Abstract
Formation of crossovers between homologous chromosomes during meiosis is positively regulated by the ZMM proteins (also known as SIC proteins). DNA damage checkpoint proteins also promote efficient formation of interhomolog crossovers. Here, we examined, in budding yeast, the meiotic role of the heterotrimeric DNA damage response clamp composed of Rad17, Ddc1 and Mec3 (known as ‘9-1-1’ in other organisms) and a component of the clamp loader, Rad24 (known as Rad17 in other organisms). Cytological analysis indicated that the 9-1-1 clamp and its loader are not required for the chromosomal loading of RecA homologs Rad51 or Dmc1, but are necessary for the efficient loading of ZMM proteins. Interestingly, the loading of ZMM proteins onto meiotic chromosomes was independent of the checkpoint kinase Mec1 (the homolog of ATR) as well as Rad51. Furthermore, the ZMM member Zip3 (also known as Cst9) bound to the 9-1-1 complex in a cell-free system. These data suggest that, in addition to promoting interhomolog bias mediated by Rad51–Dmc1, the 9-1-1 clamp promotes crossover formation through a specific role in the assembly of ZMM proteins. Thus, the 9-1-1 complex functions to promote two crucial meiotic recombination processes, the regulation of interhomolog recombination and crossover formation mediated by ZMM.
KEY WORDS: Meiosis, Recombination, ZMM, SIC, Checkpoint clamp
INTRODUCTION
Meiotic recombination, which is a highly programmed process during meiosis, is essential for the segregation of homologous chromosomes during the first round of chromosome segregation (meiosis I). Recombination generates both crossover and non-crossover (NCO) products (Heyer et al., 2010). Crossovers, together with sister chromatid cohesion, ensure the segregation of homologous chromosomes by providing the physical linkages between them. Meiotic crossover formation is initiated by the generation of DNA double-strand breaks (DSBs) (Keeney, 2001). After the ends of the DSB have undergone nucleolytic processing, the recombination machinery uses the exposed tracts of single-stranded DNA (ssDNA) to search for identical double-stranded DNA (dsDNA) sequences. It has been proposed that one of the two ends generated by a single DSB site would engage in a homology search to form homologous joint molecules, while the other end is involved in a later step called ‘second-end capture’ (Hunter and Kleckner, 2001; Lao et al., 2008; Schwacha and Kleckner, 1994). Importantly, meiotic cells are equipped with a mechanism that ensures that the exchange usually occurs at the allelic site of a homologous chromatid, rather than with a sister chromatid (Goldfarb and Lichten, 2010; Grushcow et al., 1999; Thompson and Stahl, 1999). After locating a DNA sequence match is located on the homolog, the ssDNA invades the duplex DNAs, leading to the formation of an unstable D-loop. Then, the D-loop is converted into a metastable joint molecule intermediate (Hunter and Kleckner, 2001), and further processing of joint molecules gives rise to double Holliday junctions (dHJs) (Schwacha and Kleckner, 1995). Joint molecule formation occurs preferentially between homologous chromosomes, rather than between sister chromatids (Schwacha and Kleckner, 1994); this is referred to as ‘interhomolog bias’. Finally, dHJs are specifically resolved into reciprocal crossover products. NCO recombination products are usually generated from a pathway that does not involve stable joint molecule intermediates (Allers and Lichten, 2001; Börner et al., 2004; Hunter and Kleckner, 2001).
Meiotic crossover formation is facilitated by a group of proteins called ZMM (Zip, Mer, Msh) or SIC (synaptic initiation complex) proteins, hereafter ZMM for simplicity (Agarwal and Roeder, 2000; Börner et al., 2004; Chua and Roeder, 1998; Shinohara et al., 2008). ZMMs include Zip1, Zip2, Zip3, Mer3, Msh4, Msh5, Spo22/Zip4 and Spo16. Among the ZMMs, Msh4 and Msh5 are MutS homologs (Hollingsworth et al., 1995; Snowden et al., 2004). Zip3 (also known as Cst9) is a putative SUMO or ubiquitin E3 ligase whose targets have not yet been identified (Cheng et al., 2006). ZMM proteins localize on chromosomes during meiotic prophase-I, which can be detected cytologically as immunostaining foci. The assembly of some ZMM foci depends on DSB formation and end resection (Agarwal and Roeder, 2000; Chua and Roeder, 1998; Shinohara et al., 2008; Tsubouchi et al., 2006). However, the molecular interactions leading to the assembly of ZMM proteins on chromosomes are largely unknown. Zip3 binds DNA near sites of DSBs where it promotes local assembly of other ZMM proteins (Serrentino et al., 2013). ZMM proteins also promote local initiation of the synaptonemal complex (Sym et al., 1993) and coordinate synaptonemal complex formation with recombination (Börner et al., 2004; Shinohara et al., 2008). The synaptonemal complex is a ladder-like structure consisting of two axial/lateral elements aligned in parallel by a central element (Zickler and Kleckner, 1999).
DNA damage checkpoint proteins play an important role in the cellular responses to DNA damage (Hochwagen and Amon, 2006). During the mitotic DNA damage response in Saccharomyces cerevisiae, the Rad24–RFC (replication factor C) clamp-loader complex (also called the Rad17–RFC complex in other organisms) promotes the assembly of a clamp complex composed of Ddc1, Rad17 and Mec3 at the ssDNA–dsDNA junction of tailed DNAs (Majka and Burgers, 2003). Once loaded, the clamp promotes downstream events such as Mec1 (the homolog of ATR) activation. The Ddc1–Rad17–Mec3 clamp is referred to as the ‘9-1-1’ complex (Rad9–Rad1–Hus1) in other organisms (Sancar et al., 2004); we will therefore refer to the Ddc1–Rad17–Mec3 complex in budding yeast as the 9-1-1 clamp, for simplicity. The Mec1 kinase is recruited and activated on replication protein A (RPA)-coated ssDNAs (Zou and Elledge, 2003). In meiosis, as in mitosis, DNA damage response proteins can induce delays in entry into the first meiotic division (MI) when meiotic recombination is impaired. For example, dmc1 mutants, which lack the key meiotic strand exchange protein, arrest or delay before MI (Bishop et al., 1992). This arrest/delay is alleviated by mutations in the DNA damage response genes (Lydall et al., 1996). However, mutations in the DNA damage response genes induce meiotic delay in the timing of the first meiotic division rather than permitting earlier division as would be expected if their function were limited to regulatory surveillance of recombination intermediates (Shinohara et al., 2003a). Thus, it is not clear if and how these proteins contribute to making downstream meiotic processes depend on completion of the 150–200 recombination events that occur during normal meiosis (Hochwagen and Amon, 2006). It is clear that the proteins are involved in promoting normal recombination in addition to their role in promoting arrest when recombination is defective. For example, checkpoint proteins restrict the use of sister chromatids or repeated sequences at non-allelic (ectopic) sites as recombination partners (Grushcow et al., 1999; Thompson and Stahl, 1999). Moreover, the Mec1 kinase phosphorylates a meiosis-specific chromosome component, Hop1, to promote interhomolog recombination (Carballo et al., 2008). The Hop1 activation contributes to a DSB-dependent signaling pathway that activates the kinase Mek1, a key regulator of Rad51–Dmc1 dependent interhomolog recombination (Carballo et al., 2008; Hong et al., 2013). Furthermore, one of the functions of DNA damage response proteins is the promotion of crossover formation (Grushcow et al., 1999; Shinohara et al., 2003a). The overexpression of RAD51 or RAD54 partially suppresses meiotic and mitotic DNA repair defects of checkpoint mutants (Shinohara et al., 2003a). Together these results show that the functions of the checkpoint proteins are not limited to surveillance of defective recombination events.
In this paper, we provide evidence for a role played by the DNA damage response clamp and clamp loader in the formation of crossovers during meiosis through the ZMM assembly. Importantly, the checkpoint kinase, Mec1 is not required for the ZMM assembly. Zip3 binds to the 9-1-1 clamp in vitro. These suggest that the 9-1-1 clamp promotes the loading of ZMM proteins at sites destined to form crossovers through protein–protein interactions.
RESULTS
The 9-1-1 clamp promotes the assembly of the ZMM-containing complex
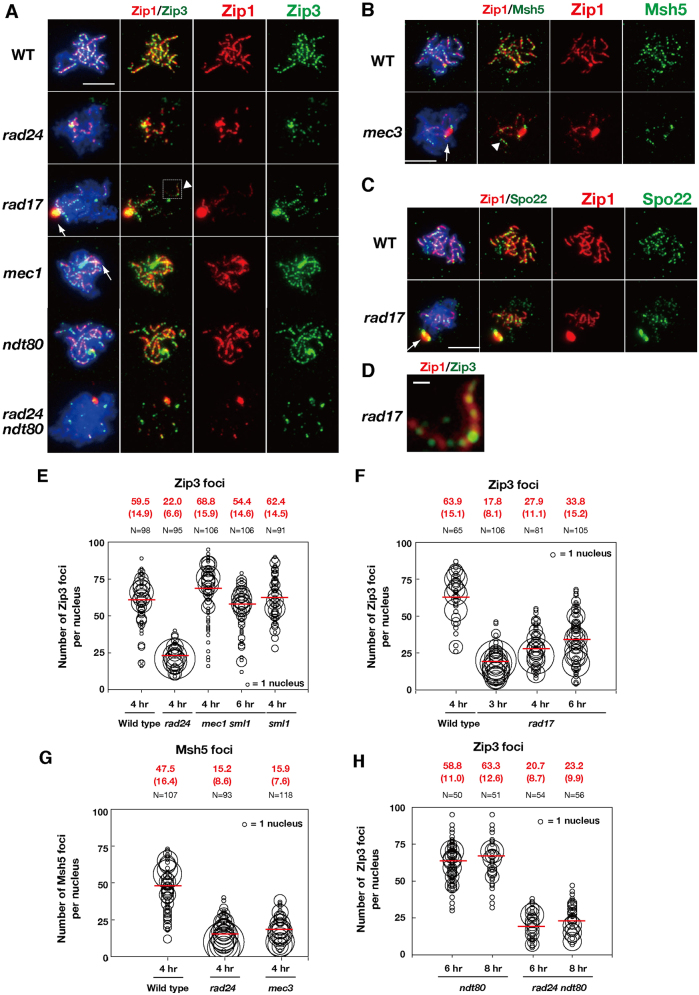
Previous studies have demonstrated that checkpoint mutants are defective in synaptonemal complex formation (Grushcow et al., 1999; Shinohara et al., 2003a). In budding yeast, synaptonemal complex assembly depends on the formation of interhomolog crossover intermediates (Börner et al., 2004); therefore, defects in mutants involved in homologous recombination might indirectly impair synaptonemal complex assembly. However, cytological analysis revealed little defects in DSB-dependent assembly of Rad51–Dmc1 in mutants lacking a 9-1-1 clamp or clamp loader component, although the turnover of Rad51 and Dmc1 foci was delayed in the checkpoint mutants (Shinohara et al., 2003a). This delay suggests that the 9-1-1 clamp and clamp loader plays a role in the process that follows Rad51–Dmc1 assembly and raised the possibility that the 9-1-1 clamp and clamp loader might also be involved in promoting synapsis during a post-strand-invasion step. It is known that ZMM proteins promote the formation of synaptonemal complexes as well as the maturation of joint molecules – processes that occur during or after the strand exchange process (Börner et al., 2004). We therefore asked whether the checkpoint clamp and clamp loader mutants affect the recruitment of ZMM proteins to meiotic chromosomes (Fig. 1). We examined the localization of ZMM proteins – Zip3, Msh5 and Spo22 – in 9-1-1 clamp/clamp loader-defective mutants (Fig. 1). The extent of synaptonemal complex elongation was also monitored using Zip1 staining. As described previously (Grushcow et al., 1999), the checkpoint clamp and clamp loader mutants such as the rad17, rad24, and mec3 were defective in Zip1 elongation (Fig. 1A–C). Consistent with a defect in synaptonemal complex assembly, the mutants formed a polycomplex, an aggregate of synaptonemal complex-associated proteins, including Zip1 (Sym et al., 1993). The rad17, rad24 and mec3 mutants showed reduced ZMM assembly on chromosomes (Fig. 1). The number of ZMM foci, such as those formed by Zip3, was greatly reduced in the rad24 clamp loader mutant compared to the wild type. In the wild type, ∼59.5±14.9 (mean±s.d.) Zip3 foci were detected on chromosomes at 4 hours (Fig. 1E). By contrast, the average number of Zip3 foci in the rad24 mutant was 22±6.6 (Fig. 1E). This difference was statistically significant as determined by a Mann–Whitney's U-test (P = 1.2×10−6). A similar defect is also observed in the 9-1-1 clamp mutant rad17 (Fig. 1F). Time course analysis indicated defective assembly of Zip3 foci in the rad17 mutant at 3 hours, 4 hours and 6 hours (Fig. 1F). At 6 hours, the number of foci was slightly higher than that observed at earlier time points (4 hours; P = 6.4×10−5), but was much lower than that of the wild type (at 4 hours, P = 9.6×10−3). The rad24 mutant also showed reduced numbers of Msh5 foci compared to the wild type (Fig. 1G). Furthermore, the number of Msh5 foci was also reduced in the other clamp mutant, mec3 (Fig. 1G). Thus, defects in the loading of two different ZMM proteins were observed in clamp and clamp loader mutants.
Fig. 1.
DNA damage clamp promotes the assembly of ZMM.DNA damage clamp promotes the assembly of ZMM. Localization of Zip3 (green), together with the synaptonemal complex component, Zip1 (red), was analyzed in various strains [wild-type (WT), and rad17, rad24, mec1 sml1, ndt80, and ndt80 rad24 mutant cells) by immunostaining. A representative image at 4 h is shown. Arrows indicate the Zip1 polycomplex. Arrowheads indicate a ‘long’ Zip1 line with multiple Zip3 foci in the mutant. (A) Msh5 staining (green) in the mec3 mutant was analyzed by Zip1-staining (red). The rectangle bordered by dotted lines is enlarged in D. (B) Staining of Spo22 (green) and Zip1 (red) in the rad17 mutant was analyzed as described above. (C) An image of Zip3 foci on a Zip1-line in the rad17 cell is enlarged (arrowhead in Fig. 1A). (E,F) Quantification of the numbers of Zip3 foci in the wild-type and checkpoint mutants. For each focus-positive chromosome spread, the numbers of Zip3 foci were counted and plotted as shown. The size of each circle represents the number of nuclei with each number of foci (i.e. the sizes of circles are proportional to the numbers of nuclei with a given number of foci). The mean number of foci per nucleus is shown in red (top), and also as a red bar in the graph. Standard deviations of focus numbers are shown in parentheses. N represents the number of nuclei analyzed for counting. (G) Quantification of the numbers of Msh5 foci in the wild type and the mec3 mutant. Msh5 focus numbers were analyzed as described above. (H) Quantification of the number of Zip3 foci in the ndt80 mutant background. Counting of Zip3 foci was studied as described above. Scale bars: 4 µm (A–C); 0.5 µm (D).
Previous studies have shown that, in response to aberrant recombination, the meiotic recombination checkpoint induces an arrest at pachytene (reviewed by Hochwagen and Amon, 2006). The pachytene arrest is mediated by the inactivation of the transcription activator Ndt80, which promotes the transcription of genes necessary for the exit from pachytene (Chu and Herskowitz, 1998). Thus, decreased numbers of ZMM foci in the checkpoint mutants may be the result of premature exit from pachytene. To exclude this possibility, we counted the numbers of Zip3 foci in the ndt80 and rad24 ndt80 mutants, both of which arrested at pachytene (Fig. 1A,H). The ndt80 single mutant maintained a similar number of Zip3 foci to that in the wild type (58.8±11.0 at 6 hours and 63.3±12.6 at 8 hours). Importantly, similar to the rad24 mutant, rad24 ndt80 double mutant cells showed reduced numbers of Zip3 foci, to 20.7±8.7 and 23.2±9.9 at 6 hours and 8 hours, respectively (versus ndt80, P = 9.6×10−3 and 9.6×10−3). Therefore, the reduced numbers of ZMM foci observed in clamp-defective mutants are not the result of accelerated exit from prophase.
Interestingly, despite the overall dramatic reduction of Zip3 foci and linear Zip1 structures, a few Zip1 linear structures that formed in the checkpoint clamp and clamp loader mutants were associated with lots of Zip3 foci (Fig. 1A,D). For example, the average number of Zip1 lines per nucleus in the rad17 mutant was lower than that of the wild type (6.3±2.2 lines per nucleus in the rad17 mutant, 14±2.2 in the wild type; Student's t-test, P = 7.3×10−6). By contrast, the density of Zip3 foci along elongated Zip1 structures was similar between the wild type (5.09±1.28 Zip3 foci/µm Zip1; n = 128) and rad17 mutant (4.68±2.10 foci/µm; n = 57). In addition, the Msh5 foci showed a similar localization pattern in the mec3 mutant (Fig. 1B). Taken together, these results suggest that there might be two mechanisms for the loading of ZMM proteins: one that is dependent on the checkpoint clamp, and a second that is associated with elongation of the synaptonemal complex and is independent of clamp function.
The mec1 mutant is proficient in ZMM focus formation
Previous analysis has shown that the mec1 deletion mutant exhibits a similar reduction in crossovers and a similar increase ectopic recombination as observed in 9-1-1 clamp mutants (Grushcow et al., 1999). Therefore, we asked whether a mec1 sml1 double mutant displays a similar defect in Zip3 focus accumulation to that seen in 9-1-1 mutants. Mutations in SML1 suppress the lethal effects of mec1 mutants allowing analysis of other mec1 functions. For simplicity the mec1 sml1 double mutant is referred to as ‘the mec1 mutant’ hereafter. We found that the number of Zip3 foci in the mec1 mutant was slightly higher than that in the wild type (68.8±15.9 at 4 hours, mean±s.d.; Fig. 1A,E). The number of Zip3 foci in the mec1 mutant is unlikely to depend on the sml1 mutation, since the number of Zip3 foci in the sml1 single mutant was analogous to that in the wild type at 4 hours (62.4±14.5). In addition, we found that the number of the foci in the rad24 sml1 double mutant was 26.9±9.1 (n = 96; supplementary material Fig. S1A), which was indistinguishable from that in the rad24 mutant (27.1±8.6). These data suggest that the function of Mec1 differs from that of the 9-1-1 clamp/clamp loader in that it exerts little or no influence on the recruitment of Zip3 to meiotic chromosomes.
9-1-1 recruitment is normal in zmm mutants
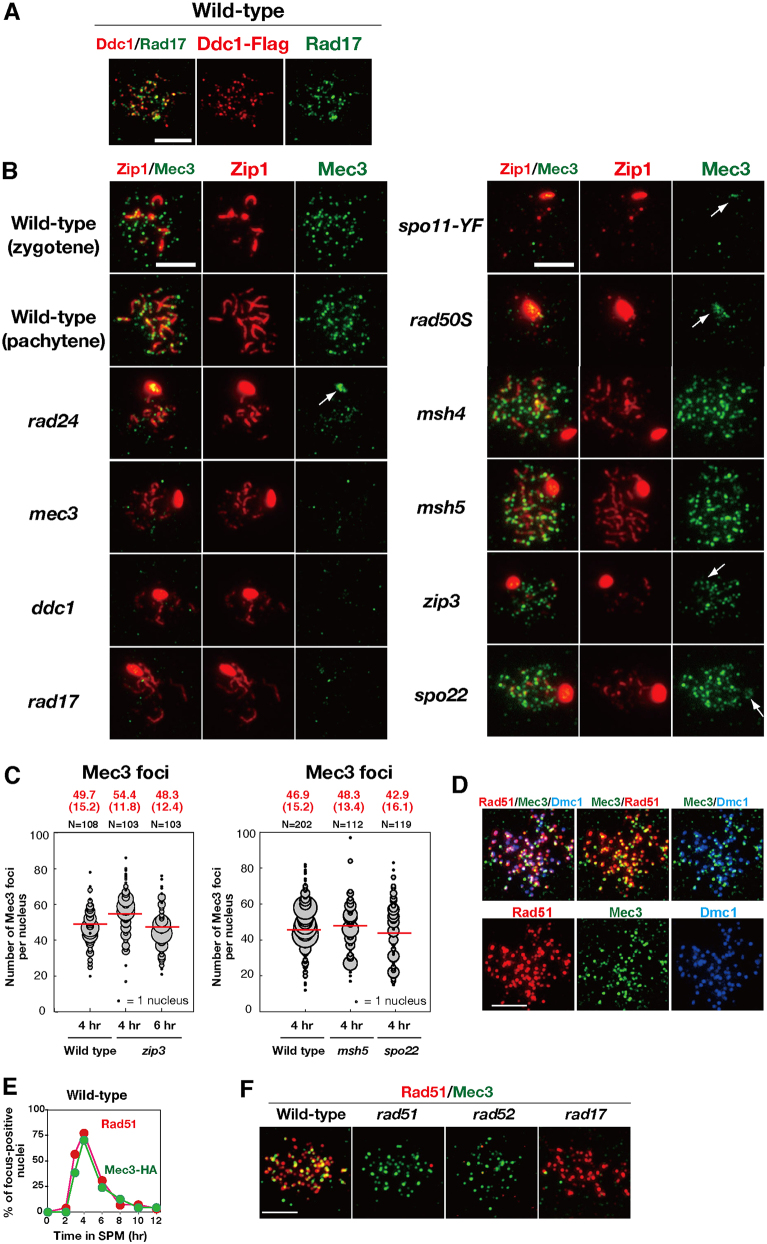
It has been previously shown that Ddc1–Myc forms foci on meiotic chromosomes (Hong and Roeder, 2002). To further study the localization of clamp components, we constructed a strain in which Mec3 and Rad17 were fused to HA and Flag epitope tags, respectively, and found that both tagged diploids form spores exhibiting wild-type viability and nearly normal DNA damage sensitivity (data not shown). For immunostaining, we also prepared antibodies against Mec3 and Rad17, which revealed a punctate staining pattern of Mec3–HA (Mec3), Ddc1–Flag, and Rad17 on meiotic chromosomes (Fig. 2A,B; supplementary material Fig. S2A). The anti-Mec3 antibody did not stain the chromosomes of a mec3 deletion mutant, indicating that the staining observed in wild-type strains was specific to Mec3 (Fig. 2B). Anti-Rad17 detected signals specific to wild-type cells, not to rad17 cells, but with a high background (Fig. 2A; supplementary material Fig. S2B). Double staining indicated that the colocalization frequency of Ddc1–Flag and Rad17 was 70% at 4 hours (n = 20; Fig. 2A), although we observed some competition between the antibodies against the clamp components; double staining weakened the signal intensity compared to single staining (data not shown). These data confirmed that the 9-1-1 clamp formed a distinct assembly on meiotic chromosomes. We opted to use the anti-Mec3 antibody in the experiments described below because it produced brighter and more specific staining patterns than the anti-Rad17 antibody.
Fig. 2.
Mec3 focus formation is independent of functions.Mec3 focus formation is independent of ZMM functions. (A) Staining of Ddc1–Flag (red; anti-Flag) and Rad17 (green; anti-Rad17) in the wild-type DDC1-Flag strain. A representative image at 4 hours is shown. (B) Mec3 (green) and Zip1 (red) staining were analyzed in the wild-type and various mutants. A representative image at 4 hours is shown. Arrows indicate Mec3 bound to a Zip1 polycomplex. (C) Quantification of the number of Mec3 foci in the wild type and zmm mutants. The numbers of Mec3 foci were analyzed as described in Fig. 1. (D) Chromosome spreads from MEC3-HA diploids were stained with anti-HA (green), anti-Rad51 (red) and anti-Dmc1 (blue) antibodies. In addition to three-color and mono-color, different two-color combinations are shown. A representative image at 4 hours is shown. (E) Kinetics of Mec3-HA (green; anti-HA) and Rad51 (red) foci in the wild type. At each time point, more than 100 spreads were examined for Rad51-foci- or Mec3–HA-foci-positive nuclei (with more than five foci). (F) Formation of Rad51 (red) and Mec3 foci (green) was analyzed in various strains (wild type, rad51, rad52 and rad17). Scale bars: 4 µm.
Previous results have shown that Ddc1 is recruited to meiotic chromosomes (Hong and Roeder, 2002). Immunostaining revealed punctate staining of Mec3 and Rad17 on meiotic chromosomes in wild-type cells (Fig. 2A,B). Consistent with previous results, Mec3 foci formation was abolished in spo11-Y135F and rad50S mutants (Alani et al., 1990; Keeney et al., 1997), which are deficient in DSB formation and resection, respectively (Fig. 2B). Moreover, the formation of Mec3 foci depended on the checkpoint clamp loader Rad24 as well as the clamp components Rad17 and Ddc1 (Fig. 2B). These data support the model in which checkpoint proteins bind ssDNA as a heterotrimeric complex in a Rad24 clamp-loader-dependent manner.
To determine the relationship between the checkpoint clamp and ZMM proteins on chromosomes, we next analyzed the localization of the 9-1-1 clamp in various zmm mutants (Fig. 2B). The number of Mec3 foci in the zip3 mutant at 4 hours was slightly higher than that in the wild type (54.4±11.8 versus 49.7±15.2, mean±s.d.; Fig. 2C), possibly due to delayed DSB turnover in the mutant (Börner et al., 2004) or increased DSB formation (Thacker et al., 2014). The number of Mec3 foci in other zmm mutants, such as msh5 and spo22/zip4, was also statistically indistinguishable from that in the wild type (P = 0.25 for wild type versus msh5, Fig. 2C). These results suggest that Mec3 loading on chromosomes is independent of ZMM function.
Mec3 colocalizes with Rad51 and Dmc1 on meiotic chromosomes
Given previous evidence for a genetic interaction of Rad17 and Rad24 with Rad51 and Rad54 in meiotic recombination (Shinohara et al., 2003a), we also determined whether the 9-1-1 clamp colocalized with recombination proteins on meiotic chromosomes. To assay for colocalization of the clamp with Rad51, spreads of meiotic chromosomes were immunostained for both Mec3–HA and Rad51 (Fig. 2D). Meiotic time-course analysis revealed that the kinetics for the appearance and disappearance of the Mec3 focus was similar to that of Rad51 (Fig. 2E). The average steady-state number of Mec3 foci was 50±15.2 (mean±s.d., n = 108) at 4 hours (also see Fig. 2C), which was similar to that of Rad51 (53.7±1.7 foci per nucleus). At 4 hours, ∼76.9±8.6% of Mec3–HA foci colocalized with Rad51 foci (n = 22), which is much higher than the proportion predicted for fortuitous colocalization (12%) (Gotta et al., 1996). We also assayed the colocalization of Mec3 with the meiosis-specific RecA homolog Dmc1 (Bishop et al., 1992). Triple staining for Rad51, Dmc1 and Mec3 (Fig. 2D; supplementary material Fig. S2C) also showed extensive colocalization, as expected given the high colocalization of Rad51 and Dmc1 (Bishop, 1994; Shinohara et al., 2000). The extensive colocalization supported the hypothesis that Mec3, and therefore the 9-1-1 clamp, functions concurrently with Rad51 and Dmc1 at sites of recombination.
Previous mutant studies have shown that Rad51 focus formation is independent of the checkpoint clamp (Lydall et al., 1996; Shinohara et al., 2003a). To determine whether the checkpoint clamp focus formation is independent of Rad51 assembly, we immunostained for Mec3 in mutants lacking Rad51 or the Rad51-assembly mediator Rad52 (Gasior et al., 1998; Miyazaki et al., 2004; Shinohara and Ogawa, 1998). These showed that the rad51 and rad52 mutants were proficient in the formation of Mec3 foci (Fig. 2F). Thus, the 9-1-1 clamp is recruited to meiotic chromosomes in a Rad51-independent manner. We also confirmed that formation of the Rad51 foci occurred normally in the rad17 mutant (Fig. 2F). Therefore, Rad51 and the 9-1-1 clamp were recruited to meiotic chromosomes independently of one another.
A subset of Mec3 foci colocalize with Msh5 foci on meiotic chromosomes
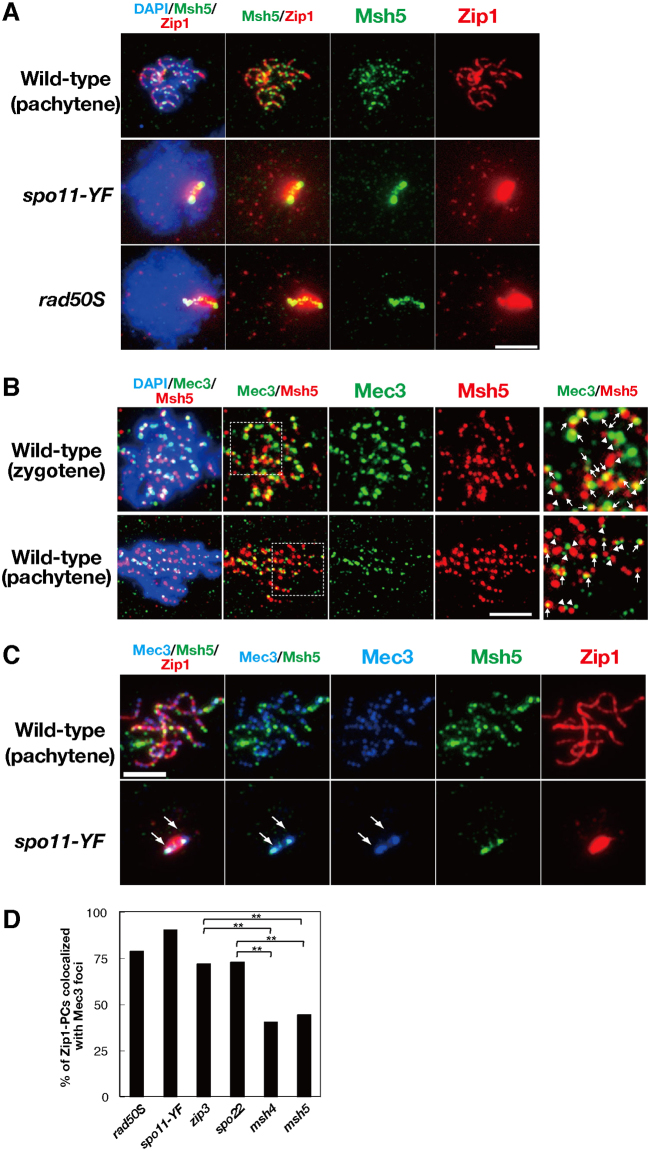
Given the dependency of ZMM protein assembly on the 9-1-1 clamp, we asked whether foci formed by the ZMM and 9-1-1 proteins colocalized. This analysis was complicated by the fact that not all the foci formed by ZMM proteins mark sites of recombination intermediates (Serrentino et al., 2013; Tsubouchi et al., 2006). Zip1 and Zip3 display high levels of colocalization to foci formed by centromeric proteins in mutants that have a block in the recombination pathway prior to formation of ssDNA (Tsubouchi and Roeder, 2005). Double staining of Zip1 and Msh5 in the spo11-Y135F and rad50S mutants showed that Zip1 forms foci on chromosomes as well as a Zip1 polycomplex (Fig. 3A), with foci likely marking centromeres (Tsubouchi and Roeder, 2005). In contrast, there was no significant focal staining pattern for Msh5 on chromosomes in either the spo11-Y135F or the rad50S mutant, with the exception of signal localizing to Zip1 polycomplex. These findings indicate that, in contrast to Zip1 and Zip3, the loading of Msh5 on chromosomes largely depends on DSB formation and DNA-end processing, and suggests that Msh5 does not bind centromeres with Zip1 and Zip3. Thus, to circumvent the complication of DSB-independent foci, we studied the localization pattern of Msh5 relative to the 9-1-1 component Mec3.
Fig. 3.
Colocalization of Mec3 and Msh5 on Zip1-polycomplex.Colocalization of Mec3 and Msh5 on Zip1-polycomplex. (A) Staining of Zip1 (red) and Msh5 (green) in the wild-type, spo11-Y135F, and rad50S strains. A representative image at 4 hours is shown. (B) Staining of Msh5 (red) and Mec3 (green) in the wild type. Representative images for zygotene (top) and pachytene (bottom) are shown. Enlarged regions are shown to the right. Arrows and arrowheads show colocalized and side-by-side staining for Mec3 and Msh5, respectively. (C) Triple staining of Zip1 (red), Msh5 (green), and Mec3 (blue) in the wild-type and the spo11-Y135F mutant. Arrows indicate bipolar localization of Mec3 at the ends of Zip1 polycomplex. Scale bars: 4 µm. (D) Colocalization frequencies of Mec3 foci or patches with Zip1 polycomplexes. Randomly selected nuclei containing Zip1 polycomplexes were assessed for colocalization with Mec3 foci or patches. More than 120 spreads were examined for each strain. **P<0.01 (chi-squared independent test). zip3 versus msh4, P = 2.7×10−5; zip3 versus msh5, P = 4.9×10−6, spo22/zip4 versus msh4, P = 2.6×10−4; spo22/zip4 versus msh5, P = 6.1×10−5.
Double staining of Msh5 and Mec3 revealed frequent overlapping or side-by-side staining of foci on wild-type chromosomes (Fig. 3B, arrows and arrowheads). We found that 80±7.3% (mean±s.d., n = 23) of Msh5 foci overlapped or lay side-by-side with foci formed by Mec3 at 4 hours. At the same time, 71±11% (n = 23) of Mec3 foci overlapped, or lay side-by-side with the Msh5 foci. However, using a more stringent criterion for colocalization, where only foci displaying more than 50% overlap in signal were counted, we found that only 20% of the Mec3 foci colocalized with Msh5 foci. This level of colocalization, although low, is nonetheless significantly higher than that predicted for fortuitous colocalization (11%; Gotta et al., 1996). It should be noted that colocalization analysis cannot provide definitive evidence for a role for direct protein–protein interaction in recruitment of foci, even when colocalization frequencies are high. At the same time, the data do not exclude the possibility that direct interactions contribute to the role of 9-1-1 in promoting ZMM focus formation because such interactions could be short-lived and/or disrupted by the spreading procedure. Indeed, there is precedent for pairs of proteins protein that are known to have functionally significant protein–protein interactions in vitro, but to display low levels of colocalization in vivo. This precedent includes are Rad51 and Rad52, which are known to bind one another directly but to display a similar low level of colocalization (25%; Gasior et al., 1998). Thus, the low level of Mec3–Msh5 colocalization observed is not inconsistent with a role of direct interactions in 9-1-1-dependent ZMM loading.
To further examine the interaction between Mec3 and ZMM proteins, we measured colocalization with Zip1 polycomplexes. Several ZMM proteins localized to Zip1 polycomplexes in mutants defective in synapsis. Colocalization of a protein with a polycomplex-associated ZMM protein is consistent with direct or indirect protein–protein interaction (Tsubouchi et al., 2006). We found that Mec3 localized to Zip1 polycomplex in mutants defective in meiotic recombination such as spo11-Y135F and rad50S (Figs 2B, 3C, arrows). Importantly, Mec3 localization to the Zip1 polycomplex was lost in the rad17 and ddc1 9-1-1 clamp mutants but not in the clamp-loader mutant rad24 (Fig. 2B). These findings suggest that Mec3 associates directly or indirectly with the Zip1 polycomplex in a manner that depends upon formation of the 9-1-1 complex. We quantified the fraction of the Mec3 foci that colocalized with Zip1 polycomplexes and found that 90% (n = 74) and 79% (n = 80) of Zip1 polycomplexes colocalized with Mec3 foci or patches in the spo11-Y135F and rad50S mutants, respectively (Fig. 3D). This localization pattern did not reflect a general tendency for recombination proteins to localize to polycomplexes, given the fact that Rad51 did not localize to polycomplexes in the spo11 mutant (supplementary material Fig. S2D). Further analysis showed that zmm mutations reduced the localization of Mec3 to the polycomplex (Fig. 2B; Fig. 3D). The msh4 and msh5 mutants exhibited significant reductions in Mec3 colocalization to polycomplexes to 40% (n = 84) and 44% (n = 83), respectively – closer to that predicted for fortuitous colocalization (35%). Thus, Mec3 localization to polycomplexes was significantly dependent on Msh4 and Msh5. In contrast, the Mec3 localization showed less dependency on Zip3 and Spo22/Zip4, with the corresponding mutants exhibiting 72% (n = 95) and 73% (n = 111) colocalization of polycomplexes with Mec3 foci, respectively. The signal intensity of Mec3 on polycomplexes was weaker in the zip3 and spo22 mutants than in the spo11 and rad50S mutants (Fig. 2B). This might be due to a lower level of free Mec3 proteins available for polycomplex localization in the zip3 and spo22 mutants, given that these mutants have more chromosome-bound Mec3 (see Fig. 2B).
The ZMM proteins, Spo22, Spo16, Msh4 and Msh5 localized to Zip1 polycomplexes in a unique bipolar manner (i.e. they form two staining foci on opposite ends of the polycomplex), whereas Zip3, Zip2 and Mer3 localized to the central region of the polycomplex (Fig. 1A–C; Shinohara et al., 2008; Tsubouchi et al., 2006). In 43% of the nuclei in the spo11-Y135F mutant, Mec3 displayed the bi-polar pattern and, importantly, colocalized with Msh5 in this localization pattern (Fig. 3C), suggesting a possible link between Msh4–Msh5 and the 9-1-1 clamp.
Interaction of Zip3 with the checkpoint clamp in vitro
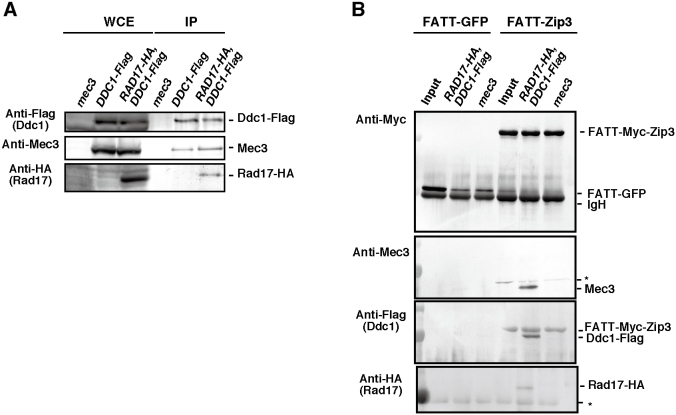
We confirmed Ddc1–Rad17–Mec3 complex formation in meiotic cell lysates of DDC1-Flag and DDC1-Flag RAD17-HA strains by immunoprecipitation. Immunoprecipitation using the anti-Flag antibody was successful in pulling down Ddc1–Flag and Mec3 from both strains, and in pulling down Rad17–HA from the DDC1-Flag RAD17-HA cells (Fig. 4A). This indicates that Ddc1, Rad17 and Mec3 bind one another during meiosis as expected.
Fig. 4.
Interaction between 9-1-1 complex and Zip3 protein.Interaction between 9-1-1 complex and Zip3 protein. (A) Immunoprecipitation (IP) of Ddc1–Flag from mec3 (non-tag), DDC1-Flag, and RAD17-HA DDC1-Flag cells. Whole-cell extracts (WCE) and immunoprecipitation products were probed with anti-Mec3, anti-Flag and anti-HA antibodies. (B) Pulldown assay of FATT(-Myc)-Zip3. E. coli lysates expressing either FATT–Zip3 or FATT–GFP were incubated with magnetic beads coated with anti-c-Myc antibodies. The beads (Input) were incubated with yeast meiotic cell lysates (at 4 hours) from the wild-type, RAD17-HA DDC1-Flag and mec3 (non-tag) deletion cells. The eluates were analyzed by western blotting using anti-Mec3, anti-c-Myc, anti-Flag or anti-HA antibodies. Anti-Flag antibody can detect both Ddc1–Flag and FATT–Zip3–Flag. The asterisk indicates a non-specific band.
Given that Zip3 functions upstream of other ZMM proteins (Shinohara et al., 2008), we examined the interaction of Zip3 with the checkpoint clamp. However, Zip3 protein, like other ZMM proteins is insoluble in yeast cell lysates and when expressed in Escherichia coli (K.H. and M.S., unpublished results). To promote efficient solubilization of the Zip3 protein, we used the extremely acidic FATT(-Myc) epitope tag (Sangawa et al., 2013) and found that the FATT-tagged Zip3 protein was soluble in E. coli lysates. The FATT–Zip3 was bound to magnetic beads coated with anti-Myc antibodies and incubated with extracts of yeast meiotic cells expressing RAD17-HA DDC1-Flag. After extensive washing, the presence of proteins was verified by western blotting of fractions eluted from the beads. Mec3 protein was eluted from FATT–Zip3-coated beads but not from the control FATT–GFP-coated beads (Fig. 4B). The signal on western blots was absent from the eluates of protein-bound beads prepared using cell extracts from a mec3 deletion strain. This control demonstrated that the western blot signal was specific for Mec3. Moreover, the eluate contained Ddc1–Flag as well as Rad17–HA. Taken together, the results provide evidence for a physical interaction between Zip3 and the 9-1-1 clamp. It is unlikely that this interaction was mediated by DNAs in the lysate because we treated both of the E. coli and yeast lysates extensively with DNase I (see Materials and Methods). In addition, the interaction between Zip3 and 9-1-1 was not due to the presence of HA and Flag tags because Mec3 bound to FATT–Zip3 in wild-type cell extracts (supplementary material Fig. S3). The ability of Zip3 to bind clamp components supports a model in which direct protein–protein interaction contributes to 9-1-1-dependent assembly of ZMM proteins at sites of meiotic recombination.
The checkpoint clamp loader promotes interhomolog bias
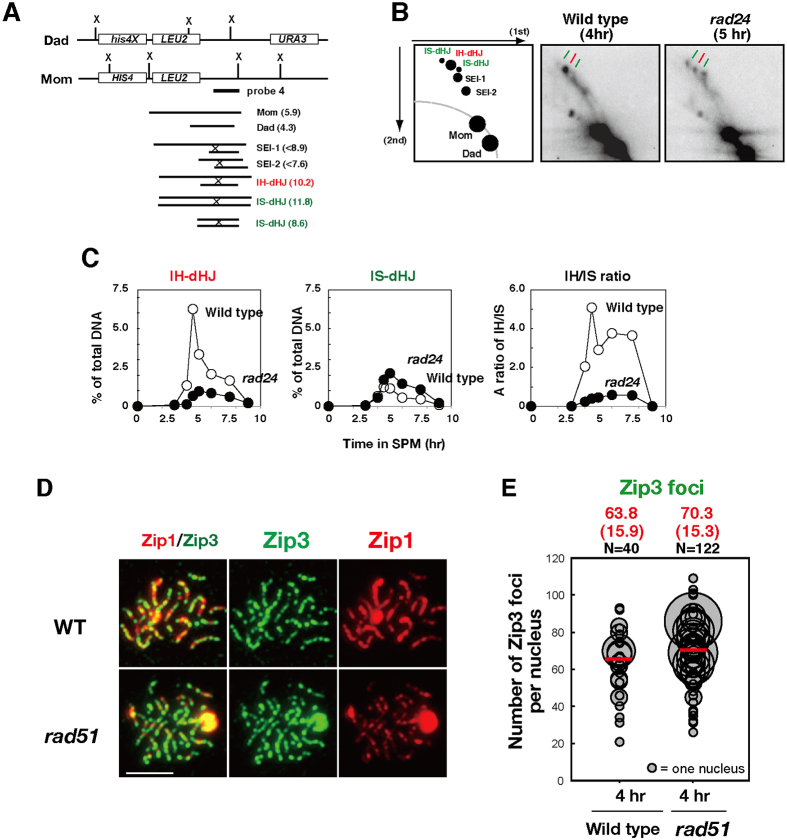
A previous study has analyzed the effect of the rad17 mutation on interhomolog bias for joint molecule formation in the background of the ndt80, which induces pachytene arrest (Ho and Burgess, 2011). We examined the effect of the clamp loader mutant rad24 on the relative abundance of interhomolog and intersister recombination intermediates at a recombination hotspot, HIS4-LEU2, using two-dimensional gel electrophoresis (Schwacha and Kleckner, 1994; Fig. 5A–C; supplementary material Fig. S4A,B). Meiotic time-course analysis of joint molecules showed that the rad24 mutant reduced steady-state levels of dHJs compared to the wild type (2.4-fold) and delayed turnover of the joint molecules. Interhomolog dHJs were decreased by 6.5-fold, whereas intersister dHJs were increased by 1.7-fold at 5 hours in the rad24 mutant relative to the wild type. These results confirmed and extended the conclusion that the 9-1-1 clamp and clamp loader influence Rad51–Dmc1-mediated partner choice.
Fig. 5.
Relationship of 9-1-1 clamp and clamp loader with Rad51 and/or Dmc1.Relationship of 9-1-1 clamp and clamp loader with Rad51 and/or Dmc1. (A) Schematic representation of the HIS4-LEU2 recombination hotspot for the analysis of recombination intermediates: single-end invasion (SEI) and interhomolog (IH; red) and intersister (IS; green) double-Holliday junctions (dHJs). Sizes of relevant joint molecule-containing fragments are indicated. (B) Southern blotting of two-dimensional gel electrophoresis for recombination intermediates. A schematic representation for a typical gel is shown on the left. DNAs from the wild type (4 hours) and the rad24 mutant (5 hours) were analyzed as described in the Materials and Methods. (C) Kinetics of IH-dHJ, IS-dHJ, and the ratio of IH to IS dHJs are shown. Wild type, open circles; the rad24 mutant, closed circles. (D) Localization of Zip3 (green), together with the synaptonemal complex component, Zip1 (red), was analyzed in wild-type and rad51 mutant cells by immunostaining. A representative image at 4 hours is shown. Scale bar: 4 µm. (E) Quantification of the numbers of Zip3 foci in the wild type and rad51 mutants. The numbers of Zip3 foci were analyzed as described in Fig. 1.
To elucidate the role of Rad51-dependent interhomolog bias in ZMM assembly, we analyzed Zip3 focus formation in the rad51 mutant. The rad51 mutant was proficient in the assembly of Zip3 foci on meiotic chromosomes (Fig. 5D). The average number of Zip3 foci in the rad51 mutant was 70.3±15.3 (mean±s.d., n = 122), which was slightly higher than that in the wild type (63.8±15.9; Fig. 5E). These results suggest that Rad51-dependent interhomolog bias is independent of the assembly of the Zip3 complex. We also analyzed the effect of the rad51 mutation on Zip3 focus formation in the absence of the RAD24 and found that an average Zip3 focus number in the rad24 rad51 mutant was 19.2±8.0 (n = 147) at 4 hours, which is indistinguishable from that in the rad24 mutant (supplementary material Fig. S1B). This suggests that Rad51 does not affect Zip3 assembly in the absence of the checkpoint clamp loader.
DISCUSSION
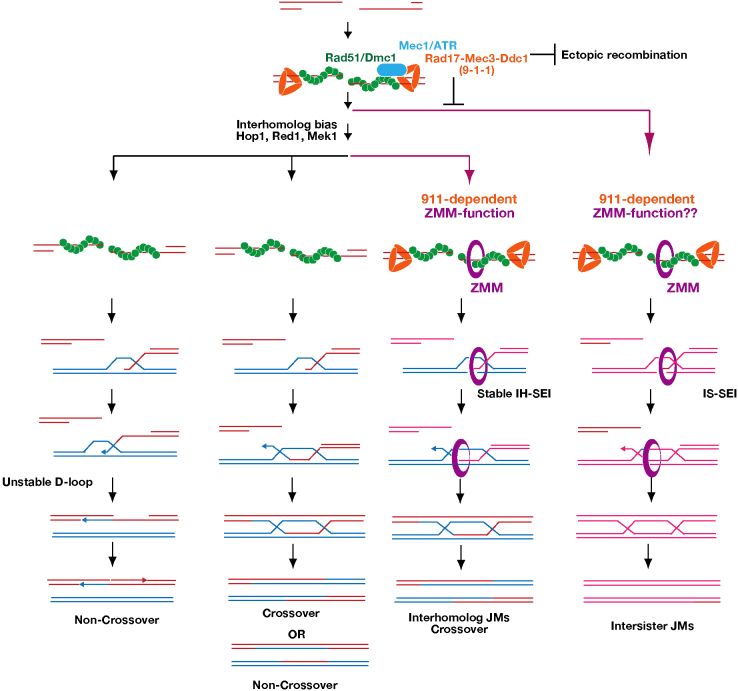
ZMM proteins play a key role in the formation of meiotic crossovers. However, the mechanism by which ZMM proteins are recruited to meiotic chromosomes and/or DSB sites remains to be elucidated. In this study, we have demonstrated that the DNA damage checkpoint clamp, 9-1-1, and its loader containing Rad24 play a role in crossover formation during meiosis by recruiting ZMM proteins. The results presented in this paper also extend data from previous studies (Ho and Burgess, 2011) by showing that the clamp is necessary for interhomolog bias during normal meiotic progression. Given that meiotic defects in the absence of the 9-1-1 clamp and its loader containing Rad24 are very similar, it is likely that meiotic role of Rad24 is to recruit the clamp to meiotic recombination intermediates. Alternatively, both the clamp and the loader might work together for these meiotic functions. Thus, the clamp plays two distinct roles in regulating the progress of meiotic recombination events, in addition to its role in DNA damage response signaling (Fig. 6).
Fig. 6.
A model of functions of the checkpoint clamp and clamp loader in crossover formation.A model of functions of the checkpoint clamp and clamp loader in crossover formation. Four possible pathways of meiotic recombination are shown. Rad51 and Dmc1 cooperate in pathways that display interhomolog bias (three leftmost pathways). Together with Rad51–Dmc1, the 9-1-1 clamp and Mec1 suppress intersister and ectopic recombination (right pathway). ZMM proteins specifically promote the ‘crossover-only’ pathway (third pathway from the left) by processing interhomolog joint molecules – this depends on Hop1–Red1–Mek1. The 9-1-1 clamp, but not Mec1, facilitates ZMM function on the crossover-only pathway. ZMM proteins might also promote intersister joint molecule formation. Non-crossover formation is independent of ZMM proteins (the leftmost pathway). Even in the absence of ZMM function, interhomolog joint molecules are formed (the second pathway from the left). In this pathway, joint molecules are resolved into either crossover or NCO.
The 9-1-1 clamp promotes ZMM loading
Given that the checkpoint proteins cooperate with Rad51–Dmc1 in crossover formation, impaired Rad51–Dmc1 function in the checkpoint mutant might underlie the defects observed in downstream events such as ZMM assembly. However, this mechanism is unlikely because, unlike clamp-defective mutants, both rad51 and dmc1 mutants are proficient in Zip3-focus formation (Fig. 5; Lao et al., 2013). These findings led us to the hypothesis that the 9-1-1 clamp, in addition to its role in homolog bias, is necessary for the efficient recruitment and/or stabilization of ZMM proteins along the developing synaptonemal complex. Consistent with this idea, we observed that the ZMM protein Zip3 bound to the clamp containing Mec3, Rad17 and Ddc1 in vitro (Fig. 4). A possible mechanism is that, during normal meiosis, the 9-1-1 complex recruits Zip3 to future crossover sites. However, the polycomplex localization results imply that there is additional complexity in the interactions between the 9-1-1 clamp and ZMMs. The localization pattern of Mec3 to Zip1 polycomplex was bipolar, a specific pattern observed for a subset of ZMM proteins, including Msh4, Msh5 and Spo22/Zip4, and was distinct from the pattern of Zip3-polycomplex colocalization (Tsubouchi et al., 2006). Furthermore, msh4 and msh5 mutants, but not spo22/zip4 or zip3 mutants, showed reduced localization of Mec3 to the Zip1 polycomplex. This indicates that Msh4–Msh5 is crucial for proper Mec3 localization on the polycomplex. These data suggest two possible modes of interaction between 9-1-1 and ZMM proteins. First, the DSB-bound 9-1-1 clamp might interact with Zip3 to stabilize ZMM proteins on meiotic chromosomes (Serrentino et al., 2013). Second, the checkpoint clamp might interact with Msh4–Msh5, a meiosis-specific MutS-γ complex, which can recognize DNA structures in meiotic recombination intermediates (Snowden et al., 2004) for the formation and/or regulation of crossover.
Most joint molecules in wild-type meiosis are formed through the ZMM-dependent pathways (Börner et al., 2004). The 9-1-1 clamp and clamp loader mutants reduced total joint molecule formation (this work; Ho and Burgess, 2011), indicating that the clamp is required to maintain normal levels of joint molecule formation. Given that mutations that disrupt the clamp function largely decreased interhomolog dHJs and slightly increased intersister dHJs, it appears that the function of the clamp is indispensible for ZMM-dependent interhomolog joint molecules rather than interhomolog bias (Fig. 6); this function can likely be explained by the role of the clamp in ZMM assembly.
Previous studies (Grushcow et al., 1999; Shinohara et al., 2003a) have shown a modest reduction of crossover at an artificial recombination hotspot, HIS4-LEU2, in the rad17 and rad24 mutants whereas the zmm mutants decreased crossovers to half of those in the wild type. The modest reduction of crossover in the checkpoint mutant can be explained by residual ZMM assembly (Fig. 1). Alternatively, this could be due to a locus-specific effect. Our recent genetic analysis showed the of the rad24 mutant decreased crossover frequencies by about half on three intervals on chromosome V (MS, unpublished result).
Mec1 is not important for ZMM assembly
The 9-1-1 clamp and clamp loader often function in concert with the effector kinase, Mec1, not only during mitosis but also in meiosis (MacQueen and Hochwagen, 2011); the mec1 and clamp mutants showed very similar defects in Hop1 phosphorylation, meiotic ectopic recombination, DSB control and recombination checkpoint control (Carballo et al., 2008; Gray et al., 2013; Grushcow et al., 1999; Lydall et al., 1996). By contrast, we found that the clamp and clamp loader mutants were phenotypically different from the mec1 mutant with respect to ZMM assembly. Although the number of Zip3 foci in the mec1 mutant was normal and similar to that of the wild type, clamp mutants showed a reduced number of Zip3 foci by about 2-fold. Thus, it appeared that one of the functions of the clamp is involvement in the formation of Zip3 foci in a Mec1-kinase-independent fashion. It should be noted, however, that yeast possesses Tel1 (the homolog of ATM), which displays partial redundancy with Mec1 in some assays (Carballo et al., 2008). Therefore, the proficiency of ZMM assembly in the mec1 mutant could be the result of redundant Tel1 activity. However, this is unlikely given that Tel1 is active in clamp mutants (Ho and Burgess, 2011), which nonetheless exhibit defective ZMM assembly. Indeed, recent analysis of the mec1 tel1 sml1 triple mutant showed the normal assembly of Zip3 in this mutant (M.S., unpublished results), supporting the ATR- and ATM-independent role of the 9-1-1 clamp and clamp loader in ZMM-dependent recombination.
As described above, interhomolog crossover formation requires the ZMM-dependent joint molecule that follows interhomolog bias (Fig. 6). The crossover defect in the mec1 mutant could be explained primarily by the defect in the bias. The same is true for the clamp and clamp loader mutants. However, the clamp and clamp loader mutants exhibit an additional defect in ZMM loading, whose effect on crossover formation might be hidden by the upstream defect in the interhomolog bias.
DNA damage response proteins versus Rad51–Dmc1 in interhomolog bias
Previous studies have demonstrated an intimate relationship between DNA damage response proteins and Rad51 in both mitotic DSB repair and meiotic recombination (Shinohara et al., 2003b). This relationship during recombination was suggested by phenotypic similarities between the rad51, dmc1 hed1 and the checkpoint mutants – reduced crossovers (decreased interhomolog joint molecules), decreased NCOs and increased ectopic recombination. In addition, we observed high colocalization frequencies of the 9-1-1 clamp components, for example, Mec3 with Rad51 and Dmc1 (Fig. 2D). Moreover, the kinetics of the appearance and disappearance of chromosomal foci of Mec3 and Rad51 were also very similar. Therefore, although the checkpoint clamp and Rad51 were recruited to single-stranded DSB ends independently of one another (Shinohara et al., 2003a), the two components function concurrently at the same sites to promote the progression of recombination intermediates during meiosis. In the absence of Rad51, intersister joint molecules were increased and interhomolog joint molecules decreased to a similar extent, suggesting a simple redistribution of joint molecule intermediates from homolog to sister (Hong et al., 2013; Lao et al., 2013; Schwacha and Kleckner, 1997). Taken together, these findings delineate a crucial role for Rad51–Dmc1 cooperation in interhomolog bias for meiotic recombination. However, Rad51–Dmc1 cooperation is not sufficient to ensure homolog bias; loading of the checkpoint clamp is also required. One key role played by 9-1-1 in partner choice likely involves the activation of the Hop1–Red1–Mek1 kinase pathway, given that the red1 and mek1 mutants have been previously shown to be defective in homolog bias (Goldfarb and Lichten, 2010; Kim et al., 2010; Schwacha and Kleckner, 1997) and that 9-1-1 signaling activates Mek1 (Carballo et al., 2008). This idea is supported by the observation that Rad17 and Ddc1 bind to Red1, an activator of Mek1 (Eichinger and Jentsch, 2010). Taken together, these results indicate that a key function of the clamp is to promote interhomolog bias through Mek1 activation and thereby regulation of Rad51–Dmc1.
The 9-1-1 clamp and clamp loaders are necessary for meiosis in other organisms. In fission yeast, which lacks ZMM proteins (and also a synaptonemal complex), the clamp and clamp loader are required for efficient intragenic recombination (Shimada et al., 2002). In mice, the clamp is localized to meiotic chromosomes as seen in the budding yeast (Freire et al., 1998; Lyndaker et al., 2013). In a conditional knockout mouse of one of 9-1-1 components, Hus1A, DSB repair is delayed with normal assembly of Rad51/Dmc1, but with abnormal synaptonemal complex formation (Lyndaker et al., 2013). These are reminiscent for the meiotic phenotypes of checkpoint mutants in budding yeast, suggesting some conservation of functions of 9-1-1 clamp and clamp loader during meiotic recombination. Further studies are necessary to reveal role of these protein complexes in crossover and synaptonemal complex formation in higher eukaryotes.
MATERIALS AND METHODS
Strains and plasmids
All strains described here are derivatives of SK1 diploids, NKY1551 and MSY831/832. MEC3-3HA, RAD17-3HA, and DDC1-3FLAG were constructed by a PCR-based tagging methodology (De Antoni and Gallwitz, 2000). Strain genotypes are given in supplementary material Table S1.
Anti-sera and antibodies
Anti-HA antibody (16B12; Covance), anti-Flag (M2, Sigma; anti-DYKDDDDK 1E6, Wako), anti-c-Myc (MC045; Nacalai Tesque), anti-GFP (8372-2; Clontech), rabbit anti-Dmc1 and guinea pig anti-Rad51 (Shinohara et al., 2000) antibodies were used for staining. Anti-Zip1, -Zip3, -Spo22/Zip4 and -Msh5 have been described previously (Shinohara et al., 2008; Zhu et al., 2010). Open-reading frames of Mec3 and Rad17 were PCR-amplified and inserted into pET21a plasmid (Novagen) resulting in the addition of hexahistidine to the C-terminus. Fusion proteins with hexahistidine were affinity-purified and used for immunization (MBL Co. Ltd). Secondary antibodies conjugated to Alexa Fluor 350, 488 and 594 dyes were used for immunostaining.
Cytology
Immunostaining of chromosome spreads was performed as described previously (Shinohara et al., 2000). Stained samples were observed using an epi-fluorescent microscope (AxioPlan; Zeiss) with a 100× objective (NA1.4). Images were captured by a CCD camera (Retiga; Qimaging) at room temperature, and then processed using IPLab (Sillicon) software. For foci counting, more than 100 nuclei were counted at each time point. Colocalization is defined as ≥50% overlap (Shinohara et al., 2000). The frequency of fortuitous colocalization was calculated by ‘Dotstat’ software (Gotta et al., 1996). In the case of fortuitous colocalization of Zip1 polycomplex with Msh5 foci, we performed manual rotation of 180° for fortuitous colocalization (Gasior et al., 1998). The length of Zip1 lines was measured using IPLab.
Analysis of meiotic recombination
Time-course analysis of events in meiosis and the cell cycle progression was performed as described previously (Shinohara et al., 1997). Two-dimensional gel analysis of branched recombination intermediates was also carried out following published methods (Lao et al., 2013).
Zip3 pulldown assay
A ZIP open reading frame was placed under the control of the T7 promoter of an expression vector, pFATT3 (Sangawa et al., 2013) by the SLIC method (Li and Elledge, 2007). FATT–Zip3-expressing E. coli cells were disrupted by sonication in lysis buffer (50 mM Hepes-KOH pH 7.5, 300 mM KCl, 20% v/v glycerol, 1 mM NaVO3, 60 mM β-glycerophosphate and 0.1% v/v NP-40) and cell lysates were applied to magnetic beads (GE Healthcare) coated with anti-c-Myc antibodies. Then, the beads were incubated with yeast extracts prepared in lysis buffer from cultures incubated in SPM medium for 4 hours with SPM medium. Yeast cell lysates were prepared as described previously (Sasanuma et al., 2013). Briefly, cells were collected and suspended in the buffer supplemented with protease inhibitor cocktail (Sigma). Cells were disrupted with glass beads (Yasui-kikai), and lysates were recovered and used for the pull-down assay. The lysates from E. coli and yeast cells were treated with DNase I (Takara, 25 units/ml for E. coli and 14 units/ml for yeast) for 0.5 hours at 37°C by adding MgCl2 to a final concentration of 5 mM. The beads were incubated with the yeast lysates for 1.5 hours at 4°C and then recovered using a magnetic stand. After extensive washing with the same buffer, bound fractions were eluted with Laemmli sample buffer.
Mec3 and Ddc1–Flag were detected using anti-Mec3 and anti-Flag antibodies, respectively. To detect Rad17–HA on membranes, we used Mouse TrueBlot(r), anti-Mouse IgG Biotin (Rockland), which was visualized with an Alexa-Fluor-680–streptoavidin conjugate.
Supplementary Material
Acknowledgements
We are grateful to Drs T. Sangawa and J. Takagi for pFATT3 and their kind guidance for the expression analysis. We also thank to Mr G. Tsuji for the construction of RAD17-3HA. We thank Drs N. Hunter, R. Weiss, and members of the Shinohara laboratory for helpful discussions and Ms A. Murakami and S. Umetani for excellent technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.S., K.H., D.K.B. and A. S. designed the experiments. M.S., K.H. and J.T.G. performed the experiments and analyzed the data. M.S., D.K.B. and A.S. prepared the manuscript.
Funding
This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture (to A.S., K.H. and M.S.); and by the National Institutes of Health (NIH) [grant number GM50936 to D.K.B.]. M.S. was supported by the Japan Society for the Promotion of Science (JSPS) through the ‘Funding Program for Next Generation World-Leading Researchers (NEXT Program). Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.161554/-/DC1
References
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/S0092--8674(00)00029--5. [DOI] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092--8674(90)90524--I. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/S0092--8674(01)00416--0. [DOI] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092--8674(94)90038--8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092--8674(92)90446--J. [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/S0092--8674(04)00292--2. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell. 1998;1:685–696. doi: 10.1016/S1097--2765(00)80068--4. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/S0092--8674(00)81164--2. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Gallwitz D. A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene. 2000;246:179–185. doi: 10.1016/S0378--1119(00)00083--4. [DOI] [PubMed] [Google Scholar]
- Eichinger CS, Jentsch S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc. Natl. Acad. Sci. USA. 2010;107:11370–11375. doi: 10.1073/pnas.1004248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire R, Murguía JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 1998;12:2560–2573. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Allison RM, Garcia V, Goldman AS, Neale MJ. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol. 2013;3:130019. doi: 10.1098/rsob.130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev--genet--051710--150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HC, Burgess SM. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 2011;7:e1002351. doi: 10.1371/journal.pgen.1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Roeder GS. A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 2002;16:363–376. doi: 10.1101/gad.938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. The logic and mechanism of homologous recombination partner choice. Mol. Cell. 2013;51:440–453. doi: 10.1016/j.molcel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/S0092--8674(01)00430--5. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/S0070--2153(01)52008--6. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/S0092--8674(00)81876--0. [DOI] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N, Bishop DK. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Lyndaker AM, Lim PX, Mleczko JM, Diggins CE, Holloway JK, Holmes RJ, Kan R, Schlafer DH, Freire R, Cohen PE. et al. Conditional inactivation of the DNA damage response gene Hus1 in mouse testis reveals separable roles for components of the RAD9-RAD1-HUS1 complex in meiotic chromosome maintenance. PLoS Genet. 2013;9:e1003320. doi: 10.1371/journal.pgen.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Hochwagen A. Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends Cell Biol. 2011;21:393–400. doi: 10.1016/j.tcb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Bressan DA, Shinohara M, Haber JE, Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sangawa T, Tabata S, Suzuki K, Saheki Y, Tanaka K, Takagi J. A multipurpose fusion tag derived from an unstructured and hyperacidic region of the amyloid precursor protein. Protein Sci. 2013;22:840–850. doi: 10.1002/pro.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A. et al. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092--8674(94)90172--4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092--8674(95)90191--4. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/S0092--8674(00)80378--5. [DOI] [PubMed] [Google Scholar]
- Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9:e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Nabeshima K, Tougan T, Nojima H. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 2002;21:2807–2818. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365--2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Ogawa T, Shinohara A. The mitotic DNA damage checkpoint proteins Rad17 and Rad24 are required for repair of double-strand breaks during meiosis in yeast. Genetics. 2003a;164:855–865. doi: 10.1093/genetics/164.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003b;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092--8674(93)90114--6. [DOI] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev. Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Mori S, Oshiumi H, Matsuzaki K, Shinohara M, Shinohara A. Cyclin-dependent kinase promotes formation of the synaptonemal complex in yeast meiosis. Genes Cells. 2010;15:1036–1050. doi: 10.1111/j.1365--2443.2010.01440.x. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.