Abstract
Chronic infections induce a complex immune response that controls pathogen replication, but also causes pathology due to sustained inflammation. Ca2+ influx mediates T cell function and immunity to infection, and patients with inherited mutations in the gene encoding the Ca2+ channel ORAI1 or its activator stromal interaction molecule 1 (STIM1) are immunodeficient and prone to chronic infection by various pathogens, including Mycobacterium tuberculosis (Mtb). Here, we demonstrate that STIM1 is required for T cell–mediated immune regulation during chronic Mtb infection. Compared with WT animals, mice with T cell–specific Stim1 deletion died prematurely during the chronic phase of infection and had increased bacterial burdens and severe pulmonary inflammation, with increased myeloid and lymphoid cell infiltration. Although STIM1-deficient T cells exhibited markedly reduced IFN-γ production during the early phase of Mtb infection, bacterial growth was not immediately exacerbated. During the chronic phase, however, STIM1-deficient T cells displayed enhanced IFN-γ production in response to elevated levels of IL-12 and IL-18. The lack of STIM1 in T cells was associated with impaired activation-induced cell death upon repeated TCR engagement and pulmonary lymphocytosis and hyperinflammation in Mtb-infected mice. Chronically Mtb-infected, STIM1-deficient mice had reduced levels of inducible regulatory T cells (iTregs) due to a T cell–intrinsic requirement for STIM1 in iTreg differentiation and excessive production of IFN-γ and IL-12, which suppress iTreg differentiation and maintenance. Thus, STIM1 controls multiple aspects of T cell–mediated immune regulation to limit injurious inflammation during chronic infection.
Keywords: Immunology, Infectious disease, Microbiology
Introduction
Modulation of intracellular Ca2+ concentrations is an important mechanism for regulating the function of many immune cells, especially T cells (1). In response to antigen binding to the T cell receptor (TCR), Ca2+ is released from ER stores. The subsequent decrease in the ER Ca2+ concentration is sensed by stromal interaction molecule 1 (STIM1) and its homologue STIM2, which translocate and bind to ORAI1 in the plasma membrane. ORAI1 is the pore-forming subunit of the Ca2+ release–activated Ca2+ (CRAC) channel. Binding of STIM1 to ORAI1 opens the CRAC channel, which mediates sustained Ca2+ influx from the extracellular space. This form of Ca2+ influx is called store-operated Ca2+ entry (SOCE) and controls numerous Ca2+-dependent signaling events, including activation of transcription factors of the nuclear factor of activated T cells (NFAT) family and expression of cytokine genes in human and murine T cells (2). Accordingly, T cells from mice with conditional deletion of Stim1 or both Stim1/Stim2 genes have severely reduced production of IFN-γ, IL-2, and other cytokines. T cell–specific deletion of Stim1/Stim2 results in increased susceptibility to acute viral infection with lymphocytic choriomeningitis virus (LCMV) (3) and impaired antitumor immunity (4) due to perturbed CD8+ T cell effector functions. Furthermore, both CD4+ and CD8+ T cells require SOCE mediated by STIM1/STIM2 to sustain memory CD8+ T cell response to acute viral infection (3). Patients (PATs) with inherited mutations in ORAI1 or STIM1 genes that abolish SOCE develop a SCID-like disease termed CRAC channelopathy (5, 6). Although T cell development is normal, PATs’ CD4+ and CD8+ T cells proliferate poorly after TCR stimulation in vitro and have reduced production of IFN-γ and other cytokines (7–9). Impaired T cell functions result in chronic bacterial and viral infections (7–13). Furthermore, several SOCE-deficient PATs vaccinated with attenuated Mycobacterium bovis (bacillus Calmette-Guerin [BCG]) displayed pathologic lymphoproliferation (ref. 7 and S. Feske, unpublished observations), suggesting that SOCE may also be required for orchestrating immune regulatory functions in response to mycobacterial infections.
Tuberculosis (TB) is a chronic infection caused by Mycobacterium tuberculosis (Mtb) and represents one of the most common causes of infection-related death worldwide. Following deposition in lung alveoli, Mtb infects alveolar macrophages (AM) and other lung myeloid cells, i.e., neutrophils, DCs, and recruited interstitial macrophages (RIM) (14). Despite active mechanisms of immune evasion deployed by Mtb, a delayed adaptive immune response is established in the lung-draining lymph node, and effector T cells migrate to the lungs (15). There, display of Mtb antigens, TCR, and costimulatory signals, together with signals received from IL-12, IL-18, and other cytokines produced by myeloid cells, results in the production of IFN-γ by T cells (14, 16–18). In turn, IFN-γ activates myeloid cells to kill intracellular mycobacteria, although additional evasion mechanisms limit the effectiveness of this response and lead to Mtb persistence (14, 19). The importance of IFN-γ for antimycobacterial immunity is emphasized by Ifng–/– mice, in which Mtb causes disseminated infection and early mortality (20, 21). PATs with mutations in IL12B, IL12RB1, IFN-GR1, IFN-GR2, or STAT1 genes that impair IL-12/IFN-γ–dependent signaling between CD4+ T cells and myeloid cells have an increased susceptibility to systemic infections with low virulence mycobacteria (17, 22). Despite the protective role of IFN-γ in early TB, PATs with high levels of IFN-γ seem more likely to progress to active disease (17), suggesting that IFN-γ levels during chronic infection correlate better with bacterial burden than with bacterial control.
During chronic infections, T cells are continuously activated by persistent pathogens (23). Mtb-infected macrophages and other myeloid cells induce T cells to continuously produce cytokines such as IFN-γ or TNF-α (19), thus contributing to chronic lung inflammation. Therefore, T cell activation is tightly regulated to prevent excessive tissue damage in the course of chronic TB (24). Mechanisms limiting sustained immune responses and immunopathology associated with chronic infection and inflammation include signaling through inhibitory receptors such as PD-1 (25), activation-induced cell death (AICD or apoptosis) in T cells (26–28), and regulatory T cells (Tregs) that express the lineage-specific transcription factor FOXP3 (29, 30). However, the ambiguous role of Tregs in delaying the onset and/or limiting the efficiency of the adaptive immune response to Mtb has attracted most of the attention in the field, and little is known about their role in controlling inflammation during chronic infection (31).
To investigate the role of SOCE in immunity to Mtb and the immune regulation of chronic infection, we studied Mtb infection in mice with conditional deletion of Stim1 in T cells. We found that, while STIM1-mediated Ca2+ influx is required for optimal production of IFN-γ in early Mtb infection, it mostly plays important immune regulatory functions in T cells during chronic Mtb infection, thereby limiting injurious pulmonary hyperinflammation. Taken together, our results show that STIM1 is a critical regulator of T cell responses in chronic infection.
Results
STIM1 in T cells is required to control chronic Mtb infection in mice.
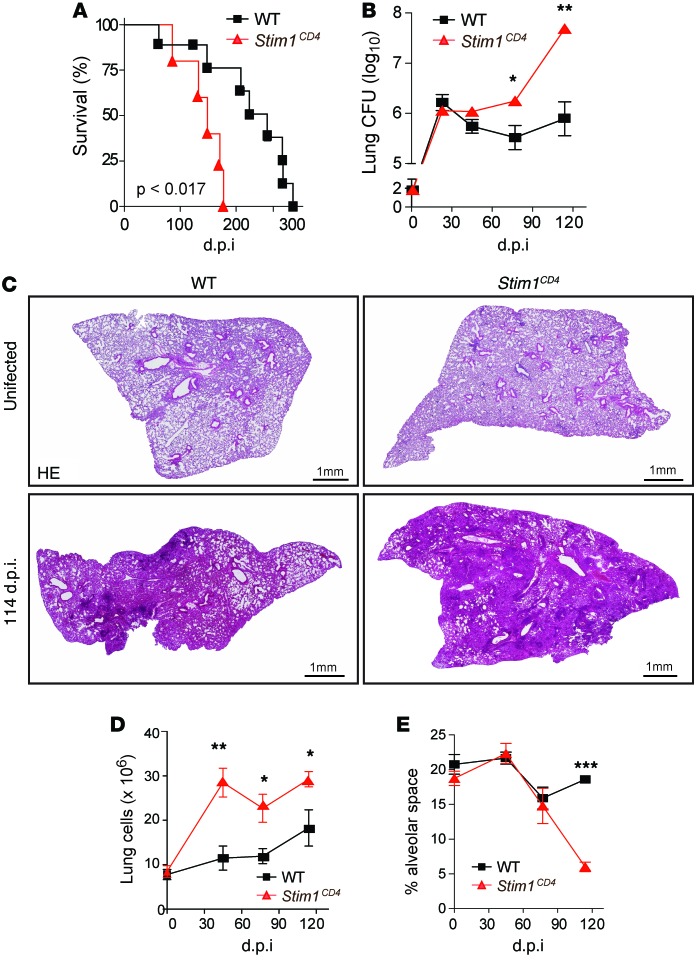
To investigate the role of STIM1 in adaptive immunity to chronic infection, we infected WT and Stim1fl/fl Cd4-Cre (Stim1CD4) mice, whose CD4+ and CD8+ T cells lack SOCE (ref. 32 and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI80273DS1), with aerosolized Mtb. Unexpectedly, Stim1CD4 mice survived the acute phase of infection, but died significantly earlier than WT littermates during chronic Mtb infection (median survival time: 149 days post infection [d.p.i.] vs. 240 d.p.i.; Figure 1A). Premature mortality of Stim1CD4 mice was accompanied by very high lung bacterial burdens at late (>70 d.p.i.) but not early (<45 d.p.i.) stages of infection when compared with WT mice (Figure 1B). By 114 d.p.i., when Stim1CD4 mice started to die, their lungs harbored 37 times more bacteria than WT mice. The lungs of chronically infected Stim1CD4 mice showed pronounced inflammation and consolidation, with increased cellularity as early as 45 d.p.i. and reduced alveolar spaces by 114 d.p.i. when compared with infected WT littermates and uninfected Stim1CD4 mice (Figure 1, C–E).
Figure 1. STIM1 in T cells is required to control chronic Mtb infection in mice.
(A and B) Survival curves (A) and lung bacterial burden (B) of Stim1CD4 and WT control mice infected with 100 CFU of aerosolized Mtb strain H37Rv. Results are representative of 3 independent experiments. (C) H&E stains of lung sections at 114 d.p.i. Pictures are representative of 5 mice per group. Original magnification, ×40. (D) Averaged total numbers of live cells isolated from the lungs of 3 to 5 mice per group and time point. (E) Open alveolar spaces quantified from histological sections in part C of 4–5 mice per group and time point using ImageJ software. Statistical significance was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
At late stages of infection (114 d.p.i.), the lungs of Mtb-infected mice contained large numbers of CD68+ monocytes/macrophages (Figure 2A). Whereas myeloid infiltrates were focal in WT mice, delineating a classic pulmonary granulomatous response, the lungs of Stim1CD4 mice were diffusely infiltrated with CD68+ cells. Flow cytometry analysis revealed that numbers of AM, neutrophils, monocytes, and RIM were already elevated by 45 d.p.i. in the lungs of Mtb-infected Stim1CD4 mice compared with infected WT littermates (Figure 2, B–D). This was in contrast to uninfected Stim1CD4 mice, which showed a size and composition of lung myeloid cell populations comparable to those of uninfected WT mice (Supplemental Figure 1B). Later in the course of Mtb infection, myeloid cells accumulated even more substantially in the lungs of Mtb-infected Stim1CD4 mice and, by 114 d.p.i., most populations, including myeloid DCs (mDC), were significantly (P < 0.05) increased. Additionally, levels of myeloid growth factors, inflammatory cytokines, and chemokines, such as IL-1β, MCP-1, MIP-1α, and RANTES, were markedly increased in the lungs of Mtb-infected Stim1CD4 mice compared with WT controls (Figure 2E). These findings demonstrate that, in the absence of STIM1 in T cells, Mtb infection results in an early onset, progressive pulmonary hyperinflammation that culminates during the chronic phase of infection.
Figure 2. STIM1 in T cells controls myeloid cell infiltration of Mtb-infected lungs.
(A) Immunohistochemistry of CD68 expression on monocytes/macrophages in lungs of Mtb-infected mice at 114 d.p.i. Images are representative of 5 mice per group. Original magnification, ×40 (left panels); ×400 (right panels). (B and C) Frequencies of lung myeloid cell populations at 114 d.p.i. Representative flow cytometry plots of 5 mice per group analyzed. Blue gate numbers represent CD11b–CD11c+Gr1– AM (no. 1), CD11b+CD11c+Gr1– mDC (no. 2), CD11b+CD11c–Gr1hi neutrophils (no. 3), CD11b+CD11c–Gr1int monocytes (no. 4), and CD11b+CD11c–Gr1– RIM (no. 5). (D) Absolute numbers of myeloid cell populations in the lungs of WT and Stim1CD4 mice (as defined in B and C) over the course of Mtb infection. Line graphs show mean ± SEM of 5 mice per group and time point. (E) Concentrations of the proinflammatory cytokine IL-1β, myeloid growth factors (GM-CSF, M-CSF), and chemokines (MCP-1/CCL2, MIP-1α/CCL3, RANTES/CCL5) in lung homogenate supernatants from Mtb-infected WT and Stim1CD4 mice analyzed at 107 d.p.i. and 114 d.p.i. by multiplex analysis. Individual values for both days are pooled and indicated by symbols. Bar graphs show the mean of 9 mice (4 on 107 d.p.i. and 5 on 114 d.p.i.) per group. Statistical significance was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
STIM1 controls AICD of T cells during chronic infection.
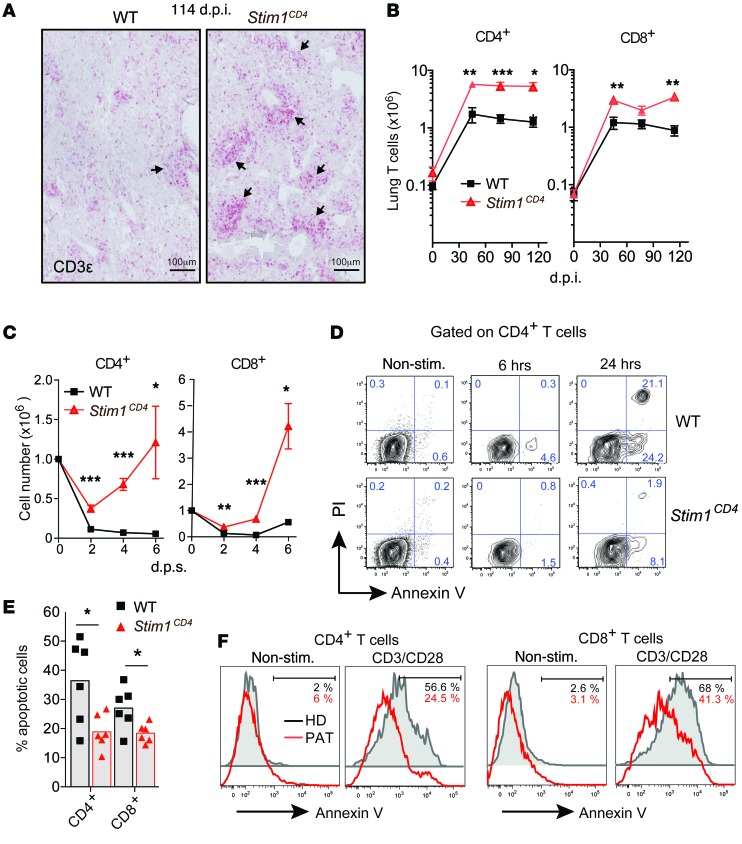
In addition to infiltration by myeloid cells, the lungs of Mtb-infected Stim1CD4 mice showed strong infiltration by CD3+ T cells compared with WT littermates (Figure 3A). Pulmonary infiltrates in Stim1CD4 mice contained significantly more CD4+ and CD8+ T cells than in WT mice as early as 45 d.p.i. and throughout Mtb infection (Figure 3B). Notably, lung T cell numbers were comparable in WT and Stim1CD4 mice before infection. These findings indicate that proliferation of T cells in vivo and their ability to home to sites of infection are independent of SOCE, confirming previous findings that SOCE is not required for T cell homing to secondary lymphoid organs or tumors (4, 33). During chronic infection, T cells continuously encounter antigen and are repeatedly stimulated through their TCR. To mimic this recurring antigen exposure and investigate the role of STIM1 in T cell homeostasis, we repeatedly stimulated CD4+ and CD8+ T cells from uninfected WT and Stim1CD4 with αCD3 in vitro. Repeated TCR stimulation induced a rapid decline in absolute numbers of CD4+ and CD8+ T cells from WT mice, whereas STIM1-deficient T cells expanded vigorously (Figure 3C). T cells continuously encountering antigen in vivo undergo AICD, thereby limiting the immune response (26). We observed significantly reduced frequencies of annexin V+ apoptotic cells among αCD3-restimulated CD4+ and CD8+ T cells from Stim1CD4 mice compared with those from WT littermates (Figure 3, D and E). To confirm that SOCE also regulates AICD in human T cells, we used CD4+ and CD8+ T cells from a PAT with an ORAI1 p.R91W loss-of-function mutation that abolishes SOCE (Supplemental Figure 2A and ref. 10). Restimulation of the PAT’s T cells with αCD3/αCD28 showed a resistance to AICD similar to that found with STIM1-deficient mouse T cells (Figure 3F). These findings demonstrate that STIM1 is required for TCR-mediated apoptosis of T cells following repeated stimulation.
Figure 3. STIM1 controls AICD of T cells during chronic infection.
(A) Infiltration of T cells into the lungs of WT and Stim1CD4 mice at 114 d.p.i. with Mtb. T cells were stained with αCD3 antibody (red); images are representative of 5 mice per group. Original magnification, ×400. Arrows indicate inflammatory infiltrates with T cell accumulation. (B) Absolute numbers of CD4+ and CD8+ T cells in lungs of WT and Stim1CD4 mice determined by flow cytometry. Line graphs show mean ± SEM for 4 to 5 mice per group and time point. (C) Absolute numbers of in vitro–expanded splenic CD4+ and CD8+ T cells from uninfected WT and Stim1CD4 mice after a second stimulation with 2 μg/ml αCD3. Line graphs show mean ± SEM for 3 mice per group and time point. d.p.s., days post stimulation. (D and E) Apoptosis of in vitro–expanded CD4+ T cells isolated from spleens of uninfected WT and Stim1CD4 mice and restimulated with αCD3 for 6 and 24 hours; cells were analyzed by flow cytometry for annexin V and PI. Representative contour plots (D) and frequencies (E) of annexin V+ apoptotic CD4+ and CD8+ T cells 24 hours after restimulation with αCD3. Each dot represents 1 mouse from 2 independent experiments with 3 mice per group. (F) Apoptosis in primary T cells from the peripheral blood of a HD and a SOCE-deficient PAT with ORAI1 p.R91W mutation (10) after restimulation for 16 hours with αCD3/αCD28-coated beads. Histograms of annexin V staining are representative of 2 independent experiments. Statistical significance in B–E was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
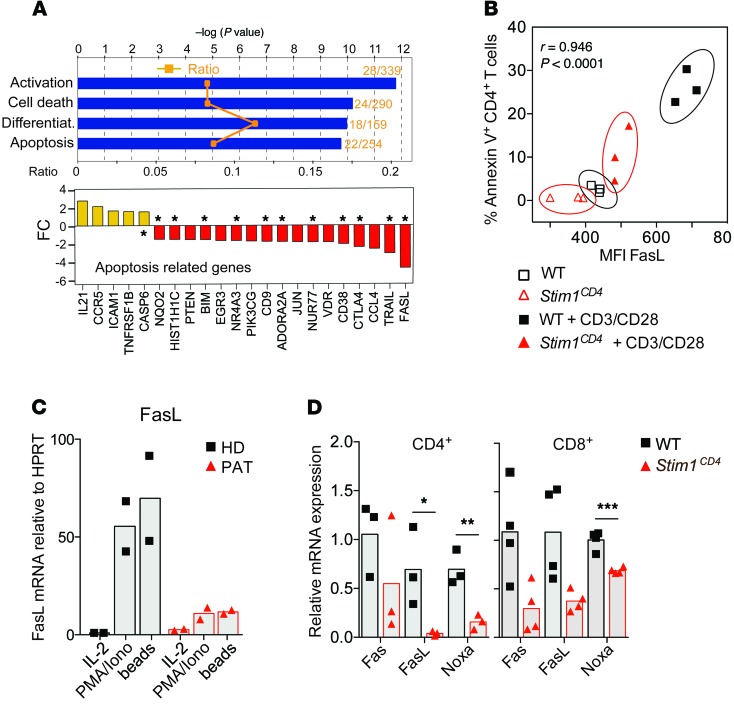
To further investigate the role of STIM1 in the regulation of AICD, we analyzed gene-expression profiles in CD4+ T cells from WT or Stim1CD4 mice after TCR stimulation. Pathway analysis of molecules differentially regulated in STIM1-deficient compared with WT CD4+ T cells revealed downregulation of numerous proapoptotic factors including Fas ligand (FasL), TRAIL, NUR77, and BIM, whereas no equivalent upregulation of proapoptotic molecules, except CASP6, was observed (Figure 4A). FasL protein expression was also reduced in STIM1-deficient T cells compared with WT controls after αCD3 restimulation and correlated strongly with decreased frequencies of annexin V+ apoptotic T cells (Figure 4B). A similar reduction of FASLG mRNA expression was observed in T cells of the ORAI1-deficient PAT after PMA/ionomycin or αCD3/αCD28 restimulation in vitro (Figure 4C), consistent with regulation of FasL expression by SOCE (4, 34). To determine whether STIM1 regulates proapoptotic factors in T cells during chronic Mtb infection in vivo, we analyzed mRNA expression of proapoptotic factors, normalized to Cd4 and Cd8 mRNA expression, in lung homogenates of WT or Stim1CD4 at 77 d.p.i. We observed significantly decreased expression of several proapoptotic factors, including Fas, FasL, and NOXA, in the lungs of Stim1CD4 mice compared with littermate controls during chronic infection (Figure 4D). Our data demonstrate that STIM1 and SOCE regulate expression of proapoptotic factors in T cells and function as inducers of AICD following repeated TCR stimulation. This mechanism may explain how, in the absence of STIM1 and thereby AICD, T cells expand unchecked in the lungs of Mtb-infected mice.
Figure 4. STIM1-dependent regulation of proapoptotic factors in T cells.
(A) Microarray analysis of mRNA expression in CD4+ cells from the spleens of WT or Stim1CD4 mice (3 mice per group) stimulated for 48 hours with αCD3/αCD28. Bar graphs show the results of a pathway analysis (IPA software) indicating the ratio of dysregulated genes per pathway (orange symbols, top panel), statistical significance (blue bars, top panel), and average fold changes (FC) in the expression of apoptosis-regulating factors (bottom panel; P < 0.05 for all genes shown). Asterisks denote known proapoptotic factors in T cells. (B) Correlation of the mean fluorescence intensity (MFI) of FasL expression on splenic CD4+ T cells and frequency of annexin V+ cells with or without restimulation with 2 μg/ml αCD3 for 16 hours. Correlation coefficient was calculated by Spearman’s rho test. (C) FasL expression by primary T cells from the peripheral blood of an HD and a PAT with ORAI1 p.R91W mutation (PAT) (10) after restimulation of cells for 16 hours with PMA/ionomycin (P+I), IL-2, or αCD3/αCD28-coated beads. Bar graphs are representative of 2 independent experiments. (D) Transcript levels of proapoptotic factors Noxa, Fas, and FasL in lungs of WT and Stim1CD4 mice 77 days after Mtb infection, as determined by quantitative RT-PCR. Gene expression was normalized to Cd4 and Cd8 mRNA levels. Each dot represents 1 mouse. Statistical significance was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Dual role of STIM1 in TCR- and IL-12/IL-18–induced IFN-γ production.
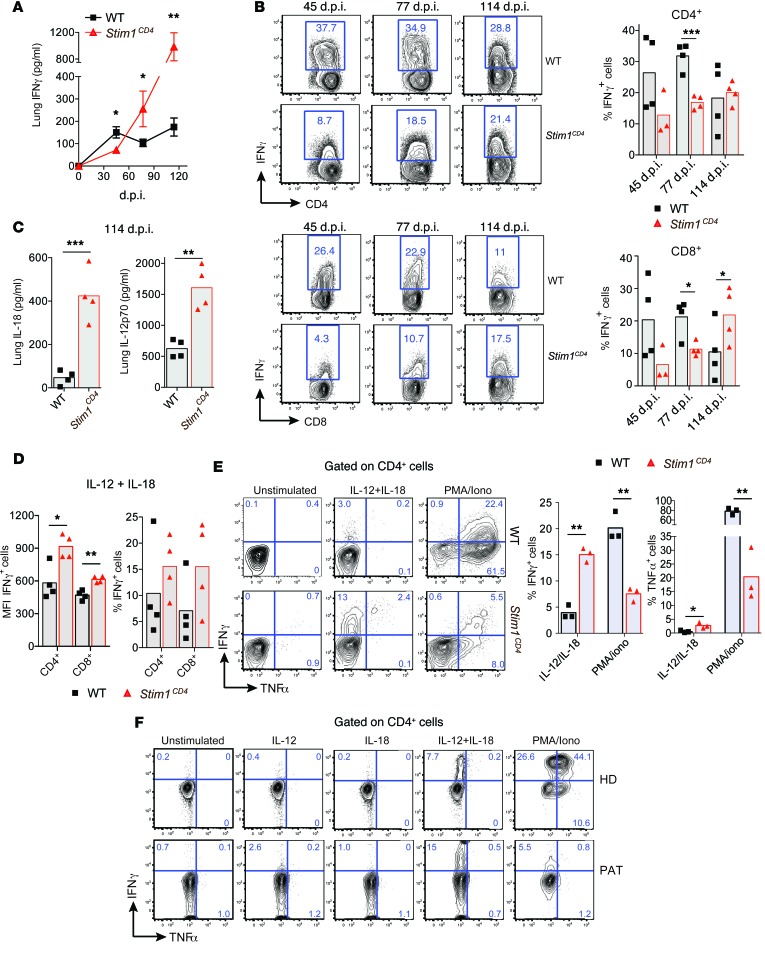
IFN-γ is critical for controlling mycobacterial infections (20, 22), notably via activation of macrophages to kill phagocytosed mycobacteria (35, 36). Early during Mtb infection (45 d.p.i.), we found significantly reduced IFN-γ levels in lung supernatants of Stim1CD4 mice (Figure 5A) and impaired IFN-γ production by isolated STIM1-deficient CD4+ and CD8+ T cells compared with WT controls (Figure 5B). This is consistent with our previous observations that SOCE is required for TCR-mediated IFN-γ production by human and mouse T cells in vitro (32, 37, 38). Surprisingly, however, lower IFN-γ levels in the lungs of Stim1CD4 mice were not accompanied by higher bacterial burdens (Figure 1B). To explain this discrepancy, we compared IFN-γ production and bacterial loads in Mtb-infected WT, Ifng heterozygous (Ifng+/–), and Ifng null (Ifng–/–) mice (Supplemental Figure 3). As expected, Ifng–/– mice failed to control bacterial growth and had approximately 10-fold increased pulmonary bacterial burdens as well as significantly (P < 0.05) larger pulmonary cellularity at 35 d.p.i compared with WT controls. Although Ifng+/– mice showed a significantly (P < 0.01) reduced frequency of IFN-γ–producing CD4+ T cells compared with WT controls, their pulmonary cellularity was comparable, and they were equally able to control Mtb in their lungs. These results indicate that, although IFN-γ is essential for protection against Mtb, it is produced in excess of what is needed for immunity and suggest that the reduced, but not absent, IFN-γ production by STIM1-deficient T cells is sufficient to control Mtb during acute infection.
Figure 5. STIM1 is required for IFN-γ production during acute but not chronic Mtb infection.
(A) IFN-γ in supernatants of lung homogenates from Mtb-infected WT and Stim1CD4 mice measured by ELISA. Line graphs show mean ± SEM for 5 mice per group and time points. (B) Frequencies of IFN-γ+ CD4+ and CD8+ T cells from the lungs of WT or Stim1CD4 mice at indicated time points p.i.; cells were restimulated ex vivo with PMA/ionomycin for 6 hours. Representative contour plots (left) and frequencies of IFN-γ+ T cells (each dot represents 1 mouse). (C) IL-12p70 and IL-18 in supernatants of lung homogenates of WT or Stim1CD4 mice measured by ELISA at 114 d.p.i. (D) IFN-γ production by lung T cells of Mtb-infected WT and Stim1CD4 mice isolated at 114 d.p.i. and restimulated ex vivo with recombinant IL-12 and IL-18 for 6 hours. Shown are MFI and frequencies of IFN-γ+ cells. Each dot represents 1 mouse. (E) Splenic CD4+ T cells from noninfected WT or Stim1CD4 mice were stimulated with αCD3/αCD28 for 6 days in vitro and restimulated with IL-12/IL-18 or PMA/ionomycin for 6 hours. Representative contour plots (left) and frequencies of IFN-γ+ and TNF-α+ T cells (right; each dot represents 1 mouse). (F) IFN-γ and TNF-α production by PBL from a PAT with ORAI1 p.R91W mutation (PAT) (10) and cord blood cells from a HD after stimulation with recombinant IL-12, IL-18, IL-12/IL-18, or PMA/ionomycin for 6 hours. Shown are representative contour plots of CD4+ T cells from 2 independent experiments (for corresponding CD8+ T cell data, see Supplemental Figure 2B). Statistical significance was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Unexpectedly, IFN-γ levels increased in the lung supernatants of Mtb-infected Stim1CD4 mice as the infection became chronic (>77 d.p.i.) and exceeded those in WT mice (Figure 5A). Additionally, STIM1-deficient CD4+ and CD8+ T cells isolated at 114 d.p.i. produced similar or increased amounts of IFN-γ compared with WT controls upon restimulation ex vivo (Figure 5B). These findings suggest that IFN-γ production by T cells becomes independent of TCR stimulation and SOCE during chronic Mtb infection. IL-12 and IL-18 produced by myeloid cells synergize to induce IFN-γ in T cells (39), and continuous production of IL-12 is required to maintain Th1 cells in the lungs of chronically Mtb-infected mice (40). We found that the lung levels of IL-12p70 and IL-18 were strongly elevated in Mtb-infected Stim1CD4 mice at 114 d.p.i. compared with those of WT controls (Figure 5C). Importantly, ex vivo restimulation of isolated CD4+ and CD8+ T cells with IL-12 and IL-18 resulted in increased IFN-γ production by STIM1-deficient T cells compared with WT controls (Figure 5D).
We next asked whether STIM1-deficient T cells might be intrinsically more responsive to IL-12/IL-18 than WT T cells. CD4+ T cells from uninfected Stim1CD4 mice are severely impaired in their ability to express IFN-γ (and TNF-α) after PMA/ionomycin treatment that mimics TCR stimulation, but unexpectedly produced significantly more IFN-γ (and TNF-α) upon IL-12/IL-18 stimulation in vitro than WT CD4+ T cells (Figure 5E). This altered response is not limited to mouse T cells, as we also found enhanced IFN-γ production by CD4+ and CD8+ T cells from the ORAI1-deficient PAT in response to IL-12p70/IL-18 stimulation (Figure 5F and Supplemental Figure 2B). As in STIM1-deficient murine T cells, PMA/ionomycin stimulation failed to induce IFN-γ expression in the PAT’s T cells, consistent with the known role of the Ca2+/NFAT pathway in TCR-mediated IFN-γ production (Figure 5F, Supplemental Figure 2B, and refs. 7, 41–43). A strong correlation between IFN-γ production and IL-12Rβ1/IL-12Rβ2 expression in T cells has been shown in human TB (44), and NFAT was reported to silence IL-12Rβ2 expression (45). We found that expression of IL-12R and IL-18R mRNA was in fact increased in T cells of the ORAI1-deficient PAT (Supplemental Figure 2C). Together, our results indicate that STIM1 and SOCE play important, albeit opposing, roles in TCR-mediated and cytokine-dependent production of IFN-γ by T cells. Impaired activation of NFAT in STIM1-deficient T cells likely accounts for the initially (<60 d.p.i.) reduced IFN-γ levels in the lungs of Mtb-infected Stim1CD4 mice but, as levels of myeloid cell–derived IL-12/IL-18 increase, STIM1-deficient T cells become hyperresponsive to cytokine stimulation and produce increased amounts of IFN-γ in a TCR-independent manner.
Selective reduction in iTregs in STIM1-deficient mice during chronic Mtb infection.
FOXP3+ Tregs play an important role in limiting immune responses and inflammation that arise during chronic infections (29, 30, 46). In PATs with chronic hepatitis C virus (HCV) infection, frequencies of virus-specific FOXP3+ Tregs inversely correlate with markers of liver damage, suggesting that Tregs contain chronic inflammation and subsequent liver damage in HCV carriers (47, 48). Two subsets of FOXP3+ Tregs can be distinguished, natural Tregs (nTreg) and inducible Tregs (iTregs), both of which are important for maintaining immunological self tolerance and restraining potentially harmful immune responses (49). Whereas nTregs develop in the thymus, iTregs convert from naive CD4+ T cells in the periphery, for instance, during chronic inflammation (50). We observed a significantly reduced frequency of STIM1-deficient CD4+CD25+FOXP3+ Tregs in the lungs of Mtb-infected mice compared with infected WT littermates at all stages of infection (Figure 6A). This is in contrast to uninfected Stim1CD4 mice that had numbers of pulmonary FOXP3+ Tregs similar to those of WT controls (Supplemental Figure 4A and ref. 32), indicating that STIM1 deficiency does not impair Treg development and/or homeostasis in uninfected mice. We hypothesized that, during Mtb infection, the lack of STIM1 might impair the expansion and/or homeostasis of nTregs or the differentiation of iTregs. nTregs can be distinguished from iTregs by the transcription factor Helios and the membrane coreceptor Neuropilin-1 (NRP-1) (51–53). Absolute Helios–NRP-1– iTreg numbers in the lungs of Mtb-infected Stim1CD4 mice were significantly decreased only at 77 d.p.i., and Helios+NRP-1+ nTreg numbers were moderately increased compared with those of control mice due to pulmonary lymphocytosis (Supplemental Figure 4B). Importantly, the frequencies of pulmonary nTregs were similar in Mtb-infected WT and Stim1CD4 mice (Figure 6B), consistent with normal numbers of thymic nTregs in uninfected Stim1CD4 mice (32). In contrast, the frequencies of pulmonary iTregs were significantly reduced in Mtb-infected Stim1CD4 mice at 77 d.p.i. and 114 d.p.i. compared with WT controls (Figure 6B). Accordingly, the ratio of iTreg to effector T cells in the lungs of STIM1-deficient mice was significantly lower than in WT mice (Supplemental Figure 4C).
Figure 6. Lack of STIM1 results in reduced iTreg numbers in chronic Mtb infection.
(A) CD4+CD25+FOXP3+ Tregs in lungs of Mtb-infected WT or Stim1CD4 mice at 77 d.p.i. Representative contour plots (left) and frequencies (mean ± SEM, right) of Tregs from 4 to 5 mice per group and time point. (B) Helios and NRP-1 expression by CD4+CD25+FOXP3+ Tregs in the lungs of mice at 77 d.p.i. Frequencies (line graphs) of Helios+NRP-1+ nTregs and Helios–NRP-1– iTregs over the course of Mtb infection are the mean ± SEM of 4 to 5 mice per group and time point. (C) Correlation of pulmonary IFN-γ levels and frequencies of iTregs in the lung of Mtb-infected WT and Stim1CD4 mice. (D) Effects of IFN-γ and IL-12 on iTreg differentiation in vitro. Naive CD4+ T cells from WT mice were stimulated with αCD3/αCD28 and IL-2/TGF-β in the presence of the indicated concentrations (in ng/ml) of IFN-γ and IL-12 for 3 days in vitro. Bar graphs show frequencies of CD25+FOXP3+ iTregs (left) and IFN-γ–producing T cells after restimulation with PMA/ionomycin for 6 hours (right) as determined by flow cytometry. (E) Effects of IFN-γ and IL-12 on the stability of iTregs. WT CD4+ T cells were differentiated into iTregs as described in D and incubated with IFN-γ, IL-12, or IFN-γ plus IL-12 for another 6 days. The frequencies of CD25+FOXP3+ cells were determined by flow cytometry. Data in D and E represent the mean ± SEM of 4 mice per group. Statistical significance was calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
IFN-γ has been shown to negatively regulate iTreg differentiation in vitro (54). The frequencies of iTregs in the lungs of Mtb-infected Stim1CD4 mice correlated negatively with pulmonary IFN-γ concentrations over the course of infection (Figure 6C), suggesting that increased IFN-γ levels in STIM1-deficient mice suppress iTreg differentiation. Indeed, we found that both IL-12 and IFN-γ significantly (P < 0.05 for IL-12; P < 0.001 for IFN-γ) compromised the differentiation of naive CD4+ T cells into FOXP3+ iTregs in vitro. Instead, both cytokines shifted the polarization of CD4+ T cells to IFN-γ–producing effector T cells despite the presence of TGF-β (Figure 6D and Supplemental Figure 5, A and B). IL-12 and IFN-γ not only prevented the de novo differentiation of iTregs, but also reduced the frequencies of established FOXP3+ iTregs, possibly by interfering with the maintenance or lineage commitment of iTregs (Figure 6E and Supplemental Figure 5C). Together, these results indicate that excessive IFN-γ and IL-12 levels in the lungs of Mtb-infected Stim1CD4 mice contribute to reduced frequencies of iTregs and thereby exacerbate pulmonary inflammation.
STIM1 has a CD4+ T cell–intrinsic role in the development of iTregs.
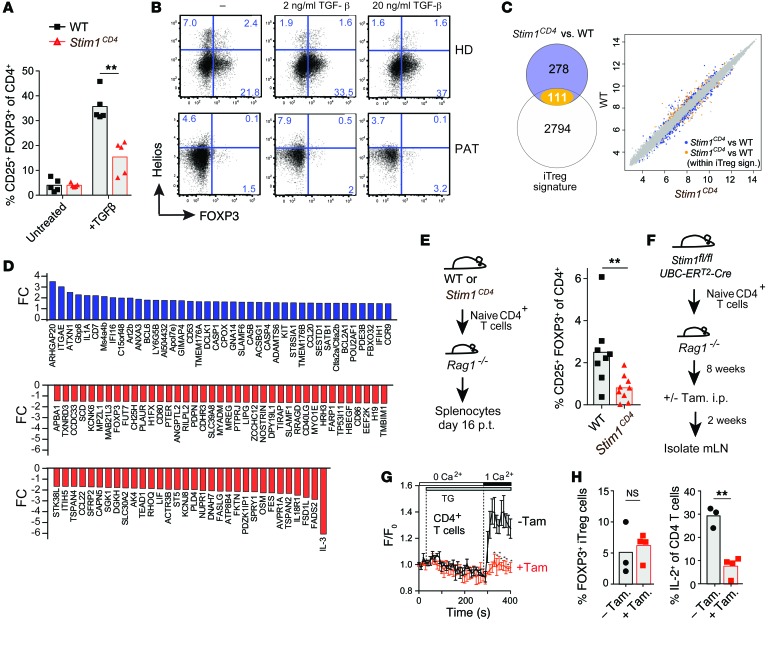
The role of STIM1 in iTreg development is not known, and we hypothesized that STIM1 might play an additional CD4+ T cell–intrinsic role, independent of the extrinsic effects of IFN-γ/IL-12. We found that the frequency of FOXP3+ iTregs was significantly reduced in the absence of STIM1 compared with WT control when we differentiated naive CD4+ T cells in the presence of TGF-β for 3 days in vitro (Figure 7A). This role of SOCE is not specific to murine iTregs because CD4+ T cells of the ORAI1-deficient PAT also failed to develop into FOXP3+ Helios– iTregs after TCR/TGF-β stimulation in vitro (Figure 7B). A global comparison of mRNA expression profiles in murine iTregs differentiated in vitro from naive WT, and STIM1-deficient CD4+ T cells revealed a significant deregulation of over 100 genes that belong to an iTreg signature (Figure 7C). Among genes up- or downregulated in the absence of STIM1 are several molecules previously linked to iTreg function and differentiation, such as FOXP3, ITGAE (CD103), SGK-1, SATB-1, and IL-3 (Figure 7D). To investigate whether STIM1 also plays a role in the differentiation of iTregs in vivo, we adoptively transferred naive CD4+ T cells from WT or Stim1CD4 mice to lymphopenic Rag1–/– mice, a commonly used model to study iTreg conversion in vivo (50). We found markedly reduced frequencies of FOXP3+ iTregs in the spleens of Rag1–/– mice that had received STIM1-deficient compared with WT CD4+ T cells (Figure 7E) despite comparable frequencies of total CD4+ T cells (data not shown). Reduced iTreg numbers in the absence of STIM1 were due to impaired differentiation, but not maintenance, of iTregs because tamoxifen-induced genetic deletion of Stim1 in iTregs after their development in vitro or in vivo did not affect their frequency compared with WT controls (Figure 7, F–H, and data not shown). We also found that STIM1 is not required for the suppressive function of iTregs because in vitro–differentiated WT and STIM1-deficient iTregs were equally able to suppress T cell proliferation and IL-12p40 production by macrophages (Supplemental Figure 6, A and B). Together, our findings reveal a CD4+ T cell–intrinsic role of STIM1 and SOCE in the development of iTregs that is likely to contribute to immunoregulation and prevention of excessive inflammation during chronic Mtb infection.
Figure 7. STIM1 regulates the development, but not the stability, of iTregs.
(A) Frequencies of CD25+FOXP3+ iTregs after in vitro αCD3/αCD28 stimulation of naive splenic CD4+ T cells from WT or Stim1CD4 mice with or without TGF-β (untreated) for 3 days. (B) iTreg differentiation of human CD4+ T cells from a HD or PAT with ORAI1 p.R91W mutation (PAT) (10) after αCD3/αCD28 stimulation with or without TGF-β for 5 days in vitro. Flow cytometry dot plots showing Helios and FOXP3 expression are representative of 3 independent experiments. (C and D) Microarray analysis of WT versus Stim1CD4 iTregs. (C) Venn diagram (left) showing more than 100 iTreg signature genes (see Methods) differentially regulated in Stim1CD4 T cells. Scatter plot (right) of robust multi-array average (RMA) expression of genes that are part (orange) or not part (blue) of the iTreg signature. (D) Bar graphs representing fold changes of iTreg genes in Stim1CD4 cells (P < 0.05 for genes shown). (E) Frequencies of splenic Tregs 16 days after adoptive transfer of naive CD4+ T cells from WT or Stim1CD4 mice. Each symbol represents 1 mouse; data are from 2 independent experiments. (F–H) Unimpaired iTreg maintenance after inducible deletion of Stim1 in vivo. (F) Experimental design (see Methods). (G) SOCE in adoptively transferred CD4+ T cells from mesenteric lymph nodes of mice injected (+Tam) or not (–Tam) with tamoxifen. Mean Fluo-4 fluorescence (F) over baseline (F0) (± SEM) measured by flow cytometry; data are representative of 3-4 mice per group. (H) Frequencies of FOXP3+ iTregs (left) and IL-2+CD4+ T cells (after PMA/ionomycin stimulation for 6 hours; right) isolated from mesenteric lymph nodes of recipient mice. Statistical significance calculated by Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
We show here that STIM1 and SOCE play multiple roles in adaptive immunity to chronic infection by controlling important regulatory mechanisms of the T cell–mediated immune response. STIM1 is thus critical to limiting the injurious pulmonary inflammation that is associated with chronic Mtb infection (see schematic in Figure 8).
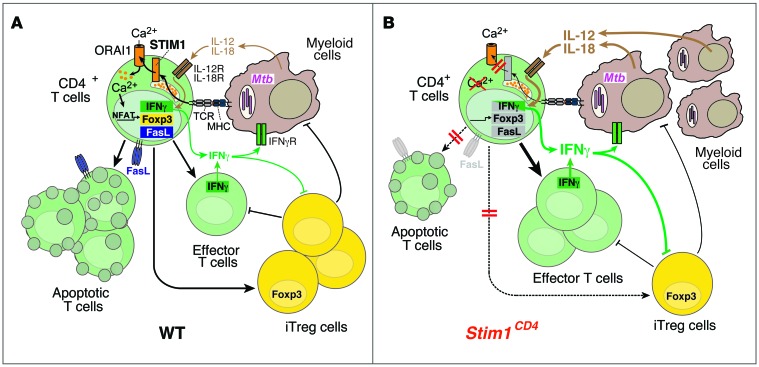
Figure 8. The role of STIM1 in chronic Mtb infection.
(A) In WT mice, Mtb-infected macrophages/DCs present mycobacterial antigens via MHC class II to CD4+ T cells, resulting in STIM1 activation, its binding to ORAI1, and Ca2+ influx (SOCE). SOCE activates NFAT and other Ca2+-dependent transcription factors, which regulate Fasl, Foxp3, and Ifng gene expression. FasL mediates AICD in T cells following repeated TCR stimulation, thereby preventing lymphoproliferation in chronic infection. FOXP3 expression is required for the differentiation of iTregs that suppress effector T cells and potentially myeloid cells. IFN-γ production after TCR, but not IL-12/IL-18, stimulation is dependent on STIM1/SOCE. IFN-γ secreted by effector T cells helps to contain Mtb infection and to recruit myeloid cells. (B) In STIM1-deficient mice, ORAI1 activation and SOCE are impaired, resulting in reduced expression of Fasl, Foxp3, and Ifng and other genes. Impaired AICD results in accumulation of effector T cells in Mtb-infected mice. Despite impaired TCR-dependent IFN-γ production, STIM1-deficient T cells respond to IL-12/IL-18 and secrete more IFN-γ during chronic Mtb infection, promoting the recruitment of myeloid cells that can be infected with Mtb. Higher bacterial burdens result in increased secretion of IL-12/IL-18 and perpetuation of pulmonary inflammation. Secretion of IFN-γ by T cells and IL-12 by myeloid cells inhibits differentiation of iTregs, adding to the STIM1-dependent, T cell–intrinsic defect in iTreg development. Together, these defects result in pulmonary hyperinflammation/consolidation, loss of respiratory function, and premature death of Stim1CD4 mice.
During acute Mtb infection, STIM1 is required for optimal IFN-γ production by T cells, which is consistent with in vitro (32, 38, 55) and acute viral infection (3) models. Although IFN-γ levels were reduced in Stim1CD4 mice early in infection, lung bacterial loads were surprisingly not increased concomitantly, as one would expect given the important role of IFN-γ in immunity to TB (20, 21). Control of Mtb growth in Stim1CD4 mice during acute infection was likely due to residual IFN-γ production by STIM1-deficient T cells, consistent with the phenotype we observed in Mtb-infected heterozygous Ifng+/– mice. In these mice, the frequency of IFN-γ–producing T cells was reduced by 50%, but lung bacterial burdens at 35 d.p.i. were similar to those in WT mice. In contrast, homozygous Ifng–/– mice have 10-fold increased bacterial burdens at this time, indicating that IFN-γ in Stim1CD4 mice and Ifng+/– is produced in excess of what is needed to control acute Mtb infection. However, we cannot rule out that the initial defect in IFN-γ production by STIM1-deficient T cells contributed to increased bacterial growth at later stages of infection, a delay that could be explained by the slow growth of Mtb in vivo.
During chronic infection with Mtb, STIM1 takes on multiple immune regulatory roles to prevent pulmonary hyperinflammation and tissue damage. First, STIM1 mediates contraction of the pool of activated T cells following repeated TCR stimulation by regulating the expression of proapoptotic molecules. In our mouse model of pulmonary Mtb infection, a reduction in the frequency of IFN-γ–producing T cells in the lungs was shown to occur 5 to 6 weeks after infection (56). In the absence of STIM1, however, CD4+ and CD8+ T cells expand dramatically at as early as 45 d.p.i. due to continuous antigen exposure and impaired AICD. Second, although STIM1-deficient T cells initially show defective TCR-dependent IFN-γ expression during acute Mtb infection, production of IFN-γ becomes independent of STIM1 during chronic TB, and high levels of IFN-γ are detectable in the lungs of Stim1CD4 mice beyond 100 d.p.i. While impaired AICD and increased responsiveness to IL-12/IL-18 in the absence of STIM1 are T cell–intrinsic defects, increased numbers of myeloid cells and approximately 40-fold higher lung bacterial burdens create an inflammatory milieu that contributes extrinsically to a positive feedback loop amplifying IL-12/IL-18 secretion, thereby fueling excessive IFN-γ production, pulmonary inflammation, and bacterial replication. Third, STIM1 controls the frequency of iTregs in the lungs of infected mice in both T cell–intrinsic and –extrinsic manners. STIM1 is required for the expression of FOXP3 in naive CD4+ T cells and therefore their conversion into iTregs. In addition, increased IFN-γ and IL-12 levels in the lungs of Mtb-infected mice further suppress iTreg differentiation and maintenance (54, 57). Thus, in the absence of STIM1, low frequencies of iTregs may fail to control the proliferation and function of effector T cells, and potentially myeloid cells, in the lungs of Mtb-infected mice. Together these immunomodulatory defects in the absence of SOCE in T cells contribute to pulmonary hyperinflammation and, eventually, to the early mortality of Mtb-infected Stim1CD4 mice.
STIM1 is essential to controlling AICD in T cells, and its deletion results in expansion of T cells in the lungs of Mtb-infected mice. This function of STIM1 is consistent with the known role of Ca2+-dependent transcription factors of the NFAT family in regulating FasL expression. Stimulation of human Jurkat T cells by CD3 crosslinking or ionomycin mediates binding of NFAT to the FASLG enhancer region, resulting in FasL expression (58). Using murine T cells, Kim et al. observed that retroviral overexpression of constitutively active NFAT1 increases apoptosis in T cells, even under resting conditions (34). In contrast, NFAT1-deficient and, even more pronouncedly, NFAT1/NFAT4 double-deficient, lymphocytes are hyperproliferative and have elevated primary and secondary immune responses (43, 59–61). This is at least partially caused by impaired lymphocyte apoptosis due to reduced FasL expression upon activation (62, 63). Fas/FasL-mediated apoptosis plays an important role in controlling Mtb infection. Mtb-infected Lpr (Fas) mutant mice showed significantly increased lung bacterial burdens compared with WT mice during chronic (40–120 d.p.i.) but not acute infection (64). This is similar to our observations in Mtb-infected Stim1CD4 mice and suggests that SOCE-dependent expression of FasL and apoptosis are required to control chronic Mtb infection. Several other proapoptotic factors expressed at lower levels in STIM1-deficient T cells were shown to be regulated by the Ca2+/calcineurin/NFAT pathway, including TRAIL (65), CD38 (66), and NUR77 (67). CD38-induced activation of T cells triggers Ca2+ influx and apoptosis in human T cells, which was inhibited by cyclosporin A (which inhibits calcineurin and NFAT activation), suggesting that CD38 mediates its proapoptotic function by regulating Ca2+ signaling (66). Expression of the steroid receptor NUR77 is regulated by Ca2+ following TCR stimulation and promotes thymocyte apoptosis by activating the transcription factor myocyte enhancer factor 2 (MEF2), which binds to the Nur77 promoter (67). Taken together, our data indicate that SOCE regulates several Ca2+-signaling pathways to control expression of proapoptotic factors, thereby limiting proliferation of effector T cells during chronic infection.
As a second important immunoregulatory mechanism, STIM1 controls differentiation of CD4+ T cells into FOXP3+ iTregs in both cell-extrinsic and cell-intrinsic manners, which accounts for the reduced frequencies of FOXP3+Helios–NRP-1– iTregs, but not of FOXP3+Helios+NRP-1+ nTregs, in the lungs of chronically Mtb-infected Stim1CD4 mice. IFN-γ and IL-12 negatively regulate conversion of naive CD4+ T cells into iTregs and instead shift the differentiation of CD4+ T cells toward Th1 cells, consistent with previous studies (54, 57). This inhibitory effect of IFN-γ on iTreg differentiation is also observed in vivo, as IFN-γ–deficient mice have elevated numbers of FOXP3+ iTregs (but not nTregs) compared with WT mice (54). In addition, we found that STIM1 and SOCE play a CD4+ T cell–intrinsic role in the differentiation of iTregs. The conversion of naive CD4+ T cells into iTregs in vitro and in vivo was impaired in the absence of STIM1. In contrast, STIM1 was not required for the maintenance or lineage commitment of iTregs, as inducible deletion of Stim1 after iTreg differentiation in vitro or in vivo did not alter the frequencies of FOXP3+ iTregs.
Our gene-expression analysis showed that STIM1 regulates expression of more than 100 iTreg signature genes, including Foxp3. This is in line with recent studies showing that members of the Ca2+-dependent NFAT transcription factor family regulate FOXP3 expression in iTregs (68, 69). NFAT binds, together with SMAD2/3, directly to the conserved noncoding sequence 1 in the Foxp3 locus and regulates FOXP3 expression in response to TCR stimulation and TGF-β (68–70). In addition to Foxp3, expression of other genes associated with iTregs was significantly deregulated in STIM1-deficient T cells. These include Itgae (Cd103) and Il3, which are also controlled by NFAT (68, 71). In contrast to reduced iTreg differentiation, nTreg numbers were normal in uninfected and chronically Mtb-infected STIM1-deficient mice, suggesting that STIM1 is more important for the development of iTregs than nTregs. Interestingly, complete deletion of SOCE in T cells from STIM1/STIM2 double-deficient mice (32, 72) or human PATs with mutations in STIM1 and ORAI1 genes (11, 73) (S. Feske, unpublished observations) also impairs nTreg development, resulting in reduced numbers of FOXP3+ Tregs. Further emphasizing the importance of SOCE in iTreg differentiation, T cells from NFAT1/NFAT2, and NFAT1/NFAT4 double-deficient mice are unable to differentiate into iTregs, whereas nTreg development and function are normal (68, 74). These findings suggest that the lymphoproliferative phenotype observed in Stim1CD4 mice chronically infected with Mtb is caused by impaired iTreg differentiation rather than defective nTreg development (38, 75, 76). Although differentiation of FOXP3+ iTregs was defective in STIM1-deficient mice, their suppressive function in vitro was intact, which is consistent with unimpaired suppressive function of Tregs of a PAT with loss-of-function STIM1 mutation (73). Similar observations were made in NFAT2-deficient CD4+ T cells, which fail to efficiently differentiate into FOXP3+ iTregs, while the remaining iTregs were functional (68). Together, our data indicate that STIM1-mediated Ca2+ signaling is essential for the conversion of naive CD4+ T cells into iTregs, but that it is largely dispensable for their stability and suppressive functions.
iTregs are essential to regulate the homeostasis of pro- and anti-inflammatory immune responses during infections (50). In Mtb infection, Tregs have been studied for their role in delaying the onset and limiting the efficiency of adaptive immune responses (31). Early during Mtb infection, pathogen-specific nTregs were shown to proliferate in lung-draining lymph nodes, where they suppress priming of effector T cells and promote bacterial dissemination. The role of iTregs in controlling inflammation during chronic Mtb infection is largely unknown. Interestingly, BCG-induced iTregs ameliorate chronic inflammation in a mouse model of atopic dermatitis (77). Our data of reduced iTreg frequencies in Mtb-infected Stim1CD4 mice indicate that STIM1 controls immune homeostasis and prevents pulmonary hyperinflammation.
The complex role of STIM1 in Mtb infection is further emphasized by its biphasic and opposing control of IFN-γ expression in the acute and chronic phases of infection. Although STIM1 is required for TCR-induced IFN-γ production early in Mtb infection, it assumes a negative regulatory role in IL-12/IL-18–mediated IFN-γ production during chronic infection. TCR stimulation results in activation of SOCE-regulated transcription factor NFAT, which cooperates with STAT4 (in naive T cells) and T-bet (in committed Th1 cells) to promote transcription of Ifng (42, 78). NFAT and T-bet bind to a highly conserved distal enhancer in the Ifng locus and synergize to enhance IFN-γ expression (79). T cells from NFAT1- or NFAT1/NFAT4-deficient mice have impaired IFN-γ production (41, 43, 63), suggesting that NFAT1 and NFAT4 promote Th1 cell differentiation. Since SOCE is required to activate NFAT, impaired IFN-γ expression in STIM1-deficient mice following TCR stimulation can be explained by defective NFAT function. In contrast, TCR-independent mechanisms of IFN-γ production in activated T cells, mediated by IL-12/IL-18, become important during chronic Mtb infection. IL-12 binds to the heterodimeric IL-12R, which consists of the constitutively expressed β1 chain and the inducible β2 chain. The IL-12Rβ2 promoter contains a NFATc2-binding site that functions as a silencer of IL-12Rβ2 expression, and preventing NFAT activation with cyclosporin A — an inhibitor of the Ca2+-dependent phosphatase calcineurin that activates NFAT — results in increased IL-12Rβ2 levels on T cells (45). Negative regulation of IL-12Rβ2 by calcineurin/NFAT is consistent with increased responsiveness of STIM1-deficient CD4+ and CD8+ T cells to IL-12/IL-18 stimulation and probably contributes to elevated IFN-γ levels in the lungs of chronically Mtb-infected Stim1CD4 mice. The proinflammatory milieu in the lungs of these mice is likely to activate other lymphoid cells, including NK cells, NKT cells, and innate lymphoid cells (ILC), to produce IFN-γ.
The initially reduced and later increased IFN-γ expression in STIM1-deficient mice reinforces the ambiguous role of IFN-γ in TB. Although IFN-γ is essential for controlling Mtb (20, 21), several lines of evidence indicate that IFN-γ levels do not ultimately correlate with protection in humans, and some studies even show positive correlation with lung disease (reviewed in ref. 80). We found that, during acute Mtb infection, lower IFN-γ levels in Stim1CD4 and Ifng+/– mice do not correlate with higher mycobacterial burdens in the lungs. But during chronic infection, high levels of IFN-γ in Stim1CD4 mice fail to limit bacterial growth and, on the contrary, are associated with pulmonary hyperinflammation, including increased recruitment of infectable myeloid cells. Similar observations were made in LCMV-infected mice, suggesting that IFN-γ is involved in the recruitment of macrophages to the site of infection (81). Continued production of IFN-γ by STIM1-deficient T cells may therefore exacerbate chronic infection with Mtb by promoting accumulation of myeloid cells in the lungs, thereby increasing the pool of cells that can be infected by Mtb and the overall pulmonary mycobacterial burden. Enhanced IFN-γ expression by STIM1-deficient T cells in response to Mtb is somewhat reminiscent of the immune reconstitution inflammatory syndrome (IRIS) in HIV-infected PATs who develop an exacerbated immune response to pathogens, including Mtb, with IFN-γ production and inflammation after initiation of antiretroviral therapy and recovery of T cell numbers (82). The role of STIM1 and SOCE in the regulation of T cell–mediated immunity likely extends beyond TB to other chronic infections in which IFN-γ serves as an important regulator of the immune response, including infections with hepatitis B and C viruses, herpes simplex virus (HSV), or Helicobacter pylori (83). Our findings in STIM1-deficient mice illuminate the ambiguous role of IFN-γ in chronic infections and may therefore have important therapeutic consequences for the assessment of IFN-γ modulators, including potential vaccines that rely on the production of IFN-γ for protection.
Taken together, our results shed light on the importance of Ca2+ signaling in T cell–mediated immune regulation and inflammation during chronic infection. They offer an explanation for the complex immunodeficiency observed in human PATs with CRAC channelopathy, who frequently suffer from chronic viral and mycobacterial infections and lymphoproliferative diseases. As Ca2+ signaling modulators are being developed for the treatment of autoimmune diseases, inflammation, and allergy (1, 84), our results underline the importance of assessing their effects on highly prevalent, chronic infections.
Methods
Mice.
WT C57BL/6 mice, B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) mice, and Rag1–/– mice were purchased from The Jackson Laboratory. Stim1CD4 mice were described previously (32). Mice with inducible deletion of the Stim1 gene (Stim1fl/flUBC-ERT2-Cre) were generated by crossing Stim1fl/fl mice with B6.Cg-Tg(UBC-Cre/ERT2)1Ejb/J (UBC-ERT2-Cre) mice (The Jackson Laboratory). Ifng+/– mice were generated by crossing Ifng–/– mice (The Jackson Laboratory) with C57BL/6 mice. For infections with Mtb, mice were housed under barrier conditions in the ABSL-3 facility at New York University Langone Medical Center (NYULMC). For tissue harvesting, mice were euthanized by CO2 asphyxiation, followed by cervical dislocation. All mice were between 8 and 12 weeks of age at the beginning of the experiment.
Bacterial infection.
Mtb laboratory strain H37Rv was grown as previously described (85). Mice were infected via the aerosol route, using an inhalation exposure system (Glas-Col) (86) calibrated to deliver approximately 100 CFU/mouse. The infectious dose was confirmed on day 1 by plating whole-lung homogenates from 5 mice on Middlebrook 7H11 agar. CFU were counted after incubation at 37°C for 2 to 3 weeks. To determine bacterial burdens throughout the infection, serial dilutions of lung homogenates were plated on Middlebrook 7H11 agar.
Histology and immunohistochemistry.
The left lungs of mice were harvested and processed as previously described (87) for histopathological examination, immunohistochemical staining, using polyclonal antibodies specific for CD68 (FA-11, AbD Serotec) and CD3ε (2GV6, Ventana), and image analysis (see below).
Image analysis.
Open source image-processing software ImageJ (http://rsbweb.nih.gov/ij/) was used for quantitation of alveolar spaces in the lungs and of specific immunohistochemical staining. Analysis was performed according to the directions of the ImageJ User Guide for sizing particles.
ELISA and multiplex assays.
Supernatants from whole-lung homogenates harvested at various time points following Mtb infection were analyzed by ELISA for detection of IFN-γ (BD Biosciences), IL12p40, IL-12p70, and IL-18 (eBioscience) and by multiplex assay for GM-CSF, M-CSF, IL-1β, IL-12p70, IL-18, MCP-1/CCL2, MIP1α/CCL3, and RANTES/CCL5 (EMD Millipore).
Quantitative real-time PCR and microarray analysis.
Total RNA was extracted from the left lungs of mice using TRIzol reagent (Invitrogen) and converted into cDNA as previously described (85). The cDNA equivalent of 50 ng of total RNA was analyzed for specific gene expression in triplicate for each sample by quantitative real-time PCR using Platinum SYBR Green qPCR SuperMix (Invitrogen) and an Opticon 2 thermocycler (Bio-Rad). Sequences of primers used for PCR can be found in Supplemental Table 1. Thermal cycling conditions were 95°C for 10 minutes followed by 40 cycles at 94°C for 45 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. For quantitation, relative Ct values were determined by normalization to Cd4, Cd8, or housekeeping gene Hrpt cDNA expression using the ΔΔCt method. For microarray analysis, CD4+ T cells from WT or Stim1CD4 mice were isolated and stimulated with functional grade αCD3ε and αCD28 (145-2C11 and 37.51, BioXCell) antibodies for 48 hours in the presence or absence of 5 ng/ml TGF-β (Peprotech) (iTreg skewing conditions). Total RNA was isolated using RNeasy Micro kit (QIAGEN). Biotin-labeled amplified aRNA was hybridized to 1.0 Eon expression chips (Affymetrix) according to the manufacturer’s protocols. The values in the sample data tables were derived from the Expression Console Software (Affymetrix) by performing a gene-level normalization and signal summarization on the raw CEL files (gene-level analysis). Obtained CHP files were subsequently analyzed and visualized using TAC software (Affymetrix) and IPA pathway analysis software (QIAGEN). Three age-matched mice per group and condition were used; no technical replicates were used. To determine the iTreg signature, gene-expression levels in WT CD4+ T cells cultured under iTreg polarizing conditions were compared with those in WT CD4+ T cells cultured under nonpolarizing (Th0) conditions (no TGF-β). Genes were included in the iTreg signature when their expression was 1.5-fold above or below mRNA levels in Th0 cells and differences were statistically significant (P < 0.05 by ANOVA using TAC software). Primary microarray data from Figures 4 and 7 were deposited in the NCBI’s Gene Expression Omnibus (GEO GS66933).
Flow cytometry.
For analysis of immune cell populations, mouse lungs were processed for flow cytometry as previously described (86). The following anti-murine fluorophore-conjugated antibodies were used for staining: CD4 (RM4-5, BioLegend), CD8α (53-6.7, BD Biosciences), CD11c (HL3, BD Biosciences), CD11b (M1/70, BD Biosciences), Gr-1 (RB6-8C5, BD Biosciences), FasL (MFL3, eBioscience), Fas (15A7, eBioscience), IFN-γ (XMG1.2, BioLegend), TNF-α (MP6-XT22, BioLegend), CD62L (MEL-14, BD Biosciences), CD44 (IM7, BioLegend), CD25 (PC61, BD Biosciences), FOXP3 (FJK-16s, eBioscience), Helios (22F6, BioLegend), and NRP-1 (biotinylated goat polyclonal antibody, catalog BAF566, R&D Systems) followed by fluorophore-conjugated streptavidin (eBioscience). For analysis of human T cells, the following fluorophore-conjugated antibodies were used for flow cytometry: CD4 (OKT4, BioLegend), CD8α (HIT8a, BioLegend), IFN-γ (4S.B3, BioLegend), TNF-α (MAb11, BioLegend), Helios (22F6, BioLegend), and FOXP3 (259D, BioLegend). For apoptotic cell staining, fluorophore-conjugated annexin V probe (BD Biosciences) and propidium iodide (PI) (BD Biosciences) were used. Samples were acquired on an LSRII flow cytometer using FACSDiva software (BD Biosciences) and further analyzed using FlowJo software (Tree Star).
In vitro murine T cell assays.
WT CD4+ and CD8+ T cells were isolated from spleen or mesenteric lymph nodes of mice using negative selection kits (STEMCELL Technologies) and stimulated with 0.5 μg/ml plate-bound αCD3 and 1 μg/ml αCD28 antibodies (BioXCell). After 48 hours, RPMI 1640 medium was supplemented with 20 U/ml recombinant human IL-2 (NIH/AIDS reagent program). After 7 days, cell numbers and frequency of apoptotic cells were determined by flow cytometry after annexin V and PI staining. For restimulation experiments, CD4+ and CD8+ T cells were incubated with 2 μg/ml αCD3 for 6, 16, or 24 hours. For cytokine production, cells were stimulated with 1 μM ionomycin plus 20 nM PMA or 20 ng/ml IL-12 and 200 ng/ml IL-18 for 6 hours in the presence of Brefeldin A. IFN-γ production was determined by intracellular cytokine staining and flow cytometry as described above. For iTreg differentiation experiments, naive CD4+CD62L+CD25– T cells were isolated from WT and Stim1CD4 mice and stimulated in vitro with αCD3/αCD28 and 2.5 ng/ml TGF-β. For some experiments, naive WT CD4+ T cells isolated from the spleen were stimulated with αCD3/αCD28 and cultured in 20 U/ml recombinant human IL-2 and 5 ng/ml TGF-β for 3 days in the presence or absence of recombinant murine IFN-γ or IL-12 (10 to 40 ng/ml; BD Biosciences). The frequency of FOXP3-expressing iTregs was determined by flow cytometry, and IFN-γ production was assessed by intracellular cytokine staining after stimulation of cells with PMA/ionomycin. To test the effects of exogenous IFN-γ/IL-12 on the maintenance of in vitro–differentiated iTregs, naive CD4+ T cells from WT mice were cultured for 3 days as described above and subsequently incubated with IFN-γ, IL-12, or IFN-γ/IL-12 for another 6 days. The frequency of FOXP3+ cells was determined by flow cytometry.
T cell transfer experiments.
Naive CD4+CD45RB+CD25– T cells were isolated from the spleens of WT, Stim1CD4, or Stim1fl/flUBC-ERT2-Cre mice using a negative isolation kit and a SONY iCyt cell sorter. 1 × 106 cells were adoptively transferred to lymphopenic recipient Rag1–/– mice by i.v. injection. To study in vivo differentiation of iTregs, spleens were harvested 16 days post transfer (p.t.) of WT or Stim1CD4 T cells and the frequencies of CD25+FOXP3+ iTregs were determined by flow cytometry. To examine the role of STIM1 in iTreg stability in vivo, mice transferred with Stim1fl/flUBC-ERT2-Cre T cells were injected i.p. daily for 5 days with 1 mg/mouse/d tamoxifen (Sigma-Aldrich) or vehicle (corn oil plus 5% ethanol) alone, starting at 8 weeks p.t. Mesenteric lymph nodes were harvested 2 weeks after the beginning of tamoxifen injections, and the frequency of FOXP3+ iTregs was determined by flow cytometry.
In vitro human T cell assays.
Cord blood cells or peripheral blood lymphocytes (PBL) from healthy donors (HD) and PBL from a SOCE-deficient PAT with ORAI1 p.R91W mutation (10) were cultured as described previously (88). Briefly, 1 × 106 cord blood cells or PBL isolated by Ficoll gradient centrifugation were stimulated using αCD3/αCD28-coated magnetic beads (Invitrogen) and cultured in RPMI 1640 medium containing 10 ng/ml IL-2 for 35 days. For apoptosis experiments, T cells were reactivated with αCD3/αCD28 beads and apoptosis was assessed 6 hours later by annexin V and PI stainings. For intracellular cytokine staining, T cells were stimulated for 16 hours with 20 ng/ml IL-12 (R&D), 200 ng/ml IL-18 (Peprotech), IL-12 plus IL-18, or PMA plus ionomycin for 4 hours or left unstimulated. GolgiStop (BD) was added for 4 hours before staining. For differentiation of iTregs, CD4+ T cells from HD and PAT were stimulated with αCD3/αCD28 beads in the presence or absence of TGF-β (R&D) at the indicated concentrations.
Statistics.
P values were calculated using a 2-tailed, unpaired Student’s t test and Prism 6 (GraphPad Software). Error bars represent mean ± SEM. P < 0.05 was considered statistically significant. Statistical correlation analyses were performed using the Spearman’s rho test. Unless otherwise stated, the data shown represent 3 to 5 biological replicates in 1 experiment.
Study approval.
All animal experiments were conducted in accordance with protocols approved by the IACUC of New York University School of Medicine. For experiments using primary human cells, informed consent for the studies was obtained from the PATs in accordance with the Declaration of Helsinki and IRB approval of the New York University School of Medicine.
Supplementary Material
Acknowledgments
This work was funded by NIH grants AI097302 (to S. Feske), AI065303 to (D. Unutmaz), AI051242 and AI084041 (to J.D. Ernst), and postdoctoral fellowships by the National Multiple Sclerosis Society (to P. Shaw) and the Deutsche Forschungsgemeinschaft (DFG) (to C. Weidinger [We 5303/1-1] and M. Vaeth [VA 882/1-1]).
Footnotes
Conflict of interest: Stefan Feske is a cofounder of CalciMedica.
Reference information:J Clin Invest. 2015;125(6):2347–2362. doi:10.1172/JCI80273.
Derya Unutmaz’s present address is: The Jackson Laboratory for Genomic Medicine, Farmington, Connecticut, USA.
Carl Weidinger’s present address is: Charité - University Medicine Berlin, Division of Gastroenterology, Rheumatology and Infection Medicine, Berlin, Germany.
References
- 1.Feske S, Wulff H, Skolnik EY. Ion channels in innate and adaptive immunity. Annu Rev Immunol. 2015;33:291–353. doi: 10.1146/annurev-immunol-032414-112212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12(7):532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw PJ, Weidinger C, Vaeth M, Luethy K, Kaech SM, Feske S. CD4+ and CD8+ T cell-dependent antiviral immunity requires STIM1 and STIM2. J Clin Invest. 2014;124(10):4549–4563. doi: 10.1172/JCI76602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidinger C, Shaw PJ, Feske S. STIM1 and STIM2-mediated Ca(2+) influx regulates antitumour immunity by CD8(+) T cells. EMBO Mol Med. 2013;5(9):1311–1321. doi: 10.1002/emmm.201302989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feske S. CRAC channelopathies. Pflugers Arch. 2010;460(2):417–435. doi: 10.1007/s00424-009-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feske S. Immunodeficiency due to defects in store-operated calcium entry. Ann N Y Acad Sci. 2011;1238:74–90. doi: 10.1111/j.1749-6632.2011.06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feske S, et al. Severe combined immunodeficiency due to defective binding of the nuclear factor of activated T cells in T lymphocytes of two male siblings. Eur J Immunol. 1996;26(9):2119–2126. doi: 10.1002/eji.1830260924. [DOI] [PubMed] [Google Scholar]
- 8.Le Deist F, et al. A primary T-cell immunodeficiency associated with defective transmembrane calcium influx. Blood. 1995;85(4):1053–1062. [PubMed] [Google Scholar]
- 9.Partiseti M, Le Deist F, Hivroz C, Fischer A, Korn H, Choquet D. The calcium current activated by T cell receptor and store depletion in human lymphocytes is absent in a primary immunodeficiency. J Biol Chem. 1994;269(51):32327–32335. [PubMed] [Google Scholar]
- 10.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441(7090):179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 11.Picard C, et al. STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity. N Engl J Med. 2009;360(19):1971–1980. doi: 10.1056/NEJMoa0900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byun M, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J Exp Med. 2010;207(11):2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarl CA, et al. ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia. J Allergy Clin Immunol. 2009;124(6):1311–1318. doi: 10.1016/j.jaci.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava S, Ernst JD, Desvignes L. Beyond macrophages: the diversity of mononuclear cells in tuberculosis. Immunol Rev. 2014;262(1):179–192. doi: 10.1111/imr.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf AJ, et al. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med. 2008;205(1):105–115. doi: 10.1084/jem.20071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urdahl KB. Understanding and overcoming the barriers to T cell mediated immunity against tuberculosis. Semin Immunol. 2014;26(6):578–587. doi: 10.1016/j.smim.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunes-Alves C, Booty MG, Carpenter SM, Jayaraman P, Rothchild AC, Behar SM. In search of a new paradigm for protective immunity to TB. Nat Rev Microbiol. 2014;12(4):289–299. doi: 10.1038/nrmicro3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bold TD, Ernst JD. CD4+ T cell-dependent IFN-γ production by CD8+ effector T cells in Mycobacterium tuberculosis infection. J Immunol. 2012;189(5):2530–2536. doi: 10.4049/jimmunol.1200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annu Rev Pathol. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 20.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon γ gene-disrupted mice. J Exp Med. 1993;178(6):2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178(6):2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupuis S, et al. Human interferon-γ-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol Rev. 2000;178:129–137. doi: 10.1034/j.1600-065x.2000.17810.x. [DOI] [PubMed] [Google Scholar]
- 23.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorhoi A, Kaufmann SH. Perspectives on host adaptation in response to Mycobacterium tuberculosis: Modulation of inflammation. Semin Immunol. 2014;26(6):533–542. doi: 10.1016/j.smim.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Barber DL, Mayer-Barber KD, Feng CG, Sharpe AH, Sher A. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J Immunol. 2011;186(3):1598–1607. doi: 10.4049/jimmunol.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenardo M, et al. Mature T lymphocyte apoptosis — immune regulation in a dynamic and unpredictable antigenic environment. Annu Rev Immunol. 1999;17:221–253. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 27.Kulinski JM, Tarakanova VL, Verbsky J. Regulation of antiviral CD8 T-cell responses. Crit Rev Immunol. 2013;33(6):477–488. doi: 10.1615/CritRevImmunol.2013007909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy FJ, Hayes I, Cotter TG. Targeting inflammatory diseases via apoptotic mechanisms. Curr Opin Pharmacol. 2003;3(4):412–419. doi: 10.1016/S1471-4892(03)00072-9. [DOI] [PubMed] [Google Scholar]
- 29.Veiga-Parga T, Sehrawat S, Rouse BT. Role of regulatory T cells during virus infection. Immunol Rev. 2013;255(1):182–196. doi: 10.1111/imr.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol Rev. 2014;259(1):40–59. doi: 10.1111/imr.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson RP, Shafiani S, Urdahl KB. Foxp3(+) regulatory T cells in tuberculosis. Adv Expe Med Biology. 2013;783:165–180. doi: 10.1007/978-1-4614-6111-1_9. [DOI] [PubMed] [Google Scholar]
- 32.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9(4):432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waite JC, et al. Interference with Ca(2+) release activated Ca(2+) (CRAC) channel function delays T-cell arrest in vivo. Eur J Immunol. 2013;43(12):3343–3354. doi: 10.1002/eji.201243255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim KD, Srikanth S, Yee MK, Mock DC, Lawson GW, Gwack Y. ORAI1 deficiency impairs activated T cell death and enhances T cell survival. J Immunol. 2011;187(7):3620–3630. doi: 10.4049/jimmunol.1100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denis M, Gregg EO, Ghandirian E. Cytokine modulation of Mycobacterium tuberculosis growth in human macrophages. Int J Immunopharmacol. 1990;12(7):721–727. doi: 10.1016/0192-0561(90)90034-K. [DOI] [PubMed] [Google Scholar]
- 36.Desvignes L, Wolf AJ, Ernst JD. Dynamic roles of Type I and Type II IFNs in early infection with Mycobacterium tuberculosis. J Immunol. 2012;188(12):6205–6215. doi: 10.4049/jimmunol.1200255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feske S, Draeger R, Peter HH, Eichmann K, Rao A. The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J Immunol. 2000;165(1):297–305. doi: 10.4049/jimmunol.165.1.297. [DOI] [PubMed] [Google Scholar]
- 38.McCarl CA, et al. Store-operated Ca2+ entry through ORAI1 is critical for T cell-mediated autoimmunity and allograft rejection. J Immunol. 2010;185(10):5845–5858. doi: 10.4049/jimmunol.1001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamura H, Kashiwamura S, Tsutsui H, Yoshimoto T, Nakanishi K. Regulation of interferon-gamma production by IL-12 and IL-18. Curr Opin Immunol. 1998;10(3):259–264. doi: 10.1016/S0952-7915(98)80163-5. [DOI] [PubMed] [Google Scholar]
- 40.Feng CG, et al. Maintenance of pulmonary Th1 effector function in chronic tuberculosis requires persistent IL-12 production. J Immunol. 2005;174(7):4185–4192. doi: 10.4049/jimmunol.174.7.4185. [DOI] [PubMed] [Google Scholar]
- 41.Kiani A, et al. Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood. 2001;98(5):1480–1488. doi: 10.1182/blood.V98.5.1480. [DOI] [PubMed] [Google Scholar]
- 42.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5(6):472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 43.Rengarajan J, Tang B, Glimcher LH. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat Immunol. 2002;3(1):48–54. doi: 10.1038/ni744. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Gong J, Presky DH, Xue W, Barnes PF. Expression of the IL-12 receptor β1 and β2 subunits in human tuberculosis. J Immunol. 1999;162(4):2441–2447. [PubMed] [Google Scholar]
- 45.van Rietschoten JG, et al. Silencer activity of NFATc2 in the interleukin-12 receptor β 2 proximal promoter in human T helper cells. J Biol Chem. 2001;276(37):34509–34516. doi: 10.1074/jbc.M102536200. [DOI] [PubMed] [Google Scholar]
- 46.Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6(4):353–360. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- 47.Bolacchi F, et al. Increased hepatitis C virus (HCV)-specific CD4+CD25+ regulatory T lymphocytes and reduced HCV-specific CD4+ T cell response in HCV-infected patients with normal versus abnormal alanine aminotransferase levels. Clin Exp Immunol. 2006;144(2):188–196. doi: 10.1111/j.1365-2249.2006.03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wohlfert E, Belkaid Y. Role of endogenous and induced regulatory T cells during infections. J Clin Immunol. 2008;28(6):707–715. doi: 10.1007/s10875-008-9248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8(7):523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30(5):626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209(10):1723–1742. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209(10):1713–1722. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang JH, Kim YJ, Han SH, Kang CY. IFN-gamma-STAT1 signal regulates the differentiation of inducible Treg: potential role for ROS-mediated apoptosis. Eur J Immunol. 2009;39(5):1241–1251. doi: 10.1002/eji.200838913. [DOI] [PubMed] [Google Scholar]
- 55.Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat Immunol. 2001;2(4):316–324. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- 56.Bold TD, Banaei N, Wolf AJ, Ernst JD. Suboptimal activation of antigen-specific CD4+ effector cells enables persistence of M. tuberculosis in vivo. PLoS Pathog. 2011;7(5):e1002063. doi: 10.1371/journal.ppat.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prochazkova J, Pokorna K, Holan V. IL-12 inhibits the TGF-beta-dependent T cell developmental programs and skews the TGF-beta-induced differentiation into a Th1-like direction. Immunobiology. 2012;217(1):74–82. doi: 10.1016/j.imbio.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 58.Holtz-Heppelmann CJ, Algeciras A, Badley AD, Paya CV. Transcriptional regulation of the human FasL promoter-enhancer region. J Biol Chem. 1998;273(8):4416–4423. doi: 10.1074/jbc.273.8.4416. [DOI] [PubMed] [Google Scholar]
- 59.Hodge S, Novembre FJ, Whetter L, Gelbard HA, Dewhurst S. Induction of fas ligand expression by an acutely lethal simian immunodeficiency virus, SIVsmmPBj14. Virology. 1998;252(2):354–363. doi: 10.1006/viro.1998.9477. [DOI] [PubMed] [Google Scholar]
- 60.Xanthoudakis S, et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272(5263):892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 61.Heyer J, et al. Inefficient termination of antigen responses in NF-ATp-deficient mice. Immunobiology. 1997;198(1-3):162–169. doi: 10.1016/S0171-2985(97)80037-X. [DOI] [PubMed] [Google Scholar]
- 62.Rengarajan J, et al. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 2000;12(3):293–300. doi: 10.1016/S1074-7613(00)80182-X. [DOI] [PubMed] [Google Scholar]
- 63.Ranger AM, Oukka M, Rengarajan J, Glimcher LH. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity. 1998;9(5):627–635. doi: 10.1016/S1074-7613(00)80660-3. [DOI] [PubMed] [Google Scholar]
- 64.Turner J, et al. CD8- and CD95/95L-dependent mechanisms of resistance in mice with chronic pulmonary tuberculosis. Am J Respir Cell Mol Biol. 2001;24(2):203–209. doi: 10.1165/ajrcmb.24.2.4370. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Zhou Y, Weiss HL, Chow CW, Evers BM. NFATc1 regulation of TRAIL expression in human intestinal cells. PLoS One. 2011;6(5):e19882. doi: 10.1371/journal.pone.0019882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morra M, Zubiaur M, Terhorst C, Sancho J, Malavasi F. CD38 is functionally dependent on the TCR/CD3 complex in human T cells. FASEB J. 1998;12(7):581–592. doi: 10.1096/fasebj.12.7.581. [DOI] [PubMed] [Google Scholar]
- 67.Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286(5440):790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 68.Vaeth M, et al. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2012;109(40):16258–16263. doi: 10.1073/pnas.1203870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9(2):194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 70.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hawwari A, Burrows J, Vadas MA, Cockerill PN. The human IL-3 locus is regulated cooperatively by two NFAT-dependent enhancers that have distinct tissue-specific activities. J Immunol. 2002;169(4):1876–1886. doi: 10.4049/jimmunol.169.4.1876. [DOI] [PubMed] [Google Scholar]
- 72.Oh-Hora M, et al. Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity. 2013;38(5):881–895. doi: 10.1016/j.immuni.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fuchs S, et al. Antiviral and regulatory T cell immunity in a patient with stromal interaction molecule 1 deficiency. J Immunol. 2012;188(3):1523–1533. doi: 10.4049/jimmunol.1102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bopp T, et al. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J Exp Med. 2005;201(2):181–187. doi: 10.1084/jem.20041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beyersdorf N, et al. STIM1-independent T cell development and effector function in vivo. J Immunol. 2009;182(6):3390–3397. doi: 10.4049/jimmunol.0802888. [DOI] [PubMed] [Google Scholar]
- 76.Gwack Y, et al. Hair loss and defective T- and B-cell function in mice lacking ORAI1. Mol Cell Biol. 2008;28(17):5209–5222. doi: 10.1128/MCB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kakeda M, et al. Heat-killed bacillus Calmette-Guerin and Mycobacterium kansasii antigen 85B combined vaccination ameliorates dermatitis in a mouse model of atopic dermatitis by inducing regulatory T cells. Br J Dermatol. 2012;166(5):953–963. doi: 10.1111/j.1365-2133.2011.10763.x. [DOI] [PubMed] [Google Scholar]
- 78.Schoenborn JR, Wilson CB.Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 200796: 41–101. [DOI] [PubMed] [Google Scholar]
- 79.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFNγ) locus revealed by genome sequence comparison. J Biol Chem. 2004;279(6):4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 80.Sakai S, Mayer-Barber KD, Barber DL. Defining features of protective CD4 T cell responses to Mycobacterium tuberculosis. Curr Opin Immunol. 2014;29:137–142. doi: 10.1016/j.coi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin AA, Tripathi PK, Sholl A, Jordan MB, Hildeman DA. Gamma interferon signaling in macrophage lineage cells regulates central nervous system inflammation and chemokine production. J Virol. 2009;83(17):8604–8615. doi: 10.1128/JVI.02477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barber DL, Andrade BB, Sereti I, Sher A. Immune reconstitution inflammatory syndrome: the trouble with immunity when you had none. Nat Rev Microbiol. 2012;10(2):150–156. doi: 10.1038/nrmicro2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shtrichman R, Samuel CE. The role of γ interferon in antimicrobial immunity. Curr Opin Microbiol. 2001;4(3):251–259. doi: 10.1016/S1369-5274(00)00199-5. [DOI] [PubMed] [Google Scholar]
- 84.Grundy S, et al. CRAC channel inhibition produces greater anti-inflammatory effects than glucocorticoids in CD8 cells from COPD patients. Clin Sci. 2014;126(3):223–232. doi: 10.1042/CS20130152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banaiee N, Kincaid EZ, Buchwald U, Jacobs WR, Jr, Ernst JD. Potent inhibition of macrophage responses to IFNγ by live virulent Mycobacterium tuberculosis is independent of mature mycobacterial lipoproteins but dependent on TLR2. J Immunol. 2006;176(5):3019–3027. doi: 10.4049/jimmunol.176.5.3019. [DOI] [PubMed] [Google Scholar]
- 86.Wolf AJ, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179(4):2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 87.Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31(6):974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wan Q, et al. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J Exp Med. 2011;208(9):1875–1887. doi: 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.