Abstract
Hypoxia-inducible factors (HIF) are a family of heterodimeric transcriptional regulators that play pivotal roles in the regulation of cellular utilization of oxygen and glucose and are essential transcriptional regulators of angiogenesis in solid tumor and ischemic disorders. The transactivation activity of HIF complexes requires the recruitment of p300/CREB-binding protein (CBP) by HIF-1α and HIF-2α that undergo oxygen-dependent degradation. HIF activation in tumors is caused by several factors including mitogen-activated protein kinase (MAPK) signaling. Here we investigated the molecular basis for HIF activation by MAPK. We show that MAPK is required for the transactivation activity of HIF-1α. Furthermore, inhibition of MAPK disrupts the HIF-p300 interaction and suppresses the transactivation activity of p300. Overexpression of MEK1, an upstream MAPK activator, stimulates the transactivation of both p300 and HIF-1α. Interestingly, the C-terminal transactivation domain of HIF-1α is not a direct substrate of MAPK, and HIF-1α phosphorylation is not required for HIF-CAD/p300 interaction. Taken together, our data suggest that MAPK signaling facilitates HIF activation through p300/CBP.
Hypoxia-inducible factors (HIF)1 consist of a family of heterodimeric transcriptional regulators that control the expression of a series of genes involved in angiogenesis, oxygen transport, and glucose metabolism (reviewed in Refs. 1–3). Each of the HIF complexes contains an α-subunit and a common dimerization partner, HIF-β, also known as aryl hydrocarbon receptor nuclear translocator. Whereas both HIF-α and HIF-β are required to form the HIF heterodimer, HIF-α is the key regulatory subunit whose transcriptional activity is indispensable for HIF complex function (1).
The activity of HIF-α is controlled at the level of protein stability (1, 2, 4, 5) and transcriptional stimulation (3, 6, 7). The degradation of HIF-α is mediated by the ubiquitin-proteasome system (5, 8) and requires the hydroxylation of prolyl residues in the conserved oxygen-dependent degradation domain (8, 9), a process carried out by the oxygen, iron, and oxoglutarate-dependent prolyl-hydroxylase enzymes (10–15). Hydroxylated oxygen-dependent degradation domain recruits the von Hippel-Lindau protein (11, 12, 16), a tumor suppressor protein that serves as a part of the E3 ubiquitin-ligase complex (17, 18). In addition to HIF-α stabilization, HIF-α activity is regulated by the functional stimulation of its transactivation domains, NAD and CAD, which are separated by a negative regulatory region (6, 7). The recruitment of p300/CBP plays an essential role in the functional activation of HIF-α (19). The interaction between HIF-α CAD and the CH1 domain of p300/CBP involves a hydrophobic interface (20–22) and thus is disrupted by the hydroxylation of the asparagine residue (Asn803) in the CAD of HIF-1α under normoxic conditions (23, 24). Hydroxylation of CAD depends on the negative regulatory region’s recruitment of factor inhibiting HIF (FIH) (24, 25), an asparagine hydroxylase that serves as an inhibitor of HIF activity (26, 27).
In addition to hypoxia, multiple oncogenic pathways including growth factor signaling or genetic loss of tumor suppressor genes, like VHL and PTEN, up-regulate HIF activity (28, 29). Particularly, ERK1/2 (also known as p44/p42), two kinases of the mitogen-activated protein kinase (MAPK) signaling pathway, have been implicated in HIF activation (30–32). ERK1/2 are activated by extracellular proliferative signaling triggered by membrane-tyrosine kinases and transduced through the Ras-Raf-MEK pathway by a cascade of phosphorylation events that can be repressed by specific kinase inhibitors (33). ERK1/2 are serine/threonine kinases that regulate gene expression by phosphorylating nuclear substrates (33). Enhanced MAPK signaling is a common event in tumors, and constitutively active MAPK signaling transforms mammalian cells (34). Therefore, up-regulation of HIF activity by MAPK signaling may play an essential role in transformation as well as, in the process of tumor growth and metastasis that depends on angiogenesis and changes in glucose metabolism (35). In this study, we examined the molecular basis by which MAPK signaling influences HIF activity. Our data indicate that MAPK signaling may affect HIF activity by promoting the formation of the HIF-p300/CBP complex and by modulating the transactivation activity of p300/CBP.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids expressing G4.H1α530–826, G4.H1α530–778, G4.H1α744–826, and G4.H1α786–826 were described previously (24). pFR-Luc is a luciferase reporter vector in which the luciferase gene is under the control of a minimal E1B promoter and upstream four copies of GAL4 binding sites (Clontech). pCMV-β-p300, pRSV-β-CBP, pCMV-G4.p300, pCMV.β-gal, and pSV.β-gal were generous gifts from Dr. Antonio Giordano (Temple University). pRL-CMV was obtained from Promega (Madison, WI). pGEX.H1α530–826, pGEX.H1α530–658, and pGEX.H1α744–826 were constructed by inserting corresponding PCR fragments into the EcoRI site of pGEX4T (Amersham Biosciences). pGEX.H1α757–826 was kindly provided by Dr. Gregg Semenza (Johns Hopkins University, Baltimore, MD) (25). pGEXp300TD (aa 1572–2370) was kindly provided by Dr. Pier Lorenzo Puri (Salk Institute). pCMV.MEK1 is a generous gift from Dr. Kun-Liang Guan (University of Michigan) (36).
Special Chemicals
Genistein and desferrioxamine (Dfx) were purchased from Sigma. PD98059 (PDx) was purchased from BioMol (Plymouth, PA). Dimethyoxalylglycine (DOG) was provided by Dr. Peter Ratcliffe (Oxford University) (12). Genistein, Dfx, and PDx were first dissolved in Me2SO and used at the final concentration as indicated in each experiment.
Cell Lines, Cell Culture, and Transfection
The establishment of the B1 cell line was described previously (5). All cells were maintained at a 37 °C humidified incubator in an atmosphere of 5% CO2. B1 cells and parental Hep3B cells were cultured in minimal essential medium, whereas HeLa cells in Dulbecco’s modified Eagle’s medium, both supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen). Transient transfection was performed with LipofectAMINE Plus reagents (Invitrogen) by following the low serum protocol provided by the manufacturer. The amount of DNA used in transfection is described in each experiment. Twenty-four hours after transfection, the cells were trypsinized, pooled, and equally divided into 12-well plates for luciferase assays or into 60-mm dishes for Western blot analysis. When needed, 0.5 μg of plasmid expressing β-galactosidase were co-transfected to normalize the transfection efficiency.
Hypoxia Treatments
Cells were flushed with a gas mixture of 0.47% O2, 5% CO2, and balanced N2 in a sealed chamber as previously described (5). Alternatively, in some experiments, cells were incubated directly in a hypoxia work station (IN VIVO2) at the O2 tension specified in each experiment.
Luciferase Assays
All cell extracts were prepared and analyzed using the luciferase assay system purchased from Promega (Madison, WI) following the manufacturer’s procedure. Luminescence was measured in a TD20/20 Luminometer (Promega), and the results were expressed as relative light units. Protein concentration was determined by the Bio-Rad method. β-Galactosidase assays were performed for normalization reasons with the β-galactosidase assay system from Promega.
GST Fusion Protein Expression, Purification, and in Vitro Pull-down Assays
Plasmids expressing the indicated GST fusion proteins were transformed into BL21-competent cells (Stratagene, La Jolla, CA), and expression was induced by the addition of isopropyl-1-thio-β-D-galacto-pyranoside (Promega) to 0.1 mM. The procedures used to perform purification and in vitro pull-down assays were in general the same as described previously (37). Briefly, cells were lysed in lysis buffer (50 mM Tris-HCl, 250 mM NaCl, 1% Triton 100, 5 mM EDTA, 50 mM NaF, 0.1 mM protease inhibitor Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1× mix, pH 7.5). GST and fusion proteins were first incubated with 3% milk in lysis buffer, washed with lysis buffer, and then incubated with cell lysates for 1 h on a roller at 4 °C, followed by three washes with lysis buffer.
In Vitro Kinase Assays
In vitro kinase assays were performed as described by Pei with minor modifications (38). Briefly, purified GST, GST fusion proteins, and commercially obtained myelin basic protein (Sigma) were incubated at 30 °C for 20 min with activated recombinant MAPK (BioMol) in the presence of 5 μCi of [γ-32P]ATP (Amersham Biosciences) in MAPK buffer (25 mM Hepes, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.5). The reaction was stopped by adding an equal volume of 2× Laemmli sample buffer and by heating at 95 °C for 3 min. After separation through a 4–20% continuous gradient SDS-PAGE (Bio-Rad), the gels were stained with Coomassie Blue R250 (Sigma), destained, and dried before autoradiography.
Antibodies, Immunoprecipitation, and Western Blot Analyses
Monoclonal antibody against HIF-1α (catalog no. 610959) and anti-p300 monoclonal antibody (NM11) were purchased from Pharmingen. Monoclonal anti-GAL4 DNA binding domain antibody was purchased from Clontech (catalog no. 5399-1). Purified polyclonal antibodies against tyrosine-phosphorylated and total MAPK and horseradish peroxidase-coupled donkey anti-rabbit polyclonal antibody were purchased from Promega. Horseradish peroxidase-coupled anti-mouse IgG (Fc fragment) was purchased from Sigma. Immunoprecipitations were carried out as described previously with minor modifications (37, 39). Briefly, cells were lysed in 1× lysis buffer supplemented with 75 μM PDx when needed. Cell lysates were first incubated with nonspecific normal mouse serum and killed protein A-positive Staphylococcus aureus cells (Roche Diagnostics). The precleared lysates were incubated with 2 μg of mono-clonal antibody on ice for 30 min followed by rocking with immobilized protein A (Pierce) at 4 °C for 45 min. For Western blot analyses, samples were separated on a 4–20% gradient SDS-PAGE (Bio-Rad), if not specified, followed by electrotransferring onto polyvinylidene difluoride membrane. The membrane was blocked with 5% milk in TBST (24), incubated with specific antibody, washed in TBST, and incubated with horseradish peroxidase-labeled secondary antibody. The membranes were finally developed with the ECL Plus system (Amersham Biosciences).
RESULTS
MAPK Signaling Enhances Both Basal and Hypoxia-stimulated HIF-1 Activity
Previously, we reported the establishment of the hepatoma-derived B1 cell line that carries a hypoxia-responsive luciferase reporter gene and showed that this cell line is responsive to hypoxia and hydroxylase inhibitors (hypoxia mimics) including transition metals, iron chelators, and oxoglutarate analogs (5). It has been observed that genistein, a tyrosine kinase inhibitor, decreased the protein levels of both HIF-1α and HIF-1β and inhibited the formation of DNA binding complex (40). Previously, we also found that in B1 cells, genistein inhibited HIF-1 activity and gene expression in response to hypoxic stimulation (30). However, PDx, a selective MEK inhibitor (41), inhibited hypoxia-stimulated gene expression but had little effect on HIF-1α level and the formation of DNA binding complex (30). Here, we investigated further the role of MAPK signaling in basal and induced activity of HIF-1α in B1 cells. The MAPK signaling pathway, the targeting sites of two kinase inhibitors, genistein (GSN) and PDx, and the experimental system are schematically represented in Fig. 1, A and B. Treatment of the B1 cells with genistein revealed a dose-dependent inhibitory effect on both the basal and hypoxia-induced HIF-1 activity (Fig. 1, C and D). Similar inhibition was observed with the use of the hypoxic mimic Dfx (an iron chelator) and DOG (an oxoglutarate analog) (not shown). We further examined the effects of PDx both on a basal level and on a stimulated transactivation activity of HIF-1. Fig. 1, E and F, shows that PDx inhibited luciferase expression in B1 cells both under normoxic and hypoxic conditions, further confirming that MAPK signaling enhances not only hypoxia-stimulated HIF-1 activity but its basal activity as well. In control experiments, PDx showed no effect on the luciferase activity from a pRL-CMV (not shown). These results indicate that MAPK signaling has a general role in promoting endogenous HIF activity, regardless of oxygen concentration.
Fig. 1. Role of MAPK signaling in endogenous HIF-1 activity.
A, schematic representation of MAPK-signaling pathway and the sites for inhibitor action. B, model of the luciferase-reporter system. C and D, effects of genistein on HIF-1 activity. B1 cells were exposed to various doses of genistein, a tyrosine kinase inhibitor, and cultured in normoxia (C) or in a hypoxic work station with 2% O2 (D) for 8 h. E and F, effects of PDx on HIF-1 activity. B1 cells were exposed to various doses of PDx, MAPK inhibitor, under normoxic or hypoxic conditions (2% O2) for 8 h. RLU, relative light units.
MAPK Signaling Promotes the Transactivation Activity of HIF-1α
We next tested the hypothesis that MAPK signaling regulates HIF-1 activity through the transactivation domains of HIF-1α. HeLa cells were transfected with plasmids expressing G4.H1α530–826 in which the HIF-1α fragment was fused with the DNA binding domain of the yeast transcription factor GAL4. A luciferase reporter driven by an E1B minimal promoter and four copies of GAL4 binding sites (pFR-luc) was co-transfected to monitor the transactivation activity. Fig. 2, A and B, shows the dose-dependent inhibitory effect of PDx on the transactivation activity of G4.H1α530–826, which contains both the NAD and the CAD, under both normoxic and hypoxic conditions. The protein levels of G4.H1α530–826 were not significantly affected by PDx treatment (Fig. 2, C and D). Taken together, these data clearly confirm that MAPK signaling up-regulates the transactivation activity of HIF-1α.
Fig. 2. Role of MAPK signaling in the transactivation activity of HIF-1α.
A and B, effects of PDx on transactivation activity of HIF-1α. HeLa cells were transfected with a plasmid expressing G4.H1α530–826 (2.5 μg) and the luciferase reporter pFR-luc (2.5 μg). Following transfection, the cells were treated with variable doses of PDx and incubated in normoxic (Nmx) on hypoxic (Hpx) conditions for 8 h before luciferase measurements. C and D, effects of PDx on protein levels of G4.H1α530–826 under normoxic and hypoxic conditions. HeLa cells cultured in 100-mm dishes were transfected with pG4.H1α530–826 (5 μg/dish). 24 h after the transfection, the cells were trypsinized, pooled, and evenly reseeded into fresh dishes and cultured for 12 h. The cells were treated with indicated doses of PDx and cultured under either normoxic on hypoxic (2% O2) conditions for 8 h before harvest. RLU, relative light units.
Direct Phosphorylation of HIF-1α by MAPK Is Not Correlated to Its Transactivation Activity
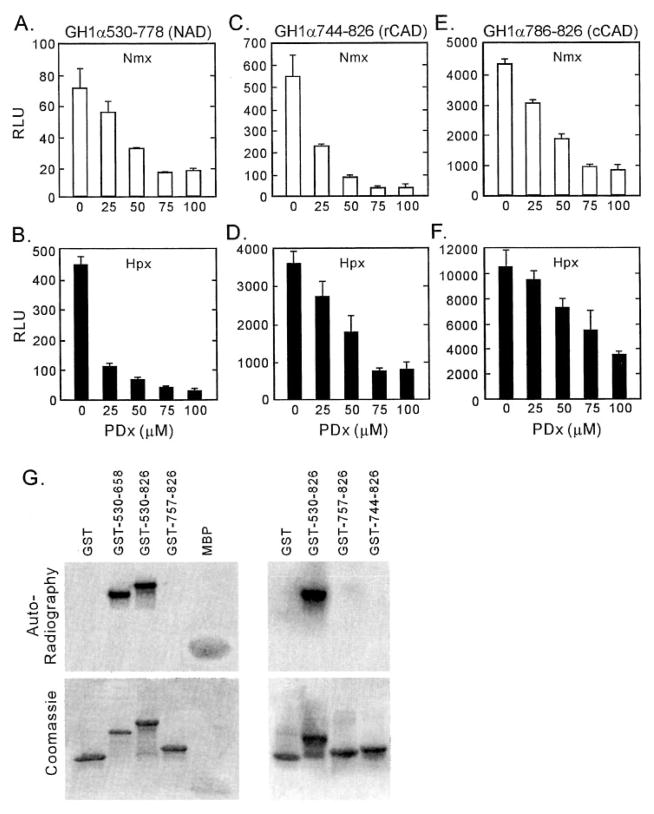
Whereas previous reports observed that HIF-1α was directly phosphorylated by MAPK (32), the exact amino acid residues involved in HIF-1α phosphorylation have not been clearly identified. Furthermore, there have been inconsistencies between results of different groups on the role of MAPK-induced direct phosphorylation of HIF-α in its transactivation activity (31, 32). To clarify further the role of MAPK in HIF activation, we first attempted to identify the functional domains that were sensitive to MAPK inhibitors. G4.HIF fusion constructs containing various segments of the carboxyl half of HIF-1α were co-transfected with the pFR-Luc reporter, and the transfected HeLa cells were treated with increasing doses of PDx. As shown in Fig. 3, A–F, all of the constructs tested, including G4.H1α530–778 (which represents the NAD), G4.H1α744–826 (which represents the hypoxia-responsive CAD), and G4.H1α786–826 (which represents the constitutive CAD), were inhibited by PDx both under normoxic and hypoxic conditions. These data suggest at least two possibilities: 1) there is more than one phosphorylation site, each of the constructs tested containing at least one site for MAPK-mediated phosphorylation, or 2) MAPK signaling affects a protein factor(s) that is required for both NAD and CAD activity. To test the first hypothesis, we expressed HIF-1α as GST fusion proteins and used them in in vitro MAP kinase assays (Fig. 3G). Whereas GST alone was not phosphorylated by active MAPK, GST.H1α530–658 and GST.H1α530–826 were readily phosphorylated. However, neither GST.H1α757–826 nor GST.H1α744–826, which corresponds to the hypoxia-responsive CAD (6), was phosphorylated, demonstrating that whereas both hypoxia-responsive CAD and constitutive CAD are sensitive to PDx treatment, they do not contain MAPK phosphorylation sites.
Fig. 3. Correlation between MAPK phosphorylation and the transactivation activity of HIF-1α.
A–F, effects of MAPK inhibitor on the transactivation activity of G4.H1α constructs. Each G4.H1α construct (3 μg) was co-transfected with the pFR-Luc reporter (2 μg), and 24 h later the cells were trypsinized and evenly divided into 12-well culture plates. Immediately before the reporter assays, the indicated doses of PDx were added, and the cells were exposed to either normoxic (A, C, and E) or hypoxic (2% O2) conditions (B, D, and F) for 8 h before luciferase assays. G, in vitro MAPK assays. HIF-1α fragments were purified as GST fusion proteins and incubated with activated MAPK and [γ-32P]ATP. The top panel shows the autoradiography, and the bottom panel shows the protein input. GST and myelin basic protein (MBP) were used as negative and positive control, respectively. RLU, relative light units.
MAPK Signaling Promotes the Transactivation Activity of p300
Since CAD is sensitive to PDx but lacks MAPK phosphorylation sites, we explored the possibility that a transcriptional coactivator might be involved in MAPK signaling to HIF. Because p300/CBP are the major coactivators involved in HIF activation, we next tested the effects of MAPK on p300 trans-activation activity. A plasmid expressing G4.p300 (aa 1–2414) was co-transfected with the pFR-Luc reporter, and the transfected HeLa cells were treated with Me2SO, PDx, or genistein and cultured under normoxic or hypoxic conditions. Fig. 4A indicates that whereas hypoxia and Me2SO had little effect on p300 transactivation activity, exposure to PDx or genistein significantly repressed this activity. The effect of PDx was dose-dependent (Fig. 4B) and affected the transactivation domain of p300 (G4.p300TD; aa 1751–2414) (Fig. 4C). Furthermore, transfection with a plasmid expressing MEK1, the upstream activator of ERK1/2 (36), enhanced the transactivation activity of p300TD (Fig. 4D). We next tested whether p300TD was a direct substrate of MAPK. We expressed and purified p300TD (aa 1572–2370) as a GST fusion protein and used it in in vitro MAPK assays. We observed that this region of p300 was readily phosphorylated by MAPK in vitro (Fig. 4E). Finally, when coexpressed with NAD (G4.H1α530–778), p300 (pCMV-β-p300) or CBP (pRSV-CBP) enhanced the transactivation activity of NAD, which was repressible by wild type E1A but not by an E1A mutant defective in targeting p300/CBP (not shown) (42), suggesting that p300 is also required for NAD activity. Therefore, the effect of PDx on both NAD and CAD activity is at least partly caused by the repression of p300/CBP.
Fig. 4. Effects of MAPK on p300 transactivation activity.
A, effect of PDx and genistein on p300 transactivation activity. HeLa cells where co-transfected with a plasmid expressing G4.p300 and with the GAL4-luciferase reporter (pFR-luc). The transfected cells were divided evenly and exposed to Me2SO (DMSO), PDx (75 μM), or genistein for 6 h in normoxia or hypoxia immediately before the luciferase assays. B, dose-dependent effect of PDx on p300 activity. HeLa cells were transfected as in B, and increasing amounts of PDx were used, as indicated (in μM). C, dose-dependent effects of PDx on the transactivation domains of p300TD. HeLa cells were transfected with plasmid G4.p300TD, expressing the transactivation domain of p300 (aa 1751–2414), and the cells were exposed to increasing concentrations of PDx. D, effects of overexpression of MEK1 on the transactivation activity of p300TD. Plasmid expressing MEK1 (2 μg) was co-transfected with pG4.p300TD (2 μg) and the luciferase-reporter (pFR-Luc; 2 μg). Luciferase activity was expressed as -fold increase over the co-transfected vector control. E, direct phosphorylation of p300TD in vitro by MAPK. GST.p300TD was expressed in and purified from BL21 cells and used in MAPK kinase assays. RLU, relative light units.
MAPK Signaling Facilitates the Interaction between p300 and HIF-1αCAD
HIF-1α transactivation activity depends on the recruitment of p300. We next investigated whether MAPK signaling affected the physical interaction between p300 and HIF-1αCAD. We exposed HeLa cells to hypoxia in the presence or absence of PDx. As shown in Fig. 5A, treatment with PDx had no effect on p300 levels. We incubated the cell lysates with bacterially expressed GST.H1α530–826 protein in pull-down assays, and the copurified p300 was detected by immunoblotting with an anti-p300 monoclonal antibody. As shown in Fig. 5B, whereas p300 in cell lysates from normal or Me2SO-treated cells readily bound to GST.H1α530–826, little p300 was copurified from PDx-treated lysates. Similar results were observed when GST.H1α757–826 was used (not shown). To further confirm that blocking MAPK signaling disrupts the HIF-1/p300 interaction in vivo, a plasmid expressing G4.H1α786–826 (constitutive CAD) was transfected into HeLa cells, and the transfected cells were treated with Me2SO or PDx and cultured under either normoxic or hypoxic conditions. Western blot analysis showed that G4.H1α786–826 was expressed in all transfected cells (Fig. 5C). Coupled immunoprecipitation and Western blot assays with either anti-p300 or anti-Gal4 antibodies confirmed that G4.H1α786–826 interacted with p300 regardless of oxygen concentration as previously demonstrated (24). This interaction, however, was significantly weakened by PDx treatment (Fig. 5C, bottom two panels). These results indicate that MAPK signaling facilitates the interaction between p300 and HIF-1αCAD in vivo.
Fig. 5. MAPK signaling regulates the interaction between p300 and HIF-1αCAD.
A, PDx treatments do not affect p300 levels. HeLa cells were incubated with PDx at 100 μM for 6 h, and the cell lysates were analyzed by immunoblotting using anti-p300 antibodies. B, GST pull-down assays. GST and GST.H1α530–826 were incubated with whole cell lysates (WCL) from HeLa that were untreated or treated with Me2SO (DMSO) or PDx (100 μM) for 6 h. Precipitates were resolved in an 8% SDS-PAGE. Top panel, Coomassie Blue staining shows GST and GST.H1α530–826 proteins used in the pull-down assays. Bottom, Western blotting with anti-p300 monoclonal antibody demonstrating that GST.H1α fails to pull down p300 from PDx-treated cell lysates. C, effects of PDx treatment on the interaction between HIF-1αCAD and p300 in vivo. HeLa cells were transfected with G4.H1α786–826 and treated with or without PDx and cultured under either normoxic or hypoxic conditions for 6 h before harvest. Whole cell lysates (WCL; top three panels) were assayed for the levels of HIF-1α, phosphorylated ERK (ERKp), and G4.H1α786–826 by Western blot (WB). Immunoprecipitations (I.P.) were performed with anti-p300 and anti-Gal4 monoclonal antibody, and the immunoprecipitates were detected with an anti-Gal4 or an anti-p300 monoclonal antibody reciprocally (bottom two panels).
MAPK Signaling Is Not a Part of the Oxygen Sensing Mechanism
Finally, we investigated whether MAPK signaling is involved in the oxygen sensing mechanism in HeLa cells. We first examined whether hypoxia or hydroxylase inhibitors might affect the levels of activated (phosphorylated) ERK. HeLa cells were cultured in normal medium with 10% fetal bovine serum, and both the total and activated ERK levels were examined by specific polyclonal antibodies in immunoblot assays. As shown in Fig. 6A, total and activated ERK1/2 levels were found at rather high levels under normal culturing conditions and were affected neither by hypoxia nor by the hypoxia mimics Dfx or DOG. The effect of further activation of MAPK on the responsiveness of HIF-1α to hypoxia and Dfx was analyzed by overexpressing MEK1, an upstream activator of ERK. Overexpression of MEK1 enhanced HIF-CAD activity under all conditions tested, whereas the addition of PDx blocked the MEK1 effect (Fig. 6B). Most importantly, variations in MAPK levels did not affect the responsiveness of HIF-1 α activity to hypoxia or Dfx.
Fig. 6. Effect of MAPK signaling in hypoxia sensing.
A, effect of hypoxia and hypoxia mimics on MAPK activity. HeLa cells were maintained in normoxia or exposed to hypoxia (2% O2), Dfx (130 μM), or DOG (1 mM) for 6 h. 50 μg of whole cell lysates were applied to an SDS-PAGE. The total and activated ERK1/2 (pERK1/2) were measured by immunoblotting. B, effect of overexpression of MEK1 on HIF-1α activity and its responsiveness to hypoxia. 2 μg of pCMV.MEK1 or vector alone were co-transfected with 2 μg of pG4.H1α744–826 and 2 μg of pRR-Luc into HeLa cells in 100-mm culture dishes. 24 h after transfection, cells were trypsinized and evenly split into 12-well culture plates. 12 h after seeding, cells were switched to the indicated culture condition or exposed to the indicated chemicals for 6 h before analyses. Luciferase activity was normalized to co-transfected 0.5 μg of pCMV.β-gal. RLU, relative light units.
DISCUSSION
The role of phosphorylation in HIF activation has become a contentious issue since it was first observed that genistein decreased the protein levels of both HIF-1α and HIF-1β and blocked the formation of DNA binding complex (40). It was later shown that PDx, a MEK-specific inhibitor, suppressed hypoxia-stimulated HIF activity but had little effect on the formation of HIF DNA-binding complex (30). These results suggested that MAPK signaling might play a role in the regulation of the transactivation activity of HIF-1 but had an insignificant effect on the protein levels of HIF-1β and hypoxia-stimulated accumulation of HIF-1α. This notion is supported by reports indicating that p44/p42 kinases (ERK1/2) directly phosphorylate HIF-1α and regulate its transcriptional activity with no effect on HIF-1α protein levels induced by hypoxia (32). This issue is complicated further by the later finding that both PTEN/phosphatidylinositol 3-kinase/Akt and MAPK pathways enhance HIF-α levels in tumor cells or cells stimulated by mitogenic signaling under normoxic conditions (43, 44) and by evidence indicating that such enhancement involves the control of HIF-1α translation (45). One recent report, however, demonstrates that the phosphatidylinositol 3-kinase/Akt pathway is neither required nor sufficient for stabilization of HIF-1α (46). The data presented here indicate that the trans-activation activity of HIF-1α is affected by MAPK signaling. Whereas at low dosages (50 μM or less), PDx was very effective in blocking the transactivation activity of HIF-1α, it showed little or no effect on the protein level. However, higher doses (100 μM or more) of PDx reduced hypoxia-accumulated HIF-1α levels (not shown). These data are consistent with the translational role of MAPK signaling and suggest that, in certain conditions, MAPK signaling may regulate HIF activity, in part, by affecting the protein levels of HIF-1α.
Although it is now clear that MAPK signaling facilitates the transactivation activity of HIF-1α and it has been suggested that MAPK directly phosphorylates HIF-α (32), there is no direct evidence for correlation of the MAPK-mediated phosphorylation to the transactivation activity of HIF-1α. Moreover, it was reported that PDx inhibited HIF-2α (EPAS1) transactivation activity but did not inhibit its phosphorylation (31). Whereas it was demonstrated recently that the C-terminal domain of HIF-α is phosphorylated at multiple sites and that mutation of a conserved threonine residue (Thr796 in HIF-1α/T844 in HIF-2α) inhibited the interactions between HIF-α and CBP/p300, such phosphorylation is not mediated by MAPK (47). We observed that whereas both responsive CAD and constitutive CAD of HIF-1α were repressed by PDx exposure, neither the aa 757–826 nor the aa 744–826 region was phosphorylated by MAPK, indicating that it is not through direct phosphorylation of HIF-1α that MAPK affects the transactivation activity of HIF-1αCAD. Instead, we observed that MAPK signaling affected the transactivation activity of p300, the co-activator required for HIF-αCAD, and that the interaction between p300 and HIF-1α was affected by MAPK signaling. Although the aa 530–743 region, encompassing NAD and part of the oxygen-dependent degradation domain, is phosphorylated by MAPK in vitro (Fig. 3G) (48), because HIF-1α530–826 and HIF-1α757–826 expressed in Escherichia coli, thus lacking posttranslational modifications, efficiently bind p300 in cell lysates, it appears that direct phosphorylation of HIF-1α is not necessary for the recruitment of p300. Moreover, whereas PDx does not affect p300 levels, HIF-1α fails to bind p300 in PDx-treated cell lysates, suggesting that phosphorylation of p300 or other cellular factor(s) by MAPK or its downstream kinases may regulate the interaction between p300 and HIF-CAD. Therefore, our data indicate that p300 and very likely CBP as well serve as integrators of MAPK signaling in HIF activation.
MAPK-dependent phosphorylation of CBP was originally reported in the regulation of Elk-1 activity (49), and later MAPK signaling through CBP was found to play a role in T-cell activation (50). Recently, phenylephrine-mediated stimulation of gene expression in cardiomyocytes has been found to involve MAPK signaling through p300 and CBP (51). Interestingly, overexpression of MEKK1 enhanced the transcriptional activity of p300 in a c-Jun N-terminal kinase-independent manner (52). Since MEKK1 is also able to activate MEK1, the MEK1-MAPK pathway may play a role in the stimulatory effect of MEKK1. How p300 and CBP integrate the MAPK signaling is currently unclear. Since p300 and CBP are co-factors for a large number of transcription factors involved in multiple cellular processes (53), signal-mediated redistribution of p300/CBP among different interacting partners may differentially regulate p300/CBP-dependent transcriptional factors (54) and the functionality of p300TD, thus reprogramming gene expression in response to the signal. Therefore, one possibility is that p300 and CBP are direct substrates of MAPK or a downstream kinase. Supporting this hypothesis, it has been reported that the C-terminal transactivation domain of CBP was phosphorylated by MAPK in vitro (49), and we now demonstrate that the C-terminal transactivation domain of p300 can be phosphorylated by MAPK in a similar manner (Fig. 4E). It is equally possible that MAPK signaling leads to phosphorylation of a pool of p300/CBP-interacting factors, as shown for several other transcriptional factors. In either case, the phosphorylation events may have differential impact on the affinity between p300 and various protein factors, thus leading to a signal-mediated redistribution of p300/CBP among interacting partners. Particularly, since MAPK signaling stimulates the transactivation activity of p300/CBP, phosphorylation events very likely enhance the interaction between p300/CBP and the basic transcriptional machinery.
The biological relevance of MAPK signaling in HIF activation is only partly understood. Previously, ERK activation upon hypoxia was observed in HMEC cells (55). In our system, however, MAPK inhibitors suppressed the transcriptional activity of HIF under both normoxic and hypoxic conditions. Therefore, the MAPK dependence is not specific for hypoxia or hypoxic mimic-stimulated HIF activity. In cells exposed to MAPK inhibitors or MEK1 overexpression, the hypoxic response remains. Furthermore, in the presence of serum, MAPK is constitutively active, and the levels of activated MAPK are not changed either by hypoxia or by hypoxia mimics in HeLa cells. Finally, we have also found that FIH is not a substrate for MAPK, ruling out the possibility that MAPK signals to HIF by modifying FIH hydroxylase activity (not shown). These results indicate that MAPK signaling is not part of the hypoxia sensing mechanism in HeLa cells. However, since MAPK signaling facilitates HIF activation, it may represent an independent pathway that links oncogenesis to HIF activation and angiogenesis and may synergize the effect of other HIF-activating pathways.
Acknowledgments
We thank Drs. A. Giordano (Temple University), P. L. Puri, K. L. Guan (University of Michigan), D. Livingston (Dana-Faber Institute), G. Semenza (Johns Hopkins University), and P. Rattcliffe (Oxford University) for providing plasmids and other reagents essential for this research. We thank Dr. S. McKenzie for support. We appreciate help from A. Likens in preparing the figures.
Footnotes
This work was supported by National Institutes of Health Grant RO1CA89212 and American Heart Association Grant 9950122N (to J. C.).
The abbreviations used are: HIF, hypoxia-inducible factor; E3, ubiquitin-protein isopeptide ligase; FIH, factor inhibiting HIF; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; CREB, cAMP-response element-binding protein; CBP, CREB-binding protein; aa, amino acids; Dfx, desferriox-amine; PDx, PD98059; DOG, dimethyoxalylglycine; GST, glutathione S-transferase.
References
- 1.Semenza GL. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Caro J. High Alt Med Biol. 2001;2:145–154. doi: 10.1089/152702901750265251. [DOI] [PubMed] [Google Scholar]
- 3.Fedele AO, Whitelaw ML, Peet DJ. Mol Interv. 2002;2:229–243. doi: 10.1124/mi.2.4.229. [DOI] [PubMed] [Google Scholar]
- 4.Huang LE, Arany Z, Livingston DM, Bunn HF. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 5.Salceda S, Caro J. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 6.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 7.Pugh CW, O’Rourke JF, Nagao M, Gleadle JM, Ratcliffe PJ. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 8.Huang LE, Gu J, Schau M, Bunn HF. Proc Natl Acad Sci U S A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivas V, Zhang LP, Zhu XH, Caro J. Biochem Biophys Res Commun. 1999;260:557–561. doi: 10.1006/bbrc.1999.0878. [DOI] [PubMed] [Google Scholar]
- 10.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 12.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim AV, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 13.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F, White SB, Zhao Q, Lee FS. Proc Natl Acad Sci U S A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruick RK, McKnight SL. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 16.Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Jr, Pavletich NP. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 17.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 18.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Proc Natl Acad Sci U S A. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. Proc Natl Acad Sci U S A. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J, Milligan J, Huang LE. J Biol Chem. 2001;276:3550–3554. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 21.Dames SA, Martinez-Yamout M, DeGuzman RN, Dyson HJ, Wright PE. Proc Natl Acad Sci U S A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman SJ, Sun ZYJ, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Proc Natl Acad Sci U S A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 24.Sang N, Fang J, Srinivas V, Leshchinsky I, Caro J. Mol Cell Biol. 2002;9:2984–2992. doi: 10.1128/MCB.22.9.2984-2992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahon PC, Hirota K, Semenza GL. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, Ratcliffe PJ, Pugh CW, Schofield CJ. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 27.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, Stokoe D, Giaccia AJ. Genes Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell PH, Pugh CW, Ratcliffe PJ. Curr Opin Genet Dev. 2001;11:293–299. doi: 10.1016/s0959-437x(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 30.Salceda S, Beck I, Srinivas V, Caro J. Kidney Int. 1997;51:556–559. doi: 10.1038/ki.1997.78. [DOI] [PubMed] [Google Scholar]
- 31.Conrad PW, Freeman TL, Beitner-Johnson D, Millhorn DE. J Biol Chem. 1999;274:33709–33713. doi: 10.1074/jbc.274.47.33709. [DOI] [PubMed] [Google Scholar]
- 32.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 33.Guan KL. Cell Signal. 1994;6:581–589. doi: 10.1016/0898-6568(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 34.Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande GF, Ahn NG. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 35.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 36.Zheng CF, Guan KL. J Biol Chem. 1993;268:11435–11439. [PubMed] [Google Scholar]
- 37.Sang N, Giordano A. J Cell Physiol. 1997;170:182–191. doi: 10.1002/(SICI)1097-4652(199702)170:2<182::AID-JCP10>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 38.Pei L. J Biol Chem. 2000;275:31191–31198. doi: 10.1074/jbc.M002451200. [DOI] [PubMed] [Google Scholar]
- 39.Sang N, Avantaggiati ML, Giordano A. J Cell Biochem. 1997;66:277–285. doi: 10.1002/(sici)1097-4644(19970901)66:3<277::aid-jcb1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 40.Wang GL, Jiang BH, Semenza GL. Biochem Biophys Res Commun. 1995;216:669–675. doi: 10.1006/bbrc.1995.2674. [DOI] [PubMed] [Google Scholar]
- 41.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 42.Sang N, Caro J, Giordano A. Front Biosci. 2002;7:407–413. doi: 10.2741/A784. [DOI] [PubMed] [Google Scholar]
- 43.Stiehl DP, Jelkmann W, Wenger RH, Hellwig-Burgel T. FEBS Lett. 2002;512:157–162. doi: 10.1016/s0014-5793(02)02247-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 45.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arsham AM, Plas DR, Thompson CB, Simon MC. J Biol Chem. 2002;277:15162–15170. doi: 10.1074/jbc.M111162200. [DOI] [PubMed] [Google Scholar]
- 47.Gradin K, Takasaki C, Fujii-Kuriyama Y, Sogawa K. J Biol Chem. 2002;277:23508–23514. doi: 10.1074/jbc.M201307200. [DOI] [PubMed] [Google Scholar]
- 48.Sodhi A, Montaner S, Miyazaki H, Gutkind JS. Biochem Biophys Res Commun. 2001;287:292–300. doi: 10.1006/bbrc.2001.5532. [DOI] [PubMed] [Google Scholar]
- 49.Janknecht R, Nordheim A. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 50.Avots A, Buttmann M, Chuvpilo S, Escher C, Smola U, Bannister AJ, Rapp UR, Kouzarides T, Serfling E. Immunity. 1999;10:515–524. doi: 10.1016/s1074-7613(00)80051-5. [DOI] [PubMed] [Google Scholar]
- 51.Gusterson R, Brar B, Faulkes D, Giordano A, Chrivia J, Latchman D. J Biol Chem. 2002;277:2517–2524. doi: 10.1074/jbc.M104626200. [DOI] [PubMed] [Google Scholar]
- 52.See RH, Calvo D, Shi Y, Kawa H, Luke MPS, Yuan Z, Shi Y. J Biol Chem. 2001;276:16310–16317. doi: 10.1074/jbc.M008113200. [DOI] [PubMed] [Google Scholar]
- 53.Vo N, Goodman RH. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 54.Giordano A, Avantaggiati ML. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 55.Minet E, Arnould T, Michel G, Roland I, Mottet D, Raes M, Remacle J, Michiels C. FEBS Lett. 2000;468:53–58. doi: 10.1016/s0014-5793(00)01181-9. [DOI] [PubMed] [Google Scholar]