the proximal promoter of the Rorc and Il17a genes.
Unphosphorylated Smad3 suppresses TH17 differentiation
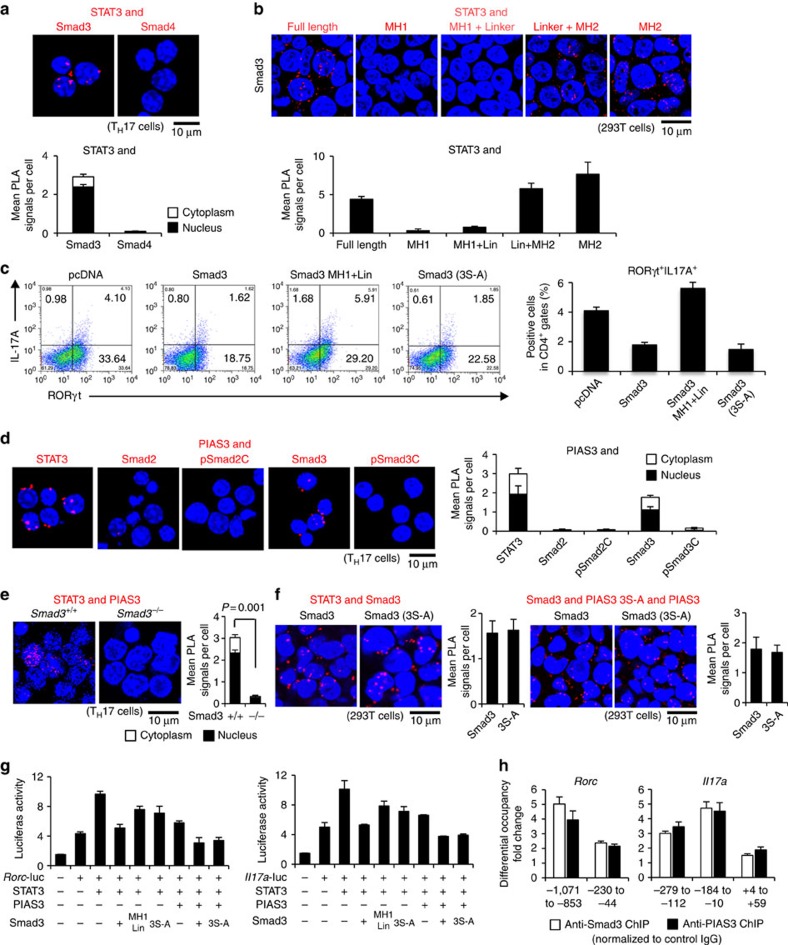
We investigated the mechanism how Smad3 functions as a transcription co-repressor of STAT3. PLA confirmed the endogenous close proximity between Smad3 and STAT3 in TH17 cells (Fig. 4a). Unlike R-Smads, Smad4 did not interact with STAT3 (Fig. 4a). Although STAT5 and STAT3 oppositely regulate TH17 differentiation by binding the multiple common sites across the locus encoding IL-17 (ref. 27), neither Smad2 nor Smad3 interacted with STAT5 (Supplementary Fig. 11). Furthermore, interactions between Smad2/3 and STAT3 were as significant as the established interaction controls: pSmad2/3C-Smad4 (refs 9, 10) and Smad2/3–RORγt18,21 (Supplementary Fig. 12). PLA and immunoprecipitation of 293T cells transfected with the various deletion mutants of Smad3 showed that Smad3 MH2 deletion mutants (MH1 and MH1+L)29 failed to bind with STAT3 (Fig. 4b and Supplementary Fig. 13). Thus, the MH2 domain is required for Smad3 to bind STAT3.
Figure 4. C-terminally unphosphorylated Smad3 recruits PIAS3 to act as a transcription co-repressor of STAT3 in TH17 differentiation.
Interactions of endogenous proteins in TH17 cells and exogenous proteins in 293T cells were determined using PLA. PLA signals (a,b,d–f) were quantified using the BlobFinder software (scale bars, 10 μm; nucleus, black; cytoplasm, white, n=10 fields). (a) Endogenous interactions between Smad3/Smad4 and STAT3 in TH17 cells. (b) Effects of truncated mutations in Smad3 on the interaction with STAT3 in 293T cells. (c) Flow cytometry analyses of IL-17A+RORγt+CD4+ T cells transduced with the indicated DNA constructs using Nucleofector (n=4). (d) Endogenous interactions between PIAS3 and STAT3/Smad2/Smad3 in TH17 cells. (e) Endogenous interaction between PIAS3 and STAT3 in Smad3+/+, −/− TH17 cells. (f) Effects of Smad3 C-terminal mutation on the interaction with STAT3 (left) or the interaction with PIAS3 (right) in 293T cells. (g) Effects of PIAS3, Smad3 MH2 deletion and Smad3 C-terminal mutation on STAT3-induced activation of the Rorc promoter and the Il17a promoter constructs transfected in TH17 cells were determined using the luciferase assay. (h) Binding of Smad3 (white) and PIAS3 (black) to the Rorc and the Il17a promoter regions in TH17 cells was determined using ChIP (differential occupancy fold changes). A representative of six (a), three (b,f,g), four (c,d) or two (e,h) independent experiments is shown. Each experiment (g,h) was performed in triplicate (n=3). Data are mean+s.d. or mean+s.d. with P values (e, unpaired Student's t-test).
PIAS3 belongs to the mammalian protein inhibitor of activated STAT (PIAS) protein family, which represses STAT3-dependent transcriptional activation by blocking the DNA-binding activity of STAT3, regardless of its small ubiquitin-like modifier-E3 ligase activity32. Overexpression of Smad3, the deletion mutant lacking MH2 domain or the C-terminal mutant in TH17 cells show that the Smad3 MH2 domain, but not the C-terminal SSXS motif, is functionally responsible for the suppression of TH17 differentiation (Fig. 4c). Because PIAS3 interacts with Smad3 at its C-terminal domain33, we examined whether Smad3 recruits PIAS3 to repress STAT3-induced transcription of the Rorc and Il17a genes. PIAS3 showed the close proximity with both STAT3 and Smad3, but not with C-terminally phosphorylated Smad3 (pSmad3C) or Smad2 in TH17 cells (Fig. 4d). STAT3–PIAS3 interaction was completely abolished in Smad3−/− TH17 cells (Fig. 4e). A mutant of serine residues to alanine in the Smad3 SSXS motif, Smad3 (3S-A), was yet capable of binding with STAT3 and PIAS3 in 293T cells (Fig. 4f). Consistently, when co-transfected with PIAS3 in TH17 cells, Smad3 (3S-A) was able to suppress STAT3-induced activation of the Rorc and Il17a reporters (Fig. 4g). ChIP revealed that PIAS3 and Smad3, but not pSmad3C, bound to the same sites in the Rorc and Il17a promoters (Fig. 4h and Supplementary Fig. 14). Thus, C-terminal phosphorylation is not required for Smad3 to bind with STAT3 and PIAS3. Overexpression of PIAS3 suppressed TH17 differentiation, whereas knockdown of PIAS3 by siRNA abolished the binding of Smad3 to the Rorc and Il17a promoter regions, although TH17 differentiation was unaltered by knockdown of PIAS3 presumably because relatively predominant binding of Smad2 over Smad3 in the absence of PIAS3 transactivated the Rorc and Il17a genes (Supplementary Fig. 15). These data indicate that unphosphorylated Smad3C in cooperation with PIAS3 represses STAT3-induced transcription of the Rorc and Il17a genes.
ERK phosphorylates Smad2 linker in TH17 differentiation
Previous studies have paid attention to C-terminal phosphorylation of R-Smads as TGF-β signalling mediators in TH17 differentiation18,19,20,21,22,23. However, pSmad2L (Ser255) and unphosphorylated Smad3 are not involved in the canonical C-terminally phosphorylated R-Smad/Smad4-mediated TGF-β signalling. Three clustered serine residues in the linker regions of Smad2 (Ser245/250/255) are the phosphorylation sites for MAPKs (ERK, JNK and p38)10,11,12,13,34. Because MAPKs are shared by TGF-β, IL-6 and TCR, we sought to identify the MAPK responsible for Smad2 linker phosphorylation in TH17 differentiation.
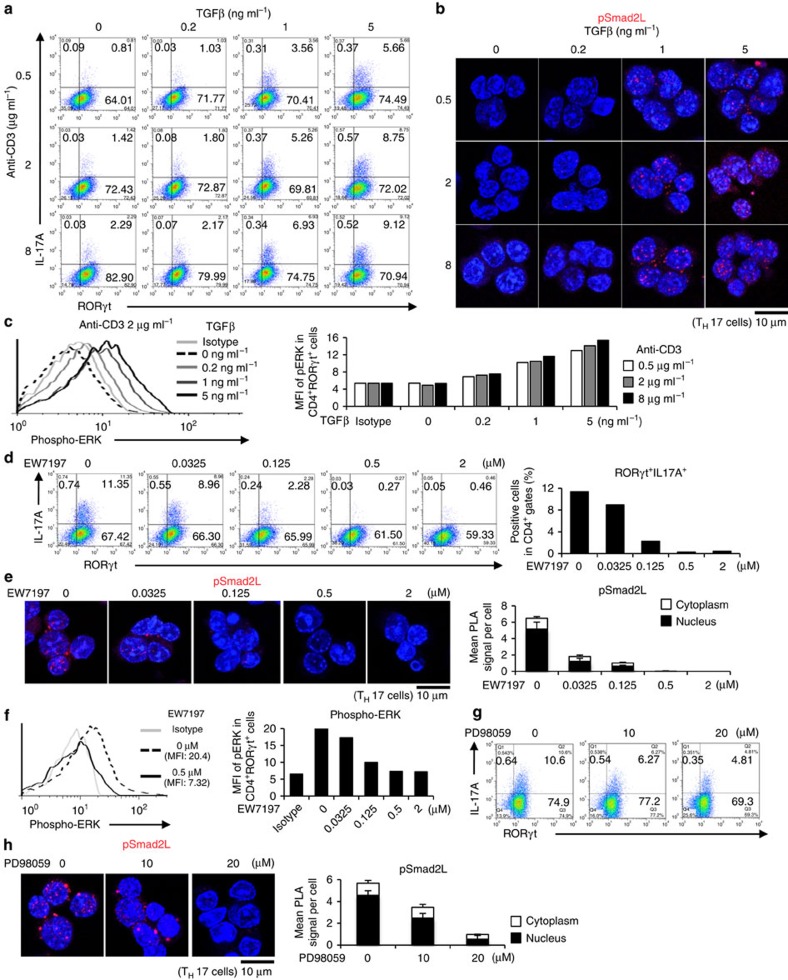
Signal intensities of TGF-β, IL-6 and TCR have been reported to correlate with the extent of TH17 differentiation3,4,16. Therefore, we treated CD4+ T cells under TH17-polarizing condition with various concentrations of TGF-β, IL-6 and anti-CD3 antibody. We confirmed that higher doses of TGF-β1, anti-CD3 antibody and IL-6 induced more TH17 differentiation (Fig. 5a and Supplementary Figs 16a and 17a). Percentages of IL-17A+RORγt+CD4+ T cells were directly proportional to phosphorylation of Smad2L (Fig. 5b and Supplementary Figs 16b and 17b) and ERK, but not to the phosphorylation of JNK or p38 (Fig. 5c and Supplementary Figs 18 and 19).
Figure 5. ERK induces Smad2 linker phosphorylation that facilitates TH17 differentiation.
Purified CD4+ T cells were activated under TH17-polarizing condition with the indicated doses of TGF-β1 and plate-coated anti-CD3 antibody, or small molecule inhibitors (EW-7197, ALK5 inhibitor; PD98059, MEK inhibitor) for 3 days. (a) Flow cytometry analyses of IL-17A+RORγt+CD4+ T cells treated with TGF-β1 and plate-coated anti-CD3 antibody. (b) Endogenous expression of pSmad2L in TH17 cells treated with TGF-β1 and plate-coated anti-CD3 antibody was determined using PLA. (c) Flow cytometry analyses of phospho-ERK in TH17 cells treated with TGF-β1 and plate-coated anti-CD3 antibody. (d) Flow cytometry analyses of IL-17A+RORγt+CD4+ T cells treated with EW-7197. (e) Endogenous expression of pSmad2L in TH17 cells treated with EW-7197 was determined using PLA. (f) Flow cytometry analyses of phospho-ERK in TH17 cells treated with EW-7197. (g) Flow cytometry analyses of IL-17A+RORγt+CD4+ T cells treated with PD98059. (h) Endogenous expression of pSmad2L in TH17 cells treated with PD98059 was determined using PLA. The values of the mean fluorescence intensity (MFI) are shown in graphs. PLA signals (b,e,h) were quantified using the BlobFinder software (scale bars, 10 μm; nucleus, black; cytoplasm, white, n=10 fields). Data are representative of two (a–h) independent experiments. Data are mean+s.d.
To confirm whether TβRI-mediated phosphorylation of Smad2L is required for TH17 differentiation, we treated CD4+ T cells under TH17-polarizing condition with specific inhibitors against TβRI35 at the doses that maintain cell viability (Supplementary Fig. 20). A potent selective ATP-competitive inhibitor of TβRI kinase (activin receptor-like kinase5: ALK5), EW-7197 (refs 35, 36) completely suppressed TH17 differentiation at the dose of 0.5 μM (Fig. 5d). Treatment with EW-7197 suppressed pSmad2L (Fig. 5e) and phosphorylation of ERK, but not phosphorylation of JNK and p38 (Fig. 5f and Supplementary Fig. 21). One of the prototype ALK5 inhibitors, SB-505124, inhibits TGF-β-induced activation of MAPKs without altering ALK5-independent MAP kinase pathways37. A more highly selective ALK5 inhibitor, EW-7197, does not directly inhibit MEK1 and ERK1 (ref. 35). Therefore, inhibitory effect of EW-7197 on ERK phosphorylation is ALK5-specific. Culture media containing IL-6, IL-23 and IL-1β is sufficient to induce TH17 in the absence of TGF-β (ref. 38). However, EW-7197 inhibited, whereas TGF-β1 enhanced TH17 differentiation along with ERK phosphorylation even under this culture condition (Supplementary Fig. 22). These results suggest that TGF-β-TβRI signal phosphorylates ERK and Smad2L in TH17 cells.
To confirm whether ERK-mediated phosphorylation of Smad2L is required for TH17 differentiation, we next treated CD4+ T cells under TH17-polarizing condition with specific inhibitors against MAPKs at the doses that maintain cell viability (Supplementary Figs 23 and 24). A MEK inhibitor PD98059 suppressed TH17 differentiation in a dose-dependent manner (Fig. 5g and Supplementary Fig. 23), whereas a JNK inhibitor SP600125 or p38 inhibitor SB203580 did not affect TH17 differentiation (Supplementary Fig. 24). PD98059 showed the similar effects with EW-7197 on pSmad2L (Fig. 5h). Specific inhibition of MAP kinase by the corresponding inhibitor was confirmed (Supplementary Fig. 25). Taken together, ERK-mediated Smad2 linker phosphorylation is responsible for TH17 differentiation and the concentrations of TGF-β, TCR and IL-6 determine the intensities of Smad2 linker phosphorylation and the extent of TH17 differentiation.
R-Smad–STAT3 interactions balance TH17 differentiation
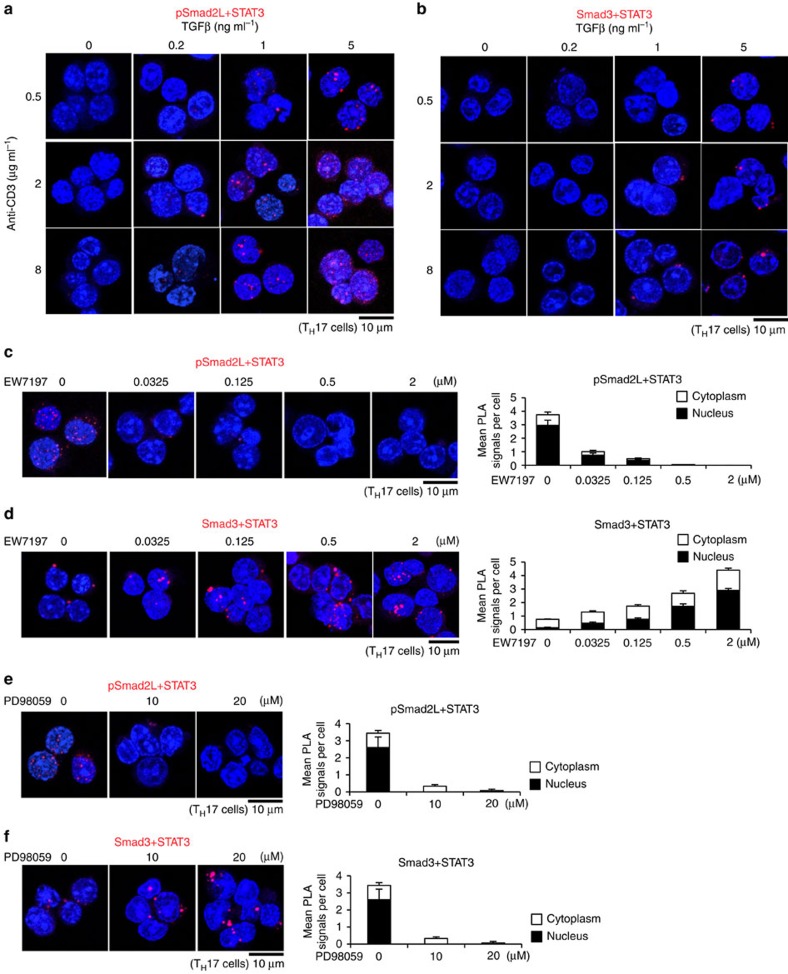
We next examined the effects of intensities and inhibitions of TGF-β/IL-6/TCR signals on the interactions of STAT3 with pSmad2L or unphosphorylated Smad3C in TH17 cells. Higher doses of TGF-β1, IL-6 and anti-CD3 antibody significantly upregulated pSmad2L–STAT3 interactions with little changes in Smad3–STAT3 interactions (Fig. 6a,b and Supplementary Fig. 26). By contrast, treatments with EW-7197 or PD98059 significantly downregulated pSmad2L–STAT3 interactions (Fig. 6c,e), whereas upregulated Smad3–STAT3 interactions (Fig. 6d,f). Interactions of pSmad2L and STAT3 were directly proportional, whereas interactions of unphosphorylated Smad3C and STAT3 were inversely proportional to TH17 differentiation of EW-7197- or PD98059-treated CD4+ T cells (Figs 5d,g and 6c–f). These data suggest that the balances between STAT3-interacting pSmad2L and STAT3-interacting unphosphorylated Smad3C determine the extent of TH17 differentiation.
Figure 6. R-Smad–STAT3 interaction balances correlate with TH17 differentiation.
Purified CD4+ T cells were activated under TH17-polarizing condition with the indicated doses of TGF-β1 and plate-coated anti-CD3 antibody, or small molecule inhibitors (EW-7197, ALK5 inhibitor; PD98059, MEK inhibitor) for 3 days. Interactions of endogenous proteins in TH17 cells were determined with PLA. PLA signals (a–f) were quantified using the BlobFinder software (scale bars, 10 μm; nucleus, black; cytoplasm, white, n=10 fields). (a) pSmad2L and STAT3, (b) Smad3 and STAT3 in TH17 cells treated with the indicated doses of TGF-β1 and plate-coated anti-CD3 antibody. (c) pSmad2L and STAT3, (d) Smad3 and STAT3 in TH17 cells treated with the indicated doses of EW-7197. (e) pSmad2L and STAT3, (f) Smad3 and STAT3 in TH17 cells treated with the indicated doses of PD98059. Data are representative of two independent experiments. Data are mean+s.d.
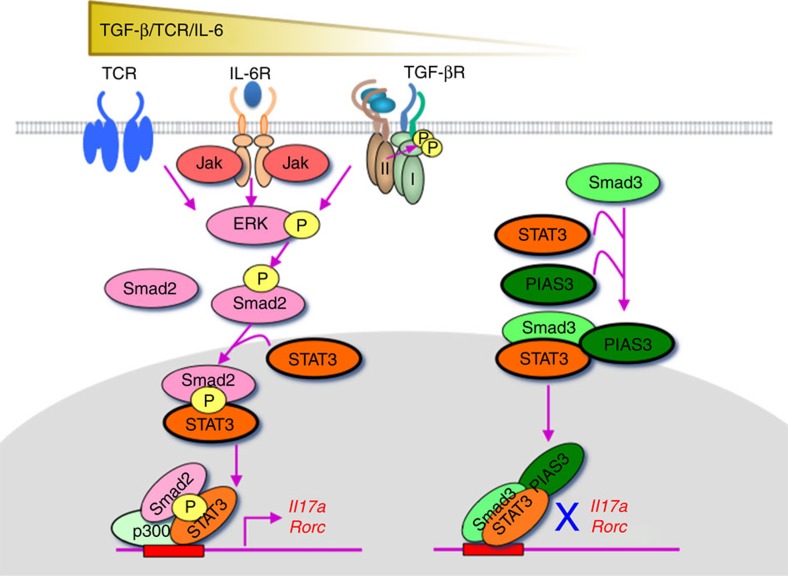
In summary, the TGF-β/IL-6/TCR–pERK–pSmad2L (Ser255) axis is the positive regulator, whereas unphosphorylated Smad3C–PIAS3 complex is the negative regulator of STAT3-induced transcriptional processes for TH17 differentiation (Fig. 7).
Figure 7. TGF-β R-Smads, Smad2 and Smad3, oppositely regulate TH17 differentiation as transcription cofactors of STAT3.
ERK-phosphorylated Smad2L (S255)/STAT3/p300 activates, whereas unphosphorylated Smad3C/STAT3/PIAS3 represses the transcription of the Rorc and Il17a genes.
Discussion
We discovered that Smad2 and Smad3 oppositely regulated STAT3-induced TH17 differentiation through the novel direct signalling networks. Transmodulation between the SMAD and STAT signalling pathways balances the interplay between TGF-β and various cytokines. Indirect crosstalk between SMAD and STAT was first reported as the inhibition of Smad3/4-mediated TGF-β signalling by Jak1-STAT1-mediated interferon (IFN)-γ signalling via induction of the inhibitory Smad, Smad7, which prevents TβRI-induced C-terminal phosphorylation of Smad3 (ref. 39). Direct crosstalk between SMAD and STAT was discovered as the synergistic signalling of leukaemia inhibitory factor and bone morphogenic protein-2, one of the TGF-β superfamily cytokines, via the STAT3–Smad1 complex bridged by p300 in fetal neural cells40. Direct crosstalk between Smad3 and STAT3 was reported as the augmentation of IL-6-STAT3-mediated transactivation by TGF-β via interaction of the STAT3–pSmad3C complex bridged by p300 in hepatoma cells41. This study clarified the mechanisms whereby R-Smads–STAT3 networks modulate TH17 differentiation; pSmad2L (Ser255) serves as STAT3 co-activator in combination with p300, a co-activator of various transcription factors including both Smads and STAT3 (refs 27, 31), whereas unphosphorylated Smad3C serves as the STAT3 co-repressor in combination with PIAS3, a negative regulator of STAT3-induced transcription32. The preceding reports and our findings indicate that SMAD–STAT signalling networks are highly cell-type-specific and context-dependent. Because of the relatively low DNA-binding affinity of Smad3 and lack of DNA-binding ability of Smad2, they interact with a wide variety of DNA-binding proteins to co-regulate the target genes. Recently, genome-wide transcriptome analyses have elucidated the diverse regulatory networks of Smad2/3 with cell-type-specific master transcription factors and/or DNA-binding cofactors in variety of cells42. The thorough iterative approach to delineate the TH17 global transcriptional regulatory network shows that STAT3 works as one of the key activators of the initial transcriptional programme, RORγt works as an expression modulator and Smad3 is the negative regulators43. It is noteworthy that a histone demethylase, JMJD3 (KDM6B) regulates the expression of numerous targets of RORγt and STAT3 (ref. 43) because JMJD3 causes a loss of the H3K27me3-repressive epigenetic mark by interacting with R-Smads at their target sites42. Therefore, it is possible that Smad2 may interact with JMJD3 to induce active chromatin state for TH17 regulation in the same manner with Nodal-Smad2/3 signalling in embryonic development44 (Supplementary Fig. 8). Further studies are required to elucidate the details of divergent context-dependent SMAD–STAT signalling networks implicated by genome-wide transcriptome analyses.
We further uncovered the novel roles of R-Smads with noncanonical phosphorylation status in networking with STAT3: linker phosphorylated Smad2 as a STAT3 co-activator and unphosphorylated Smad3 as a STAT3 co-repressor. Serine/threonine-rich R-Smad linker regions contain multiple phosphorylation sites by proline-directed protein kinases such as MAPKs, glycogen synthase kinase 3 and cyclin-dependent kinase family11,12,13. Linker residues Ser245/250/255, Thr220 in Smad2 and Ser204/208/213, Thr179 in Smad3 are the sites for phosphorylation12,13. Three clustered serine residues are preferred phosphorylation sites for ERK, JNK and p38 in response to receptor tyrosine kinases and proinflammatory cytokines, whereas threonine residues are preferred phosphorylation sites for cyclin-dependent kinase family in response to TGF-β. TβRI possesses the intrinsic tyrosine kinase activity to directly induce activation of MAPK pathways and subsequent phosphorylation of R-Smad linker residues in addition to the serine/threonine kinase activity to phosphorylate R-Smads in their conserved C-terminal SSXS motif5,11. Mitogens and hyperactive Ras induce ERK-mediated linker phosphorylation of Smad2 at Ser245/250/255/Thr220 and Smad3 at Ser204/208/Thr179 (refs 12, 13). Therefore, roles of R-Smad linker phosphorylation in carcinogenesis have been investigated intensively11,12,13. Central role of ERK in TCR signals15,24 suggests the important roles of R-Smad linker phosphorylation in T-cell signalling network. Thus far, MEKK2/3-ERK1/2 signalling has been reported to induce pSmad3L, which negatively regulates canonical TGF-β signalling for TH17 differentiation45. Because we found that Smad3 linker region was not involved in STAT3-induced TH17 differentiation (Fig. 4b), the mechanisms how pSmad3L regulates TH17 differentiation are independent of STAT3. By contrast to phosphorylated Smads, very little has been known about physiological functions of unphosphorylated R-Smads. It has been reported that PIAS3 enhances TGF-β-induced transcriptional activity of C-terminally phosphorylated Smad3 by recruiting p300 and CBP in COS and 293T cells33. By contrast, we discovered that unphosphorylated Smad3C due to less TGF-β signalling was required for PIAS3 to function as a co-repressor of STAT3 (ref. 32). Recent genome-wide studies implicate that unphosphorylated Smad3 may bind to some cell-type-specific transcription factors in both TGF-β-dependent and TGF-β-independent manners42,46,47. Our finding shed light on as-yet-unrecognized functions of unphosphorylated Smad3 as a transcription cofactor.
The discovery of a new proinflammatory effector T-cell subset, TH17, revised the functions of TGF-β, which had been long considered as the most potent immunosuppressive cytokine. TGF-β has been identified as the requisite factor for TH17 differentiation in combination with IL-6 and other proinflammatory cytokines such as IL-21, IL-23, IL-1β and tumour-necrosis factor-α (TNF-α)3,4,6. However, as functions of TGF-β have been frequently described as dual, bidirectional, pleiotropic, complex or contextual5,9,10,11,12,13, the roles of TGF-β in TH17 differentiation have become controversial18,19,20,21,22,23. Requirement of TGF-β for TH17 differentiation remains contradictory, indispensable4 or dispensable38. Our results provide explanation for these conflicting reports. TGF-β ligand-independent TH17 differentiation is possible because Smad2 linker phosphorylation could be induced by ERK signals downstream of the IL-6 receptor and TCR. However, significantly more effective inhibitory effect of the ALK5 inhibitor than that of a MEK inhibitor suggests that the TβRI–pERK–pSmad2L axis is more efficient than the non-TGF-β growth-stimulatory signal-pERK-pSmad2L axis for TH17 differentiation. Nonetheless, the report showing the dispensability of TGF-β demonstrates that TGF-β induces significantly more TH17 differentiation38. Likewise, our results provide explanations for the discrepancies in the reported roles of Smads in TH17 differentiation18,19,20,21,22,23. Our data are consistent with the previous reports showing that Smad2 is a positive regulator and Smad3 is a negative regulator of TH17 differentiation, although the mechanisms of actions are distinct18,19,20,21. It has been reported that TGF-β signalling via Smad2 indirectly induced STAT3 phosphorylation by inducing the expression of mRNA and protein of IL-6Rα (ref. 19); however, we could not confirm the differences in IL-6Rα mRNA expression in our systems (Supplementary Fig. 7). It has been reported that Smad3 interacted with RORγt and decreased its transcriptional activity21. We confirmed that not only Smad3 but also Smad2 interacted with RORγt (Supplementary Fig. 12). Whether RORγt forms the complex with STAT3 and R-Smads remains to be determined. Our data also suggest that the signalling intensity balances of TCR, co-stimulation, IL-6, TGF-β and other cytokines could yield the seeming dispensability of R-Smads22,23 because of their opposing effects. The signalling balances between TβRI-PKCα-mediated C-terminal phosphorylation of R-Smads48 and pERK-pSmad2L may be also crucial for TH17 differentiation.
An ALK5 inhibitor is efficacious against a mouse type II collagen antibody-induced arthritis model49. Our results of a CIA model showed the promoting role of pSmad2L at Ser255 and the suppressive role of unphosphorylated Smad3 in the arthritogenic TH17 differentiation. Therefore, inhibiting phosphorylation of Smad2 linker or Smad3 C-terminus may have therapeutic utility for RA and various TH17-related inflammatory diseases. Considering the crucial roles of SMADs and STATs in cell regulation, homeostasis and the pathogenesis of various diseases such as infection, cancers, fibrosis and inflammation, our findings will lead to the elucidation of cytokine signalling networks in various settings. In summary, we show the novel signalling networks of R-Smads and STAT3 for TH17 differentiation, which revise the classical linear signalling cascades50.
Methods
Mice
Smad23loxp/3loxp mice targeting exons 9 and 10 (ref. 51), Smad3ex8/ex8 mice targeting exon 8 (ref. 52) were generated as described on Sv129 × C57BL/6J background and backcrossed to C57BL/6J mice (Nihon SLC) for eight generations. For in vitro experiments, Smad3ex8/ex8 mice were backcrossed to C57BL/6J background for four generations. We used Smad3+/− mice because Smad3−/− mice develop osteoarthritis, bone malformation53 and impaired mucosal immunity52, and the embryonic lethality of Smad3−/− mice in the C57BL/6 background was extremely high (Supplementary Fig. 27), similarly with Tgf-β1−/− mice54. Cd4Cre transgenic mice55 were purchased from Jackson Laboratories. Mx-1Cre transgenic mice56 were kindly provided by Dr Masayuki Yamamoto (Tohoku University, Japan). For Mx-1Cre mice at 2–3 weeks of age, gene deletion was induced by intraperitoneal injections of 250 μg polyI:C dissolved in sterile saline at 2-day intervals for a total of three injections56. For immunophenotyping, spleens and superficial lymph nodes (cervical, axillary, brachial and inguinal) from the female mice aged between 12 and 16 weeks of age were used (age- and sex-matched, no randomized). All animals were maintained and used for experiments according to the ethical guidelines for animal experiments and the safety guidelines for gene manipulation experiments at the Konkuk University, Republic of Korea, University of Tsukuba, Japan, Tokyo Medical University, Japan under approved animal study protocols.
CIA
For induction of CIA, we used the immunization protocol for C57BL/6 strain (H-2b)57. Briefly, mice aged between 8 and 10 weeks of age were injected intradermally at several sites into the base of the tail and back with type II collagen (Sigma-Aldrich, Cat. no. C9301) emulsified in complete Freund adjuvant: incomplete Freund adjuvant (GIBCO), heat-killed mycobacterium tuberculosis (Difco Laboratories) on day −21 and the same injection was repeated on day 0 (Fig. 1a and Supplementary Fig. 2). Arthritis development in each paw was scored by macroscopic evaluation58 as follows: (0) no change, (1) erythema and mild swelling confined to the ankle, (2) erythema and mild swelling from the ankle to midfoot, (3) moderate swelling and (4) severe swelling. The maximum score per mouse is 16. The investigators (M.M., K.S., S.N. and J.S.H.) were blinded to the genotypes. Ten to twenty mice/genotype were used (Fig. 1 legend and Supplementary Fig. 2 legend). Mice were dissected 2 weeks after the second immunization to evaluate the draining lymph nodes (popliteal, inguinal, axillary and brachial).
Histological analysis
Paws from collagen-immunized mice were harvested, fixed in 10% neutral-buffered formalin, decalcified, dehydrated with 70% ethanol, embedded in paraffin and sectioned at 3 μm. Sections were stained with haematoxylin and eosin, toluidine blue or safranin O. For immunohistochemistry, sections were stained with rat anti-CD4 (Abcam, Cat. no. ab25475, 1:50), rabbit anti-RORγt (Abcam, Cat. no. ab78007, 1:50) and rabbit anti-IL-17A (Abcam, Cat. no. ab79056, 1:50) antibodies. Slides were observed using an optical microscope, DM5000B (Leica).
Flow cytometry analyses
Fluorophore-conjugated antibodies were purchased from BD Pharmingen and eBioscience. CD16/32 were blocked by Fc-Block (BD Pharmingen, Cat. no. 553142) and isotype-matched control antibodies were used in each experiment. For cytokine intracellular staining, cultured cells or freshly isolated cells from CIA mice were treated with 5 ng ml−1 of phorbol-12-myristate 13-acetate (Sigma-Aldrich) and 500 ng ml−1 of ionomycin (Sigma-Aldrich) in the presence of GolgiPlug (BD Pharmingen) for the last 4 h of culture. For intracellular staining, cultured cells were fixed with the Cytoperm/Cytofix kit (BD Pharmingen). For Foxp3 staining, the Foxp3 staining kit (eBioscience, Cat. no. 00-5523-00) was used. Stained cells were acquired and analysed using LSR II (BD) and FlowJo software (Tree Star Inc.).
T-cell stimulation in vitro
CD4+ T cells from superficial lymph nodes and spleens were enriched using T-cell enrichment columns (R&D Systems) and MACS system (Miltenyi Biotech). Purity was confirmed as >90% using LSR II. In some experiments, CD44lowCD62LhighCD4+ T cells were sorted with FACSAria (BD) and the purity was >98%. Purified CD4+ T cells were stimulated by plate-coated anti-CD3 (2.0 μg ml−1; BD Pharmingen, Cat. no. 553057) and soluble anti-CD28 antibodies (5.0 μg ml−1; BD Pharmingen, Cat. no. 553294) with mIL-6 (50 ng ml−1), TGF-β1 (1 ng ml−1; Peprotech), anti-mouse IL-4 (10 μg ml−1; Biolegend, Cat. no. 504108) and anti-mouse IFN-γ antibodies (10 μg ml−1; BioLegend, Cat. no. 505812) in 10% FCS RPMI 1640 media supplemented with penicillin and streptomycin (HyClone) for TH17 differentiation for 3–4 days3. In some experiments, Purified CD4+ T cells were stimulated by the indicated doses of plate-coated anti-CD3, TGF-β1 and mIL-6, or by plate-coated anti-CD3 (10.0 μg ml−1; BD Pharmingen, Cat. no. 553057), soluble anti-CD28 antibodies (5.0 μg ml−1; BD Pharmingen, Cat. no. 553294), IL-1β (10 ng ml−1; Peprotech), IL-23 (10 ng ml−1; Peprotech) and the neutralizing antibodies described above, or by the indicated doses of various small molecule inhibitors: EW-7197 (ALK5 inhibitor) from Dr Dae-Kee Kim, PD98059 (MEK inhibitor), SP600125 (JNK inhibitor) and SB203580 (p38 inhibitor; Sigma-Aldrich). STAT3 siRNA (Dharmacon), PIAS3 siRNA (Santa Cruz), DNA constructs: Smad2, Smad2 (S255A), Smad3, Smad3 (3S-A) and Smad3 (MH1+L) from Dr Koichi Matsuzaki and Dr Takeshi Imamura, PIAS3 (Addgene, submitted by Shuai) and pcDNA or control RNA were transfected to purified CD4+ T cells using the 4D-Nucleofector and Amaxa Mouse T-cell Nucleofector kit (Lonza) before the cell culture.
RNA isolation and quantitation of mRNA using real-time RT–PCR
Total RNA was extracted using Trizol according to the manufacturer's instructions (Invitrogen). RNA was reverse-transcribed with the cDNA reverse transcription kit (Invitrogen). Amount of cDNA was quantitated with SYBR green (Applied Biosystems) real-time PCR using ABI 7900 and ABI 7300 machines (Applied Biosystems). The primers are described in Supplementary Table 1.
Western blot analysis and immunoprecipitation
293T cells (ATCC-CRL-3216) were transfected using PEI with STAT3 (Addgene, submitted by J. Darnell), FLAG-tagged Smad2 (full length, MH1, MH1+Linker, MH2+Linker, MH2, Y220V, S245A, S250A, S255A) and FLAG-tagged Smad3 (full length, MH1, MH1+Linker, MH2+Linker, MH2, 3S-A) from Dr Koichi Matsuzaki and Dr Takeshi Imamura. Cells were lysed with lysis buffer (PBS containing 0.5% Triton X-100, 20 mM HEPES (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerol phosphate, 1.5 mM MgCl2, 10 mM NaF, 2 mM dithiothreitol, 1 mM NaOV, 2 mM EGTA, 1 mM phenylmethylsulphonyl fluoride (PMSF) and protease inhibitor cocktail) were electrophoresed on 10% SDS–polyacrylamide gel and transferred to the polyvinylidene difluoride (PVDF) membrane, and probed with antibodies against phospho-Smad2 (Abcam, Cat. no. ab53100, 1:1,000 dilution), phospho-Smad3 (Abcam, Cat. no. ab51451, 1:1,000 dilution), Smad2 (Santa Cruz, Cat. no. sc-101153, 1:1,000 dilution), Smad3 (Santa Cruz, Cat. no. sc-101154, 1:1,000 dilution), Smad4 (Santa Cruz, Cat. no. sc-7966, 1:1,000 dilution) and β-actin (Santa Cruz, Cat. no. sc-7210, 1:10,000 dilution). Blots were visualized using an electrochemiluminescence kit (GE Healthcare).
For immunoprecipitation, the lysates were cleared using centrifugation at 16,000g for 10 min, incubated with protein A/G agarose beads and with anti-STAT3 antibody (Santa Cruz, Cat. no. sc-7179, 2.0 μg per 1 ml of cell lysate) at 4 °C for 12–16 h. The beads were washed three times with lysis buffer and immunoprecipitates were separated from the beads by adding 2 × sample buffer and boiled. SDS–PAGE-separated immunoprecipitates were transferred on PVDF membranes. The membranes were denatured with denaturation buffer containing 6 M guanidine chloride, 20 mM Tris (pH 7.5), 100 mM PMSF and 5 mM β-mercaptoethanol at 4 °C for 30 min and washed three times with TBST. The membranes were blocked with 5% BSA and incubated with anti-FLAG antibody (Biomol, Cat. no. ADI-SAB-410-0100, 1:1,000 dilution). 293T cells (ATCC-CRL-3216) were confirmed to be mycoplasma-negative using the e-Myco plus Mycoplasma PCR Detection Kit (iNtRON Biotechnology, Cat. no. 25237).
PLA
CD4+ T cells cultured in TH17 condition for 3–5 days or 293T cells (ATCC-CRL-3216) transfected with various constructs were fixed on the slides by 3.7% formaldehyde in PBS. The slides were washed, permeabilized by 0.1% Triton X-100 in TBS and blocked by 0.5% BSA. PLA was performed using the Duolink II Fluorescence kit (OLINK) using the rabbit antibodies (1:50 dilution) against: STAT3 (Cell Signaling Technology, Cat. no. 12640), phospho-STAT3 Y705 (Cell Signaling Technology, Cat. no. 9145), phospho-STAT3 S727 (Cell Signaling Technology, Cat. no. 9134), Smad2 (Cell Signaling Technology, Cat. no. 5339), phospho-Smad2C S465/467 (Cell Signaling Technology, Cat. no. 3101), phospho-Smad2L S245/250/255 (Cell Signaling Technology, Cat. no. 3104), Smad3 (Cell Signaling Technology, Cat. no. 9523), phospho-Smad3C S423/425 (Cell Signaling Technology, Cat. no. 9520), RORγt (Abcam, Cat. no. ab78007), PIAS3 (Santa Cruz, Cat. no. sc-14017), Flag (Biomol, Cat. no. ADI-SAB-410-0100) and phospho-Smad3L S208/213 (IBL, Cat. no. JP28029), mouse antibodies (1:50 dilution) against: Smad2/3 (Santa Cruz, Cat. no. sc-133098), Smad4 (Santa Cruz, Cat. no. sc-7966), STAT3 (Santa Cruz, Cat. no. sc-8019) and p300 (Santa Cruz, Cat. no. sc-48343). Target-specific rabbit primary antibodies and the secondary antibodies conjugated with oligonucleotides: PLA probe anti-rabbit PLUS (Sigma-Aldrich, Cat. no. DUO92002) and PLA probe anti-rabbit MINUS (Sigma-Aldrich, Cat. no. DUO92005) or PLA probe anti-mouse PLUS (Sigma-Aldrich, Cat. no. DUO92001) and PLA probe anti-mouse MINUS (Sigma-Aldrich, Cat. no. DUO92004) were used for single recognitions. Two primary antibodies raised in different species and the secondary antibodies conjugated with oligonucleotides: PLA probe anti-rabbit PLUS and PLA probe anti-mouse MINUS were used for double recognitions. After incubation of the slides with Blocking Solution for 30 min at 37 °C, they were incubated with primary antibodies diluted in the Antibody Diluent overnight at 4 °C, in PLA probe solution for 1 h at 37 °C and in Ligation-Ligase solution for 30 min at 37 °C with washing with Wash Buffer A in the interim of each step. The slides were incubated in Amplification-Polymerase solution for 100 min at 37 °C and then washed in Wash Buffer B. The nucleus was stained with DAPI. Then, the slides were dried at room temperature in the dark. Slides were observed using a confocal microscope, LSM700 (Carl Zeiss). PLA signals were quantified using the BlobFinder software (Centre for Image Analysis, Uppsala University).
Luciferase assay
The 2,000-bp promoter region of RORγt was generated using PCR from genomic C57BL/6 DNA using primers described in Supplementary Table 2. Products were verified by sequencing and were subcloned into pGL4 firefly luciferase construct (Promega) using NheI, EcoRV sites and XhoI, HindIII sites, respectively. The pGL4 mIL-17 2-kb promoter construct was from Addgene (submitted by W. Strober). The promoter constructs in various combinations with Flag-tagged STAT3 (Addgene, submitted by J. Darnell), Flag-tagged Smads, Flag-tagged Smad mutants, haemagglutinin-tagged p300, Flag-tagged PIAS3 (Addgene, submitted by Shuai) or empty pcDNA3 plasmid were co-transfected with control TK-pRL Renilla plasmid using PEI for 293T cells or using the 4D-Nucleofector and Amaxa Mouse T-cell Nucleofector kit (Lonza) for TH17 cells. Six hours after transfection, 293T cells (ATCC-CRL-3216) were lysed for the measurement using luminometer. CD4+ T cells were transfected before the cell culture under TH17-polarizing condition for 4 days.
ChIP
Chromatin was prepared from 1 × 107 CD4+ T cells isolated from C57BL/6 mice, Cd4Cre;Smad2fl/fl mice, Smad3−/− mice and the littermate control mice under TH17-polarizing condition for 3–4 days. Immunoprecipitation was performed with antibodies (1:50 dilution) against Smad2 (Cell Signaling Technology, Cat. no. 5339), Smad3 (Cell Signaling Technology, Cat. no. 9523), phospho-Smad3C S423/425 (Cell Signaling Technology, Cat. no. 9520), Smad4 (Santa Cruz, Cat. no. sc-7966), STAT3 (Santa Cruz, Cat. no. sc-7179), tri-methyl histone H3 Lys4 (Cell Signaling Technology, Cat. no. 9751), tri-methyl histone H3 Lys27 (Cell Signaling Technology, Cat. no. 9733), acetyl histone H3 Lys23 (Millipore, Cat. no. 17–10112) and PIAS3 (Santa Cruz) using ChIP kit (Cell Signaling Technology) according to the manufacturer's protocol. Immunoprecipitated DNA released from the crosslinked proteins was quantitated with real-time PCR using the primers (Supplementary Table 3) and was normalized to input DNA.
Statistical analyses
Statistical analysis was performed using analysis tools on the VassarStats Statistical Computation site (http://vassarstats.net/) and Excel. Data were analysed using the parametric unpaired Student t-test, or two-way analysis of variance test for CIA scoring.
Additional information
How to cite this article: Yoon, J.-H. et al. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat. Commun. 6:7600 doi: 10.1038/ncomms8600 (2015).
Supplementary Material
Supplementary Figures 1-27 and Supplementary Tables 1-3
Acknowledgments
We thank C. Deng (National Institutes of Health, USA) for Smad3+/− mice; M. Yamamoto (Tohoku University, Japan) for Mx-1Cre Tg mice; A. Nakao (University of Yamanashi, Japan), L. Wakefield R. Horai, H. Yamane (National Institutes of Health, USA), H. Yanai, H. Negishi, J. Nishio, H. Ikushima, T. Taniguchi (University of Tokyo, Japan) and M. Sporn (Dartmouth Medical School, USA) for helpful discussions and critical reading of the manuscript; B. Oh (Gachon University, Korea) for assistance in designing some luciferase constructs. This work was supported by Mochida Memorial Foundation for Medical and Pharmaceutical Research, Research Resident Grant by The Association for Preventive Medicine of Japan, Research Grant by Japan Rheumatism Foundation, a grant from the Kwang-dong Pharmaceutical Co., Bio Technology R&D Program, Republic of Korea (20090081756), Basic Science Research Program through the National Research Foundation of Korea (NRF-2012R1A1A3015334 and NRF-2015R1A1A3A04001051), a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A092258), a grant of the Korea Health technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea. (Grant Number: A111345) and the Pioneer Research Center Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2015-001937 and 2015-001923).
Footnotes
Author contributions J.-H.Y. designed and performed experiments, analysed data and prepared the manuscript. K.S., I.L., J.S.H., S.N., I.M. and T.S. contributed to the animal experiments including CIA and contributed to drafting the manuscript. Ma.K., Mi.K. and J.H.J. performed histological analyses and contributed to drafting the manuscript. J.K. and J.H.J. contributed to the experiments and analyses using flowcytometry and confocal microscopy. D.-K.K. developed EW-7197. K.M. and T.I. contributed to the experiments using various Smad2/3 mutants. M.W. generated Smad2loxp/loxp mice and contributed to the animal experiments. M.M. conceived the project idea, designed and performed the experiments, analysed data, supervised the project and wrote the manuscript.
References
- Li M. O., Wan Y. Y., Sanjabi S., Robertson A. K. & Flavell R. A. Transforming growth factor-beta regulation of immune responses. Ann. Rev. Immunol. 24, 99–146 (2006) . [DOI] [PubMed] [Google Scholar]
- Travis M. A. & Sheppard D. TGF-beta activation and function in immunity. Ann. Rev. Immunol. 32, 51–82 (2014) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R. J., Atkins C. J., Locksley R. M. & Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006) . [DOI] [PubMed] [Google Scholar]
- Manel N., Unutmaz D. & Littman D. R. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9, 641–649 (2008) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M. & Kuchroo V. K. IL-17 and Th17 cells. Ann. Rev. Immunol. 27, 485–517 (2009) . [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishigame H., Saijo S. & Nakae S. Functional specialization of interleukin-17 family members. Immunity 34, 149–162 (2011) . [DOI] [PubMed] [Google Scholar]
- Nakae S., Nambu A., Sudo K. & Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J. Immunol. 171, 6173–6177 (2003) . [DOI] [PubMed] [Google Scholar]
- Massague J., Seoane J. & Wotton D. Smad transcription factors. Genes Dev. 19, 2783–2810 (2005) . [DOI] [PubMed] [Google Scholar]
- Heldin C. H. & Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 347, 21–36 (2012) . [DOI] [PubMed] [Google Scholar]
- Massague J. Integration of Smad and MAPK pathways: a link and a linker revisited. Genes Dev. 17, 2993–2997 (2003) . [DOI] [PubMed] [Google Scholar]
- Wrighton K. H., Lin X. & Feng X. H. Phospho-control of TGF-beta superfamily signaling. Cell Res. 19, 8–20 (2009) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Smad phospho-isoforms direct context-dependent TGF-beta signaling. Cytokine Growth Factor Rev. 24, 385–399 (2013) . [DOI] [PubMed] [Google Scholar]
- Neurath M. F. & Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 22, 83–89 (2011) . [DOI] [PubMed] [Google Scholar]
- Liu H. et al. ERK differentially regulates Th17- and Treg-cell development and contributes to the pathogenesis of colitis. Eur. J. Immunol. 43, 1716–1726 (2013) . [DOI] [PubMed] [Google Scholar]
- Hirahara K. et al. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine Growth Factor Rev. 21, 425–434 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A., Pietenpol J. A. & Moses H. L. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J. Cell. Biochem. 101, 9–33 (2007) . [DOI] [PubMed] [Google Scholar]
- Martinez G. J. et al. Smad2 positively regulates the generation of Th17 cells. J. Biol. Chem. 285, 29039–29043 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N., Robertson E. & Kang J. SMAD2 is essential for TGF beta-mediated Th17 cell generation. J. Biol. Chem. 285, 29044–29048 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S. et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J. Immunol. 181, 2277–2284 (2008) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G. J. et al. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J. Biol. Chem. 284, 35283–35286 (2009) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T. et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185, 842–855 (2010) . [DOI] [PubMed] [Google Scholar]
- Lu L. et al. Role of SMAD and non-SMAD signals in the development of Th17 and regulatory T cells. J. Immunol. 184, 4295–4306 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Davis R. J. & Flavell R. A. MAP kinases in the immune response. Ann. Rev. Immunol. 20, 55–72 (2002) . [DOI] [PubMed] [Google Scholar]
- Kimura A. & Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur. J. Immunol. 40, 1830–1835 (2010) . [DOI] [PubMed] [Google Scholar]
- Durant L. et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. P. et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12, 247–254 (2011) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaarenstroom T. & Hill C. S. TGF-beta signaling to chromatin: How Smads regulate transcription during self-renewal and differentiation. Semin. Cell Dev. Biol. 32C, 107–118 (2014) . [DOI] [PubMed] [Google Scholar]
- Kawabata M. et al. Intracellular signaling of the TGF-beta superfamily by Smad proteins. Ann. N. Y. Acad. Sci. 886, 73–82 (1999) . [DOI] [PubMed] [Google Scholar]
- Sekimoto G. et al. Reversible Smad-dependent signaling between tumor suppression and oncogenesis. Cancer Res. 67, 5090–5096 (2007) . [DOI] [PubMed] [Google Scholar]
- Janknecht R., Wells N. J. & Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 12, 2114–2119 (1998) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. D. et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 (1997) . [DOI] [PubMed] [Google Scholar]
- Long J., Wang G., Matsuura I., He D. & Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3). Proc. Natl Acad. Sci. USA 101, 99–104 (2004) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M., Doody J., Timokhina I. & Massague J. A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes Dev. 13, 804–816 (1999) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C. H. et al. Discovery of N-((4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-5-(6-methylpyridin-2-yl)-1H-imidazol-2 -yl)methyl)-2-fluoroaniline (EW-7197): a highly potent, selective, and orally bioavailable inhibitor of TGF-beta type I receptor kinase as cancer immunotherapeutic/antifibrotic agent. J. Med. Chem. 57, 4213–4238 (2014) . [DOI] [PubMed] [Google Scholar]
- Yoon J. H. et al. Activin receptor-like kinase5 inhibition suppresses mouse melanoma by ubiquitin degradation of Smad4, thereby derepressing eomesodermin in cytotoxic T lymphocytes. EMBO Mol. Med. 5, 1720–1739 (2013) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCosta Byfield S., Major C., Laping N. J. & Roberts A. B. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 65, 744–752 (2004) . [DOI] [PubMed] [Google Scholar]
- Ghoreschi K. et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa L., Doody J. & Massague J. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature 397, 710–713 (1999) . [DOI] [PubMed] [Google Scholar]
- Nakashima K. et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284, 479–482 (1999) . [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Matsuda T., Muraguchi A., Miyazono K. & Kawabata M. Cross-talk between IL-6 and TGF-beta signaling in hepatoma cells. FEBS Lett. 492, 247–253 (2001) . [DOI] [PubMed] [Google Scholar]
- Morikawa M., Koinuma D., Miyazono K. & Heldin C. H. Genome-wide mechanisms of Smad binding. Oncogene 32, 1609–1615 (2013) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle O., Kumar A. & Kuehn M. R. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 3, ra48 (2010) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X., Liu F., Wang X., Lin A., Zhao H. & Su B. The kinases MEKK2 and MEKK3 regulate transforming growth factor-beta-mediated helper T cell differentiation. Immunity 34, 201–212 (2011) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A. C. et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576 (2011) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogaya K. et al. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res. 24, 994–1008 (2014) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel M. et al. The kinase PKCalpha selectively upregulates interleukin-17A during Th17 cell immune responses. Immunity 38, 41–52 (2013) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma M. et al. TGF-beta type I receptor kinase inhibitor down-regulates rheumatoid synoviocytes and prevents the arthritis induced by type II collagen antibody. Int. Immunol. 19, 117–126 (2007) . [DOI] [PubMed] [Google Scholar]
- Levy E. D., Landry C. R. & Michnick S. W. Cell signaling. Signaling through cooperation. Science 328, 983–984 (2010) . [DOI] [PubMed] [Google Scholar]
- Liu Y., Festing M. H., Hester M., Thompson J. C. & Weinstein M. Generation of novel conditional and hypomorphic alleles of the Smad2 gene. Genesis 40, 118–123 (2004) . [DOI] [PubMed] [Google Scholar]
- Yang X. et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 18, 1280–1291 (1999) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Chen L., Xu X., Li C., Huang C. & Deng C. X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 153, 35–46 (2001) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyadi M. et al. Mapping of a major genetic modifier of embryonic lethality in TGF beta 1 knockout mice. Nat. Genet. 15, 207–211 (1997) . [DOI] [PubMed] [Google Scholar]
- Lee P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001) . [DOI] [PubMed] [Google Scholar]
- Kuhn R., Schwenk F., Aguet M. & Rajewsky K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995) . [DOI] [PubMed] [Google Scholar]
- Campbell I. K., Hamilton J. A. & Wicks I. P. Collagen-induced arthritis in C57BL/6 (H-2b) mice: new insights into an important disease model of rheumatoid arthritis. Eur. J. Immunol. 30, 1568–1575 (2000) . [DOI] [PubMed] [Google Scholar]
- Rosloniec E. F., Cremer M., Kang A. H., Myers L. K. & Brand D. D. Collagen-induced arthritis Curr. Protoc. Immunol. Chapter 15, Unit 15 15, 11–25 (2010) . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-27 and Supplementary Tables 1-3