Abstract
Objectives:
The present study was conducted for isolation, screening, and identification of Azotobacter and Trichoderma from different soil samples.
Methods:
A total of 10 isolates of Azotobacter and Trichoderma were isolated from rhizospheric soils. The test isolates were biochemically characterized and screened in in-vitro conditions for their plant growth promoting properties. DNA polymorphism of isolates was studied using randomly amplified polymorphic DNA analysis.
Results:
A total of 41 bands were scored, out of which 35 bands were found to be 85.59% polymorphic in Azotobacter and in Trichoderma among total 37 bands scored of which 29 were found to be 78.37% polymorphic. The influence of isolated plant growth promoting rhizobacteria (PGPR) strains on plant growth was studied using different parameters such as height of the plant, number of leaves, and number of branches, and bio-control activity was studied.
Conclusion:
The present results concluded that the multiple beneficial activities of PGPR traits increase the plant growth and bio-control activity.
Keywords: Azotobacter, bio-control activity, plant growth, plant growth promoting rhizobacteria, polymerase chain reaction-randomly amplified polymorphic DNA, trichoderma
INTRODUCTION
Plant growth promoting rhizobacteria (PGPR) are soil inhabitants that are able to colonize plant roots, stimulate plant growth, and increase crop yields.[1] The bacteria from soil aggressively colonize the root zone and promote plant growth are generally termed as PGPR.[2] PGPR directly enhance plant growth by a variety of mechanisms like atmospheric nitrogen fixation, siderophores production that chelates iron and make it available to the plant roots, solubilization of minerals such as phosphorus, increased uptake of nutrients such as nitrogen, phosphorus, potassium, and synthesis of phytohormones, indole acetic acid, and gibberlic acid and antifungal activity. PGPR are diverse, complex, and important assemblages in the biosphere,[3] they are considered as a group of beneficial free-living soil bacteria for sustainable agriculture and environment.[4] Along with this, they are also involved suppressing the root pathogenicity.[5] Several studies were reported the inoculation of bacteria to the plants enhances plant growth.[6,7,8] Different strains of PGPR genera exhibiting plant growth promoting activity are Azoarcus, Pseudomonas, Azospirillum, Azotobacter, Arthrobacter, Bacillus, Clostridium, Burkholdaria, Enterobacter, Gluconacetobacter, Rhizobium, Erwinia, Mycobacterium, Mesorhizobium, Flavobacterium, etc. have been reported.[1,9,10,11,12] Rhizosphere bacteria Azotobacter and fungus Trichoderma are the well-known predominant microorganisms having novel features in improving plant growth and are the most extensively studied. Azotobacter is a free-living aerobic, nonsymbiotic nitrogen fixer and acts as PGPR helps in root expansion, improve uptake of plant nutrients, protects plants from root diseases and most important improves biomass production in the rhizosphere of almost all crops. Trichoderma spp. is one of the efficient bio-control agents against to several plant and soil pathogenic fungi, used for plant disease control.[13,14,15,16,17] Inoculation of these bacteria competitively colonizes the roots of the plant and can act as biofertilizers and/or antagonists (biopesticides) or simultaneously both. In view of this, the present study was undertaken to investigate the PGPR activity of strains Azotobacter and Trichoderma, isolated from different crop soils and used as biofertilizers for stimulation of plant growth and bio-control without damaging the environment.
MATERIALS AND METHODS
Collection of soil samples
A total of 10 isolates were isolated from rhizospheric soils. Plant was gently and carefully uprooted, soil tightly adhering the root was collected, randomly selected, mixed and ¼ parts was used as composite rhizospheric soil sample of the crops. The pH of soil was determined in 1:2 (soil:Water) ratio, keeping 30 min as equilibration time.
Isolation and identification of isolates
Collected soil samples were air-dried for 4 h, and isolation was done by serial dilution technique.[18] Selective N-free mannitol agar for Azotobacter and Trichoderma selective medium for Trichoderma were used for isolation of the strains.[19] One ml of soil suspension was taken with the help of sterilized pipette and poured on the petri plate seeded with selective mediums. The plates were incubated at 37°C ± 1°C for Azotobacter and 28°C ± 1°C for Trichoderma for 5 days. Appearance of colonies was recorded, and individual colonies selected and maintained as a pure culture for further study.
The isolated colonies were sub-cultured and identified by using morphological and biochemical tests. The various tests were performed for Azotobacter like, catalase activity, motility test, oxidase activity, citrate test, indole test, methyl red test, Voges–Proskauer test, triple sugar ion test, nitrate reduction test.[20] Production of chlamydospores, conidial diameter, hydrolysis of gelatin, growth on glucose, nutrient agar, citric acid, lactic acid, ammonium oxalate, 4°C, 37°C, 40°C[21] tests were conducted for Trichoderma.
Extraction of DNA from isolates
Total genomic DNA was extracted from the isolated samples using Zymo-Research fungal/bacterial DNA kit. Bacterial isolates were grown in luria broth and incubated at 33°C for overnight under shaking. About 1.5 ml of culture was taken in a microcentrifuge tube, spin for 7 min and supernatant was decanted. To the pellet, 567 μl of Tris-EDTA (TE) buffer, 3 μl of 20 mg/ml proteinase-k, 30 μl of 10% sodium dodecyl sulfate were added and incubated for 1 h at 37°C. Again 100 μl of 5M NaCl and 80 μl of cetyltrimethylammonium bromide solution were added and incubated for 10 min at 65°C. Further it was extracted with equal volume of chloroform:Isoamyl alcohol and the aqueous phase was transferred to the fresh tube and to this equal volume of phenol:Chloroform:Isoamyl alcohol was added and subjected to centrifugation at 8000 rpm for 5 min at 4°C. It was washed with chloroform:Isoamyl alcohol until the clear supernatant was obtained. Then equal volume of chilled propanol was added, mixed gently and kept at −20°C overnight for precipitation of DNA. To pellet the DNA, centrifuged at 10,000 rpm for 20 min at 4°C then, pellet was washed with 70% ethanol and air-dried and DNA was dissolved in TE buffer.
Polymerase chain reaction amplification
Polymerase chain reaction (PCR) was performed using 16s recombinant DNA (rDNA) primers forward primer (5’-AGA GTT TGA TCC TGG CTC AG-3’) and reverse primer (5’-TGA CTG ACT GAG GCT ACC TG-3’). PCR reactions were performed in a final volume of 25 μl containing 30 ηg of template DNA, 0.75 μl of 2 mM deoxynucleotide triphosphates each, 2.5 μl of 10X Taq buffer,0.36 μl 1 unit of Taq DNA polymerase, 3 ml of 10 pico mole primer. Amplifications were achieved in eppendrof thermocycler with the program consisting initial denaturation of 94°C for 3 min followed by 45 cycles each consisting denaturation at 94°C for 1 min, primer annealing temperature at 37°C for 1 min, primer extension at 72°C for 3 min, and a final extension of 72°C for 10 min. These reactions were repeated to check the reproducibility of the amplification. The banding pattern was visualized on ultraviolet transilluminator and documented by Alpha image analyzer.
Effect of isolated strains on selected plants
The isolated 10 samples were inoculated to Amaranthus spinosus and Stevia rebaudiana plants at 1:4 ratios. The plant growth parameters such as plant height, number of leaves, and number of branches were recorded at 15 days interval till the time of harvest.
Bio-control activity
Bio-control activity was performed using dual plate technique. Potato dextrose agar plates were inoculated with 5-day-old cultures of the Fusarium. After 2 days, a 5 mm disc of the Trichoderma culture is placed in the same plate at a distance of 55 mm from the phytopathogen (Fusarium) disc.
RESULTS AND DISCUSSION
Isolated Azotobacter and Trichoderma colonies were sub-cultured on selective media; morphological and biochemical identification tests are tabulated [Tables 1 and 2]. Total genomic DNA was extracted, and PCR-randomly amplified polymorphic DNA (RAPD) analyses were performed using 16s rDNA forward and reverse primers for 10 strains. A total of 41 bands were scored, out of which 35 bands were found to be 85.59% polymorphic in Azotobacter and in Trichoderma among total 37 bands scored of which 29 were found to be 78.37% polymorphic [Figure 1]. In other studies, RAPD analysis for molecular variability in Azospirillum lipoferum found 93.2%,[22] in Azotobacter chroococcum 84.4%[23] polymorphism were observed which were isolated from different agroclimatic zones of Karnataka,[24] found 55.5% and[25] found 87% intra-specific genetic variation in Trichoderma isolates. In the present study, the growth parameters were significantly increased in PGPR inoculated plants when compared to control plants. Increases in the biomass in the inoculated plants with 10 strains of both Azotobacter and Trichoderma were found to be significantly higher growth parameters like plant height, number of leaves, and number of branches as compared to uninoculated plant of A. spinosus [Figure 2] and S. rebaudiana [Figure 3]. Similar studies were conducted with combined inoculation of a phosphate solubilizing Bacillus megaterium sub sp, and a bio-control fungus Trichoderma spp.[26] and with the tetra-inoculants of R. leguminosarum + A. chroococcum + P. aeruginosa + T. harzianum on chickpea and observed increased germination, nutrient uptake, height of the plant, number of leaves, number of branches, pea yield, nodulation, and total biomass and less infection with pathogens.[27]
Table 1.
Biochemical characteristics for the identification of Azotobacter species
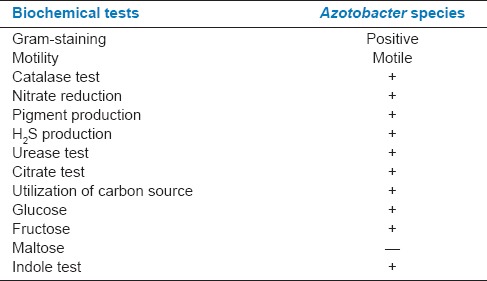
Table 2.
Biochemical characteristics for the identification of Trichoderma species
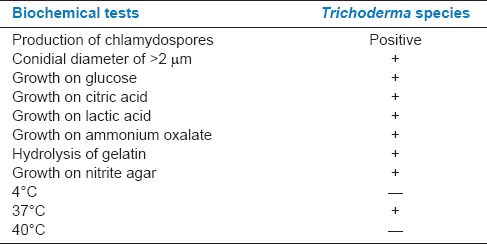
Figure 1.
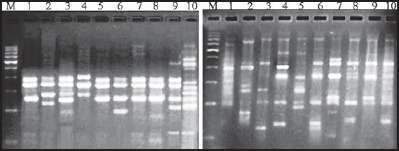
PCR-RAPD profiles obtained with forward and reverse primers (a) Azatobacter, (b) Trichoderma and markers on left side
Figure 2.
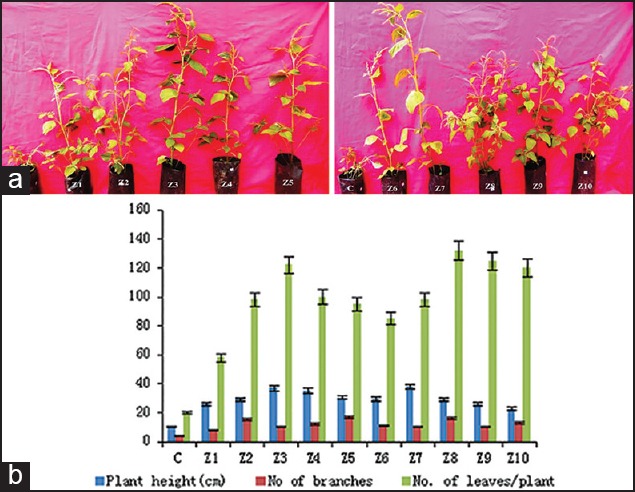
(a) Amaranthus spinosus plants inoculated with the combination of both Azotobacter and Trichoderma showing growth after 45 days C: control, Z1–Z10, (b) Effect of isolated strains of Azotobacter and Trichoderma on growth parameters of Amaranthus spinosus plants
Figure 3.
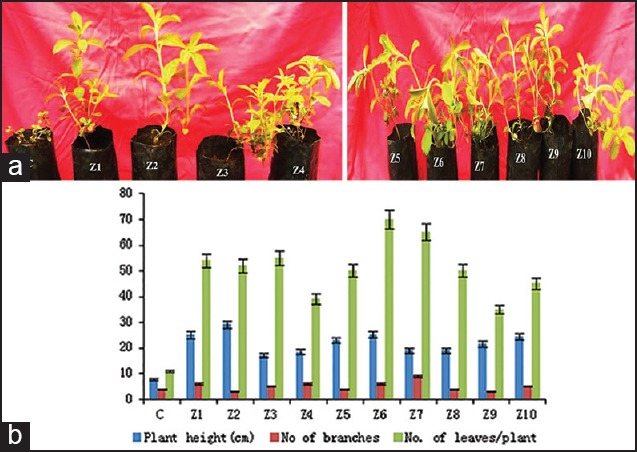
(a) Stevia rebaudiana plants inoculated with the combination of both Azotobacter and Trichoderma showing growth after 45 days. C: control, Z1–Z10, (b) Effect of isolated strains of Azotobacter and Trichoderma on growth parameters of Stevia rebaudiana plants
In the present study, bio-control activity of Trichoderma was observed against to fungal pathogen Fusarium using dual plate technique [Figure 4]. Several studies reported the isolates of Trichoderma spp. showed antifungal activity against to different fungal spp. Rhizoctonia solani, Sclerotium rolfsii, Pythium sp., Fusarium graminearum, Fusarium oxysporum f. phaseoli sp. and F. oxysporum f. Lycopersici sp.[22,24,25,28,29,30,31,32,33] In the present study, growth of the Stevia and Amaranthus plants was observed by treatment with Azotobacter and Trichoderma inoculation by direct mechanism of PGPR by the nutrients uptake and production of phytohormone (indole-3-acetic acid) and indirect mechanism of PGPR has suppressed plant diseases like wilt disease (Fusarium oxysporumi) by Trichoderma.
Figure 4.
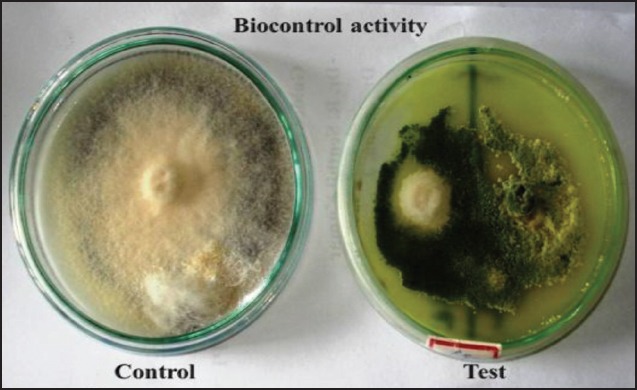
Bio-control activity of Trichoderma on Fusarium oxysporum. Sp. by dual plate technique
CONCLUSION
The results concluded that the combination of both isolated strains of Azotobacter and Trichoderma promotes plant growth and not infected with fungal pathogens. These strains may be used simultaneously as a biofertilizer and bio-control agent. These are eco-friendly in nature and cost effective, so PGPR help in improving profitability in agriculture and improve livelihoods of small and marginal farmers.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Kloepper JW, Lifshiftz R, Zablotowicz RM. Free living bacterial inoculation for enhancing crop productivity. Trends Biotechnol. 1989;7:39–44. [Google Scholar]
- 2.Tiwari PK, Thrimurthy VS. Isolation and characterization of the Pseudomonas fluorescens from rhizosphere of different crops. J Mycol Plant Pathol. 2007;37:231–4. [Google Scholar]
- 3.Khan AG. Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J Trace Elem Med Biol. 2005;18:355–64. doi: 10.1016/j.jtemb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Babalola OO. Beneficial bacteria of agricultural importance. Biotechnol Lett. 2010;32:1559–70. doi: 10.1007/s10529-010-0347-0. [DOI] [PubMed] [Google Scholar]
- 5.Fitte AH, Garbaye J. Interactions between mycorrhizal fungi and other organisms. Plant Soil. 1994;159:123–32. [Google Scholar]
- 6.Sakthivel N, Sivamani E, Unnamalai N, Ganamanickam SS. Plant growth promoting rhizobacteria in enhancing plant growth and suppressing plant pathogens. Curr Sci. 1986;55:22–5. [Google Scholar]
- 7.Kloepper JW, Lifshitz R, Schroth MN. Pseudomonas inoculants to benefit plant production. Anim Plant Sci. 1988;1:60–4. [Google Scholar]
- 8.Mroz A, Martiniuk S, Kus J. Response of winter wheat to seed applied microorganisms. Phytopathol Pol. 1994;19:15–20. [Google Scholar]
- 9.Okon Y, Labandera-Gonzalez CA. Agronomic applications of Azospirillum. In: Ryder MH, Stephens PM, Bowen GD, editors. Improving Plant Productivity with Rhizosphere Bacteria. Adelaide, Australia: Common Wealth Scientific and Industrial Research Organization; 1994. pp. 274–8. [Google Scholar]
- 10.Glick BR. The enhancement of plant growth by free living bacteria. Can J Microbiol. 1995;41:109–14. [Google Scholar]
- 11.Hurek T, Reinhold-Hurek B. Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes. J Biotechnol. 2003;106:169–78. doi: 10.1016/j.jbiotec.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Joseph B, Patra RR, Lawrence R. Characterization of plant growth promoting rhizobacteria associated with chickpea (Cicer arietinum L) Int J Plant Prod. 2007;1:141–52. [Google Scholar]
- 13.Whipps JM, McQquilken MP, Budge SS. Use of fungal antagonists for biocontrol of damping-off and Sclerotinia disease. Pestic Sci. 1993;37:309–13. [Google Scholar]
- 14.van Loon LC, Bakker PA, Pieterse CM. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–83. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 15.Weller DM. Biological control of soil borne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 16.Elad Y. Biological control foliar pathogens by means of Trichoderma harzianum and potential modes of action. Crop Prot. 2000;19:709–14. [Google Scholar]
- 17.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species — Opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 18.Don Brenner J, Krieg JR, Staley JT. 1st ed. United States: Michigan State University Publishers; 2005. Manual of Systematic Bacteriology; pp. 384–402. [Google Scholar]
- 19.Elad Y, Chet I, Henis Y. A selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica. 1981;9:59–67. [Google Scholar]
- 20.Kreig NR, Holf JG. Vol. 1. Baltimore, USA: William and Wilkins; 1984. Bergeys Manual of Systematic Bacteriology; pp. 459–67. [Google Scholar]
- 21.Bridge PD. An evaluation of some physiological and biochemical methods as an aid to the characterisation of species of Penicillium subsection fasciculata. J Gen Microbiol. 1985;131:1887–95. [Google Scholar]
- 22.Sumesh MK. Molecular and Physiological Characterization of Azospirillum lipoferum Isolated from Different Agroclimatic Zones of Karnatak, Thesis, University Agricultural Science, Bangalore. 2006 [Google Scholar]
- 23.Ananthnaik T. Biological and Molecular Characterization of Azotobacter chroococcum Isolated from Different Agroclimatic Zones of Karnataka and Their Influence on Growth and Biomass of Adhatoda vasica, Thesis, University Agricultural Science, Bengaluru. 2006 [Google Scholar]
- 24.Goes LB, Cost AB, Freire LL, Oliveria NT. Randomly amplified polymorphic DNA of Trichoderma isolates and antagonism against Rhizoctonia solani. Braz Arch Biol Technol. 2002;45:151–60. [Google Scholar]
- 25.Zaki A, El-Fiky, Osama Y Shalaby, Nada F. Ahmed, Biochemical and Molecular Characterization of some Trichoderma Isolates Antagonistic to Rhizoctonia solani the Causal of Bean Root-Rot. The Second Conference on Farm Integrated Pest Management. 2006 [Google Scholar]
- 26.El-Fiky A, Zaki Shalaby Y, Osama F, Ahamed, Nada Biochemical and Molecular Characterization of some Trichoderma Isolates Antagonistic to Rhizoctonia solani the Causal of Bean Root-Rot. The Second Conference on Farm Integrated Pest Management. 2006 [Google Scholar]
- 27.Rudresh DL, Shivaprakash MK, Prasad RD. Effect of a combined inoculation of Rhizobium, a phosphate solubilizing bacterium and Trichoderma spp. on growth, nutrient uptake and yield of chickpea. Appl Soil Ecol. 2005;28:139–46. [Google Scholar]
- 28.Verma JP, Yadav J. Evaluation of plant growth promoting rhizobacteria and their Effect on plant growth and grain yield of chickpea (cicer arietinum L.) Under sustainable agriculture production. Recent Trends Agric Water Environ Res. 2012;1:118–22. [Google Scholar]
- 29.Elad Y, Chet I, Katan Y. Trichoderma harzianum a biocontrol agent effective against Sclerotium rolfsii and Rhizoctonia solani. Phytopathology. 1980;70:119–21. [Google Scholar]
- 30.Sivan A, Elad Y, Chet I. Biological control effects of a new isolate of Trichoderma harzianum on Pythium aphanidermatum. Phytopathology. 1984;74:498–501. [Google Scholar]
- 31.Tianhui Z, Dexun Q. Antagonism of Trichoderma harzianum to Rhizoctonia solani. J Sichuan Agric Univ. 1994;12:11–11. [Google Scholar]
- 32.Kucuk C, Kivanc M. Isolation of Trichoderma Spp. and determination of their antifungal, biochemical and physiological features. Turk J Biol. 2003;27:247–53. [Google Scholar]
- 33.Dubey SC, Suresh M. Randomly amplified polymorphic DNA markers for Trichoderma species and antagonism against Fusarium oxysporum f. sp. ciceris causing chickpea wilt. J Phytopathol. 2006;154:663–9. [Google Scholar]
- 34.Siameto EN, Okoth S, Amugune NO, Chege NC. Molecular characterization and identification of biocontrol isolates of Trichoderma harzianum from Embu district, Kenya. Trop Subtrop Agroecosys. 2011;13:81–90. [Google Scholar]


