Abstract
The sialyl Lewis a and x (sLea/x) antigens frequently displayed on the surface of tumor cells are involved in metastasis. Their synthesis has been attributed to altered expression of selective glycosyltransferases. Identification of these glycosyltransferases and the glycoproteins that carry these carbohydrate antigens should help advance our understanding of selectin-mediated cancer metastasis. In this study, quantitative real-time polymerase chain reaction analysis coupled with in situ proximity ligation assay and small interference RNA treatment shows involvement of β3galactosyltransferase-V in the synthesis of MUC16-associated sLea in H292 cells. Also, α3fucosyltransferase-V, which is absent in BEAS-2B human immortalized bronchial epithelial cells and A549 lung carcinoma cells, participates in the synthesis of MUC1-associated sLex in CFT1 human immortalized bronchial epithelial cells and H292 lung carcinoma cells. Neither selectin ligand is found on MUC1 in BEAS-2B and A549 cells. Knockdown of either enzyme suppresses migration, and selectin tethering and rolling properties of H292 cells under dynamic flow as determined by wound healing and parallel plate flow chamber assays, respectively. These results provide insights into how the synthesis of mucin-associated selectin ligands and the metastatic properties of cancer cells can be regulated by selective glycosyltransferases that work on mucins. They may help develop novel anticancer drugs.
Keywords: cell adhesion, glycosylation, glycosyltransferases, lung cancer, metastasis, sialyl Lewis antigens
Introduction
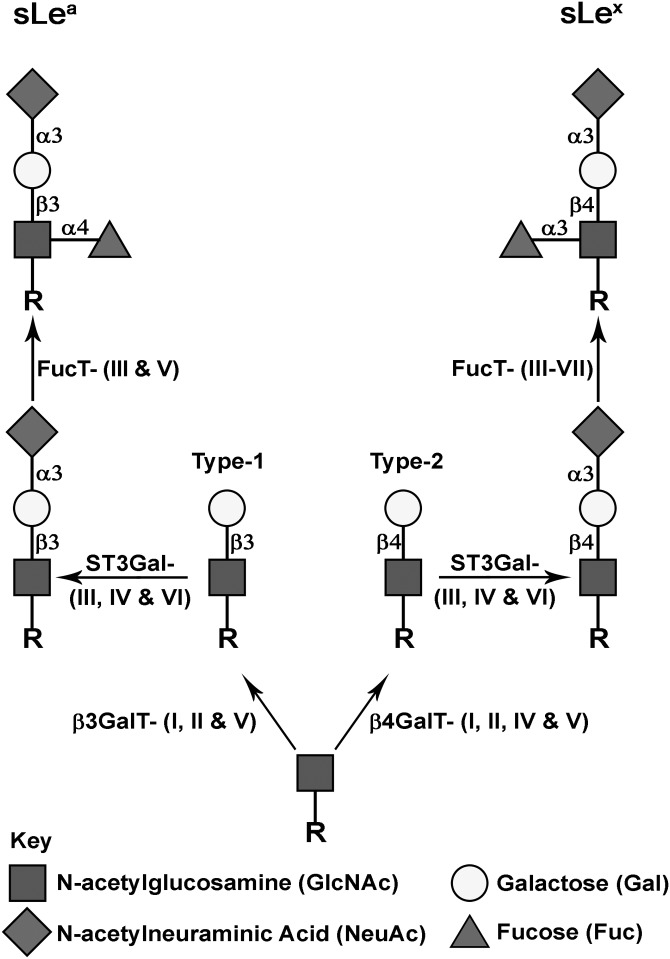
Around 90% of cancer deaths are caused by tumor metastasis. Metastatic cancer cells employ the trafficking mechanisms utilized by leukocytes to target distant sites (Laubli and Borsig 2010). Following dissemination from the primary tumor into the blood stream, the circulating cancer cells are tethered to and roll on the endothelium at distant sites to establish firm adhesion prior to extravasation (Chambers et al. 2001). Tethering and rolling are mediated by interaction of selectins with their cognate carbohydrate ligands, Sialyl Lewis a (sLea) and sialyl Lewis × (sLex). These selectin ligands expressed on various cell types, including lymphocytes, monocytes, neutrophils and advanced cancers (Kannagi et al. 2004), play a key role in hematogenous cancer metastasis (Fukuda 1996; Kim et al. 1999; Kawarada et al. 2000; Ugorski and Laskowska 2002). They are also used as diagnostic and prognostic markers for cancer (Nakayama et al. 1995; Paganuzzi et al. 2003; Sumikura et al. 2003; Kannagi 2007). Both sialyl Lewis antigens are found on a variety of cell surface glycoproteins, including mucins (Fernandez-Rodriguez et al. 2001; Barthel et al. 2009; Remmers et al. 2013). Biosynthesis of sLea or sLex (Figure 1) is initiated by addition of Gal in a β1,3 or β1,4 linkage, respectively, to a N-acetylglucosamine (GlcNAc) residue at the non-reducing terminus of an oligosaccharide chain. The structures of these two glycan products, which are designated as type-1 and type-2 lactosamine chains, are generated by β3GalT and β4GalT enzymes, respectively. Then, sialic acid is added to the Gal residue in an α2,3 linkage as catalyzed by ST3Gal (Ellies et al. 2002). Finally, fucose is added to the GlcNAc residue in an α1,3/1,4 linkage to complete the synthesis of these glycans (Brockhausen 2006). The isozymes within these four groups of glycosyltransferases (GTs) involved in the synthesis of these two sialyl Lewis antigens are: β3GalT-I, II and V (Hennet et al. 1998; Isshiki et al. 1999), β4GalT-I, II, IV and V (Hennet 2002), ST3Gal-III, IV and VI (Ellies et al. 2002) and α3/4FucT-III–VII (de Vries et al. 1995; Oriol et al. 1999; Norden et al. 2009). One major limitation in this area of glycobiology is a lack of a clear understanding of specific GT isozymes that work on a specific glycoprotein. This is the void current study is intended to fill.
Fig. 1.
Biosynthetic pathways for glycoprotein-associated sLea and sLex epitopes. Synthesis of sLea is initiated by formation of type-1 lactosamine chain by addition of Gal to GlcNAc in a β1,3-linkage catalyzed by β3GalT-I, II and V. sLex synthesis is initiated by formation of type-2 lactosamine by addition of Gal to GlcNAc in a β1,4-linkage catalyzed by β4GalT-I, II, IV and V. These two lactosamines are further elongated by a sialic acid-linked α2,3 to Gal to generate sialyl lactosamine as catalyzed by ST3GalT-III, IV and VI. Subsequent addition of Fuc to GlcNAc in an α1,4-linkage catalyzed by FucT-III and V produces sLea, and addition of Fuc to GlcNAc in an α1,3-linkage catalyzed by FucT-III–VII produces sLex. R represents a glycoprotein containing glycans terminated with GlcNAc.
MUC1, MUC4 and MUC16 (also called CA125) are the most widely studied membrane-bound mucins (Bafna et al. 2010). All three mucins have been shown to be decorated with various pancarcinoma antigens including sLea and sLex (Fernandez-Rodriguez et al. 2001; Croce et al. 2007; Groux-Degroote et al. 2008; Barthel et al. 2009; Remmers et al. 2013). Although some of the isozymes that participate in the synthesis of these two selectin ligands have been identified, specific isozymes of these four groups of enzymes that synthesize these glycotopes on a given glycoprotein are largely not known. Recently, we showed that β3GalT-I and ST3Gal-VI are involved in the synthesis of sLea and sLex on MUC1 in prostate and colon cancer cells, respectively (Chachadi et al. 2011; Chachadi et al. 2013). These studies also show that these two enzymes control the metastatic properties of these two cancer cells in an enzyme, mucin and cancer-specific fashion. Identification of additional glycoprotein-specific glycosyltransferase isozymes that are involved in the synthesis of these two selectin ligands is needed in order to gain a comprehensive understanding of the selectin-mediated cancer metastasis and develop a novel therapy.
In the present study, we have shown that β3GalT-V is involved in the synthesis of MUC16-associated sLea in H292 cells while FucT-V participates in the synthesis of sLex associated with MUC1 in CFT1 and H292 cells. Depletion of β3GalT-V or FucT-V in H292 cells results in suppression of their metastatic potential by reducing cell migration, and suppression of tethering to and rolling on immobilized E- and P-selectin under dynamic flow. The results further support the concept that glycosylation of a given glycoprotein is GT isozyme specific. However, this phenomenon may be cell type specific.
Results
Expression of sLea and sLex selectin ligands in human immortalized bronchial epithelial and lung carcinoma cells
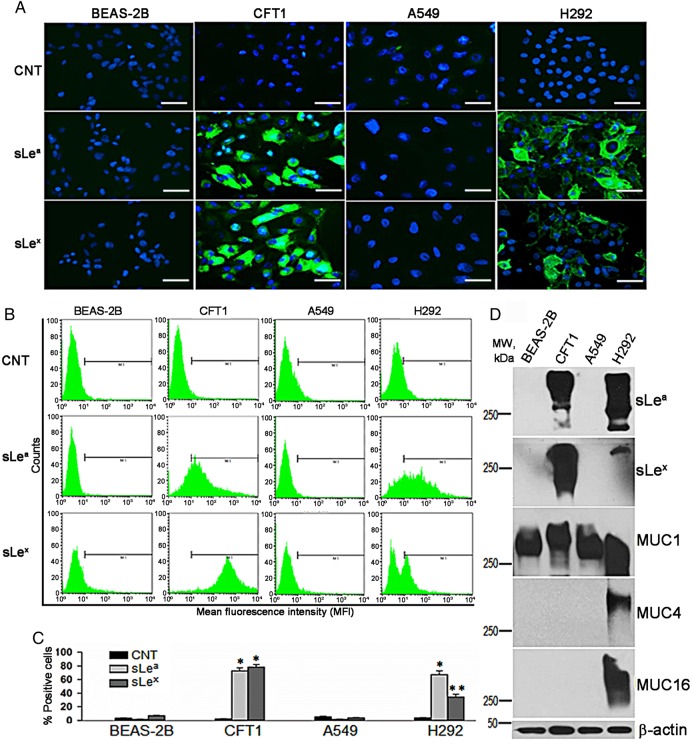
Given the important role sLea and sLex play in selectin-mediated cancer metastasis, we set out to identify key GTs that are involved in the synthesis of these glycotopes, and the membrane-bound mucins that carry them in lung carcinoma A549 and H292 cells. For comparison, we also performed same analysis in two human lung immortalized epithelial cells, BEAS-2B and CFT1. Confocal immunofluorescence microscopic analysis showed that CFT1 and H292 cells but not BEAS-2B and A549 cells were labeled with KM231 and CSLEX antibodies (Abs), which recognize sLea and sLex, respectively (Figure 2A). Next, we employed flow cytometric analysis to quantify % of cells which displayed these two selectin ligands. sLea- and sLex were detected in CFT1 and H292 but not BEAS-2B and A549 cells (Figure 2B and C). In CFT1 cells, sLea- and sLex-positive cells were 72.9 ± 4.1 and 78.4 ± 3.6%, respectively, when compared with 2.27 ± 0.29% in control (CNT) cells. In H292 cells, sLea- and sLex-positive cells were 67.2 ± 5.7 and 34.4 ± 3.6%, respectively, when compared with 3.26 ± 0.84% in CNT cells. sLea- and sLex-positive cells were 1.87 ± 0.52 and 3.89 ± 0.29%, in BEAS-2B cells, respectively, and 0.94 ± 0.04 and 1.59 ± 0.84%, in A549 cells, respectively. To determine the membrane-bound mucins which carry these two glycotopes, we performed western blot analysis of sLea, sLex and membrane-bound mucins. As shown in Figure 2D, protein-associated sLea and sLex were detected only in the lysates of CFT1 and H292 cells. The high-molecular weight (≥250 kDa) of these bands suggested that these two glycotopes were associated with mucins. To test this idea, western blotting of MUC1, MUC4 and MUC16 was performed. We detected MUC1 core protein in all four cell types while MUC4 and MUC16 in only H292 cells. The results suggested that sLea and sLex were associated with MUC1 in CFT1 cells but could be with any one of these three membrane-bound mucins in H292 cells.
Fig. 2.
Detection of sLea and sLex antigens in human lung immortalized epithelial cells and carcinoma cells. (A) Confocal immunofluorescence microscopic images of BEAS-2B, CFT1, A549 and H292 cells treated with control IgG/M (CNT), KM231 (sLea) and CSLEX (sLex) Abs followed by HRP-labeled secondary Ab (bar = 20 µm). (B) Flow cytometric analysis of these four cells treated as described in (A). (C) Quantitation of % sLea- and sLex-positive cells described in (B) (n = 3). Error bars indicate SD (*P < 0.05, **P < 0.01). (D) Western analysis of sLea, sLex, MUC1, MUC4 and MUC16 in the lysates of these four cells. β-Actin was used as a loading control. Data shown are a representative of three independent experiments. This figure is available in black and white in print and in color at Glycobiology online.
sLea and sLex are associated with MUC1 in CFT1 and H292 cells, and sLea with MUC16 in H292 cells
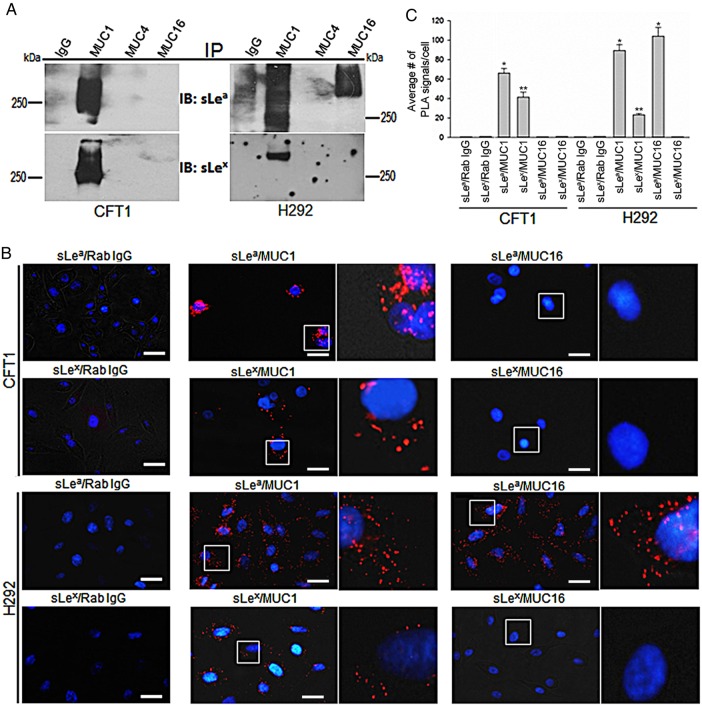
To identify the membrane-bound mucins that carry sLea and sLex, we performed Co-immunoprecipitation (Co-IP) and in situ proximity ligation assays (PLA). In the Co-IP assay, MUC1, MUC4 and MUC16 immunoprecipitates were subjected to sLea and sLex western blot analysis. As shown in Figure 3A, sLea and sLex were detected in MUC1 immunoprecipitates from the lysates of CFT1 and H292 cells. sLea but not sLex was detected in MUC16 immunoprecipitate from the lysate of H292 cells. Further, these two selectin ligands were not found in MUC4 immunoprecipitates from the lysates of CFT1 and H292 cells. The results were confirmed by in situ PLA. As shown in Figure 3B and C, strong PLA signals of sLea/MUC1 were detected in CFT1 (68.3 ± 7.3) and H292 (89.1 ± 13.4) cells. Moderate PLA signals of sLex/MUC1 were detected in CFT1 (40.1 ± 5.4) and H292 (23.2 ± 3.14) cells. Further, the PLA signal of sLea/MUC16 was very strong in H292 (105 ± 16.2) but not detectable in CFT1 cells. The PLA results confirm the Co-IP results that sLea and sLex are associated with MUC1 in CFT1 and H292 cells while sLea but not sLex is associated with MUC16 in H292 cells.
Fig. 3.
Association of sLea and sLex with MUC1 and MUC16 mucins in CFT1 and H292 cells. (A) sLea and sLex western blots of the MUC1, MUC4 and MUC16 immunoprecipitates from the lysates of CFT1 and H292 cells. Mouse IgG was used as a negative control. (B) Fluorescence microscopic images of proximity ligation assay. Positive PLA signals were detected for sLea/MUC1 and sLex/MUC1 in CFT1 and H292 cells, and sLea/MUC16 in H292 cells. Areas marked with white boxes were enlarged (4×) and shown to the right side of the original images. As the controls, one of the paired Abs was replaced with non-specific IgG (bar = 20 µm). (C) Quantification of PLA signals measured in 250 cells (n = 4) and expressed as average number of PLA signals. Error bars indicate SD (*P < 0.05, **P < 0.01). This figure is available in black and white in print and in color at Glycobiology online.
sLea and sLex are associated with mucin-type O-glycans but not N-glycans
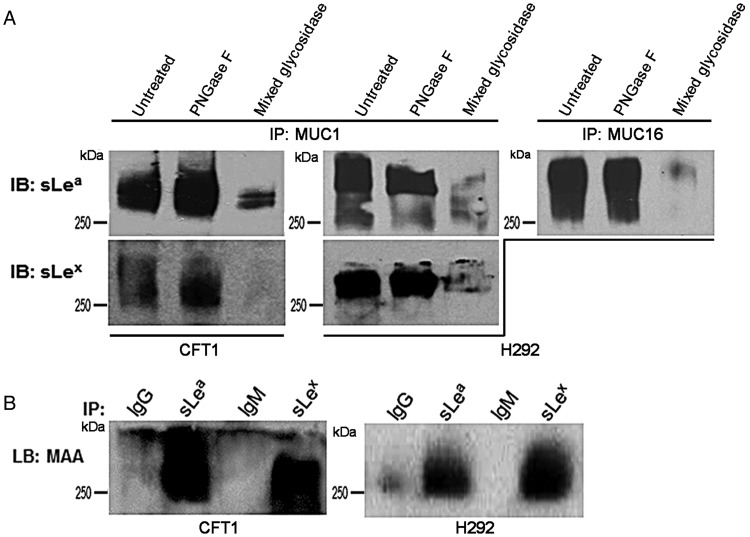
Since MUC1 and MUC16 contain both asparagine (N)-linked and Ser/Thr-linked mucin-type O-glycans, we set out to determine which of these two glycans carries sLea and sLex. The peptide N-glycosidase (PNGase) F resistance of sLea and sLex in MUC1 immunoprecipitates from the lysates of CFT1 and H292 cells suggested that these two glycotopes were associated with mucin-type O-glycans (Figure 4A). The susceptibility of these glycotopes to mixed glycosidases, which contains neuraminidase, showed the congruence of the chemical nature of these glycotopes with those of the selectin ligands. Same result was obtained for MUC16-associated sLea in H292 cells (Figure 4A). To confirm the presence of terminal α2,3 sialic acid, the sLea and sLex immunoprecipitates were analyzed by Maackia amurensis lectin blotting. Figure 4B clearly shows that sLea and sLex from CFT1 and H292 cells contain terminal α2,3 sialic acid, which further confirms the chemical nature of these two glycotopes.
Fig. 4.
Characterization of the type of glycan in MUC1 and MUC16 with which sLea and sLex are associated. (A) sLea and sLex western blots of MUC1 immunoprecipitates from the lysates of CFT1 and H292 cells, and sLea western blot of MUC16 immunoprecipitate from the lysate of H292 cells. The immunoprecipitates were treated with PGNase F or mixed glycosidases containing neuraminidase prior to western analysis. Untreated controls were similarly processed without enzymes. Proteins were subjected to western blot analysis and developed with HRP-labeled anti-sLea and anti-sLex antibodies. Both sLea and sLex pulled down by MUC1 and MUC16 antibodies were resistant to PNGase F suggesting that both glycotopes are associated with mucin-type O-glycan. (B) MAA lectin blot of IgG, sLea, IgM or sLex immunoprecipitates from the lysates of CFT1 and H292 cells. Result indicates the presence of α2–3 sialic acid in the immunoprecipitates which supports the presence of sLea and sLex in them.
Expression profile of B3GALT, B4GALT, ST3GAL and FUT genes, which participate in the synthesis of glycoprotein-associated sLea/sLex, in BEAS-2B, CFT1, H292 and A549 cells
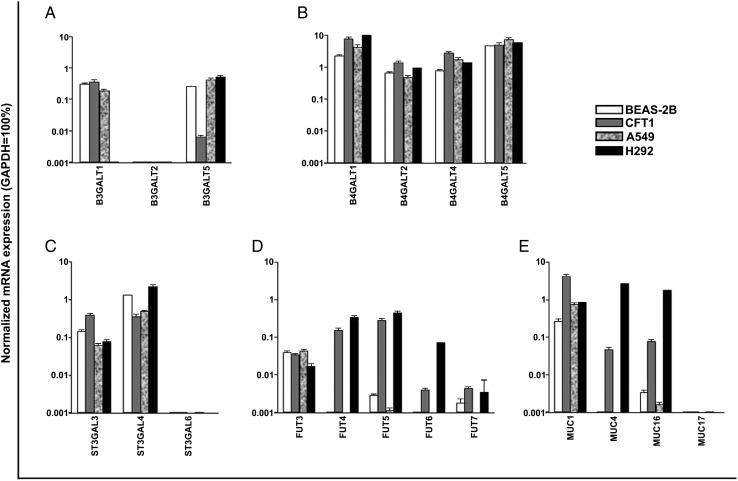
To identify the GT isozymes involved in the synthesis of sLea and sLex associated with mucin O-glycans, we analyzed by quantitative real-time PCR (qRT-PCR) the expression profile of four groups of mucin O-glycan-specific GT genes, including B3GALT, B4GALT, ST3GALT and FUT, in all four lung cell types. The data are expressed as % of GAPDH. We detected moderate expression levels of B3GALT1 in BEAS-2B, CFT1 and A549 cells, very low expression levels of B3GALT1 in H292 cells and B3GALT2 in all four cell types, and moderate expression levels of B3GALT5 in BEAS-2B, A549 and H292 cells but very low level in CFT1 cells (Figure 5A). Moderate-to-high levels of expression of B4GALT1, 2, 4 and 5 were detected in all four cell types (Figure 5B). Low-to-moderate expression levels of ST3GAL3, moderate-to-high expression levels of ST3GAL4 and no expression of ST3GAL6 were found in all four cell types (Figure 5C). Also, low FUT3 expression levels were found in all four cell types and moderate expression levels of FUT4 & 5 were detected in CFT1 and H292 cells but very low expression levels in the other two cell types. Low level of FUT6 expression was found in H292 cells and very low to no expression in the other three cell types (Figure 5D). To confirm the western blotting results of the membrane-bound mucins (Figure 2D), we measured the mRNA levels of MUC1, 4, 16 and 17 genes. Moderate-to-high levels of MUC1 gene expression were found in all four cell types (Figure 5E). High levels of MUC4 & 16 gene expression were detected in H292 cells but only low levels of expression in CFT1 cells. MUC4 and MUC16 gene expression levels in BEAS-2B and A549 cells and MUC17 gene expression level in all four cells were extremely low. The relative gene expression levels of MUC1, 4 and 16 among these cells matched the western blotting results (Figure 2D).
Fig. 5.
Gene expression profiles of glycosyltransferases and membrane-bound mucins in BEAS-2B, CFT1, A549 and H292 cells, which are involved in the biosynthesis of sLea and sLex. (A–D) Gene expression profiles of glycosyltransferases involved in the biosynthesis of sialyl Lewis antigens analyzed by qRT-PCR. (A) B3GALT1, 2 and 5: type-1 β1,3 Gal transferases (B) B4GALT1, 2, 4 and 5: type-2 β1,4 Gal transferases, (C) ST3GAL3, 4 and 6 and (D) FUT3–7. (E) Expression profile of MUC1, 4, 16 and 17 genes as measured by qRT-PCR. Gene expression levels were sorted according to ΔΔCt method (see Methods for details), normalized with GAPDH (=100%) in same cell preparation, expressed as mean ± SD and plotted in a log scale (n = 3). Gene encoding β3GalT-V involved in the synthesis of sLea was expressed at moderate levels in all but CFT1 cells, which expressed this gene at a very low level, while high level of MUC16 gene expression was detected only in H292 cells. Similarly, moderate levels of FUT4 and FUT5 genes were expressed in CFT1 and H292 cells.
Silencing of B3GALT5 gene decreases sLea in H292 cells, and silencing of FUT5 decreases sLex in CFT1 and H292 cells
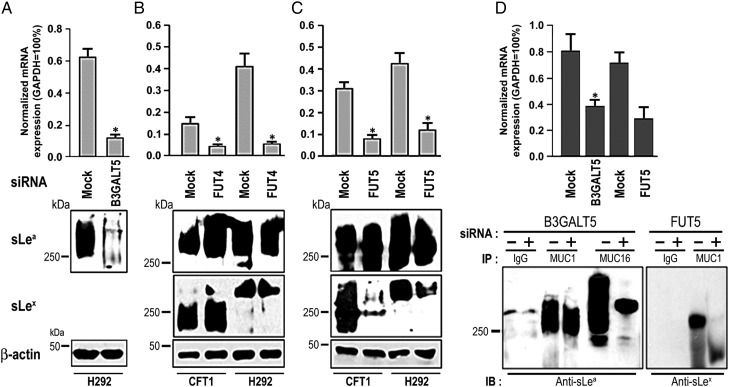
Although highly expressed in all four cell types, MUC1 in only CFT1 and H292 cells was decorated with sLea and sLex, suggesting expression of all GTs needed for the synthesis of these two glycotopes in these two cell types. Since the expression levels of B4GALT1, 2, 4 and 5 and ST3GAL3 and 4 genes were adequate for producing the expected glycan structures, they were not considered crucial in determining the differential expression of these two selectin ligands between these two and the other two lung cell types. Therefore, analysis of the differential expression of B3GALT and FUT genes among these cells could help identify specific β3GalT and α1,3/4FucT isozymes involved in the synthesis of MUC-associated sLea and sLex. Further, presence of sLea and absence of sLex on MUC16 in H292 cells suggested that the differential expression of B3GALT5 could be the key determinant. Also, cell-type-specific differences in the expression of glycoprotein-specific FUT genes suggested that FUT4 and 5 were the candidate genes, because these two genes were expressed in CFT1 and H292 but not the other two cell types. To test these ideas, series of siRNA knockdown experiments were carried out to identify the specific GT isozymes involved in the synthesis of sLea and sLex. Figure 6A shows that knockdown of B3GALT5 gene in H292 cells by 80.5% decreased sLea by 87.2%. To investigate the possible involvement of FucT-IV and FucT-V in the synthesis of sLea and sLex, these genes were individually silenced. Knockdown of FUT4 mRNA by 68.8 and 86.2% in CFT1 and H292 cells, respectively, did not affect the production of either sLea or sLex (Figure 6B). However, knockdown of FUT5 gene expression in CFT1 and H292 cells by 74.0 and 71.3% reduced sLex by 85 and 61%, respectively (Figure 6C), suggesting involvement of FucT-V in the synthesis of MUC1-associated sLex in CFT1 and H292 cells. This effect is FUT5-specific because knockdown of FUT5 mRNA did not affect the expression levels of FUT3 and FUT6 (data not shown), which exhibit very high sequence identity with FUT5 (de Vries et al. 2001), Furthermore, FucT-V did not participate in the synthesis of MUC1 and MUC16-associated sLea in CFT1 and H292 cells because knockdown of FUT5 did not affect sLea detected in these cells by western blotting. Additionally, knockdown of B3GALT5 mRNA (by 52%) in H292 cells decreased sLea (by 73%) associated with MUC16 but not MUC1 (Figure 6D), indicating that B3GALT5 participates in the synthesis of sLea associated with MUC16 but not MUC1 in this cell type. Knockdown of FUT5 mRNA (by 54%) in H292 cells decreased MUC1-associated sLea to below detectable level (Figure 6D), indicating that FucT-V is involved in the synthesis of MUC1-associated sLea in this cell type.
Fig. 6.
B3GALT5 is involved in the synthesis of sLea in H292 cells, and FUT5 is involved in the synthesis of sLex in CFT1 and H292 cells. (A) sLea western blot (bottom panel) of the lysates of H292 cells treated with either B3GALT5 or scrambled siRNAs (Mock). mRNA levels were measured by qRT-PCR (upper panel). Reduction of B3GALT5 mRNA level greatly reduced sLea level as measured by western blotting. (B and C) sLea and sLex western blots of the lysates of CFT1 and H292 cells treated with scrambled (Mock), (B) FUT4 and (C) FUT5 siRNAs. Reduction of FUT4 mRNA levels in CFT1 and H292 cells did not affect sLea or sLex level. Reduction of FUT5 mRNA levels in CFT1 and H292 cells greatly reduced sLex levels in these two cells. β-Actin was used as equal protein-loading control. (D) B3GALT5 and FUT5 gene knockdown experiments were repeated in H292 cells, and MUC1 and MUC16 were immunoprecipitated followed by western blotting for sLea and sLex. Efficiency of gene knockdown was determined by measuring the mRNA level by qRT-PCR. All the experiments were performed in triplicates and data were expressed as mean values ± SD (*P < 0.05).
Knockdown of either B3GALT5 or FUT5 gene decreases in vitro migratory and metastatic properties of H292 cells
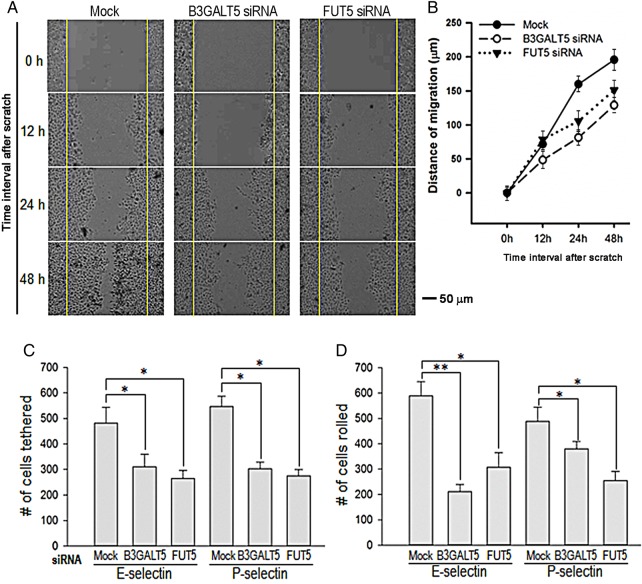
To investigate the functional consequences of the aforementioned molecular findings, in vitro cell migration and cell binding were measured by wound healing and parallel plate flow chamber assays under dynamic flow, respectively. Knockdown of either B3GALT5 or FUT5 mRNA in H292 cells by siRNA treatment suppressed cell migration (Figure 7A). The rate of cell migration towards the scratch wound was significantly reduced when compared with that of mock-treated CNT cells (Figure 7B). The difference still existed even at 48 h post scratch. Since sLea and sLex are carbohydrate ligands involved in selectin-mediated hematogenous metastasis of cancer cells (Kannagi et al. 2004), we proceeded to examine the effect of knocking down either B3GALT5 or FUT5 mRNA in H292 cells on their in vitro interaction with immobilized P- or E-selectin under dynamic flow. We found that knockdown of B3GALT5 mRNA in H292 cells significantly reduced their tethering to and rolling on immobilized E- and P-selectins. When compared with mock-treated cells, cells treated with B3GALT5 siRNAs showed a reduction in the number of cells tethered to E-selectin by 35.6% and P-selectin by 44.7% (Figure 7C), and rolled on E-selectin by 64.2% and P-selectin by 22.1% (Figure 7D). Also, when compared with mock-treated cells, cells treated with FUT5 siRNAs exhibited a reduction in the number of cells tethered to E-selectin by 44.9% and P-selectin by 49.6% (Figure 7C), and rolled on E-selectin by 47.9% and P-selectin by 48.0% (Figure 7D). The results indicate that both β3GalT-V and FucT-V play an important role in selectin-mediated tethering and rolling of H292 cells by participating in the synthesis of sLea and sLex in a MUC-specific fashion.
Fig. 7.
Knockdown of B3GALT5 or FUT5 in H292 cells decreases migration, and tethering to and rolling on E- and P-selectins. (A) Wound healing migration assay of H292 cells after treatment with B3GALT5- or FUT5-specific siRNAs. Phase contrast images were obtained at 0, 12, 24 and 48 h after scratching following 72 h siRNA treatment. (B) Quantification of the migration distance in cultures after the scratch as shown in (A). Data were obtained from three independent experiments (n = 3, *P < 0.05 vs. mock). (C) No. of cells tethered to (D) and rolled on E- or P-selectin-coated cover slip inside a parallel plate flow chamber. H292 cells (2.5 × 105 cells/mL) that had been treated with scrambled (Mock), B3GALT5 or FUT5 siRNAs were perfused at a constant wall shear stress of 1.0 dyne/cm2. Tethering and rolling were visualized with a phase-contrast microscope at ×40 magnification and videotaped. B3GALT5 or FUT5 siRNA-treated cells show reduced in vitro migration, and selectin-adhesion properties. (*P < 0.05, **P < 0.001 for specific siRNA-treated vs. scrambled siRNA-treated cells). This figure is available in black and white in print and in color at Glycobiology online.
Discussion
Cell surface sialyl Lewis antigens play an important role in selectin-mediated leukocyte trafficking (Kawarada et al. 2000; Ugorski and Laskowska 2002) and cancer metastasis (Shimodaira et al. 1997; Dimitroff et al. 2004). Synthesis of these two glycotopes requires participation of four groups of GTs, β3GalT, β4GalT, ST3Gal and FucT. Each of these GT gene families contains several isozymes. The involvement of specific isozymes of each GT family in the synthesis of these two selectin ligands in a given glycoprotein is largely unknown. In this study, we have identified β3GalT-V as the isozyme involved in the synthesis of sLea associated with MUC16 in H292 cells. Also, we have identified FucT-V as the isozyme involved in the synthesis of sLex associated with MUC1 in CFT1 and H292 cells. Further, we have also discovered that by participating in the synthesis of sLea and sLex, β3GalT-V and FucT-V play a critical role in the in vitro migratory and metastatic properties of H292 cells.
Biosynthesis of mucin glycans takes place exclusively in the Golgi apparatus in a template-independent process. The glycan structure is determined primarily by (a) the expression levels of GT genes, (b) the substrate specificity of GTs, (c) the Golgi localization of the isozymes of each GT and (d) the intra-Golgi transport route of the glycoprotein substrates in the Golgi in relationship to the Golgi localization of their cognate GTs. Since most isozymes within a GT gene family have over-lapping substrate specificities and can produce same product when their enzymatic activities are measured in vitro, it is not possible to attribute a glycan structure generated on a glycoprotein to a specific isozyme based on the enzyme activity measured in cell homogenate. Therefore, to understand the biological function of a GT isozyme, the glycoprotein substrate that carries the glycotope generated by each isozyme in vivo needs to be identified.
It should be pointed out that only few isozymes within each of these four enzyme groups are involved in the synthesis of glycoprotein-associated sLea and sLex. For example, there are five β3GalT isozymes which can synthesize β3Gal structure, but only three, including β3GalT-I, II and V, can generate type-1 lactosamine structure (Hennet et al. 1998; Isshiki et al. 1999; Hennet 2002). There are seven β4GalT isozymes which can synthesize β4Gal structure, but only four, including β4GalT-I, II, IV and V can produce type-2 lactosamine chain (Amado et al. 1999; Furukawa and Sato 1999). There are six ST3Gal isozymes which can synthesize N-acetylneuraminic acid (NeuAc) α2,3Gal structure, but only three, including ST3Gal-III, IV and VI, can generate NeuAcα2,3Galβ3/4GlcNAc, the precursors of sLea and sLex (Sasaki et al. 1993; Okajima et al. 1999). Further, among the isozymes within each enzyme family, differences in substrate specificity also exist. For example, the preference of ST3Gal-III for type-1 over type-2 lactosamine acceptor suggests that sialylated type-1 chain is its predominant product (Wen et al. 1992; Kitagawa and Paulson 1993). Also, ST3Gal-IV and VI prefer type-2 over type-1 lactosamine as their acceptor, which makes synthesis of sialylated type-2 chain their primary function (Rohfritsch et al. 2006; Carvalho et al. 2010). There are 10 α3/4FucT isozymes which can synthesize α3/4Fuc structure, but only five, including FucT-III–VII, can make sLea and sLex in glycoproteins (de Vries et al. 2001). Further, there are differences in the specificities toward four different acceptors for these enzymes. FucT-III and V can work on both type-1 and type-2 acceptors, and FucT-III prefers type-1 over type-2 and vice versa for FucT-V. These enzymes can work on their respective lactosamine acceptors with or without sialic acid. However, FucT-VII is only active with α2,3 sialylated type-2 lactosamine acceptor (Shinoda et al. 1998). Table I summarizes the carbohydrate acceptor specificities of the GT isozymes involved in the synthesis of glycoprotein-associated sLea and sLex.
Table I.
Carbohydrate and glycoprotein acceptor specificity of the glycosyltransferase isozymes involved in the synthesis of glycoprotein-associated sLea and sLex
| Isozymes | Glycan acceptors and preference | Glycan products | Glycoprotein substrates in vivo |
|---|---|---|---|
| B3GALT1 | GlcNAcβ-saccharide (Amado et al. 1998; Holgersson and Lofling 2006) | Galβ1,3GlcNAc | MUC1 (Chachadi et al. 2013) |
| B3GALT5 | GlcNAcβ-saccharide (Holgersson and Lofling 2006) | Galβ1,3GlcNAc | CD43 (Holgersson and Lofling 2006); MUC16a |
| B4GALT1b | GlcNAcβ-saccharide (Hennet 2002) | Galβ1,4GlcNAc | PSGL1 (Nakayama et al. 2000) |
| ST3GAL4c | Type-2>Type-1 (Sasaki et al. 1993) | NeuAcα2,3Galβ1,4/3GlcNAc | PSGL1 (Harduin-Lepers et al. 2001) |
| ST3GAL6c | Type-2>Type-1 (Okajima et al. 1999) | NeuAcα2,3Galβ1,4/3GlcNAc | MUC1 (Chachadi et al. 2011) |
| FUT3 | (±Sialyl)Type1>Type-2 (de Vries et al. 1995) | ±NeuAcα2,3Galβ1,3/4(Fucα1,4/3)GlcNAc | CD44 (Padro et al. 2011) |
| FUT4 | (±Sialyl)Type-2 (Niemela et al. 1998) | ±NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAc | ESL-1, PSGL1 (Homeister et al. 2001; Martinez et al. 2005) |
| FUT5 | (±Sialyl)Type-2>Type-1 (Okajima et al. 1999) | ±NeuAcα2,3Galβ1,4/3(Fucα1,3/4)GlcNAc | CD44 (Padro et al. 2011); MUC1a |
| FUT6 | (±Sialyl)Type-2 (de Vries et al. 1995) | ±NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAc | CEA (Barthel et al. 2009) |
| FUT7 | Sialyl Type-2 (Shinoda et al. 1998) | NeuAcα2,3Galβ1,4(Fucα1,3)GlcNAc | CD24, GlyCAM-1, ESL-1, PSGL1, MCAM (Barthel et al. 2009; Homeister et al. 2001; Huang et al. 2000; Li et al. 2005; Martinez et al. 2005) |
aIdentified in this study.
bGlycoprotein substrates for B4GALT2, 4 and 5, which exhibit same carbohydrate acceptor specificity and generate same glycan product as B4GALT1, have not been identified.
cGlycoprotein substrates for ST3GAL3, which acts on NeuAcα2,3Galβ1,3/4GlcNAc and has higher specificity towards type-1 over type-2 glycans (Weinstein et al. 1982), have not been identified.
CEA, carcinoembryonic antigen; MCAM, melanoma cell adhesion molecule; ESL, E-selectin ligand; PSGL1, P-selectin glycoprotein ligand-1; GlyCAM-1, glycosylation-dependent cell adhesion molecule 1.
Altered tumor-associated carbohydrate antigens have been identified in many solid tumors (Cazet et al. 2010). Tissue and tumor-specific expression of GT genes also has been reported (Tsuji 1996). But, very little is known about the specific GT isozymes that are responsible for the synthesis of these altered glycans. Even much less is known about the glycoproteins that carry these tumor-associated carbohydrate antigens in vivo. As shown in Table I, the glycan synthesis in a given glycoprotein is GT isozyme specific. Recently, our lab has shown that β3GalT-I and ST3Gal-VI are involved in the synthesis of MUC1-associated sLea in human normal prostatic RWPE-1 cells (Chachadi et al. 2013) and sLex in human colonic adenocarcinoma HCT15 cells (Chachadi et al. 2011), respectively. In the absence of β3GalT-I, MUC1-associated sLea was not produced despite expression of high level of B3GALT5 gene in RWPE-1 cells. sLea was generated only after induction of the expression of B3GALT1 gene by treatment with suberoylanilide hydroxamic acid, a histone deacetylase inhibitor (Chachadi et al. 2013). Also, sLex was not detected in HCT15 cells which express very low level of ST3GAL6 but moderate-to-high levels of ST3GAL3, ST3GAL4 and ST3GALT5. Following treatment with 5-aza-2′-deoxycytidine, a DNA methylase inhibitor, sLex was generated following a 754-fold increase in the expression of ST3GAL6 gene. The data support the concept that in vivo, each glycoprotein is glycosylated by its cognate GTs in an isozyme-specific fashion.
Current study provides further evidence to support the concept that in vivo glycosylation of a given glycoprotein is GT isozyme specific. In this study, we have identified specific isozymes of two GTs which participate in the synthesis of MUC-associated sLea and sLex. We have found that β3GalT-V is involved in the synthesis of MUC16-associated sLea in H292 cells. Further, FucT-V is found to be involved in the synthesis of MUC1-associated sLex but not sLea in this type cell. Interestingly, sLex in H292 cells is greatly reduced after knockdown of FUT5 gene even though the moderate expression levels of FUT3, FUT4 and FUT6 are sufficient for generation of sLex if these enzymes have access to MUC1-associated sialylated type-2 lactosamine. FucT-IV is not involved in the synthesis of sLea and sLex because knockdown of mRNA of this gene in CFT1 and H292 cells did not affect synthesis of these selectin ligands. FucT-III appears to be the likely candidate for the synthesis of sLea in H292 cells because FucT-III but not FucT-VI has the affinity for sialylated type-1 lactosamine. Further, lack of FucT-V is the likely explanation for the inability of BEAS-2B and A549 cells to generate MUC1-associatedsLex. The low level of MUC16 gene expression in BEAS-2B, CFT1 and A549 cells may further contribute to the absence of MUC16-associated selectin ligands in these cell types. These predictions remain to be verified. These results clearly show that synthesis of MUC-associated glycans is GT isozyme-specific. However, this phenomenon may be cell type specific because of the following reasons. First, ST3Gal6 is required for synthesis of sLex associated with MUC1 in HCT15 cells (Chachadi et al. 2011) but not H292 cells as shown in current study. Second, β3GalT-1 is involved in the synthesis of MUC1-associated sLea in normal prostatic RWPE-1 cells (Chachadi et al. 2013) but not H292 cells. In this cell type, we show that β3GalT-V is the isozyme which participates in the synthesis of this selectin ligand on MUC1. To gain more insight into this phenomenon, it is necessary to identify GTs required for the synthesis of MUC1-associated sLea and sLex in more cell types. Further, given that Golgi targeting of GTs is altered in aggressive cancer cells, which leads to altered glycosylation (Petrosyan et al. 2014), it is imperative to compare the results between normal and cancerous cells and between same cancer cells with normal and altered Golgi targeting of GTs to determine if the Golgi targeting of a GT remains the same between the above-mentioned cell pair because differences in mucin substrate specificity of ST3Gal6 and β3GalT-1 in different carcinoma cells may be caused by altered Golgi targeting of GTs.
Another interesting finding of current study is that β3GalT-V and FucT-V play an important role in the in vitro metastatic property of H292 cells because knockdown of either gene reduces the migratory and selectin-mediated adhesion properties of these cells. These enzymes and/or the mucins which they work on may serve as potential targets for developing therapeutic strategy to inhibit the metastatic properties of the primary tumors.
Current work provides an additional insight into our concept that each GT isozyme acts only on a subset of glycoproteins even though they show similar sugar substrate specificity and can produce same carbohydrate structure in vitro. Further, it will be of interest to elucidate the mechanism by which GT isozymes meet up with their glycoprotein substrates in cancer cells in vivo, which may provide an opportunity for developing cancer therapy.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM), F-12K, penicillin–streptomycin, fetal bovine serum (FBS), bovine serum albumin (BSA) and trypsin/EDTA were procured from Life Technologies Inc. (Grand Island, NY). Bronchial epithelial growth medium (BEGM) was obtained from Lonza (Walkersville, MD). Solutions of SDS–PAGE and protein estimation kit were purchased from Bio-Rad (Hercules, CA). A mixture of prestained protein molecular weight standard marker was from Fermentas (Glen Burnie, MD). ECL reagent kit was obtained from Pierce Biotechnology (Rockford, IL). Nuclear stain DAPI was from Life Technologies Inc. (Grand Island, NY). KM231 mouse monoclonal Abs to sLea was from EMD Millipore Corporation (Billerica, MA) and anti-sLex (CSLEX1) antibody was procured from BD Biosciences (San Jose, CA). Mouse monoclonal Ab to β-actin was from Sigma (St. Louis, MO). Anti-MUC1 mouse Ab was obtained from Life Span Biosciences (Seattle, WA) while rabbit polyclonal Abs to MUC1 was from Abcam (Cambridge, MA). Anti-MUC4 (8G-7) Ab was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and anti-MUC16 Ab was from Abcam (Cambridge, MA). Horseradish peroxidase, DyLight® 488 (green) and DyLight® 595 (red) conjugated secondary Abs (donkey anti-rabbit, donkey anti-mouse) were obtained from Jackson ImmunoResearch (West Grove, PA). Biotinylated Maackia amurensis lectin (MAA) was purchased from EY laboratories (San Mateo, CA).
Cell culture
Human immortalized bronchial epithelial cells (BEAS-2B) or lung carcinoma cells, including A549 and H292, were obtained from the American Type Culture Collection (Rockville, MD), whereas immortalized CFT1-C2 cells (derived from human tracheal epithelial cells with the CF ΔF508 mutation) were provided by Dr. James Yankaskas (The University of North Carolina at Chapel Hill, NC) (Yankaskas et al. 1993). CFT1 and BEAS-2B cells were grown in BEGM supplemented with hormones and growth factor while A549 and H292 cells were grown in DMEM high glucose medium supplemented with 10% FBS and antibiotics (50 units/mL penicillin and 50 μg/mL streptomycin) at 37°C under 5% CO2 and water saturated environment.
Gel electrophoresis and immunoblotting
Cell homogenate proteins (50 µg) were separated on 2% SDS–agarose gel as well as SDS–4% polyacrylamide gels under reducing conditions and transferred to PVDF membranes overnight at 4°C. The blotted membranes were blocked at RT for 1 h with a blocking solution [tris buffered saline (TBS), pH 7.4, 5% nonfat dried milk, 0.1% Tween-20] and incubated at 4°C overnight in a blocking solution containing primary Abs with appropriate dilutions. After washing with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary Ab for 1 h. Immunoreactive bands were detected by Thermo Scientific SuperSignal West Pico Chemo-luminescent substrate reagents and exposed to BioExpress Blue Basic Autorad chemo-luminescence film (Kaysville, UT). Prestained protein ladder was used as the molecular-weight markers. Immunodetected β-actin was used to ensure equal protein loading after separation by SDS–12% polyacrylamide gel electrophoresis.
Fluorescence microscopy
BEAS-2B, CFT1, A549 and H292 were grown overnight on cover slips and fixed in 4% paraformaldehyde/PBS at RT for 30 min (Chachadi et al. 2013). After treated with primary Ab (1 : 50) at 37°C for 1 h, the cells were stained with dyLight-488 conjugated donkey anti-mouse IgG or IgM Ab and mounted in ProLong Gold antifade reagent with DAPI. Cells processed without primary antibodies served as negative control. Finally, cells stained with sLea and sLex Abs were examined under EVOS® FL digital inverted fluorescence microscope from AMG (Bothell, WA) and photographed at 60× magnification.
Flow cytometric analysis
Quantitative and comparative assessment of cell surface sLea and sLex were performed by flow cytometry using KM231 (sLea) and CSLEX1 (sLex) Abs as described (Chachadi et al. 2011; Chachadi et al. 2013). Finally, cells were re-suspended in 500 µL of 0.5% paraformaldehyde and analyzed using a FACS Vantage (Becton Dickson San Jose, CA) equipped with 488 nm argon laser and Cell Quest-pro software. The unstained cells and cells incubated with secondary Ab alone served as negative and antibody controls, respectively.
Immunoprecipitation
For immunoprecipitation assays, cell lysates containing equal amounts of protein (500 μg) were precleared with 20 μL of protein A/L-coupled sepharose beads (50%, v/v) (Amersham, Little Chalfont, Buckinghamshire, UK) for 1 h at 4°C. The precleared samples were incubated with 5 μg of anti-MUC1, anti-MUC4, anti-MUC16, anti-sLea or anti-sLex Ab overnight at 4°C. The immune complexes were mixed with 20 μL of protein A-coupled (protein l-coupled in case of sLex) sepharose beads (50% v/v) and rotated overnight at 4°C. The trapped immune complexes were subjected to western blot analysis with sLea, sLex or MAA staining. Immunoprecipitates obtained with non-specific mouse IgG and IgM Abs served as negative controls.
In situ PLA
In situ PLA was performed in CFT1 and H292 cells to analyze association of sLea and sLex with MUC1 and MUC16 according to the previously described protocol with minor modifications (Chachadi et al. 2013). In brief, cells were grown on cover slip for 48 h and fixed with 4% cold paraformaldehyde/PBS at RT for 30 min, washed thrice with PBS and blocked for 1 h in 3% BSA. Cells were incubated with the indicated Ab pairs (Figure 2B) (1 : 50 in PBS with 3% BSA) at 37°C for 1 h and washed thrice (5 min each) with PBST. Oligonucleotide-conjugated donkey anti-mouse IgG minus (for sLea), donkey anti-mouse IgM minus (for sLex) and donkey anti-rabbit plus (for MUC1 and MUC16) PLA secondary probes were added at appropriate dilutions prepared in PBS with 3% BSA and the cells were incubated in a humidified chamber for 1 h at 37°C. PLA secondary donkey anti-mouse IgM minus probe was prepared using Duolink® In Situ Probemaker Minus kit (DUO92010) from Sigma-Aldrich (St. Louis, MO) while other probes were directly purchased. The PLA assay was performed using the Duolink PLA kit (Olink Bioscience cat# LNK-92101-KI01, Uppsala, Sweden) according to manufacturer's instruction. Briefly, connector oligonucleotides were hybridized and circularized by ligation for 30 min at 37°C. After thorough washing, the cells were incubated with DNA polymerase for 100 min at 37°C to produce rolling circle amplification products tagged with a red fluorescence probe. Cells were stained with DAPI for nuclei and images were obtained from EVOS® FL digital inverted fluorescence microscope and photographed at 60× magnification. PLA signals (red fluorescent spots) were quantified by counting all signals obtained from three different images divided by the number of cells in those three images to obtain average signals/cell.
PNGase F or mixed glycosidase treatments
The MUC1 and MUC16 immunoprecipitates were boiled (100°C) for 10 min in 0.5% SDS and 1% β-mercaptoethanol and then treated with PNGase F (Sigma) (5 U/mg protein) in 50 mM phosphate, pH 7.5 containing 1% NP-40 at 37°C for 16 h. The PNGase F activity was validated using transferrin as previously described (Chachadi et al. 2011). The immunoprecipitate was also treated with mixed glycosidases containing neuraminidase, β1-4galactosidase/β-N-acetylglucosaminidase and O-glycanase (ProZyme, Inc., San Leandro, CA). The samples treated with or without glycosidases were analyzed by sLea and sLex western blotting. Untreated samples served as the negative controls. For MAA detection, glycoproteins in the sLea and sLex immunoprecipitates were resolved by SDS–PAGE and blotted on to PVDF membrane as described earlier. Then, membranes were blocked with TBS containing 3% BSA (60 min), followed by incubation with 1 µg/mL of biotin-conjugated MAA (in TBS containing 1 mM MgCl2, 1 mM MnCl2 and 1 mM CaCl2, pH 7.5) for 60 min. The treated membranes were washed with TBS containing 0.05% Tween 20 and then incubated with streptavidin-HRP (Pierce) (1 : 20,000) for 30 min. Finally, the blots were washed (5×) with TBS containing 0.05% Tween 20 and then developed using ECL.
Quantitative gene expression analysis of glycosyltransferases and mucins by qRT-PCR
qRT-PCR analyses of the mRNAs of the genes encoding the GTs involved in the synthesis of sialyl Lewis antigens and genes encoding membrane-bound mucins were performed in BEAS-2B, CFT1, A549 and H292 cell types according to the protocol described previously (Chachadi et al. 2011; Chachadi et al. 2013). The results were expressed as % of the target gene relative to that (100%) of GAPDH and plotted as mean expression ± SD. Primer sequences used for the study can be found in earlier publications (Chachadi et al. 2011, 2013; Radhakrishnan et al. 2011).
siRNA transient transfection
Pool of 3 siRNAs targeting each of B3GALT5, FUT4 and FUT5, and scrambled siRNA were purchased from Santa Cruz Biotechnology. CFT1 and H292 cells at 50% confluence were transfected for 8 h in OPTI-MEM® reduced serum medium containing 50 nM siRNAs using Lipofectamine RNAi MAX reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendation. The medium was then replaced with a fresh DMEM medium containing 10% FBS for H292 and BEGM supplemented with hormones and growth factor for CFT1 as mentioned in previous section. After cultured for 72 h, the transfected cells were analyzed for B3GALT5, FUT4 and FUT5 mRNA by qRT-PCR for successful knockdown as mentioned above. Effects of gene knockdown on the expression of sLea and sLex were determined by western blotting as described in previous section.
Wound healing assay
A monolayer scratch assay as described previously (Cory 2011) was used to compare the migratory ability of H292 cells after knocking down the B3GALT5 or FUT5 gene. Knockdown of B3GALT5 and FUT5 was performed. After 72 h of siRNA treatment, H292 cells were dislodged from T25 culture flasks with a non-enzymatic cell stripper and seeded to six-well tissue culture dishes and grown to confluence. Each confluent monolayer was then wounded linearly using a sterile 200 µL pipette tip and washed three times with PBS. Thereafter, migration was observed and photographed at regular intervals of 0, 12, 24 and 48 h. The distance of cells migrating into the cell-free zone was acquired under a phase contrast microscope and analyzed by ImageJ software. The distances of cell migration were calculated by subtracting the distance between the scratch's edges (wound closure) at indicated time from the distance measured at 0 h.
Parallel plate flow chamber adhesion assay
Recombinant human E- and P-selectins/IgG Fc chimeras were purified and coated to cover slips as described (Chachadi et al. 2011; Ali et al. 2012). The siRNA-treated H292 cells that tethered to and rolled on E- and P-selectin-coated cover slips were measured and compared with mock-treated cells by a parallel plate flow chamber assay as described (Wiese et al. 2009; Chachadi et al. 2011) and evaluated according to Krull et al. (1999). Tethering was determined by counting the number of cells that adhered to slides over the first 2–3 s of continuous shear flow at a given field of view and remained tethered for at least 3 s while rolling was defined as five times cell diameters of forward movement which was assessed at 1.0 dyne/cm2. Tethering or rolling was expressed as number of tethered or rolled cells/high-power field during a 10-m observation period.
Statistical analysis
Student's t-test was used for statistical analysis; P-values of <0.05 were considered statistically significant. SigmaPlot software (Systat Software, San Jose, CA) was used for all the graphing and statistical analyses.
Funding
This work is supported in part by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (VA 1I1BX000985), the National Institutes of Health (2RO1HL48282), the State of Nebraska (LB506) (to P.W.C.) and the Alcohol & Education Research Council (hfygr667789).
Conflict of interest statement
None declared.
Abbreviations
Abs, antibodies; BEGM, bronchial epithelial growth medium; BSA, bovine serum albumin; CNT, control; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; GT, glycosyltransferase; HRP, horseradish peroxidase; MAA, Maackia amurensis lectin; NeuAc, N-acetylneuraminic acid; PLA, proximity ligation assay; PNGase, peptide N-glycosidase; qRT-PCR, quantitative real-time polymerase chain reaction; sLea, sialyl Lewis a; sLex, sialyl Lewis x; TBS, tris buffered saline.
Acknowledgements
We thank the Flow Cytometry Research Facility Core supported by the Nebraska Research Initiatives and the Eppley Cancer Center.
References
- Ali MF, Chachadi VB, Petrosyan A, Cheng PW. 2012. Golgi phosphoprotein 3 determines cell binding properties under dynamic flow by controlling Golgi localization of core 2 N-acetylglucosaminyltransferase 1. J Biol Chem. 287(47):39564–39577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado M, Almeida R, Carneiro F, Levery SB, Holmes EH, Nomoto M, Hollingsworth MA, Hassan H, Schwientek T, Nielsen PA, et al. 1998. A family of human β3-galactosyltransferases: Characterization of four members of a udp-galactose:β-n-acetyl-glucosamine/β-nacetyl-galactosamine β-1,3-galactosyltransferase family. J Biol Chem. 273:12770–12778. [DOI] [PubMed] [Google Scholar]
- Amado M, Almeida R, Schwientek T, Clausen H. 1999. Identification and characterization of large galactosyltransferase gene families: Galactosyltransferases for all functions. Biochim Biophys Acta. 1473(1):35–53. [DOI] [PubMed] [Google Scholar]
- Bafna S, Kaur S, Batra SK. 2010. Membrane-bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 29(20):2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel SR, Wiese GK, Cho J, Opperman MJ, Hays DL, Siddiqui J, Pienta KJ, Furie B, Dimitroff CJ. 2009. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc Natl Acad Sci USA. 106(46):19491–19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I. 2006. Mucin-type O-glycans in human colon and breast cancer: Glycodynamics and functions. EMBO Rep. 7(6):599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AS, Harduin-Lepers A, Magalhaes A, Machado E, Mendes N, Costa LT, Matthiesen R, Almeida R, Costa J, Reis CA. 2010. Differential expression of alpha-2,3-sialyltransferases and alpha-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int J Biochem Cell Biol. 42(1):80–89. [DOI] [PubMed] [Google Scholar]
- Cazet A, Julien S, Bobowski M, Burchell J, Delannoy P. 2010. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 12(3):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachadi VB, Ali MF, Cheng PW. 2013. Prostatic cell-specific regulation of the synthesis of MUC1-associated sialyl Lewis a. PLoS ONE. 8(2):e57416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachadi VB, Cheng H, Klinkebiel D, Christman JK, Cheng PW. 2011. 5-Aza-2′-deoxycytidine increases sialyl Lewis X on MUC1 by stimulating beta-galactoside:alpha2,3-sialyltransferase 6 gene. Int J Biochem Cell Biol. 43(4):586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AF, Naumov GN, Varghese HJ, Nadkarni KV, MacDonald IC, Groom AC. 2001. Critical steps in hematogenous metastasis: An overview. Surg Oncol Clin N Am. 10(2):243–255, vii. [PubMed] [Google Scholar]
- Cory G. 2011. Scratch-wound assay. Methods Mol Biol. 769:25–30. [DOI] [PubMed] [Google Scholar]
- Croce MV, Isla-Larrain M, Rabassa ME, Demichelis S, Colussi AG, Crespo M, Lacunza E, Segal-Eiras A. 2007. Lewis x is highly expressed in normal tissues: A comparative immunohistochemical study and literature revision. Pathol Oncol Res. 13(2):130–138. [DOI] [PubMed] [Google Scholar]
- de Vries T, Knegtel RM, Holmes EH, Macher BA. 2001. Fucosyltransferases: Structure/function studies. Glycobiology. 11(10):119R–128R. [DOI] [PubMed] [Google Scholar]
- de Vries T, Srnka CA, Palcic MM, Swiedler SJ, van den Eijnden DH, Macher BA. 1995. Acceptor specificity of different length constructs of human recombinant alpha 1,3/4-fucosyltransferases. Replacement of the stem region and the transmembrane domain of fucosyltransferase V by protein A results in an enzyme with GDP-fucose hydrolyzing activity. J Biol Chem. 270(15):8712–8722. [DOI] [PubMed] [Google Scholar]
- Dimitroff CJ, Lechpammer M, Long-Woodward D, Kutok JL. 2004. Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin. Cancer Res. 64(15):5261–5269. [DOI] [PubMed] [Google Scholar]
- Ellies LG, Sperandio M, Underhill GH, Yousif J, Smith M, Priatel JJ, Kansas GS, Ley K, Marth JD. 2002. Sialyltransferase specificity in selectin ligand formation. Blood. 100(10):3618–3625. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rodriguez J, Dwir O, Alon R, Hansson GC. 2001. Tumor cell MUC1 and CD43 are glycosylated differently with sialyl-Lewis a and x epitopes and show variable interactions with E-selectin under physiological flow conditions. Glycoconj J. 18(11–12):925–930. [DOI] [PubMed] [Google Scholar]
- Fukuda M. 1996. Possible roles of tumor-associated carbohydrate antigens. Cancer Res. 56(10):2237–2244. [PubMed] [Google Scholar]
- Furukawa K, Sato T. 1999. Beta-1,4-galactosylation of N-glycans is a complex process. Biochim Biophys Acta. 1473(1):54–66. [DOI] [PubMed] [Google Scholar]
- Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Vincent A, Lehoux S, Lafitte JJ, Van Seuningen I, Delannoy P. 2008. IL-6 and IL-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated Lewisx epitopes in the human bronchial mucosa. Biochem J. 410(1):213–223. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. 2001. The human sialyltransferase family. Biochimie. 83(8):727–737. [DOI] [PubMed] [Google Scholar]
- Hennet T. 2002. The galactosyltransferase family. Cell Mol Life Sci. 59(7):1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T, Dinter A, Kuhnert P, Mattu TS, Rudd PM, Berger EG. 1998. Genomic cloning and expression of three murine UDP-galactose: Beta-N-acetylglucosamine beta1,3-galactosyltransferase genes. J Biol Chem. 273(1):58–65. [DOI] [PubMed] [Google Scholar]
- Holgersson J, Löfling J. 2006. Glycosyltransferases involved in type 1 chain and Lewis antigen biosynthesis exhibit glycan and core chain specificity. Glycobiology. 16(7):584–593. [DOI] [PubMed] [Google Scholar]
- Homeister JW, Thall AD, Petryniak B, Maly P, Rogers CE, Smith PL, Kelly RJ, Gersten KM, Askari SW, Cheng G, et al. 2001. The alpha(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity. 15:115–126. [DOI] [PubMed] [Google Scholar]
- Huang MC, Zollner O, Moll T, Maly P, Thall AD, Lowe JB, Vestweber D. 2000. P-selectin glycoprotein ligand-1 and E-selectin ligand-1 are differentially modified by fucosyltransferases Fuc-TIV and Fuc-TVII in mouse neutrophils. J Biol Chem. 275:31353–31360. [DOI] [PubMed] [Google Scholar]
- Isshiki S, Togayachi A, Kudo T, Nishihara S, Watanabe M, Kubota T, Kitajima M, Shiraishi N, Sasaki K, Andoh T, et al. 1999. Cloning, expression, and characterization of a novel UDP-galactose:beta-N-acetylglucosamine beta1,3-galactosyltransferase (beta3Gal-T5) responsible for synthesis of type 1 chain in colorectal and pancreatic epithelia and tumor cells derived therefrom. J Biol Chem. 274(18):12499–12507. [DOI] [PubMed] [Google Scholar]
- Kannagi R. 2007. Carbohydrate antigen sialyl Lewis a – its pathophysiological significance and induction mechanism in cancer progression. Chang Gung Med J. 30(3):189–209. [PubMed] [Google Scholar]
- Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. 2004. Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci. 95(5):377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarada Y, Ishikura H, Kishimoto T, Kato H, Yano T, Kato H, Yoshiki T. 2000. The role of sialylated Lewis antigens on hematogenous metastases of human pancreas carcinoma cell lines in vivo. Pathol Res Pract. 196(4):259–263. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Borsig L, Han HL, Varki NM, Varki A. 1999. Distinct selectin ligands on colon carcinoma mucins can mediate pathological interactions among platelets, leukocytes, and endothelium. Am J Pathol. 155(2):461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Paulson JC. 1993. Cloning and expression of human Gal beta 1,3(4)GlcNAc alpha 2,3-sialyltransferase. Biochem Biophys Res Commun. 194(1):375–382. [DOI] [PubMed] [Google Scholar]
- Krull M, Klucken AC, Wuppermann FN, Fuhrmann O, Magerl C, Seybold J, Hippenstiel S, Hegemann JH, Jantos CA, Suttorp N. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J Immunol. 162(8):4834–4841. [PubMed] [Google Scholar]
- Laubli H, Borsig L. 2010. Selectins promote tumor metastasis. Semin Cancer Biol. 20(3):169–177. [DOI] [PubMed] [Google Scholar]
- Li G, Sanders JM, Phan ET, Ley K, Sarembock IJ. 2005. Arterial macrophages and regenerating endothelial cells express P-selectin in atherosclerosis-prone apolipoprotein E-deficient mice. Am J Pathol. 167(6):1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Joffraud M, Giraud S, Baisse B, Bernimoulin MP, Schapira M, Spertini O. 2005. Regulation of PSGL-1 interactions with L-selectin, P-selectin, and E-selectin: Role of human fucosyltransferase-IV and -VII. J Biol Chem. 280:5378–5390. [DOI] [PubMed] [Google Scholar]
- Nakayama F, Teraki Y, Kudo T, Togayachi A, Iwasaki H, Tamatani T, Nishihara S, Mizukawa Y, Shiohara T, Narimatsu H. 2000. Expression of cutaneous lymphocyte-associated antigen regulated by a set of glycosyltransferases in human T cells: Involvement of alpha1, 3-fucosyltransferase VII and beta1,4-galactosyltransferase I. J Invest Dermatol. 115(2):299–306. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Watanabe M, Katsumata T, Teramoto T, Kitajima M. 1995. Expression of sialyl Lewis(a) as a new prognostic factor for patients with advanced colorectal carcinoma. Cancer. 75(8):2051–2056. [DOI] [PubMed] [Google Scholar]
- Niemelä R, Natunen J, Majuri ML, Maaheimo H, Helin J, Lowe JB, Renkonen O, Renkonen R. 1998. Complementary acceptor and site specificities of Fuc-TIV and Fuc-TVII allow effective biosynthesis of Sialyl-TriLex and related polylactosamines present on glycoprotein counterreceptors of selectins, J Biol Chem. 273:4021–4026. [DOI] [PubMed] [Google Scholar]
- Norden R, Nystrom K, Olofsson S. 2009. Activation of host antiviral RNA-sensing factors necessary for herpes simplex virus type 1-activated transcription of host cell fucosyltransferase genes FUT3, FUT5, and FUT6 and subsequent expression of sLe(x) in virus-infected cells. Glycobiology. 19(7):776–788. [DOI] [PubMed] [Google Scholar]
- Okajima T, Fukumoto S, Miyazaki H, Ishida H, Kiso M, Furukawa K, Urano T, Furukawa K. 1999. Molecular cloning of a novel alpha2,3-sialyltransferase (ST3Gal VI) that sialylates type II lactosamine structures on glycoproteins and glycolipids. J Biol Chem. 274(17):11479–11486. [DOI] [PubMed] [Google Scholar]
- Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C. 1999. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology. 9(4):323–334. [DOI] [PubMed] [Google Scholar]
- Padró M, Cobler L, Garrido M, de Bolós C. 2011. Down-regulation of FUT3 and FUT5 by shRNA alters Lewis antigens expression and reduces the adhesion capacities of gastric cancer cells. Biochim Biophys Acta. 1810(12):1141–1149. [DOI] [PubMed] [Google Scholar]
- Paganuzzi M, Bobbio B, Marroni P, Filiberti R, Secco GB, Grossi CE. 2003. Prognostic role of serum sialyl Lewisx (CD15s) in colorectal cancer. Oncology. 65(1):52–59. [DOI] [PubMed] [Google Scholar]
- Petrosyan A, Holzapfel MS, Muirhead DE, Cheng P-W. 2014. Restoration of compact Golgi morphology in advanced prostate cancer enhances susceptibility to galectin 1-induced apoptosis by modifying mucin O-glycan synthesis. Mol Cancer Res. 12(12):1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan P, Chachadi V, Lin MF, Singh R, Kannagi R, Cheng PW. 2011. TNFalpha enhances the motility and invasiveness of prostatic cancer cells by stimulating the expression of selective glycosyl- and sulfotransferase genes involved in the synthesis of selectin ligands. Biochem Biophys Res Commun. 409(3):436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F, Hollingsworth MA. 2013. Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 19(8):1981–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohfritsch PF, Joosten JA, Krzewinski-Recchi MA, Harduin-Lepers A, Laporte B, Juliant S, Cerutti M, Delannoy P, Vliegenthart JF, Kamerling JP. 2006. Probing the substrate specificity of four different sialyltransferases using synthetic beta-D-Galp-(1–>4)-beta-D-GlcpNAc-(1–>2)-alpha-D-Manp-(1–>O) (CH(2))7CH3 analogues general activating effect of replacing N-acetylglucosamine by N-propionylglucosamine. Biochim Biophys Acta. 1760(4):685–692. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Watanabe E, Kawashima K, Sekine S, Dohi T, Oshima M, Hanai N, Nishi T, Hasegawa M. 1993. Expression cloning of a novel Gal beta (1–3/1–4) GlcNAc alpha 2,3-sialyltransferase using lectin resistance selection. J Biol Chem. 268(30):22782–22787. [PubMed] [Google Scholar]
- Shimodaira K, Nakayama J, Nakamura N, Hasebe O, Katsuyama T, Fukuda M. 1997. Carcinoma-associated expression of core 2 beta-1,6-N-acetylglucosaminyltransferase gene in human colorectal cancer: Role of O-glycans in tumor progression. Cancer Res. 57(23):5201–5206. [PubMed] [Google Scholar]
- Shinoda K, Tanahashi E, Fukunaga K, Ishida H, Kiso M. 1998. Detailed acceptor specificities of human alpha1,3-fucosyltransferases, Fuc-TVII and Fuc-TVI. Glycoconj J. 15(10):969–974. [DOI] [PubMed] [Google Scholar]
- Sumikura S, Ishigami S, Natsugoe S, Miyazono F, Tokuda K, Nakajo A, Okumura H, Matsumoto M, Hokita S, Aikou T. 2003. Disseminated cancer cells in the blood and expression of sialylated antigen in gastric cancer. Cancer Lett. 200(1):77–83. [DOI] [PubMed] [Google Scholar]
- Tsuji S. 1996. Molecular cloning and functional analysis of sialyltransferases. J Biochem. 120(1):1–13. [DOI] [PubMed] [Google Scholar]
- Ugorski M, Laskowska A. 2002. Sialyl Lewis(a): A tumor-associated carbohydrate antigen involved in adhesion and metastatic potential of cancer cells. Acta Biochim Pol. 49(2):303–311. [PubMed] [Google Scholar]
- Weinstein J, de Souza-e-Silva U, Paulson JC. 1982. Purification of a Gal beta 1 to 4GlcNAc alpha 2 to 6 sialyltransferase and a Gal beta 1 to 3(4)GlcNAc alpha 2 to 3 sialyltransferase to homogeneity from rat liver. J Biol Chem. 257(22):13835–13844. [PubMed] [Google Scholar]
- Wen DX, Livingston BD, Medzihradszky KF, Kelm S, Burlingame AL, Paulson JC. 1992. Primary structure of Gal beta 1,3(4)GlcNAc alpha 2,3-sialyltransferase determined by mass spectrometry sequence analysis and molecular cloning. Evidence for a protein motif in the sialyltransferase gene family. J Biol Chem. 267(29):21011–21019. [PubMed] [Google Scholar]
- Wiese G, Barthel SR, Dimitroff CJ. 2009. Analysis of physiologic E-selectin-mediated leukocyte rolling on microvascular endothelium. J Vis Exp. (24). pii: 1009 doi:10.3791/1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankaskas JR, Haizlip JE, Conrad M, Koval D, Lazarowski E, Paradiso AM, Rinehart CA, Jr, Sarkadi B, Schlegel R, Boucher RC. 1993. Papilloma virus immortalized tracheal epithelial cells retain a well-differentiated phenotype. Am J Physiol. 264(5 Pt 1):C1219–C1230. [DOI] [PubMed] [Google Scholar]