ABSTRACT
Serratia marcescens generates secondary metabolites and secreted enzymes, and it causes hospital infections and community-acquired ocular infections. Previous studies identified cyclic AMP (cAMP) receptor protein (CRP) as an indirect inhibitor of antimicrobial secondary metabolites. Here, we identified a putative two-component regulator that suppressed crp mutant phenotypes. Evidence supports that the putative response regulator eepR was directly transcriptionally inhibited by cAMP-CRP. EepR and the putative sensor kinase EepS were necessary for the biosynthesis of secondary metabolites, including prodigiosin- and serratamolide-dependent phenotypes, swarming motility, and hemolysis. Recombinant EepR bound to the prodigiosin and serratamolide promoters in vitro. Together, these data introduce a novel regulator of secondary metabolites that directly connects the broadly conserved metabolism regulator CRP with biosynthetic genes that may contribute to competition with other microbes.
IMPORTANCE This study identifies a new transcription factor that is directly controlled by a broadly conserved transcription factor, CRP. CRP is well studied in its role to help bacteria respond to the amount of nutrients in their environment. The new transcription factor EepR is essential for the bacterium Serratia marcescens to produce two biologically active compounds, prodigiosin and serratamolide. These two compounds are antimicrobial and may allow S. marcescens to compete for limited nutrients with other microorganisms. Results from this study tie together the CRP environmental nutrient sensor with a new regulator of antimicrobial compounds. Beyond microbial ecology, prodigiosin and serratamolide have therapeutic potential; therefore, understanding their regulation is important for both applied and basic science.
INTRODUCTION
In order for organisms to survive and prosper, they must be able to sense their environment and effectively compete with other organisms. To respond to these environmental changes, bacteria have developed elaborate transcriptional regulatory systems that enable fine-tuning of factors that allow for their adaptation and proliferation. One of the most studied signaling systems involved in adaptation to the nutritive status of the environment is the cyclic AMP (cAMP)-associated catabolite repression system (1–4). The second messenger cAMP has been classified as an alarmone that induces positive regulation of alternative carbon transport systems in times of carbon/fuel deprivation (5). In addition to catabolite repression control, this system also can positively regulate flagellum production in unfavorable conditions (6) and activate attachment factors in nutrient-rich conditions (7).
Evidence suggests that cAMP-cAMP receptor protein (CRP) can directly bind to and promote expression of secondary metabolite genes involved in antibiotic production in Streptomyces coelicolor (2). A positive or negative role for cAMP has been suggested for control of antimicrobial production in other organisms, including fungi, although direct or indirect control of gene expression has not been determined (8–11). In general, cAMP-associated transcriptional circuits that regulate secondary metabolism are poorly understood.
The Gram-negative bacterium S. marcescens is known for its ability to produce numerous secondary metabolites (12–14). These include the surfactant serratamolide and the red pigment prodigiosin, which are broad-spectrum antibiotics that may aid the bacterium in competition, as well as having therapeutic potential for initiating apoptosis in cancer cells (15–17).
Mutation of genes involved in 3′-5′-cAMP production (cyaA) and the transcription factor that responds to cAMP (crp) confers robust phenotypes beyond catabolite repression, including increased prodigiosin production (18), elevated serratamolide production (19), increased biofilm formation through increased type I pili production (20), enhanced extracellular protease production (21), and a loss of flagellum-based motility (22). CRP similarly regulates motility, adhesion, and secondary metabolite production by such diverse bacteria as Salmonella enterica serovar Typhimurium and Streptomyces coelicolor (1–4).
Whereas recombinant S. marcescens CRP bound directly to the promoter of flhDC to regulate flagellum production (18), interactions were not detected between recombinant CRP and promoters of the prodigiosin biosynthetic operon (pigA-N) or the swrW gene, required for serratamolide production (18, 21, 23). Based on these observations, we hypothesized an intermediate regulatory protein, regulated by cAMP-CRP, that in turn regulates expression of pigA-N and swrW. The purpose of this study was to identify this theoretical intermediate regulatory protein. Using a genetic approach, suppressor mutations of the hyperhemolysis and pigment phenotypes of the crp mutant strain were generated and mapped to an uncharacterized putative two-component transcriptional regulator locus. These genes, named eepR and eepS, were found to be a novel regulatory system for control of secondary metabolites by S. marcescens. The data further support that EepR is an intermediate transcriptional regulator between cAMP-CRP and secondary metabolite biosynthetic genes.
MATERIALS AND METHODS
Microbial strains, media, and growth.
S. marcescens strains are listed in Table 1. Human keratitis isolate K949 was isolated at the Charles T. Campbell Laboratory Ophthalmic Microbiology Laboratory. Bacteria were grown with aeration in lysogeny broth (LB) medium (26) (0.5% yeast extract, 1% tryptone, 0.5% NaCl) with or without 1.5% agar, tryptic soy agar supplemented with 5% sheep erythrocytes (blood agar), or M9 minimal medium (27) supplemented with glucose (0.4%) and casein amino acids (0.06%). Swimming agar and swarming agar used in this study were LB medium with agar concentrations at 0.3% and 0.6%, respectively. Escherichia coli strains used were the EC100D pir-116 (Epicentre), SM10 λpir, and S17-1 λpir strains (28). Saccharomyces cerevisiae strain InvSc1 (Invitrogen) was grown with either yeast extract-peptone-dextrose (YPD) or synthetic complete (SC)-uracil medium (29). Antibiotics used in this study include gentamicin (10 μg ml−1), kanamycin (100 μg ml−1), and tetracycline (10 μg ml−1).
TABLE 1.
S. marcescens strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| CMS376 | WT strain PIC3611 | Presque Isle Cultures |
| CMS531 | K949, clinical keratitis isolate | This study |
| CMS534 | K949 with transposon upstream of eepR | This study |
| CMS592B | CMS376 with pigB::Tn (transposon insertion) | 18 |
| CMS613 | CMS376 with crp-1 null mutation | 7 |
| CMS635 | CMS376 with swrW::Tn | 23 |
| CMS786 | crp-23 transposon null mutation | 7 |
| CMS794 | CMS592B with crp-1 null mutation | 18 |
| CMS795 | CMS613 with transposon upstream of eepR | This study |
| CMS827 | CMS376 with pMQ178 inserted in the eepR ORF | This study |
| CMS853 | K904 clinical keratitis isolate | 18 |
| CMS1075 | CMS376 with crp-1 eepS::Tn | This study |
| CMS1076 | CMS1075 with restored crp | This study |
| CMS1464 | CMS376 with crp-23 eepS::Tn | This study |
| CMS1687 | Δcrp-4 deletion null mutation | 18 |
| CMS1787 | Nima pigmented environmental isolate | 24 |
| CMS2089 | Nima with ΔeepR | This study |
| CMS2091 | Nima with ΔeepS | This study |
| CMS2093 | Nima with ΔeepR ΔeepS | This study |
| CMS2096 | CMS376 ΔpigP | 25 |
| CMS2097 | CMS376 ΔeepR | This study |
| CMS2157 | CMS376 Δcrp-4 ΔeepR | This study |
| CMS2395 | CMS376 Δcrp-4 eepS::Tn | This study |
| CMS2701 | CMS376 ΔeepS | This study |
| CMS2881 | CMS376 Δcrp-4 swrW::Tn | 15 |
| CMS2904 | K904 ΔeepR | This study |
| CMS2924 | K904 with ∆eepS | This study |
| CMS2921 | CMS2097 with ΔeepR replaced by wild-type eepR | This study |
| CMS2032 | CMS1076 with eepS::Tn replaced with wild-type eepS | This study |
Mutagenesis and plasmid construction.
Transposons were introduced into S. marcescens by conjugation as previously described (20) using mariner-based transposon delivery plasmids pBT20 (30) and pSC189 (31). Tetracycline (10 μg ml−1) was used to eliminate donor E. coli growth, and kanamycin (100 μg ml−1) or gentamicin (10 μg ml−1) was used to select for S. marcescens with transposon mutations. These were performed on blood agar plates to screen for pigment- and hemolysis-defective mutants as described below.
Cloning was performed using in vivo recombination (32) of PCR-generated amplicons or using T4 DNA ligase (New England BioLabs). PCR of amplicons used for cloning was performed using a high-fidelity polymerase, Phusion (New England BioLabs). Cloned genes were verified by diagnostic PCR and DNA sequencing (University of Pittsburgh Genomic and Proteomic Core). Plasmids are listed in Table S1 in the supplemental material. Directed mutagenesis was achieved by two-step allelic replacement or insertional mutagenesis as noted in the text and as previously described (21, 32). Mutations were verified using PCR primers outside the cloned region on the mutagenesis plasmid.
Allelic replacement of eepR, eepS, and eepRS.
To generate the eepS deletion strain, we cloned eepR, including 458 bp upstream of eepR and the entire eepS open reading frame (ORF), here referred to as eepRS, in pMQ236 to generate pMQ289. Primers 1577 and 1578 were used to clone eepRS. Primers are listed in Table S2 in the supplemental material.
To delete the eepR ORF, pMQ289 was digested with MluI and SalI, the ends were blunted with a multiple enzyme mixture (End-It kit; Epicentre), and the plasmid was recircularized using T4 DNA ligase. The resulting plasmid has an in-frame deletion of 67 out of 283 amino acids from E126-V192. The plasmid was named pMQ318z and used for allelic replacement. To make an insertion mutation in eepR at base pair 400 with respect to the translational start, a 359-bp internal fragment was amplified with primers 1234 and 1235 and cloned in pMQ118.
To mutate eepS, pMQ289 was digested with ApaLI, which cuts twice in eepS. The plasmid was recircularized using T4 DNA ligase, yielding the eepS deletion allele and plasmid pMQ308. This deletion is in frame and removes S337-H422 out of the total of 594 amino acids in EepS.
To generate the double eepR eepS mutation, pMQ289 was digested with AatII, which has sites in both eepR and eepS. The plasmid was recircularized using T4 DNA ligase, yielding the eepRS deletion allele. The resulting plasmid, pMQ291, has an in-frame deletion of the last 93 amino acids of eepR and an in-frame mutation of the last 79 amino acids of eepS.
To generate complementation vectors, the eepR open reading frame was cloned with primers that changed the start codon from ATG to TTG using primers 1222 and 2552 and placed the gene under the control of the E. coli Plac promoter in plasmid pMQ132, yielding pMQ364, or in plasmid pMQ131, yielding pMQ432. A similar plasmid with a C-terminal polyhistidine (His8) tag was generated with plasmid pMQ132, yielding pMQ369 using primers 2552 and 2698.
For purification of EepR, a maltose-binding protein (MBP)-EepR fusion construct was made with pMal-C2 (New England BioLabs). The eepR ORF was amplified with primers containing EcoRI and HindIII sites, and the restriction-digested amplicon was introduced into pMal-C2, which also was digested with EcoRI and HindIII. Primers used to amplify eepR were MBP-R-R1 and MBP-R-H3. T4 DNA ligase was used to recombine the amplicon and vector to generate pMQ403.
To ensure that the MalE-EepR (MBP-EepR) fusion was functional, EepR and MalE ORFs were amplified from pMQ403 with primers 919 and 2948, and the amplicon was recombined into expression vector pMQ124. The resulting plasmid, pMQ438, has the MBP-EepR fusion under transcriptional control of the PBAD promoter.
Prodigiosin production assays.
Single colonies were inoculated in 5 ml of LB medium and incubated at 30°C for 16 to 18 h with aeration as noted above. Culture optical density was recorded, 1 ml of culture was transferred to a microcentrifuge tube, and the cells were pelleted. Prodigiosin was extracted from centrifuged cell pellets with 1 ml acidified ethanol (2 ml of 2 N HCl added to 98 ml of 95% ethanol), and pigment levels were measured by absorbance at 534 nm based on the method of Slater et al. (33), as previously described (18).
Transcriptional analysis.
For β-galactosidase (β-gal) assays, cultures were grown overnight in LB medium with antibiotics at 30°C, subcultured (1:100) two times, and grown to an optical density at 600 nm (OD600) of 0.1 in order to synchronize cultures in the early exponential growth phase. After growth to the desired optical density, culture aliquots were pelleted and washed with Z-buffer, and β-gal activity was determined (34). Lysates were prepared by sonication in Z-buffer and were clarified by centrifugation at 16,100 × g for 5 min. The supernatant protein concentration was determined by Bradford analysis, and the same amount of protein (0.2 mg) from each sample in a given experiment was added to microtiter plate wells. The volume was adjusted to 100 μl with Z-buffer. ONPG (o-nitrophenyl-β-d-galactopyranoside; 25 μl at 4 mg ml−1) was added as a colorimetric substrate, and A410 readings were taken with a plate reader over a 30-min period (Biotek Synergy 2; Winooski, VT).
For eepR analysis, a transcriptional lacZ reporter construct was targeted into the chromosome of the wild-type and the crp mutant strains using a 263-bp region of DNA upstream of the eepR ORF containing the predicted CRP binding site in a lacZ-containing suicide vector (pMQ254). When this construct integrates, the promoter region is duplicated such that the lacZ gene becomes a reporter for eepR expression, and the native eepR gene comes under the control of the regulatory elements in the 263 bp of DNA upstream of the ORF, maintaining EepR, which may be necessary for eepR expression. For pigA and swrW promoter analysis, transcriptional lacZ fusions to internal fragments of the pigB and swrW genes (plasmids pMQ268 and pMQ223) were targeted to the chromosome by homologous recombination, verified by PCR, and used as previously described (18, 21, 23).
For quantitative PCR (qPCR) analysis, cultures for RNA extraction were grown at 30°C with aeration in 5 ml of LB broth following inoculation from a single colony. The overnight cultures were diluted to an OD600 of 0.1 in fresh LB medium, grown as described above, and harvested at the desired optical density. Bacterial aliquots were treated with RNAprotect bacterial reagent (Qiagen) by following the manufacturer's protocol and stored at −80°C for a maximum of 1 week. RNA was extracted (Qiagen RNeasy kit) using two rounds of DNase treatment (one Qiagen on-column DNase treatment and one Promega RQ1 DNase treatment) by following the manufacturer's protocols. RNA was concentrated using a Zymo Research RNA Clean & Concentrator-5 kit (R1015) according to the manufacturer's protocol. The RNA concentration was measured using a Thermo Scientific NanoDrop (model 2000) and normalized to 50 ng μl−1 using nuclease-free water. cDNA synthesis was performed using Superscript III reverse transcriptase (Invitrogen) as specified by the manufacturer using 250 ng of RNA. The cDNA was diluted 1:5 in DNase-free water and tested for chromosomal DNA contamination by PCR using a thermal cycler (2720; Applied Biosystems) with oligonucleotide primers for the 16S rRNA gene (see Table S2 in the supplemental material) with amplification for 26 rounds, and any samples with a band on an agarose gel, indicating contamination, were discarded. Negative-control reactions without reverse transcriptase and without RNA were included and failed to produce an amplicon. Quantitative reverse transcription-PCR (qRT-PCR) was performed using Sybr green reagent (Applied Biosystems) according to the manufacturer's protocol using an Applied Biosystems Step One real-time PCR system with oligonucleotide primers listed in Table S2. The primers were 2638 and 2639 for the 16S rRNA gene, 1471 and 1472 for eepR, 2911 and 2912 for pigA, and 1786 and 2919 for swrW. qRT-PCR analysis was determined using the ΔΔCT method (where CT is threshold cycle).
Protein purification, electrophoretic mobility shift analysis (EMSA), and chromatin affinity precipitation (ChAP) assays.
An MBP fusion to EepR (MBP-EepR) and MBP alone were generated for affinity purification of EepR using pMal-C2 (New England BioLabs) as previously described (25). His8-CRP purification was described previously (18).
To perform EMSA, labeled DNA amplicons were made with a 5′-biotinylated oligonucleotide primer (Integrated DNA Technologies, Skokie, IL), gel purified, and verified by sequencing. A commercial EMSA kit was employed as specified by the manufacturer (LightShift chemiluminescent EMSA kit; Pierce, Rockford IL), using biotinylated target DNA (1 to 3 ng), purified His8-CRP (≥50 ng), or MBP-EepR (≥50 ng) and poly(dI-dC) (500 ng), cAMP where indicated, and nonlabeled competitor DNA (20 to 600 ng) as specified, in a 20-μl reaction mixture. A 10-μl aliquot of the reaction mix was separated on a 5% PAGE, Tris-borate-EDTA (TBE) gel (Bio-Rad) with a running buffer containing 500 μM cAMP when His8-CRP was used. EMSAs were repeated at least three times. Primers and amplified regions for the oxyR, pigA, and swrW promoter regions used in EMSAs have been described previously (25). Primers for the eepR promoter are 1346 and 1884, or a biotinylated version of primer 1884, which amplify a 263-bp region of DNA just upstream of the eepR start codon, using pMQ254 as a template. A 359-bp internal region of eepR was used to test the specificity of CRP binding to the eepR promoter interaction; this region was amplified using primers 1234 and 1235.
ChAP assays were performed as previously described (25), except using pMQ242 (His8-CRP) and the pMQ124 vector as a negative control. Primer pairs for flhDC, eepR, and oxyR promoters were 1670 and 1671, 1667 and 1668, and 1432 and 1433, respectively. The experiment was repeated twice, yielding similar results.
Serratamolide measurement.
Zones of biosurfactant around colonies on swarming agar plates were measured 18 to 20 h after inoculation of the bacteria onto the surface of the agar as previously reported (23, 25). Quantitative analysis of serratamolide from culture supernatants by high-performance liquid chromatography-mass spectrometry (HPLC-MS) was carried out as previously described (25). Bacterial cultures were grown in LB (10 5-ml cultures per genotype) for 20 h at 30°C. Bacteria were pelleted by centrifugation from pooled cultures, and the supernatant was extracted three times with equal volumes of ethyl acetate (30 ml). The ethyl acetate layers were dried over sodium sulfate and evaporated in vacuo. The dried residue was dissolved in methanol and analyzed by HPLC-MS (Shimadzu LCMS-2020) using a Dionex Acclaim 120 C18 column (3-μm particle size, 120-Å pore size; dimensions, 2.1 by 150 mm). A mobile-phase gradient, 40% acetonitrile (AcCN)–60% H2O (0 min), 40% AcCN–60% H2O (1 min), 90% AcCN–10% H2O (15 min), 90% AcCN–10% H2O (35 min), 40% AcCN–60% H2O (40 min), and 40% AcCN–60% H2O (45 min), was used for this analysis. The column flow rate was set to 0.2 ml min−1, and the column oven temperature was set at 40°C. Serratamolide was monitored at m/z = 515 (for [M+H]+) using an electrospray ionization-mass spectrometry (ESI-MS) detector in positive mode. Previously purified serratamolide (23) was used as a positive control.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software. One-way analysis of variance (ANOVA) with Tukey's posttest and the two-tailed Student's t tests were used with significance set at P < 0.05.
Nucleotide sequence accession number.
The sequence of the eepR gene from strain CMS376 was deposited in GenBank under accession number JQ914138.
RESULTS
Identification of eepR and eepS.
Previous studies demonstrated cAMP-CRP regulation of flagellum and secondary metabolite production by S. marcescens strain PIC3611 (18, 21, 23). Direct binding of cAMP-CRP to the promoter of the flagellar master regulator operon, flhDC, was observed, but a positive interaction between cAMP-CRP and promoters of the genes required for biosynthesis of the secondary metabolites prodigiosin (pigA-N) or serratamolide (swrW) was not (18). Because the regulation of flagella by cAMP-CRP involves intermediate regulators FlhD and FlhC (22, 35), we predicted an analogous intermediate regulator functions between CRP and pigA-N and swrW.
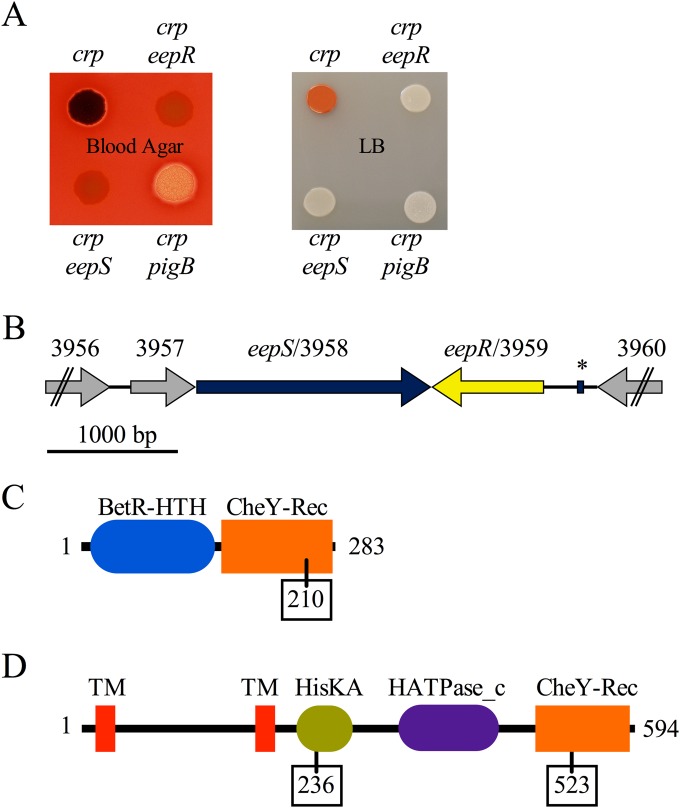
To find the predicted intermediate regulator downstream of cAMP-CRP, suppressor analysis was performed. Specifically, random mutations were introduced using a mariner transposon in a crp mutant background, and the mutant colonies were screened for suppression of the crp hyperprodigiosin phenotype. The mutants with reduced or eliminated prodigiosin then were screened for hyperhemolysis suppression phenotypes; an example is shown in Fig. 1A. A progidiosin-defective mutant with an insertion in the pigB pigment biosynthetic gene was included to demonstrate a strain defective in the pigment phenotype is not necessary for the hemolysis phenotypes (Fig. 1A). Hemolysis under the crp pigB colony is evident in Fig. 1A and shared by the crp mutant (not evident in Fig. 1A) but was absent from crp eepR and crp eepS mutants.
FIG 1.
Hyperhemolysis phenotype of crp mutants and genetic analysis. (A) Photograph demonstrating suppression of the crp hemolysis (left) and pigment (right) phenotypes by mutation of eepR and eepS. The eepR and eepS colonies appear red because of the blood agar (left), but their severe lack of pigment phenotype is clear when grown on LB agar (right). The crp pigB mutant is included as a control and only has a defect in pigment production. The crp mutant is strain number CMS613, the crp eepR mutant is CMS795, the crp eepS mutant is CMS1075, and the crp pigB mutant is CMS794. (B) Map of the genetic context of eepR and eepS; ORF numbers are from the Db11 genome, and “SMDB11_” was removed from each ORF number to save space. The asterisk indicates a predicted CRP binding site. (C) Predicted protein domains and amino acid length of EepR. The amino acid location of the predicted phosphorylation site is boxed. (D) Predicted protein domains of EepS. The amino acid locations of predicted phosphorylation sites are boxed.
Mutations that suppressed both phenotypes of the crp mutant were mapped to one of two adjacent and convergently transcribed uncharacterized ORFs (Fig. 1B; also see Tables S3 and S4 in the supplemental material), corresponding to SMDB11_3958 and SMDB11_3959, respectively (ORF designations are based on the DB11 genome [36]), that are predicted to code for a two-component histidine kinase and a response regulator based on sequence.
Other transposon-based genetic screens that provided impetus to analyze these two ORFs included the following: (i) a mutation in SMDB11_3959 eliminated production of an antistaphylococcal compound produced by S. marcescens (15), (ii) multiple mutations of SMDB11_3958 eliminated secreted hemolysis activity by a clinical isolate, K904, and (iii) a mutation upstream of SMDB11_3959 eliminated protease secretion by a nonpigmented ocular clinical isolate, K949 (data not shown). Altogether, 12 independent mutations in SMDB11_3958 and SMDB11_3959 have been identified, and all of these mutant strains were found to be deficient in hemolysis (serratamolide) and, if the strain was pigmented, defective in prodigiosin production. Transposon insertion sites are listed in Tables S1 and S2 in the supplemental material. Based on these phenotypes and subsequent data, we are naming these genes exoenzyme and pigment response regulator and sensor kinase, i.e., eepR for the putative response regulator (SMDB11_3959) and eepS for the sensor histidine kinase (SMDB11_3958). The role of these genes in exoenzyme regulation will be described elsewhere. This study focuses on determining whether the eepR or eepS genes are CRP regulated and provides a primary characterization of this novel locus.
Analysis of EepR and EepS sequences.
The eepR gene codes for a predicted 283-amino-acid response regulator transcription factor with an N-terminal helix-turn-helix domain of the BetR family and a C-terminal CheY receiver domain with a predicted phosphorylation site at D210 (Fig. 1C). The eepR gene from strain CMS376 was sequenced (GenBank accession number JQ914138) and shares 94.8% (DNA) identity with the corresponding DNA of a sequenced S. marcescens strain, Db11 (36); the resulting proteins from each strain are predicted to be 100% identical. Beyond other strains of S. marcescens, the predicted EepR protein is most similar to predicted response regulators in S. plymuthica (86% amino acid identity), other Serratia species (up to 79% amino acid identity), and various Burkholderia species (up to 50% amino acid identity).
The predicted EepS protein (Fig. 1D) is a 594-residue hybrid histidine kinase with two N-terminal transmembrane domains, a dimerization-photoreceptor domain (HisKA) with a predicted phosphorylation site at reside 236, a histidine kinase ATPase domain (HATPase_c), and a C-terminal CheY receiver domain with a predicted phosphorylation site at residue 523 (CheY-Rec). The organization of EepS suggests the existence of an intermediate phosphate carrier protein(s) between EepR and EepS (see Fig. S1 in the supplemental material). The AtsR protein from B. cenocepacia is 56.9% identical at the amino acid level and was found to mediate bacterial attachment to abiotic and biotic surfaces and host-pathogen interactions in that opportunistic pathogen (37, 38).
The cAMP receptor protein directly regulates eepR expression.
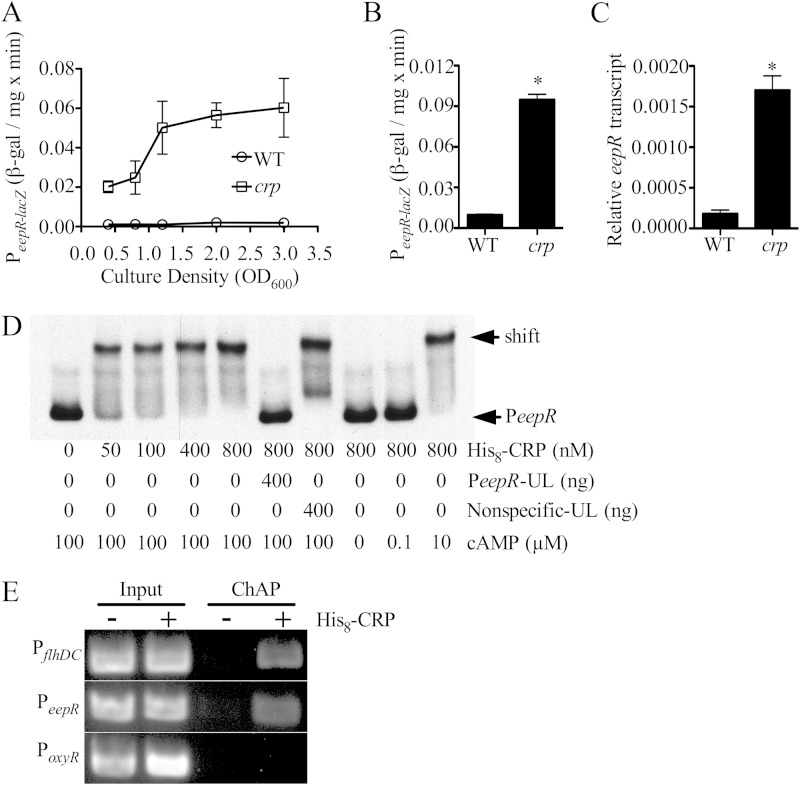
A predicted CRP-binding site was observed in the DNA sequence 241 to 257 bp upstream of the eepR ORF (TGAGACGATGATCACA) (Fig. 1B, asterisk; also see Fig. S2 in the supplemental material), but none were noted upstream of the SMDB11_3957-eepS predicted operon. To test transcriptional regulation of eepR by CRP, a chromosomal lacZ fusion was used. In the WT strain, eepR-lacZ expression was low throughout growth but was highly elevated in the crp mutant strain (Fig. 2A). In separate experiments with cells grown for 20 h in LB medium with aeration (OD600 = ∼4), β-galactosidase activity was 9.7-fold higher in the crp mutant than in the WT (n = 9) (P < 0.01) (Fig. 2B). qRT-PCR analysis of the native eepR gene agreed with the lacZ reporter data that there is a higher level of eepR transcript (9.5-fold) in the crp mutant than in the WT at an OD600 of 1.5 (Fig. 2C). As a third level of confirmation, the tdtomato fluorescent reporter gene was placed under the control of the eepR promoter (412 bp upstream of the eepR ORF) on a pBBR1-based plasmid. Higher levels of fluorescence were measured in the crp mutant (30,062 ± 1,186 relative fluorescence units [RFU]) cultures than in wild-type (11,812 ± 464 RFU) cultures grown overnight (P < 0.01 by Student's t test). Together, these data suggest a negative regulatory role for CRP on the eepR promoter.
FIG 2.
Direct regulation of eepR by cAMP-CRP. (A) Expression of the eepR promoter measured from a chromosomal lacZ reporter integrated at eepR in a WT and crp mutant background. The WT strain is CMS376, and the crp mutant is CMS786. (B) As described for panel A but at an OD600 of 4.0. (C) qPCR analysis of eepR expression from the WT and crp mutant measured at an OD600 of 1.5. The WT strain is CMS376, and the crp mutant is CMS1687. (D) EMSA of His8-CRP interaction with the biotin-labeled eepR predicted promoter (PeepR; 2 ng) in vitro. His8-CRP produced a gel shift of labeled PeepR that could be inhibited by an excess of unlabeled PeepR (PeepR-UL) but not by a nonspecific unlabeled amplicon (Nonspecific-UL), a 360-bp internal region of eepR. The gel shift required cAMP; whereas 0 and 0.1 μM did not support binding, 10 μM was sufficient. (E) Chromatin affinity purification of His8-CRP suggests binding of the eepR promoter in vivo. PCR amplification of the eepR promoter was elevated from ChAP purification of CRP-bound DNA in the WT strain containing a plasmid expressing His8-CRP (+CRP) compared to the WT strain with the empty vector (−CRP). The flhDC promoter was included as a positive control and the oxyR promoter as a negative control.
EMSA was performed to test whether CRP protein binds to the eepR promoter in vitro. We observed that purified His8-tagged recombinant CRP (His8-CRP) bound to the eepR promoter in a dose-dependent and cAMP-dependent manner, and that CRP-eepR promoter interactions could be titrated with excess unlabeled eepR promoter DNA but not with unlabeled DNA internal to the eepR ORF, supporting that the interaction was specific (Fig. 2D). Furthermore, ChAP analysis was performed to test whether CRP binds to the eepR promoter in vivo. ChAP analysis was performed three times, and a semiquantitative analysis of PCR amplicon density indicates a 10.1- ± 1.8-fold increase in detection of the eepR promoter in the CRP pulldown samples (pMQ124+His8-crp) compared to the level for the negative control (pMQ124) (Fig. 2E). A similar enrichment was found for the positive-control flhDC promoter but not the oxyR promoter, which serves as a negative control (Fig. 2E). Together, these data support the model that cAMP-CRP directly binds to the promoter of eepR and inhibits its expression (see Fig. S1A in the supplemental material).
EepR directly regulates prodigiosin biosynthesis.
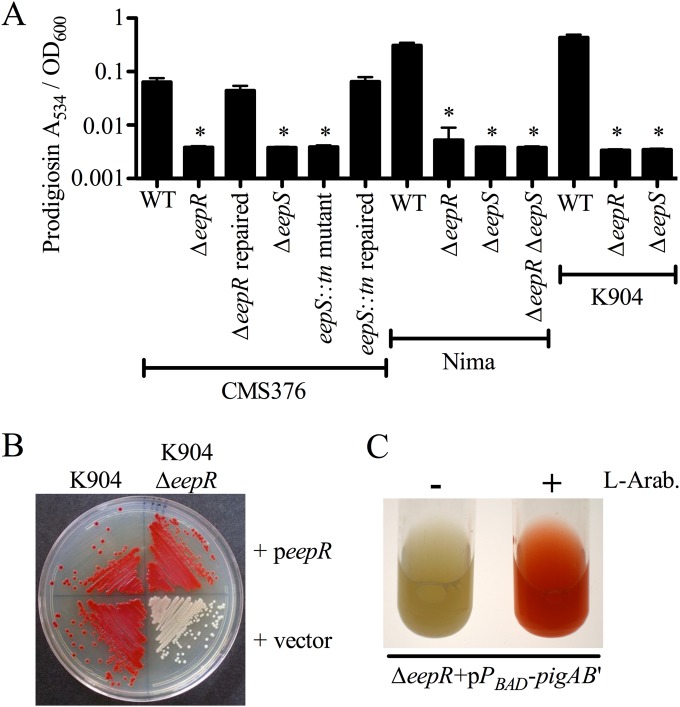
To further confirm that EepR and EepS play a role in prodigiosin production, we generated in-frame deletion mutations of eepR and eepS in WT strain CMS376. Both the eepR (CMS2097) and the eepS (CMS2701) deletion strains were defective in pigment production (Fig. 3A). Importantly, the eepR deletion mutation strains grown in M9 minimal medium were not defective in growth compared to the WT strain (see Fig. S1B in the supplemental material), indicating that the eepR mutant phenotypes are not due to reduced growth. Similar results were measured for growth of the CMS376 strain with an eepS transposon mutant, an eepR insertion mutant, and the parental strain grown in LB medium (see Fig. S1C). Furthermore, there were no detected differences between eepR and eepS mutants for the assays listed below; therefore, eepR mutant phenotypes alone generally will be shown for the sake of brevity.
FIG 3.
Growth and complementation analysis of eepR mutant pigment phenotypes. (A) Prodigiosin extracted and measured from stationary-phase bacteria. WT, CMS376; ΔeepR strain, CMS2097; ΔeepR repaired strain, CMS2921; ΔeepS strain, CMS2701; eepS::Tn strain, CMS1076; eepS::Tn repaired strain, CMS2032; Nima ΔeepR strain, CMS2089; Nima ΔeepS strain, CMS2091; Nima ΔeepR ΔeepS strain, CMS2093; K904 ΔeepR strain, CMS2094; K904 ΔeepS strain, CMS2924. Means and standard deviations are shown (n = 8). An asterisk indicates P < 0.05 by ANOVA with Tukey's posttest. (B) Complementation of the ΔeepR mutant phenotype in K904 and K904 ΔeepR strains using plasmid pMQ369 (peepR) and a vector negative control (pMQ132). (C) Culture pigmentation in the ΔeepR strain (CMS2097) without (left) and with (right) l-arabinose (L-Arab.)-induced expression of the prodigiosin biosynthetic operon.
Given the phenotypic variation caused by mutation of crp in S. marcescens and other Enterobacteriaceae (39, 40), we assessed whether eepR and eepS have a conserved role in other S. marcescens strains. The eepR and eepS genes were deleted from another laboratory strain, Nima, and from a contact lens-associated keratitis isolate, K904. Nima eepR and eepS and K904 eepR and eepS strains also lost pigment production (Fig. 3A and B). The K904 mutant differs from the other eepR mutants in that it eventually gained partial pigmentation when grown on plates for 2 days (data not shown). The K904 ΔeepR mutant was not growth defective (see Fig. S1B in the supplemental material). Strain Nima with deletion of both eepR and eepS was as defective in prodigiosin biosynthesis as either single mutant (Fig. 3A), suggesting a single pathway rather than EepR and EepS acting independently to promote pigmentation.
Complementation of the eepR mutant defect was performed to determine whether the deletion mutation rather than an unknown mutation elsewhere in the genome or a polar effect upon expression of adjacent genes was responsible for the observed phenotypes. In trans complementation of ΔeepR mutant phenotypes by expression of the wild-type eepR gene from the Plac promoter on a pBBR1-based plasmid was performed (Fig. 3B; also see Fig. S1D in the supplemental material). In addition, allelic replacement of the eepR and eepS mutant alleles with the CMS376 wild-type (WT) genes restored pigment production (Fig. 3A and data not shown). This allelic replacement approach indicates that there is not a mutation elsewhere on the chromosome that caused the mutant phenotype, but it does not tell us whether the mutant phenotype was due to a polar effect. The similar phenotype conferred by multiple independent insertion mutations, in-frame deletion mutations in different strains, restoration of phenotypes by replacement of the mutant alleles with the wild-type genes, and expression of the wild-type eepR gene on a plasmid together support that mutation of the eepR gene rather than an unknown mutation or a polar effect confers the mutant phenotypes.
Consistent with EepR being a positive regulator of prodigiosin production, multicopy expression of the wild-type eepR gene from the Plac promoter (pMQ369) in the WT strain (CMS376) increased prodigiosin production above the level of the vector-alone control (see Fig. S1D in the supplemental material). When prodigiosin was quantified from stationary-phase cultures there was almost twice as much isolated from the WT with multicopy eepR (A534/OD600, 0.23 ± 0.05) as there was for the WT with the vector control (0.12 ± 0.02; n = 6 per group; P < 0.05 by Student's t test).
The chromosomal prodigiosin biosynthetic operon was placed under the control of an arabinose-inducible promoter through integration of pMQ262 (15) in the ΔeepR strain (CMS2097) to test whether expression of pigA-N alone is sufficient to restore prodigiosin production. Red pigment production was restored when the strain was grown with the addition of the inducer l-arabinose but not when treated with glucose or water (Fig. 3C and data not shown). This observation supports that (i) induced expression of pigA-N is sufficient to restore pigmentation to an eepR mutant, (ii) the requirement of EepR in prodigiosin production is not downstream of pigA-N expression, and (iii) EepR has a regulatory rather than biochemical role in prodigiosin production. As a control for unintended effects of the l-arabinose sugar, we confirmed that the l-arabinose concentration used did not affect pigmentation of the eepR mutant without the pMQ262 plasmid (data not shown).
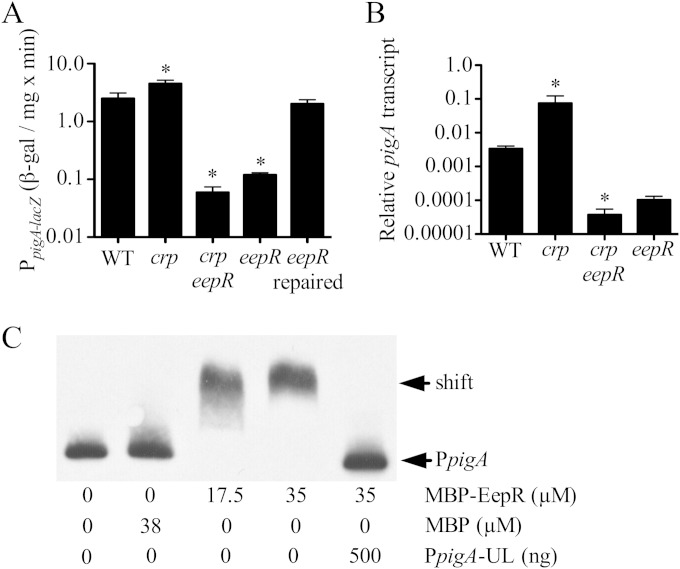
A chromosomal lacZ fusion and qPCR analysis both indicated that eepR is required for wild-type levels of expression from the prodigiosin biosynthetic operon pigA-N (Fig. 4A and B). Expression levels were more than 10-fold lower in the ΔeepR mutant than in the WT, with similar but slightly lower levels found in crp eepR double mutants compared to those of the eepR mutants (Fig. 4A and B). This lack of pig operon expression correlates with the lack of pigmentation in the eepR and crp eepR double mutant strains and are consistent with EepR and CRP functioning in a linear regulatory pathway (see Fig. S1A in the supplemental material).
FIG 4.
Regulation of pigA by EepR and epistasis analysis. (A) β-Galactosidase-based expression from the pigA promoter at an OD600 of 4. WT, CMS376; crp strain, CMS1687; crp eepR strain, CMS2157; eepR strain, CMS2097; eepR repaired strain, CMS2921. (B) qPCR analysis of the pigA promoter at an OD600 of 2. WT, CMS376; crp strain, CMS1687; crp eepR strain, CMS795; eepR strain, CMS2097. (C) EMSA analysis of MBP-EepR interaction with biotin-labeled pigA promoter (PpigA; 2 ng) in vitro. MBP-EepR produced a gel shift of labeled PpigA that could be inhibited by an excess of unlabeled PpigA (PpigA-UL). Recombinant MBP was not sufficient to produce a gel shift of PpigA. An asterisk indicates significant difference from the WT by ANOVA with Tukey's posttest.
A maltose-binding protein (MBP) fusion to EepR (MBP-EepR) was generated and found to be functional by complementation of the eepR mutant pigment phenotype (see Fig. S1D in the supplemental material). MBP-EepR bound to the pigA-N promoter in vitro in EMSAs, whereas purified MBP, by itself and at similar concentrations, was unable to bind to the pigA promoter (Fig. 4C). Furthermore, the MBP-EepR interaction with the labeled pigA-N promoter could be outcompeted using an excess of unlabeled pigA-N promoter DNA (Fig. 4C). We observed that MBP-EepR did not bind to other candidate promoters, such as the gdhS glucose-dehydrogenase and pigP transcription factor genes, lending additional evidence to the idea that the MBP-EepR binding to the pigA-N promoter is specific (see Fig. S3 in the supplemental material). Together, these data support that EepR directly and positively regulates pigA-N.
EepR/S is required for serratamolide production, hemolysis, and swarming motility.
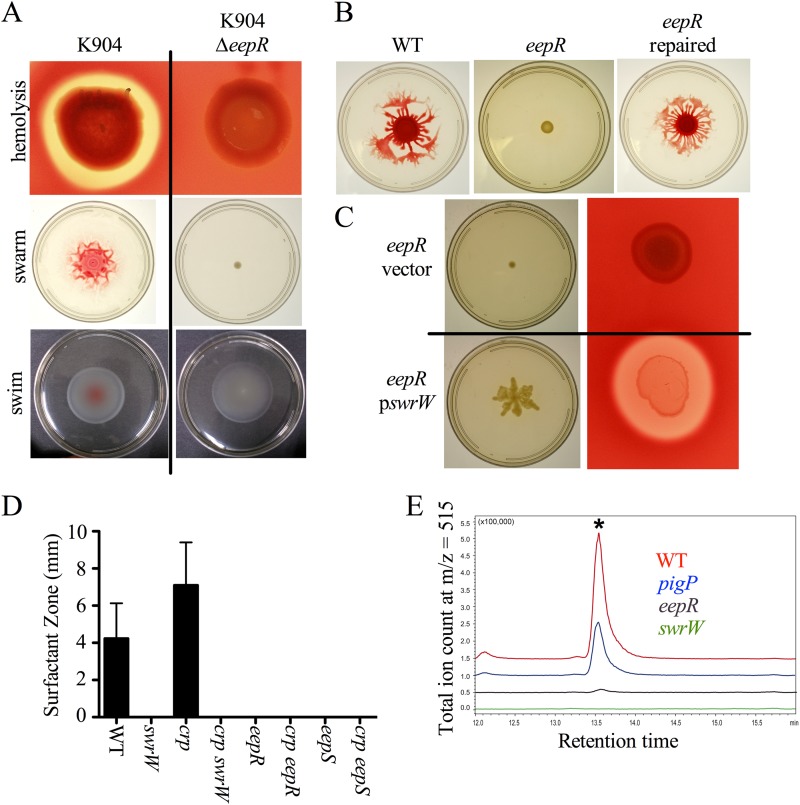
Mutation of crp in CMS376 results in higher levels of hemolysis than that of the isogenic parental strain due to increased production of the biosurfactant serratamolide (23), also known as serrawettin W1 (41). Since EepR/S appears to function with CRP in a regulatory pathway that controls prodigiosin production and EepR and EepS are required for the hemolysis phenotype of crp mutants (Fig. 1B), we tested whether EepR played a role in serratamolide production. Mutation of eepR and eepS in other isolates, such as K904 and Nima, eliminated hemolysis as measured from clearing zones on blood agar plates (Fig. 5A and data not shown).
FIG 5.
EepR is necessary for serratamolide and serratamolide-dependent phenotypes. (A) Hemolysis and swarming are EepR dependent in strain K904, but swimming is not. K904 ΔeepR mutant, CMS2904. (B) Swarming motility is defective in the ΔeepR (CMS2097) mutant and could be restored when the chromosomal eepR deletion allele was replaced by the wild-type eepR gene (CMS2921). (C) Hemolysis and swarming phenotypes of the ΔeepR mutant (CMS2097) can be rescued by induced expression of swrW on a plasmid (pswrW/pMQ367) but not by the vector control (pMQ125). (D) Means and standard deviations from surfactant radii around colonies on a swarming agar plate (n ≥ 5 independent isolates). WT, CMS376; swrW strain, CMS635; crp strain, CMS1687; crp swrW strain, CMS2281; eepR strain, CMS2097; crp eepR strain, CMS2701; crp eepS strain, CMS2395. (E) HPLC-MS analysis of serratamolide levels in supernatants from stationary-phase cultures. An asterisk indicates the serratamolide peak. WT, CMS376; pigP strain, CMS2096; eepR strain, CMS2097; swrW strain, CMS635).
Serratamolide also is required for swarming motility in many strains of S. marcescens (25, 42). We observed that eepR and eepS mutants were defective in swarming in the wild-type (CMS376) and K904 strain backgrounds (Fig. 5A and B). The eepR mutant swarming defect was complemented when the wild-type eepR gene was used to replace the deletion allele on the chromosome (Fig. 5B). Whereas swarming motility was defective, swimming motility was equivalent to that of the wild type (Fig. 5A). This result was consistent with the swarming defect resulting from reduced surfactant production rather than a deficiency in functional flagella. Expression of the serratamolide biosynthetic gene, swrW, from a multicopy plasmid restored swarming motility and hemolysis to tested eepR and eepS mutants (Fig. 5C and data not shown), suggesting that a loss of swrW expression was the mechanism underlying the eepR hemolysis and swarming defects.
Similar to swrW mutants, strains defective in eepR and eepS exhibited no zones of surfactant around colonies, whereas WT colonies produced surfactant zones extending about 4 mm beyond the colony edge by 24 h (Fig. 5D). The crp mutant was previously shown to generate zones larger than those of the WT (23). Here, we observed that the crp mutant produced an average zone of 7 mm, whereas the crp eepR and crp eepS double mutants, as well as the crp swrW strain, produced no zone of surfactant (Fig. 5D).
HPLC-MS analysis verified that serratamolide is reduced in spent supernatants produced by eepR mutants (CMS2097) grown to saturation (Fig. 5E). Unlike the WT, which exhibits a large serratamolide peak, the eepR mutant extracts were almost completely devoid of detectable serratamolide, much like the negative-control swrW mutant (CMS635) that is unable to generate serratamolide (Fig. 5E). Supernatants from the K904 strain and its isogenic eepR mutant were similarly defective in serratamolide when assessed by HPLC-MS (data not shown). Interestingly, the eepR mutant was more defective than a pigP deletion mutant (Fig. 5E). PigP is a previously described positive regulator of serratamolide production in S. marcescens (25).
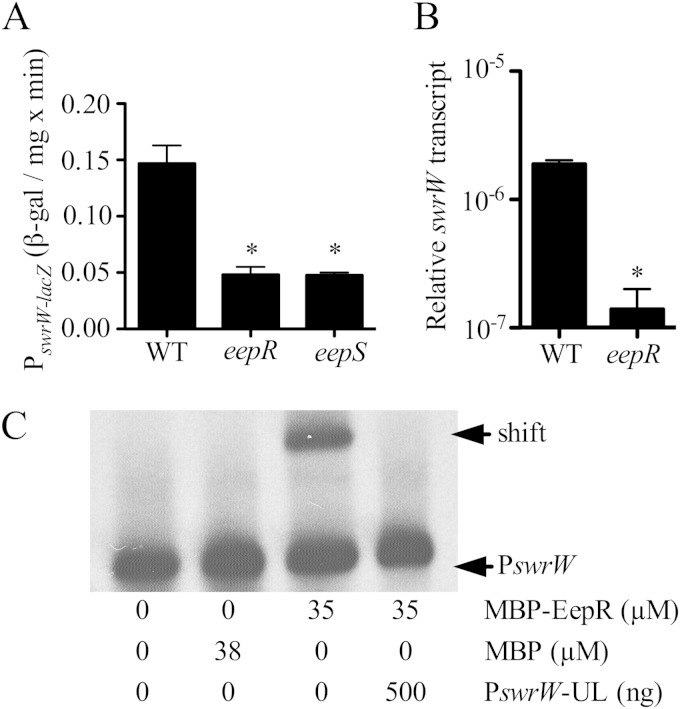
To test the prediction that EepR regulates expression of the swrW gene, a chromosomal lacZ reporter and qPCR were used to measure swrW expression from cells grown for 18 h at 30°C. Expression was significantly higher when measured from the wild-type strain compared to that of the ΔeepR mutant and ΔeepS mutant in the CMS376 strain background (Fig. 6A). Results from qPCR indicate the WT had 21- ± 6-fold higher swrW expression than the ΔeepR mutant in stationary-phase cells (OD600 of 3.0) (Fig. 6B). EMSA was used to assess whether recombinant EepR (MBP-EepR) was able to bind directly to the swrW promoter in vitro, unlike the MBP negative control (Fig. 6C). As with the pigA-N promoter described above, an MBP-EepR–swrW promoter interaction was observed, suggesting that EepR positively and directly regulates the serratamolide biosynthetic gene.
FIG 6.
Positive regulation of serratamolide biosynthetic gene, swrW, by EepR. (A) β-Galactosidase analysis of swrW expression from cultures at an OD600 of 4. Means and standard deviations are shown. An asterisk indicates significant difference from results for the WT by ANOVA with Tukey's posttest. WT, CMS376; eepR strain, CMS2097; eepS strain, CMS1076. (B) Quantitative RT-PCR analysis of swrW expression from cultures at an OD600 of 3. Means and standard deviations are shown. An asterisk indicates significant differences by Student's t test. WT, CMS376; eepR strain, CMS2097. (C) EMSA analysis of MBP-EepR interaction with biotin-labeled swrW promoter (PswrW; 2 ng) in vitro. MBP-EepR produced a gel shift of labeled PswrW that could be inhibited by an excess of unlabeled PswrW (PswrW-UL). Recombinant MBP was not sufficient to produce a gel shift of PswrW.
DISCUSSION
The previously undescribed eepR and eepS genes were identified in a number of genetic screens in different strain backgrounds that were focused on determining regulators of secondary metabolites. Evidence from this study suggests that EepR directly and positively regulates the pigA and swrW promoters, making EepR the only described direct and positive regulator of swrW transcription. Furthermore, the data presented here support the model that EepR is the hypothetical intermediate regulator in the cAMP-CRP-mediated pathway postulated at the onset of the project (see Fig. S1A in the supplemental material). To our knowledge, this is the first example of a CRP family protein regulating production of antimicrobial secondary metabolites through an intermediate regulator. This multiregulator model may be a common theme. Consistent with this, of the eight secondary metabolite gene clusters regulated by CRP in Streptomyces coelicolor, only six were directly regulated (2). This outcome ties a major metabolic regulator, the catabolite repression system (cAMP-CRP), to the regulation of factors that likely aid in competition. Serratamolide and prodigiosin both are reported to kill or inhibit a wide range of microbes in order to limit competition for nutrients. In addition, serratamolide and prodigiosin can be toxic to eukaryotic cells, which can provide an additional source of factors, such as iron, important for bacterial growth. Since cAMP is reduced under high nutrient conditions, it is expected that these EepR levels would increase, leading to greater production of competition factors in a favorable niche.
Another potential benefit for secondary metabolite regulation to be tied to a central metabolic regulator, cAMP-CRP, is suggested by Haddix et al.; prodigiosin was shown to have a role in energy spilling that may protect cells at high culture density (43). Furthermore, recent work supports that prodigiosin has a positive effect on cell yield (P. Haddix, personal communication). It is possible that prodigiosin, like other bacterial pigments, protects the bacterium from metabolism-derived oxidative stress fitting, with a model where low cAMP levels derepressing eepR expression should lead to increased prodigiosin production.
cAMP-CRP directly and positively regulates transcription of the flagellum master regulator in S. marcescens, flhDC (22), and negatively regulates biofilm formation through regulation of type I pili (20). Together these data lead to a simplified model in which cAMP levels are elevated under low-nutrient conditions, shutting off biofilm formation and eepR expression and turning on FlhDC, so that the bacterium can seek a more favorable environment. Under ideal conditions, such as a glucose-rich environment, cAMP levels will be low, deactivating flagellum production and turning on biofilm formation and EepR-regulated nutrient acquisition enzymes and competition factors.
The sequences and phenotypes of mutant strains suggest that EepS and EepR are a histidine sensor kinase and response regulator pair; however, further biochemical and/or directed mutagenesis analysis will be required to formally demonstrate that EepS and EepR together form a two-component regulatory system. Moreover, the amino acid sequence of EepS suggests that, rather than being part of a simple two-component system, it will require an intermediate phosphocarrier protein to phosphorylate EepR. Although we demonstrated here that cAMP-CRP negatively regulates eepR transcription, the impact of EepS on EepR activity is less clear. The signal for most histidine kinases is unknown and notoriously difficult to determine. Nevertheless, as a sensor kinase, EepS is predicted to regulate EepR activity in response to some additional signal, for example, through an unknown intermediate phosphocarrier protein (see Fig. S1 in the supplemental material). This may fine-tune or serve as an override switch of cAMP-CRP regulation of EepR, when, for example, EepR-regulated genes are necessary or deleterious. Interestingly, recombinant EepR bound to the pigA and swrW promoters in vitro in the absence of a phosphodonor such as carbamoyl phosphate. One possible model is that EepR is active in a nonphosphorylated form; therefore, EepS may promote secondary metabolite production by removing phosphate from EepR rather than acting as a kinase.
We recently reported that the S. marcescens PigP transcription factor positively regulates pigA-N directly and swrW indirectly. PigP was necessary for the hyperprodigiosin and hyperserratamolide phenotypes of crp mutants, and pigP expression was regulated by cAMP-CRP. However, CRP did not directly regulate transcription of pigP, suggesting that PigP, while involved in the same pathway, was not the missing regulator directly controlled by CRP. Extensive work with another Serratia species, ATCC 39006, demonstrated that secondary metabolism biosynthetic genes are regulated by quorum-sensing, cyclic-di-GMP, gluconate, phosphate, temperature, and other signals (17, 44–50). The existence of multiple regulators involved in controlling these factors (secondary metabolites, secreted enzymes, flagella, and adhesins) underscores the complexity of the regulation, the large number of external stimuli that must be coordinated, and the energy investment involved in making these factors.
In conclusion, this study introduces two genes predicted to code for a two-component transcriptional regulatory system composed of EepR as a putative response regulator and EepS as a putative sensor kinase. These two genes have an important impact on a variety of processes by S. marcescens, including motility, hemolysis, and production of antimicrobial compounds that likely play a role in bacterium-bacterium interactions and successful colonization of environmental niches.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kristin Arena, Marissa Aston, James Fender, and Nikolai Klena for outstanding technical assistance.
This work was supported by the Charles T. Campbell Laboratory of Ophthalmic Microbiology, the Eye and Ear Foundation of Pittsburgh, a career development award from Research to Prevent Blindness (R.M.Q.S.), NIH grant AI085570, and an NEI Core Grant for Vision Research, EY08098. K.M.B. was supported by NIH training grant 2T32 EY017271.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00136-15.
REFERENCES
- 1.Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol Rev 56:100–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao C, Hindra Mulder D, Yin C, Elliot MA. 2012. Crp is a global regulator of antibiotic production in streptomyces. mBio 3:e00407–12. doi: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komeda Y, Suzuki H, Ishidsu I, Iino T. 1976. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet 142:289–298. [DOI] [PubMed] [Google Scholar]
- 4.Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog 5:e1000303. doi: 10.1371/journal.ppat.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alper MD, Ames BN. 1978. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol 133:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to “foraging”-like behavior in E. coli. Nucleic Acids Res 35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalivoda EJ, Stella NA, O'Dee DM, Nau GJ, Shanks RM. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol 74:3461–3470. doi: 10.1128/AEM.02733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fomenko D, Veselovskii A, Khmel I. 2001. Regulation of microcin C51 operon expression: the role of global regulators of transcription. Res Microbiol 152:469–479. doi: 10.1016/S0923-2508(01)01220-7. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Martinez J, Adam AL, Avalos J. 2012. Adenylyl cyclase plays a regulatory role in development, stress resistance and secondary metabolism in Fusarium fujikuroi. PLoS One 7:e28849. doi: 10.1371/journal.pone.0028849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai Y, Wang L, Qing L, Chen F. 2011. Effects of cyclic AMP on development and secondary metabolites of Monascus ruber M-7. Lett Appl Microbiol 52:420–426. doi: 10.1111/j.1472-765X.2011.03022.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Zhao Y, Zhang J, Shen Y, Su Z, Xu G, Du L, Huffman JM, Venturi V, Qian G, Liu F. 2014. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 98:9009–9020. doi: 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binet R, Letoffe S, Ghigo JM, Delepelaire P, Wandersman C. 1997. Protein secretion by Gram-negative bacterial ABC exporters–a review. Gene 192:7–11. doi: 10.1016/S0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 13.Hines DA, Saurugger PN, Ihler GM, Benedik MJ. 1988. Genetic analysis of extracellular proteins of Serratia marcescens. J Bacteriol 170:4141–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadouri DE, Shanks RM. 2013. Identification of a methicillin-resistant Staphylococcus aureus inhibitory compound isolated from Serratia marcescens. Res Microbiol 164:821–826. doi: 10.1016/j.resmic.2013.06.002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Tomas R, Vinas M. 2010. New insights on the antitumoral properties of prodiginines. Curr Med Chem 17:2222–2231. doi: 10.2174/092986710791331103. [DOI] [PubMed] [Google Scholar]
- 17.Williamson NR, Fineran PC, Leeper FJ, Salmond GP. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- 18.Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, Kowalski RP, Shanks RM. 2010. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol 161:158–167. doi: 10.1016/j.resmic.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fender JE, Bender CM, Stella NA, Lahr RM, Kalivoda EJ, Shanks RMQ. 2012. Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl Environ Microbiol 78:6225–6235. doi: 10.1128/AEM.01778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanks RM, Stella NA, Kalivoda EJ, Doe MR, O'Dee DM, Lathrop KL, Guo FL, Nau GJ. 2007. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol 189:7262–7272. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanks RM, Stella NA, Arena KE, Fender JE. 2013. Mutation of crp mediates Serratia marcescens serralysin and global secreted protein production. Res Microbiol 164:38–45. doi: 10.1016/j.resmic.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stella NA, Kalivoda EJ, O'Dee DM, Nau GJ, Shanks RM. 2008. Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol 159:562–568. doi: 10.1016/j.resmic.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanks RM, Stella NA, Lahr RM, Wang S, Veverka TI, Kowalski RP, Liu X. 2012. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7:e36398. doi: 10.1371/journal.pone.0036398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams RP, Green JA, Rappo-Port DA. 1956. Studies on pigmentation of Serratia marcescens. I. Spectral and paper chromatographic properties of prodigiosin. J Bacteriol 71:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, Kwak DH, Liu X, Kalivoda EJ. 2013. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams MH. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY. [Google Scholar]
- 28.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke D, Stearns DDT. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 30.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system contols the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol 55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 31.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. doi: 10.1016/S0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 32.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slater H, Crow M, Everson L, Salmond GP. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol Microbiol 47:303–320. doi: 10.1046/j.1365-2958.2003.03295.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 35.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aubert DF, Flannagan RS, Valvano MA. 2008. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun 76:1979–1991. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aubert DF, O'Grady EP, Hamad MA, Sokol PA, Valvano MA. 2013. The Burkholderia cenocepacia sensor kinase hybrid AtsR is a global regulator modulating quorum-sensing signalling. Environ Microbiol 15:372–385. doi: 10.1111/j.1462-2920.2012.02828.x. [DOI] [PubMed] [Google Scholar]
- 39.Eisenstein BI, Beachey EH, Solomon SS. 1981. Divergent effects of cyclic adenosine 3′,5′-monophosphate on formation of type 1 fimbriae in different K-12 strains of Escherichia coli. J Bacteriol 145:620–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stella NA, Shanks RM. 2014. Cyclic-AMP inhibition of fimbriae and prodigiosin production by Serratia marcescens is strain-dependent. Arch Microbiol 196:323–330. doi: 10.1007/s00203-014-0970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. 1992. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol 174:1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuyama T, Sogawa M, Nakagawa Y. 1989. Fractal spreading growth of Serratia marcescens which produces surface active exolipids. FEMS Microbiol Lett 52:243–246. [DOI] [PubMed] [Google Scholar]
- 43.Haddix PL, Jones S, Patel P, Burnham S, Knights K, Powell JN, LaForm A. 2008. Kinetic analysis of growth rate, ATP, and pigmentation suggests an energy-spilling function for the pigment prodigiosin of Serratia marcescens. J Bacteriol 190:7453–7463. doi: 10.1128/JB.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fineran PC, Williamson NR, Lilley KS, Salmond GP. 2007. Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J Bacteriol 189:7653–7662. doi: 10.1128/JB.00671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gristwood T, Fineran PC, Everson L, Salmond GP. 2008. PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol Microbiol 69:418–435. doi: 10.1111/j.1365-2958.2008.06291.x. [DOI] [PubMed] [Google Scholar]
- 46.Gristwood T, Fineran PC, Everson L, Williamson NR, Salmond GP. 2009. The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol 9:112. doi: 10.1186/1471-2180-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gristwood T, McNeil MB, Clulow JS, Salmond GP, Fineran PC. 2011. PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J Bacteriol 193:1076–1085. doi: 10.1128/JB.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilf NM, Salmond GP. 2012. The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology 158:648–658. doi: 10.1099/mic.0.055780-0. [DOI] [PubMed] [Google Scholar]
- 49.Wilf NM, Williamson NR, Ramsay JP, Poulter S, Bandyra KJ, Salmond GP. 2011. The RNA chaperone, Hfq, controls two luxR-type regulators and plays a key role in pathogenesis and production of antibiotics in Serratia sp. ATCC 39006. Environ Microbiol 13:2649–2666. doi: 10.1111/j.1462-2920.2011.02532.x. [DOI] [PubMed] [Google Scholar]
- 50.Williamson NR, Fineran PC, Ogawa W, Woodley LR, Salmond GP. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a posttranscriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ Microbiol 10:1202–1217. doi: 10.1111/j.1462-2920.2007.01536.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.