Abstract
Background
Neuroblastoma is a neural crest-derived tumor and is the most common cancer in children less than 1 year of age. We hypothesized that aberrations in genes that control the cell cycle could play an important role in the pathogenesis of neuroblastoma and could provide a tractable therapeutic target.
Methods
In this study, we screened 131 genes involved in cell cycle regulation at different levels by analyzing the effect of siRNA-mediated gene silencing on the proliferation of neuroblastoma cells.
Results
Marked reductions in neuroblastoma cellular proliferation were recorded after knockdown of CCND1 or PLK1. We next showed that pharmacological inhibition of cyclin D1 dependent kinases 4/6 (CDK4/6) with PD 0332991 (palbociclib) reduced the growth of neuroblastoma cell lines, induced G1 cell cycle arrest, and inhibited the cyclin D1-Rb pathway.
Conclusion
Selective inhibition of CDK4/6 using palbociclib may provide a new therapeutic option for treating neuroblastoma.
Electronic supplementary material
The online version of this article (doi:10.1186/s12935-015-0224-y) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Neuroblastoma, Palbociclib, Cyclin D1, Targeted therapy
Background
Neuroblastoma is a solid childhood tumor that arises from the sympathoadrenal lineage of the neural crest during development [1]. It represents the most common cancer in children younger than 1 year [2] and is a heterogeneous disease that may either have a very good prognosis (e.g., stage 4S neuroblastoma) or a dismal outlook (e.g., MYCN-amplified neuroblastoma) [1]. Neural crest-derived malignancies such as neuroblastoma are believed to have defects in the regulatory circuits that are essential for embryonic development and regulation of cellular differentiation, in particular at the G1-S transition [1, 3]. The involvement of cell cycle regulators such as PLK1 [4], TRIM16 [5], WEE1 [6], CDK4/6 [7], and CCND1 [8] in various aspects of neuroblastoma pathogenesis suggests that deregulation of the normal cell cycle could be an important factor in driving neuroblastoma tumorigenesis and opens the possibility that more oncogenic cell cycle regulators could be identified by screening a large set of cell cycle regulators.
In this study, we used an siRNA library targeting 131 cell cycle regulators that belong to different gene families involved in cell cycle regulation. The siRNA library was used to assess the effect of knockdown of cell cycle regulators on the proliferation of neuroblastoma cells. This approach is expected to result in the identification of genes that are required for proliferation or survival of neuroblastoma cells, and therefore may serve as new therapeutic targets. Our screen showed that several cell cycle regulators are critical for neuroblastoma cellular proliferation. The strongest reductions in cellular proliferation were observed after knockdown of CCND1 or PLK1. To translate these findings into a pharmacologically useful approach, we explored the potential therapeutic utility of palbociclib, a small-molecule inhibitor of cyclin D1 associated kinases, mainly CDK4 and CDK6. We showed that palbociclib, which is widely used in clinical trials, inhibits the growth of neuroblastoma cells in vitro. Moreover, cell cycle analysis demonstrated that palbociclib induces cell cycle arrest exclusively in G1. In addition, we showed that inhibiting CDK4/6 by palbociclib suppresses the cyclin D1-pRb pathway by inhibiting the phosphorylation of Rb and the expression of E2F target genes. Our data suggest that the CDK4/CDK6 inhibitor palbociclib may offer therapeutic benefit for the treatment of neuroblastoma.
Results
Screening of neuroblastoma cellular proliferation using an siRNA library of 131 cell cycle regulators
We used an siRNA library to target 131 cell cycle regulators in two neuroblastoma cell lines (NGP and IMR-32) followed by monitoring of neuroblastoma cell growth in real time. The siRNA library included siRNAs that target genes which belong to several gene families involved in cell cycle regulation at different levels, such as cyclin dependent kinases (CDKs), members of the retinoblastoma protein family, DNA replication factors like the cell division cycle proteins (CDCs), members of the CIP/KIP family, and the INK4 family of cell cycle inhibitors. Our results showed that several genes of these families are required for the growth of both NGP and IMR-32 cells. In particular, silencing of CCND1 and PLK1 showed the most pronounced effects in reducing neuroblastoma cellular proliferation (Fig. 1; Additional file 1: Table S1).
Fig. 1.
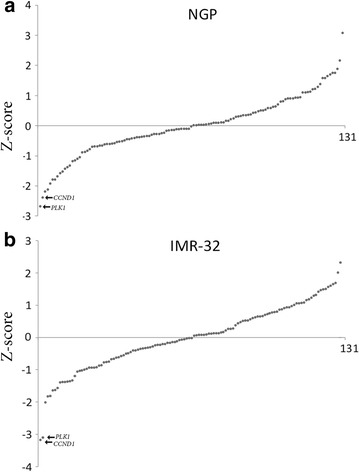
siRNA library screening of cell cycle regulators. NGP cells (a) and IMR-32 cells (b) were transfected with the siRNA library including negative controls (scrambled siRNA) and their growth was assessed using the xCELLigence system. The data shown in the graphs represent the cellular proliferation measurements recorded at 48 h post transfection. The data are depicted as Z score values (Additional file 1: Table S1), which represent the number of standard deviations from the average for every measurement. Positive Z score values indicate an increase in cell growth, whereas negative values represent a decrease in cell growth.
Knockdown of CCND1 in neuroblastoma cell lines
CCND1 and PLK1 were the primary hits of our siRNA library screen. As inhibition of PLK1 using small molecule compounds has already been extensively studied in neuroblastoma [4, 9], we decided to select CCND1 for further experiments. We first measured the mRNA expression levels of CCND1 in a panel of 31 neuroblastoma cell lines (Additional file 1: Figure S1) and evaluated the effects of CCND1 knockdown on the proliferation of eight neuroblastoma cell lines in real time using the xCELLigence system. The cell lines had various levels of CCND1 expression and were either MYCN amplified or MYCN nonamplified and either mutant or wild-type for TP53 (Additional file 1: Table S2). Our results showed that knockdown of CCND1 induces a reduction in cellular proliferation in the majority of the neuroblastoma cell lines in comparison to the scrambled siRNA negative control (Fig. 2). The real-time data is shown in Additional file 1: Figure S2.
Fig. 2.
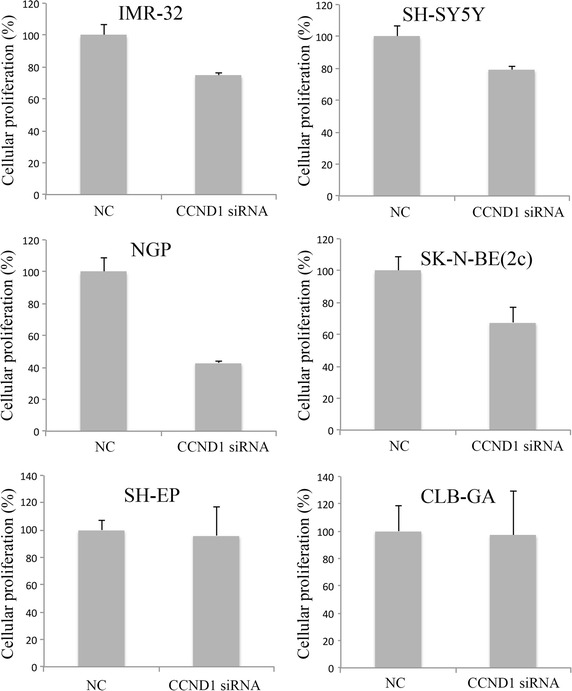
Assessment of cellular proliferation after knockdown of CCND1. The cellular proliferation was measured in real time using the xCELLigence system. The data shown represents the percentage of cellular proliferation after 72 h of knockdown of CCND1. NC scrambled siRNA negative control. Error bars represent the standard deviation (n = 2).
Targeting the cyclin D1-CDK4/CDK6 complex with palbociclib
Targeting cyclin D1 with small molecule compounds is currently not possible. However, an alternative approach to inhibit its activity is by inhibiting the associated kinases, CDK4 and CDK6. Indeed, a small molecule compound known as PD 0332991 or palbociclib has been reported to be a potent and highly selective inhibitor of CDK4 and CDK6 [10]. We treated neuroblastoma cells with this compound and evaluated the effect on cellular proliferation using xCELLigence. Our results showed that palbociclib inhibits the growth of IMR-32, SH-SY5Y, and NGP cells in a time- and dose-dependent manner. In contrast, other neuroblastoma cell lines such as SK-N-SH and CLB-GA were relatively resistant to the treatment. Other cell lines such as SH-EP responded at relatively high concentrations (Fig. 3). IC50 values are shown in Table 1 and the real-time data is shown in Additional file 1: Figure S3. The effect of palbociclib on the proliferation of neuroblastoma cells is very similar to the effect induced by the knockdown of CCND1, which suggests that both treatments induce their effects by targeting the same pathway.
Fig. 3.
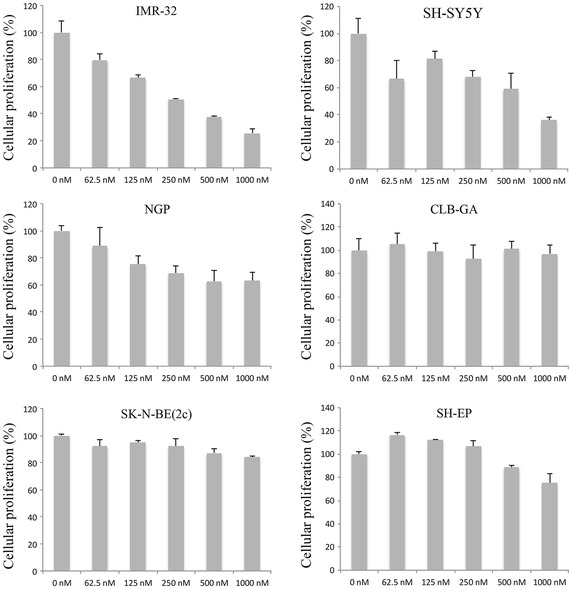
Assessment of cellular proliferation after treatment with palbociclib. The cellular proliferation data was measured in real time using the xCELLigence system. The data shown represents the percentage of cellular proliferation 48 h after treatment with palbociclib. Error bars represent the standard deviation (n = 2).
Table 1.
IC50 values of neuroblastoma cell lines treated with palbociclib
| Cell line | IC50 (nM) |
|---|---|
| IMR-32 | 261 |
| SH-SY5Y | 676 |
| NGP | 2077 |
| SH-EP | 2211 |
| SK-N-SH | 7,482 × 105 |
| NLF | 1,109 × 104 |
| CLB-GA | 2,579 × 105 |
| SK-N-BE(2c) | 1,285 × 104 |
Palbociclib induces G1 arrest in neuroblastoma cells
We evaluated the effect of palbociclib on the progression of the cell cycle in the neuroblastoma cell lines. The cells were exposed to vehicle control (0 µM) or 1 µM of palbociclib for 24 h. Cell cycle analysis showed that palbociclib induces an increase in the percentage of cells in G1 and a decrease in the other phases of the cell cycle in the neuroblastoma cell lines (Fig. 4). This induction of a G1 cell cycle arrest is consistent with what is expected after selective inhibition of CDK4 and CDK6 [10].
Fig. 4.
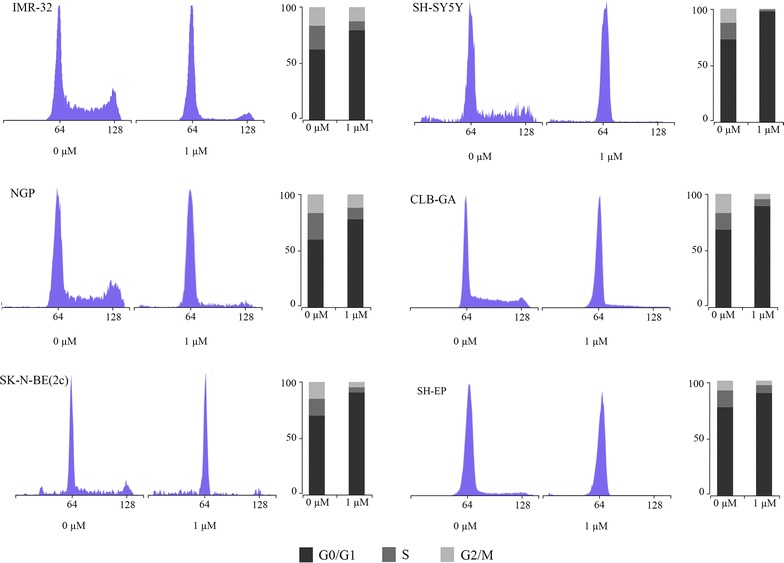
Cell cycle analysis after treatment with palbociclib. DNA histograms generated by flow cytometry for neuroblastoma cell lines treated with palbociclib for 24 h. The DNA histograms show the distribution of cell populations in each phase of the cell cycle. The percentages of cells are shown to the right of each histogram.
Palbociclib inhibits Rb phosphorylation and the Rb/E2F pathway in neuroblastoma cells
The cyclin D1-CDK4/6 complex specifically phosphorylates Rb at Ser780 [11]. Reduction in Rb phosphorylation at Ser780 is indicative of CDK4 and CDK6 inhibition. Upon treatment of neuroblastoma cells with 0, 250, 500, or 1,000 nM of palbociclib, we evaluated the degree of Rb phosphorylation by Western blot analysis. Treatment with palbociclib inhibited the phosphorylation of Rb in a dose-dependent manner only in the cells that showed a decrease in cellular proliferation after palbociclib treatment (Fig. 5a). Phosphorylation of Rb by the cyclin D1-CDK4/6 complex releases E2F from the inhibitory grip of Rb [12]. Therefore, hypophosphorylation of Rb due to inhibition of CDK4/6 should lead to the inhibition of E2F activity. We evaluated the effect of palbociclib on the Rb/E2F pathway in neuroblastoma cells by measuring the mRNA expression levels of direct E2F target genes, topoisomerase 2A (TOP2A), cyclin E2 (CCNE2), and thymidine kinase (TK1). IMR-32 and NGP cells were treated with 0, 250, 500, or 1,000 nM of palbociclib for 24 h. Treatment with palbociclib reduced the expression of TOP2A, CCNE2, and TK1 in a dose-dependent manner, as expected (Fig. 5b).
Fig. 5.

Palbociclib inhibits Rb Ser780 phosphorylation and reduces the expression of E2F target genes. Neuroblastoma cell lines were treated with different concentrations of palbociclib for 24 h. Western blot analysis was performed using anti-Rb Ser780 to detect specific phosphorylation of Rb at Ser780. Anti-Rb was used to detect total Rb, and anti-β actin as a loading control (a). RT-qPCR data of NGP and IMR-32 cells showing the expression levels of E2F target genes after 24 h of treatment with palbociclib. Error bars represent the standard error of the mean (n = 2) (b).
Discussion
Several cell cycle regulators, such as PLK1 [4], TRIM16 [5], WEE1 [2, 6], CDK4/6 [1, 7], and CCND1 [1, 3, 8], have been shown to be involved in the tumorigenesis of neuroblastoma. This suggests that neuroblastoma tumors might be addicted to individual activated oncogenes in the process of cell cycle regulation and that additional oncogenic cell cycle regulators could play a role in neuroblastoma tumorigenesis. We screened two neuroblastoma cell lines using a commercially available arrayed siRNA library representing 131 cell cycle regulators from different gene families to identify genes that are required for the proliferation or survival of neuroblastoma cells and that might serve as new therapeutic targets. The gene families included cyclin dependent kinases (CDKs), members of the retinoblastoma protein family, DNA replication factors such as the cell division cycle proteins (CDCs), members of the CIP/KIP family, and the INK4 family of cell cycle inhibitors. Real-time monitoring of cell growth showed that knockdown of CCND1 and PLK1 had the strongest effects in reducing the proliferation of neuroblastoma cells. PLK1 has previously been extensively studied in neuroblastoma and has been shown to inhibit the transactivation activity of p53 and to promote cell survival [4, 13, 14]. Moreover, targeting PLK1 with small molecule compounds induced apoptosis and growth arrest in neuroblastoma tumor-initiating cells [5, 9] and reduced the growth of neuroblastoma xenografts in nude mice [4, 6]. Cyclin D1 has also been studied to some extent in neuroblastoma and has previously been shown to be highly expressed in a subset of neuroblastoma cell lines and tumors due to gene amplification or GATA3 binding [7, 8, 15, 16]. Cyclin D1 belongs to a group of proteins known as cyclins, which are involved in the temporal coordination of each mitotic event during the cell cycle. Cyclins exhibit a cell-cycle dependent pattern of expression and degradation, and they activate the CDKs. Cyclin-CDK complexes phosphorylate target proteins such as Rb and coordinate the progression into the next phase of the cell cycle [8, 12]. Cyclin D1 activates CDK4 and CDK6 and this initiates the phosphorylation of the tumor suppressor protein Rb resulting in the activation of E2F. The activity of E2F leads to the transcriptional activation of E2F-responsive genes, such as TOP2A, CCNE2, and TK1, that are essential for DNA synthesis and cell cycle progression [4, 9, 10, 17]. Despite being an attractive therapeutic target, small molecule compounds that can directly and specifically inhibit cyclin D1 do not exist at present. It is possible, however, to inhibit cyclin D1 indirectly by several means. For example, inhibition of CCND1 can be achieved by mTOR inhibitors, since CCND1 mRNA translation is mTOR-dependent [10, 18], or by inhibitors of glycogen synthase kinase 3 β (GSK3β), which phosphorylates cyclin D1 at Thr-286 suggested to regulate the turnover and the intracellular distribution of cyclin D1 [10, 19]. The most effective approach to inhibit cyclin D1 activity is by inhibiting its associated kinases CDK4 and CDK6 [11, 12]. Pan-CDK inhibitors can inhibit CDKs but induce also considerable toxicity due to off-target effects [12, 20]. In contrast, early phase clinical trials have shown that specific and potent inhibition of CDK4/6 by small molecule drugs, such as palbociclib [4, 21–24] and R547 [5, 25], can result in antitumor activity with tolerable side effects. Recently, it has been shown that inhibition of CDK4/6 by LEE011 reduces the growth of neuroblastoma tumors with MYCN amplification in murine xenograft models [7]. In this study, we used palbociclib, a selective CDK4/6 inhibitor that has already shown promising antitumor activity in several clinical trials in other cancer types [12]. Treatment of neuroblastoma cells in vitro with palbociclib resulted in variable effects: some cell lines, such as IMR-32, SH-SY5Y, and NGP, were sensitive, whereas other cell lines, such as SK-N-SH and CLB-GA, were relatively resistant. It is worth mentioning that NGP cells have CDK4 amplification. Additional studies are needed to explain the different sensitivity to this compound.
Conclusions
We show that palbociclib induces a G1 cell cycle arrest in neuroblastoma cells, which is in line with previous reports in other cancer types [10]. Palbociclib also induces hypophosphorylation of Rb at Ser780, indicative of CDK4/6 inhibition, in those neuroblastoma cell lines that show a reduction in cellular proliferation after treatment. Upon hypophosphorylation, Rb forms a complex with E2F and we show that treatment with palbociclib reduces the expression levels of E2F target genes involved in cell cycle progression, such as TOP2A, CCNE2, and TK1. Taken together, our results suggest that inhibition of CDK4/6 may provide an effective strategy to treat a subset of neuroblastoma tumors.
Methods
Cell lines and palbociclib treatment
Neuroblastoma cell lines were grown as monolayer cultures at 37°C and 5% CO2 in a humid atmosphere. The culture medium was RPMI 1640 (GIBCO, Life Technologies) containing 10% Fetal Calf Serum (FCS), 2 mmol/l glutamine, and the following antibiotics: Penicillin (1%), Kanamycin (1%), and Streptomycin (1%). Palbociclib (Selleck Chemicals) was dissolved in DMSO and stored as a 10 mmol/l stock solution at −20°C. Keeping the final concentration of DMSO constant, cells were treated with palbociclib ranging from 0 to 2 µmol/l for the time periods indicated.
siRNA transfection
Neuroblastoma cells were seeded in RPMI 1640 (GIBCO, Life Technologies) with 10% FCS and without antibiotics and transfected with the siRNA library against 131 cell cycle regulators (Dharmacon Cell Cycle ON-TARGETplus, Thermo Scientific) at a concentration of 100 nM using Dharmafect 2 transfection reagent (Thermo Scientific). The genes targeted by the siRNA library are listed in Additional file 1: Table S3. Transfections with siRNA against CCND1 and scrambled negative control siRNA were done using smart pools of siRNAs (Dharmacon, Thermo Scientific).
Cell growth assessment
xCELLigence MP (Roche Diagnostics) was used to monitor cell proliferation in real time. Background impedance was measured before seeding the cells using 40 µl of RPMI 1640 containing 10% FCS and always subtracted as blank value. 1 × 104 cells in 50 µl of RPMI containing 10% FCS were added. Cell proliferation was measured with a programmed signal detection every 1 h and the signal was normalized to the transfection or palbociclib treatment time point where the cell index at every time point was divided by the cell index at the time of transfection. Data acquisition and analysis was performed with the RTCA software (version 1.2, Roche Diagnostics). IC50 values were defined as the drug concentration that inhibited cell proliferation by 50% at 48 h of treatment and were calculated using GraphPad Prism (version 6).
RNA extraction and RT-qPCR (reverse transcription-quantitative polymerase chain reaction)
Analysis of gene expression was performed by RT-qPCR according to MIQE guidelines [26]. RNA extraction, cDNA synthesis and RT-qPCR were performed as described previously [27]. The primers were designed using an in-house developed web tool (http://www.primerXL.org) and the sequences can be found in Additional file 1: Table S4. HMBS, HPRT1, SDHA, TBP, and YWHAZ were used for normalization. Biogazelle’s qbase + qPCR data-analysis software version 3.0 [28] was used to quantify the relative expression of the genes.
Western blot
Neuroblastoma cells treated with palbociclib (Selleck Chemicals) for 24 h were harvested and washed using ice-cold PBS, centrifuged and the supernatant discarded. The pellet was solubilized in RIPA lysis buffer (Pierce) containing protease and phosphatase inhibitor mixture (Roche). Cell lysates were placed on ice for 30 min and centrifuged for 10 min at 12,000 rpm at 4°C. The protein concentration was measured using the Bio-Rad Protein Assay (Bio-Rad). Protein samples were mixed at 1:1 ratio with Laemmli denaturation buffer (Bio-Rad) and β-mercaptoethanol (Sigma Aldrich) at a final dilution of 1/40 and boiled for 10 min at 95°C. Approximately 25 µg of protein was loaded and fractionated using a 10% SDS-PAGE gel (Bio-Rad). The protein was transferred onto a nitrocellulose membrane (Bio-Rad) and immunoblotted with rabbit monoclonal antibody against phospho-Rb (Ser780) (Cell Signaling Technology), rabbit polyclonal antibody against total Rb (Abcam), and mouse monoclonal antibody against β-actin (Sigma Aldrich) as a loading control. Secondary antibody was anti-mouse HRP-linked (Cell Signaling Technology) and anti-rabbit HRP-linked (Cell Signaling Technology). Visualization of the proteins was done using ChemiDoc-It2 Imaging System (UVP, Sopachem).
Cell cycle analysis
Neuroblastoma cells were treated with palbociclib for 24 h, harvested, washed with PBS and resuspended in 100 µl of PBS. 100 µl of DNA Prep lysis buffer (Beckman Coulter) was added to the cells and the suspension was vortexed and incubated for 5 min. The DNA was stained using 2 ml DNA Prep stain (Propidium iodide) and the suspension was placed in the dark for 30 min at room temperature. Cellular DNA content was analyzed using the S3 cell sorter (Bio-Rad).
Authors’ contributions
AR participated in the intellectual design of the study, carried out the transfection of neuroblastoma cell lines with the siRNA library, treated the cells with palbociclib performed the viability experiments, gene expression and western blot, and drafted the manuscript. FSP and JV participated in the intellectual design of the study. TVM participated in the intellectual design of the study, critically revised the manuscript and gave the final approval for submission. All authors read and approved the final manuscript.
Acknowledgements
We thank Nurten Yigit for technical assistance. AR is supported by a PhD fellowship from the Ghent University research fund (BOF; 01D02210). TVM is a senior clinical investigator of the Research Foundation—Flanders (FWO; 1803115N) and a recipient of a research grant of the FWO (1510813N). This work was supported by the BOF, the FWO (G022509N), the Concerted Research Actions of Ghent University (GOA), the Childhood Cancer Fund, and the National Cancer Plan of the Belgian State (Action 29).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interest.
Ethics The manuscript does not contain any human data or samples. All the experiments were performed on neuroblastoma cell lines.
Abbreviations
- CDK4/6
cyclin D1 dependent kinases 4/6
- CDCs
cell division cycle proteins
- TOP2A
topoisomerase 2A
- CCNE2
cyclin E2
- TK1
thymidine kinase
- GSK3β
glycogen synthase kinase 3 β
- FCS
fetal calf serum
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
Additional file
Real-time viability data, expression data and primers sequences.
Contributor Information
Ali Rihani, Email: Rihani_Ali@hotmail.com.
Jo Vandesompele, Email: Joke.Vandesompele@UGent.be.
Frank Speleman, Email: Franki.Speleman@UGent.be.
Tom Van Maerken, Email: Tom.VanMaerken@UGent.be.
References
- 1.Cheung NKV, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simões-Costa M, Bronner ME. Insights into neural crest development and evolution from genomic analysis. Genome Res. 2013;23:1069–1080. doi: 10.1101/gr.157586.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackermann S, Goeser F, Schulte JH, Schramm A, Ehemann V, Hero B, et al. Polo-like kinase 1 is a therapeutic target in high-risk neuroblastoma. Clin Cancer Res. 2011;17:731–741. doi: 10.1158/1078-0432.CCR-10-1129. [DOI] [PubMed] [Google Scholar]
- 5.Bell JL, Malyukova A, Kavallaris M, Marshall GM, Cheung BB. TRIM16 inhibits neuroblastoma cell proliferation through cell cycle regulation and dynamic nuclear localization. Cell Cycle. 2013;12:889–898. doi: 10.4161/cc.23825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creevey L, Ryan J, Harvey H, Bray IM, Meehan M, Khan AR, et al. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol Cancer. 2013;12:23. doi: 10.1186/1476-4598-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rader J, Russell MR, Hart LS, Nakazawa MS, Belcastro LT, Martinez D, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molenaar JJ, van Sluis P, Boon K, Versteeg R, Caron HN. Rearrangements and increased expression of cyclin D1 (CCND1) in neuroblastoma. Genes Chromosom Cancer. 2003;36:242–249. doi: 10.1002/gcc.10166. [DOI] [PubMed] [Google Scholar]
- 9.Grinshtein N, Datti A, Fujitani M, Uehling D, Prakesch M, Isaac M, et al. Small molecule kinase inhibitor screen identifies polo-like kinase 1 as a target for neuroblastoma tumor-initiating cells. Cancer Res. 2011;71:1385–1395. doi: 10.1158/0008-5472.CAN-10-2484. [DOI] [PubMed] [Google Scholar]
- 10.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 11.Connell-Crowley L, Harper JW, Goodrich DW. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 13.Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- 14.Ismail IA, Kang K-S, Lee HA, Kim J-W, Sohn Y-K. Genistein-induced neuronal apoptosis and G2/M cell cycle arrest is associated with MDC1 up-regulation and PLK1 down-regulation. Eur J Pharmacol. 2007;575:12–20. doi: 10.1016/j.ejphar.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Molenaar JJ, Ebus ME, Koster J, van Sluis P, van Noesel CJM, Versteeg R, et al. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008;68:2599–2609. doi: 10.1158/0008-5472.CAN-07-5032. [DOI] [PubMed] [Google Scholar]
- 16.Molenaar JJ, Ebus ME, Koster J, Santo E, Geerts D, Versteeg R, et al. Cyclin D1 is a direct transcriptional target of GATA3 in neuroblastoma tumor cells. Oncogene. 2010;29:2739–2745. doi: 10.1038/onc.2010.21. [DOI] [PubMed] [Google Scholar]
- 17.Grant GD, Brooks LI, Zhang X, Mahoney JM, Martyanov V, Wood TA, et al. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell. 2013;24:3634–3650. doi: 10.1091/mbc.E13-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musgrove EA. Cyclins: roles in mitogenic signaling and oncogenic transformation. Growth Factors. 2006;24:13–19. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- 19.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 21.O’Dwyer PJ, LoRusso P, DeMichele A, Gupta V. A phase I dose escalation trial of a daily oral CDK 4/6 inhibitor PD-0332991. J Clin Oncol. 2007;25:3550. [Google Scholar]
- 22.Niesvizky R, Lentzsch S, Badros AZ, Chanan-Khan AA (2010) A phase I study of PD 0332991: complete CDK4/6 inhibition and tumor response in sequential combination with bortezomib and dexamethasone for relapsed and refractory multiple myeloma. https://ash.confex.com/ash/2010/webprogram/Paper26540.html
- 23.Flaherty KT, LoRusso PM, DeMichele A (2012) Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin Cancer [DOI] [PubMed]
- 24.Schwartz GK, LoRusso PM, Dickson MA, Randolph SS, Shaik MN, Wilner KD, et al. Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1) Br J Cancer. 2011;104:1862–1868. doi: 10.1038/bjc.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diab S, Eckhardt S, Tan A, Frenette G, Gore L. A phase I study of R547, a novel, selective inhibitor of cell cycle and transcriptional cyclin dependent kinases (CDKs)—Diab et al. 25 (Suppl 18):3528—ASCO Meeting Abstracts. J Clin Oncol. 2007;25:3528. [Google Scholar]
- 26.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 27.Rihani A, Van Maerken T, Pattyn F, Van Peer G, Beckers A, De Brouwer S, et al. Effective Alu repeat based RT-Qpcr normalization in cancer cell perturbation experiments. PLoS One. 2013;8:e71776. doi: 10.1371/journal.pone.0071776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]


