Abstract
We studied the use of peak deviations for application in phosphoproteomics. Due to the differences in the mass defects, the peak deviations of samples containing mixtures of phosphorylated and nonphosphorylated peptides show bimodal distributions. The ratios of peak heights accurately predict the phosphoproteome content of a sample. In this work we apply a signal-processing tool, singular value decomposition (SVD), to reveal characteristic features of the phosphorylated, nonphosphorylated and mixed samples. We show that a simple application of SVD to the peak deviation (PD) matrix 1) detects transitions from mostly phosphorylated samples to mostly nonphosphorylated samples, 2) reveals modes of low-abundance species in the presence of the high-abundance species (e.g., phosphorylated peptides), and 3) simplifies the interpretation of the clustering of a covariance matrix obtained from PDs.
As the eigenfunctions of the inner-product of the data matrix (made from the PDs) are Hermite functions, we observe a change of sign in the transition from samples enriched in phosphorylated peptides to samples containing fewer phosphorylated peptides. The ordering of the singular values of the data matrix points in the direction of changes to the phosphorylation content. No peptide identifications from a database were used for this study.
Keywords: peak deviation, mass defect, singular value decomposition, phosphopeptides, phosphopeptide enrichment
1 Introduction
Mass distributions of peptides are structured with well-defined peaks and troughs, sometimes referred to as quiet zones (very few peptide populations) or forbidden zones (no peptide populations). The distinct distributions of peptide masses are due to the discrete nature of the masses of the amino acids that make up the peptides. The peak widths vary, but normally are within a few tenths of a Dalton (for peptides limited to 3.5 kDa in mass), which makes them detectable by high-resolution/ high-accuracy mass spectrometers [1]. Chemical modifications of amino acids, including post-translational modifications, e.g. phosphorylations or glycosylations, change the peak distributions. [2-4] Most notably, the modifications shift the peak centers. This shifting stems from the distinct differences between the mass defects (MDs) of the amino acids and those of the chemical modifications. Several studies have looked into using structured MDs in the context of various practical proteomic workflows – noise filtering, differentiating between modified and unmodified peptides, improving peptide identifications, etc. [5;6]. The most interest has stemmed from the potential use of MDs in workflows for phosphopeptide identification, with the goal of improving phosphopeptide analyses [3, 7]. In our most recent study [8], we introduced a new concept, peak deviation (PD), which is the difference between a precursor mass and a dynamic peak center mass. We have shown that PDs model data-dependent acquisition with a well-defined, unimodal distribution. This is contrasted with the traditional concept, MD. In data-dependent acquisition, the MD exhibits a bimodal distribution, which was explained by uncertainty stemming from undetermined sequence information. Peak deviation distributions are constructed solely on the bases of precursor monoisotopic masses; no amino acid sequence or composition information of the precursor peptide sequence is used. We have also found that unlike the MDs, PDs can be uniquely separated into those of phosphorylated and unmodified peptides. Further studies are needed to determine effective ways of incorporating PD distributions in proteomic workflows to improve quality of peptide spectrum matching. In this work, we examined the singular value decompositions (SVDs) of PDs computed from monoisotopic masses of experimental precursors in large-scale proteomic experiments. The analyses were applied to freely available datasets [9] that have a mixture of phosphorylated and nonphosphorylated peptides.
2 Materials and Methods
We used a freely available murine brain phosphoproteomics dataset [9] for our study. From this dataset we used all 10 strong cation exchange (SCX) fractions that were analyzed using collision-induced dissociations (CID). These data were acquired on an LTQ-Orbitrap Velos mass spectrometer. The dataset was enriched for phosphopeptides, and was downloaded from the Tranche data repository [10]. We used the msconvert tool of ProteoWizard [11] to convert the raw data into the mzML file format. Precursor mass-to-charge ratios and charge states were extracted from mzML files using in-house developed software [12]. No database searches to identify peptides were done in this study. We used only monoisotopic precursor masses. Below we will show how the experimental monoisotopic masses, and the SVDs of their peak deviations, can be combined to estimate phosphoproteome content in order to detect its dynamics along the fractionation steps, and to uncover modes of distribution by noise removal.
Of importance for this work is our newly introduced concept of the peak deviation [8]. We have defined the PD as the monoisotopic mass of the experimental precursor minus the mass of the nearest peak center of the theoretical peptides’ mass distributions. The range of the PD is between ∼-0.5 and 0.5 Da. Theoretical peptide distributions were computed from amino acid compositions, as previously described [1, 13]. We have shown that unlike the traditionally used concept of the mass defect, PDs from a single experiment form a well-defined, unimodal distribution, which is also characteristic of the modification state of peptides in a phosphoproteome sample [8]. Here, we have applied the signal-processing technique SVD to detect differences between phosphorylated and nonphosphorylated peptides based on their PDs. For the SVD analyses (described below), we combine the PDs from all 10 fractions into a single matrix, A, which has 10 columns (corresponding to the number of fractions/experiments) and 1000 rows (the PDs for each experiment). The number of rows is determined by the bin size, 0.001 Da, that is used to create PDs. The accuracy of PDs is dependent on the accuracy of the measurements of the precursor masses and positions of the peaks of the theoretical peptides. The standard deviation of the precursor mass measurements in these experiments was about 5 ppm. The theoretical peptide distribution was computed using the masses of all theoretically possible tryptic peptides [14]. Since the distribution was obtained from a complete sampling of all theoretically possible peptides (limited only by the peptide mass of 3.5 kDa), the peak center positions were assumed to be deterministic values with no associated error. Thus the standard deviation of PDs is about 5 ppm. The peak deviation modes of the phosphorylated and nonphosphorylated peptides are separated by more than 0.15 Da [8]. The bin size of 0.001 Da used for the PDs in this work is better than the standard deviation of the experimental measurements (5 ppm), and they are both substantially smaller than the distance between the masses of the modes corresponding to phosphorylated and nonphosphorylated peptide distributions (which makes it possible to extract major differences between the distributions). The rows of A are the “variables” (peak deviations for each bin), and its columns are the instances of these variables (as measured in each LC-MS run from the monoisotopic masses of the experimental precursors). Every column of A is essentially a histogram of PDs obtained for the monoisotopic masses of all precursors fragmented in the experiment. We used PD data from all experimental precursors, including singly and multiply phosphorylated peptides.
Singular Value Decomposition
Any non-zero, rectangular matrix A with m rows and n columns, A ∈ Rm × n, and rank r, can be factored into a product of three matrices:
where U is a matrix, U∈ Rm × r, whose columns consist of r orthonormal vectors, , in the column space of A, ui ∈ Rm; V is a matrix consisting of r orthonormal vectors, , in the row space of A, vi ∈ Rn; and Σ is the diagonal matrix, Σ ∈ Rr × r. The positive diagonal elements of Σ are called singular values, and are the square roots of the non-zero eigenvalues of both the outer product (ATA) and inner product (AAT) of matrix. The columns of the U and V matrices are eigenvectors, corresponding to the non-zero eigenvalues of ATA and AAT, respectively. There are other forms of SVD, but for our purposes the above form, termed compact SVD, is satisfactory. We will use the decomposition of the PD matrix into the sum of dyadic products, Σiiui ⊗vi,:
where each term is an outer product of ui and vi, weighed by the corresponding singular value, Σii As seen from this decomposition, the left and right singular vectors corresponding to large singular values capture the most important features of matrix. This property of the SVD will be used in our analysis of the PDs and phosphoproteome content changes along the fractionation steps. It is relevant to note here that a similar strategy for decomposing data matrix A into the sum of dyads has previously been used for differentiating letters in handwriting [15]. It has been determined that the first dyads closely represent the major features of a number as inferred from a set of handwriting samples.
Hermite Functions
A Hermite function of order n, hn(√kx), is defined as the product of an exponential function with a Hermite polynomial of order n, Hn(√kx):
where k is a Hooke's constant, and x is a variable, which in this study is the PD. The order of the Hermite function n takes on non-negative integer values, n=0, 1, 2, … Hermite polynomials are symmetric (with respect to the variable x) for the even- numbered orders, and antisymmetric for the-odd numbered orders. Since the other term in the Hermite functions, the Gaussian, is a symmetric function with respect to the variable x, the Hermite functions are either symmetric (for even orders) or antisymmetric (for odd orders). This property of the Hermite functions will be exploited below in the discussions on clustering phosphorylated and nonphosphorylated peptides. Note that there is an n versus (n+1) relationship between the Hermite functions and the corresponding singular values. Thus, the zeroth order Hermite function corresponds to the first singular value, and the first-order Hermite function corresponds to the second singular value, etc.
Our approach in this work was inspired by the recent applications of SVD to improve clustering, [16] to reduce data dimensionality [17], and to model transcript length distribution functions from DNA microarray data [18]. SVD analysis is an efficient choice for an initial state in k-means clustering. In the representation given by SVD, the clustered structure of the data appears naturally and leads to simplifications in the interpretation of clusters [16]. SVD has also been used for missing data imputation from DNA microarrays [19] and gene expression profiles [20]. When the number of missing values is relatively low, the results of SVD will change the values of only the smallest singular value. The average row method can be used, which is sufficiently precise in most cases [19].
3 Results
SVD dyads corresponding to the second singular value differentiate between phosphorylated and nonphosphorylated samples
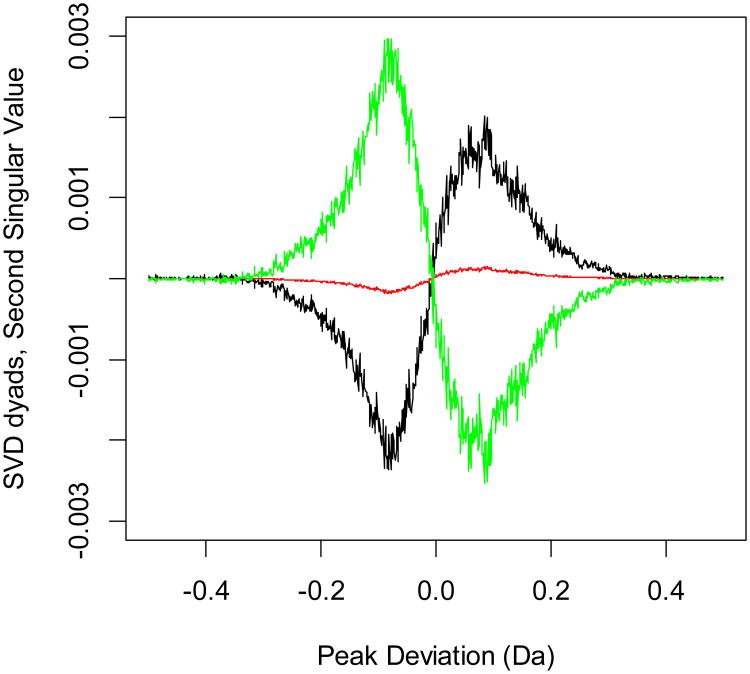
It has recently been reported that the dyads of data matrices in large-scale experiments exhibit patterns that are characteristic of the underlying changes in sample properties, and that the dyads of SVD decomposition behave as Hermite functions [18]. We examined the distributions of the SVD dyads from peak deviations for patterns that differentiate between phosphorylated and nonphosphorylated samples and demonstrate transition between them. The dyads of the first singular value (highest singular value) capture the most important features in the PD distributions (Figure S1, Supporting Information). These are even functions, and they align the PDs from all fractions. The true differences between the PDs are obtained from the second singular value. Figure 1 shows the dyad functions for the first (black), fifth (red), and tenth (green) fractions. The dyads for several other fractions are shown in Figure S2 of the Supporting Information. As is seen from the figures, dyads of the second singular value change sign along the fractionation steps, in parallel with the changes of the phosphopeptide content of the samples. The smallest absolute values of the dyads are observed for the 5th fraction. This is the transition fractionation step, where the relative proportions of the phosphorylated and nonphosphorylated peptides are found to comparable.
Figure 1.
SVD dyads corresponding to the second most important singular value. The black curve denotes the dyad of the 1st fraction, the red curve that for the 5th fraction, and the green curve that for the 10th fraction. The figure was generated using raw data – no database search or information about the sequence identity of a peptide was used. The only information used was the precursor monoisotopic mass values (neutral peptide plus proton) and peak center information from the mass distributions of theoretical peptides. The dyads clearly exhibit a transition from the phosphopeptide-rich samples (fractions 1 through 4) to nonphosphorylated peptide-rich samples (fractions 6 through 10). The transition is exhibited via changes in the coefficients of dyads along the peak deviations. Note that the absolute sign of the coefficients is not as important as the relative sign (change of the sign). This phenomenon is due to the different mass defects of phosphorylated and nonphosphorylated peptides. The dyads of several other fractionation steps are shown in Figure S2 of Supporting Information.
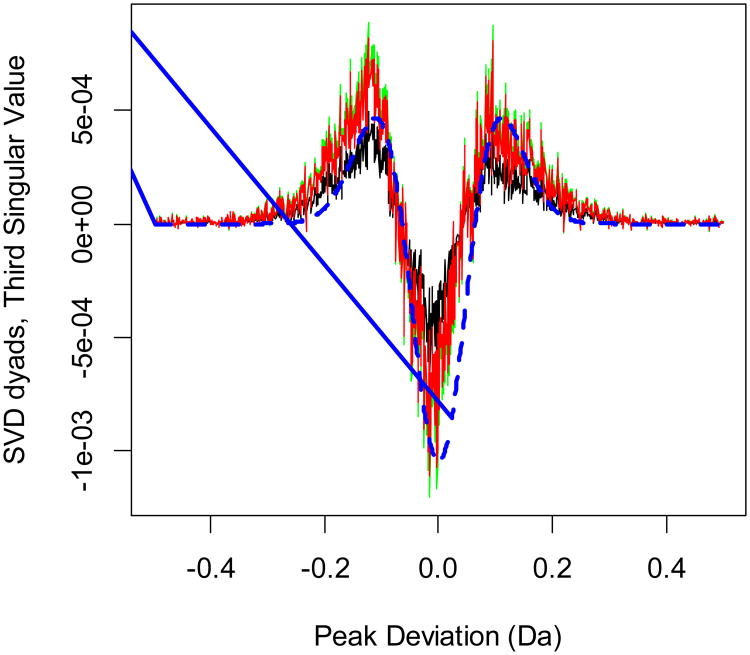
The second dyads, or the first-order Hermite functions, obtained in SVD are antisymmetric. This reflects the fact that the distributions of nonphosphorylated (mode at negative PDs) and phosphorylated peptides (mode at positive PDs) have different profiles. The profiles generate different dyads depending on the fractionation step. As the phosphopeptide content of the samples changes, the dyads of the second singular value go through a sign change. These are the most important singular value dyads to exhibit changes, as is seen from the comparison of different dyads in Figure S1 (the most important singular value dyads), Figure 1 (the second most important singular value dyads) and Figure 2 (the third most important singular value dyads). Only the dyads of the second singular value show a dramatic change in the form of a sign change. The first and third singular value dyads are symmetric, and their absolute values are comparable for samples from various fractions. Therefore, the dyads of the second largest singular value determine the degree to which the symmetry of PD distributions is distorted. The columns corresponding to experiments with large relative content of phosphorylated or nonphosphorylated peptides will have strong amplitudes.
Figure 2.
Dyads of the third singular value for the first (black), fifth (green) and tenth (red) fractions. The dyads corresponding to the symmetric eigenfunctions do not distinguish between the phosphorylated and nonphosphorylated samples. The shape of the functions is similar to that of the second-order Hermite functions. The broken line (blue) is the Hermite function (of the second-order) approximation.
4 Discussion
We have studied the distributions of peak deviations for separating phosphorylated and nonphosphorylated peptides using the monoisotopic masses of experimental precursors. Our ultimate goal is the application of precursor monoisotopic mass information and theoretical peptide distributions to improve phosphopeptide identification, validation and characterization in mass spectral data. We have previously shown that distributions of phosphorylated and nonphosphorylated peptides can be distinguished using a k-means clustering approach [8]. However, when one of the species (either phosphorylated or nonphosphorylated peptides) is present in relatively small amounts, the clustering algorithm is often unable to locate the low-abundance peak. The relative phosphopeptide content in such cases is difficult to estimate, as one needs to examine the distributions manually. Here we show that by using the SVD of the peak deviations and examining dyads corresponding to the most important singular values we can identify the modes of low-abundance distributions. The dyads of the first singular value, which are symmetric, capture the most prominent features of the peak deviations, but are not informative for differentiating purposes (Figure S1 of the Supporting Information). The dyads of the second singular value, which are antisymmetric, identify the patterns of the most prominent difference within the sample (Figure 1). In our example, these second dyads clearly show the transition from phosphopeptide-enriched to nonphosphorylated peptide samples. The dyads of the third singular value, which are symmetric, do not distinguish between the samples that are differentially enriched in their phosphopeptide content (see Figure 2). When summarized, the dyads of the three most important singular values reveal additional features that are not detectable in the raw PD distribution function. Note that the dyads of the fourth singular value (Figure S3 of the Supporting Information) are antisymmetric as well, and can in principle be used for evaluating the phosphoproteome content. However, the fourth singular value is five times smaller than the second singular value (in the present dataset). Therefore, the dyads of the second value contribute more to the SVD expansion.
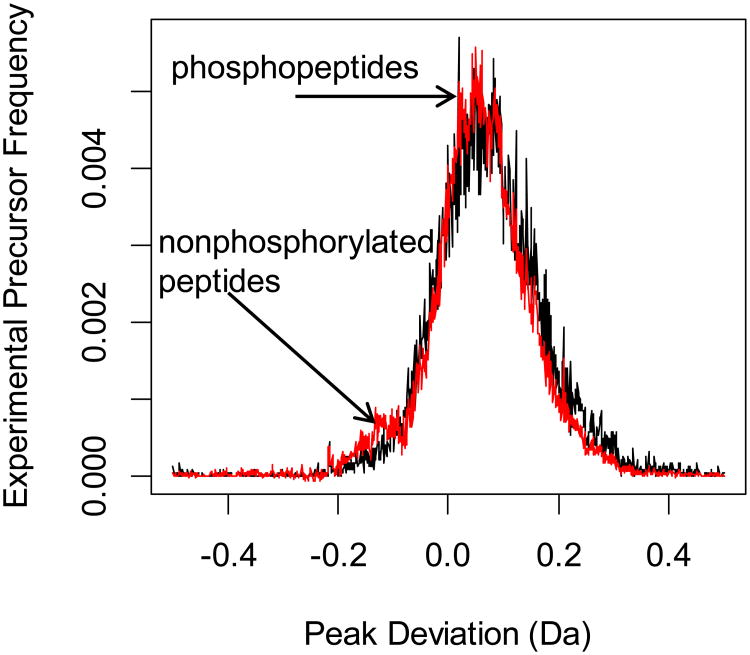
Figure 3 shows the PD distribution functions of the first SCX fractionation sample. The sample is highly enriched in phosphopeptides, and the raw PD distribution function (black line) shows only one mode, which is that of the phosphopeptides. k-means clustering is not able to detect the peak center for the nonphosphorylated peptides [8]. However, as is seen from the figure, by retaining the dyads of the first three most important singular values, we can effectively locate the mode of the distribution corresponding to the nonphosphorylated peptides (red line). Thus, processing and data reduction by the SVD (retaining only the dyads of the first three most important singular values) uncovers the peak mode of the nonphosphorylated peptides.
Figure 3.
Distribution functions of the experimental precursors from raw data (black line) and the sum of the first three dyads (red line) from the first fraction of the murine brain phosphoproteome [9]. The sum of the SVD dyads clearly identifies the peak associated with the nonphosphorylated peptides.
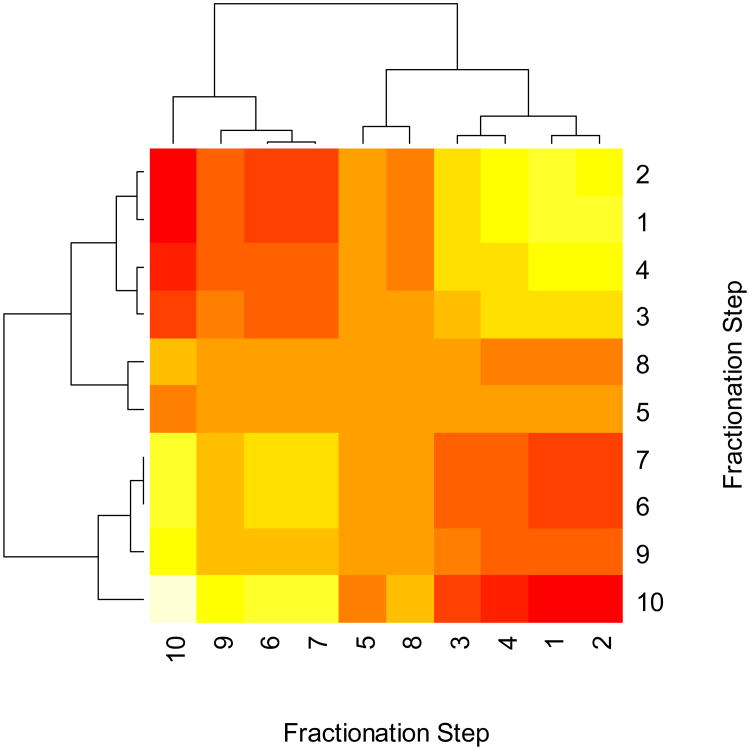
As the preceding analyses showed, the dyads corresponding to the second singular value play the most important role in exhibiting the transition in enrichments from phosphorylated to nonphosphorylated peptides (Figure 1). This observation is further validated by examining the clustering of dyads of this singular value for the ten fractionation steps. Figure 4 shows the heat map and clusters obtained from a covariance matrix of the PD for ten SCX fractions. The light colors indicate high covariance, and red indicates weak covariance. The clustering of the covariance matrix between the fractionation steps groups together the SCX fractions 1 through 4 and 6,7,9 and 10. Fractions 5 and 8 are grouped into a separate cluster. This is an important detail which is not transparent from the raw data of PDs. It is expected that the first four fractions are rich in phosphopeptides, and fractions 6, 7, 9 and 10 hold less phosphopeptides. However, the clustering analysis indicates that the eighth fraction is grouped with the fifth fractions. This is a departure from a straightforward expectation of a linear increase in nonphosphorylated peptides starting with the fifth fraction, and suggests that the eighth fractions holds more phosphopeptides than the sixth or seventh fractions – an observation confirmed by database searches [9]. Note that if we do a similar clustering for unprocessed PD, the results are more complex, and the specific observation (about the grouping of the fifth and eighth fractions) is missed (Figure S4, Supporting Information).
Figure 4.
Heat map and clustering of the covariance matrix of the dyads of the second most important singular value for 10 fractionation steps. High covariance values are shown in yellow and light colors, low covariance values are shown in red colors. The functions corresponding to fractionation steps one through four and six through ten (except eight) are clustered separately. This is partly expected, as the earlier fractionation steps are enriched in phosphopeptides and the later fractionation steps contain mostly nonphosphorylated peptides. It is interesting to note the clustering of the fifth and eighth fractions. As an exception from the linear progression towards nonphosphorylated peptide enrichment, the eighth fractionation is correctly identified as holding more phosphorylated peptides than the sixth or seventh fractions [9].
Determining phosphoproteome content of a sample is important for the development and optimization of workflows in phosphoproteomics [21, 22]. It is known that different workflows may be optimal, depending, for example, on the starting amount of available sample [22]. The phosphoproteome content is normally estimated using peptide identification from tandem mass spectra [9, 21]. Our approach is an alternative, as we do not use database searching, so the results are not affected by potential artifacts stemming from the procedures used for identifying peptides from tandem mass spectra and protein sequence databases. Our future goal is to combine the peak deviation distribution functions with the database search results to improve the efficiency of phosphoproteome research. We note that the SVD analysis of PDs used in this work is applicable to other types of PTMs, which have MDs different from those of the amino acids. In particular, we expect a similar approach will be appropriate for the differentiation of glycopeptides.
In summary, our analysis has found three advantages in the application of SVD analysis to peak deviations of samples enriched in phosphopeptides. First, the SVD detects the changes in the sample content via changes in the dyads corresponding to the second singular value. As the peptide content changes, the dyads (antisymmetric, first-order Hermite functions) change their sign, and go through an inflection point (via a fractionation step). The result is similar to that from the recent application of SVD to the transcript length distribution function of DNA microarray data [18]. In our opinion, the role of the evolutionary forces identified in this DNA microarray study is played in our dataset by the phosphoproteome gradient, which results from the SCX fractionations. Second, SVD simplifies the interpretation of clustering in the PD matrix. The covariance matrix of the dyads of the second most important singular value naturally clusters into phosphopeptide-enriched and nonphosphorylated peptide-enriched clusters. There is a third cluster which combines samples in “transition.” The identified sample clustering fully correlates with the corresponding results from database searching [9]. Our third conclusion is that SVD reveals modes of low-abundance species in the presence of the high-abundance species, Figure 3. Again, this is in accord with a recent study showing that the results of SVD were effective inputs as starting points for the k-means clustering algorithm [16]. Our data matrix, A, did not have any missing values, and there was no relevant analysis to check performance of SVD for missing value imputations [19] with the peak deviations.
Supplementary Material
Acknowledgments
This work was supported, in part, by UL1TR000071, the UTMB Clinical and Translational Science Award; HHSN272200800048C, the NIAID Clinical Proteomics Center; and NIH-NLBIHHSN268201000037C, the NHLBI Proteomics Center for Airway Inflammation. We thank Dr. David Konkel for critically editing the manuscript.
Abbreviations
- CID
collision-induced dissociation
- Eq
equation
- LC
liquid chromatography
- LTQ
Linear trap quadrupole
- MD
Mass defect
- MS
mass spectrometry
- ppm
parts per million
- PD
peak deviation
- SCX
strong cation exchange
- SVD
singular value decomposition
Footnotes
Conflict of Interest Statement. The author has no financial/commercial conflict of interest.
References
- 1.Nefedov AV, Mitra I, Brasier AR, Sadygov RG. Examining troughs in the mass distribution of all theoretically possible tryptic peptides. J Proteome Res. 2011;10:4150–4157. doi: 10.1021/pr2003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann WD, Bohne A, von Der Lieth CW. The information encrypted in accurate peptide masses-improved protein identification and assistance in glycopeptide identification and characterization. J Mass Spectrom. 2000;35:1335–1341. doi: 10.1002/1096-9888(200011)35:11<1335::AID-JMS70>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Bruce C, Shifman MA, Miller P, Gulcicek EE. Probabilistic enrichment of phosphopeptides by their mass defect. Anal Chem. 2006;78:4374–4382. doi: 10.1021/ac060046w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Froehlich JW, Dodds ED, Wilhelm M, Serang O, Steen JA, Lee RS. A classifier based on accurate mass measurements to aid large scale, unbiased glycoproteomics. Mol Cell Proteomics. 2013;12:1017–1025. doi: 10.1074/mcp.M112.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez H, Niehauser S, Boltz SA, Gawandi V, Phillips RS, Amster IJ. Mass defect labeling of cysteine for improving peptide assignment in shotgun proteomic analyses. Anal Chem. 2006;78:3417–3423. doi: 10.1021/ac0600407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X, Diego P, Ramos AA, Shi Y. Averagine-scaling analysis and fragment ion mass defect labeling in peptide mass spectrometry. Anal Chem. 2008;80:7383–7391. doi: 10.1021/ac801096e. [DOI] [PubMed] [Google Scholar]
- 7.Spengler B, Hester A. Mass-based classification (MBC) of peptides: highly accurate precursor ion mass values can be used to directly recognize peptide phosphorylation. J Am Soc Mass Spectrom. 2008;19:1808–1812. doi: 10.1016/j.jasms.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Kalita M, Kasumov T, Brasier AR, Sadygov RG. Use of theoretical Peptide distributions in phosphoproteome analysis. J Proteome Res. 2013;12:3207–3214. doi: 10.1021/pr4003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jedrychowski MP, Huttlin EL, Haas W, Sowa ME, Rad R, Gygi SP. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkner JA, Andrews PC. P6-T Tranche: Secure Decentralized Data Storage for the Proteomics Community. Journal of Biomolecular Techniques. 2007;18:1. [Google Scholar]
- 11.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilski MJ, Sadygov RG. Comparison of Programmatic Approaches for Efficient Accessing to mzML Files. J Data Mining Genomics Proteomics. 2011;2 doi: 10.4172/2153-0602.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nefedov AV, Sadygov RG. A parallel method for enumerating amino acid compositions and masses of all theoretical peptides. BMC Bioinformatics. 2011;12:432. doi: 10.1186/1471-2105-12-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra I, Nefedov AV, Brasier AR, Sadygov RG. Improved mass defect model for theoretical tryptic peptides. Anal Chem. 2012;84:3026–3032. doi: 10.1021/ac203255e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning data mining, inference, and prediction. Springer; New York: 2009. [Google Scholar]
- 16.Ding C, He X. Proceedings of the 21 st International Conference on Machine Learning. ACM Press; 2004. K-means clustering via principal component analysis; pp. 225–232. Ref Type: Conference Proceeding. [Google Scholar]
- 17.Becavin C, Tchitchek N, Mintsa-Eya C, Lesne A, Benecke A. Improving the efficiency of multidimensional scaling in the analysis of high-dimensional data using singular value decomposition. Bioinformatics. 2011;27:1413–1421. doi: 10.1093/bioinformatics/btr143. [DOI] [PubMed] [Google Scholar]
- 18.Bertagnolli NM, Drake JA, Tennessen JM, Alter O. SVD Identifies Transcript Length Distribution Functions from DNA Microarray Data and Reveals Evolutionary Forces Globally Affecting GBM Metabolism. PLoS ONE. 2013;8:e78913. doi: 10.1371/journal.pone.0078913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 20.Brock GN, Shaffer JR, Blakesley RE, Lotz MJ, Tseng GC. Which missing value imputation method to use in expression profiles: a comparative study and two selection schemes. BMC Bioinformatics. 2008;9:12. doi: 10.1186/1471-2105-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonslow BR, Niessen SM, Singh M, Wong CC, Xu T, Carvalho PC, Choi J, Park SK, Yates JR., III Single-Step Inline Hydroxyapatite Enrichment Facilitates Identification and Quantitation of Phosphopeptides from Mass-Limited Proteomes with MudPIT. J Proteome Res. 2012;11:2697–2709. doi: 10.1021/pr300200x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, Di PS, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.