Abstract
The asymmetric unit of the title compound, C18H16O4, contains two crystallographically independent molecules. The anthraquinone ring systems are slightly bent with dihedral angles of 2.33 (8) and 13.31 (9)° between the two terminal benzene rings. In the crystal, the two independent molecules adopt slipped-parallel π-overlap with an average interplanar distance of 3.45 Å, forming a dimer; the centroid–centroid distances of the π–π interactions are 3.6659 (15)–3.8987 (15) Å. The molecules are also linked by C—H⋯O interactions, forming a tape structure along the a-axis direction. The crystal packing is characterized by a dimer-herringbone pattern.
Keywords: crystal structure; 9,10-anthraquinone; crystallographically independent molecules; π–π interactions; C—H⋯O interactions
Related literature
For synthesis of alkoxy-substituted 9,10-anthraquinones, see: Kitamura et al. (2004 ▸). For background information on substitution effects of alkoxy-substituted 9,10-anthraquinones, see; Ohta et al. (2012 ▸). For related structures of 1,4-dipropoxy-9,10-anthraquinone polymorphs, see: Kitamura et al. (2015 ▸).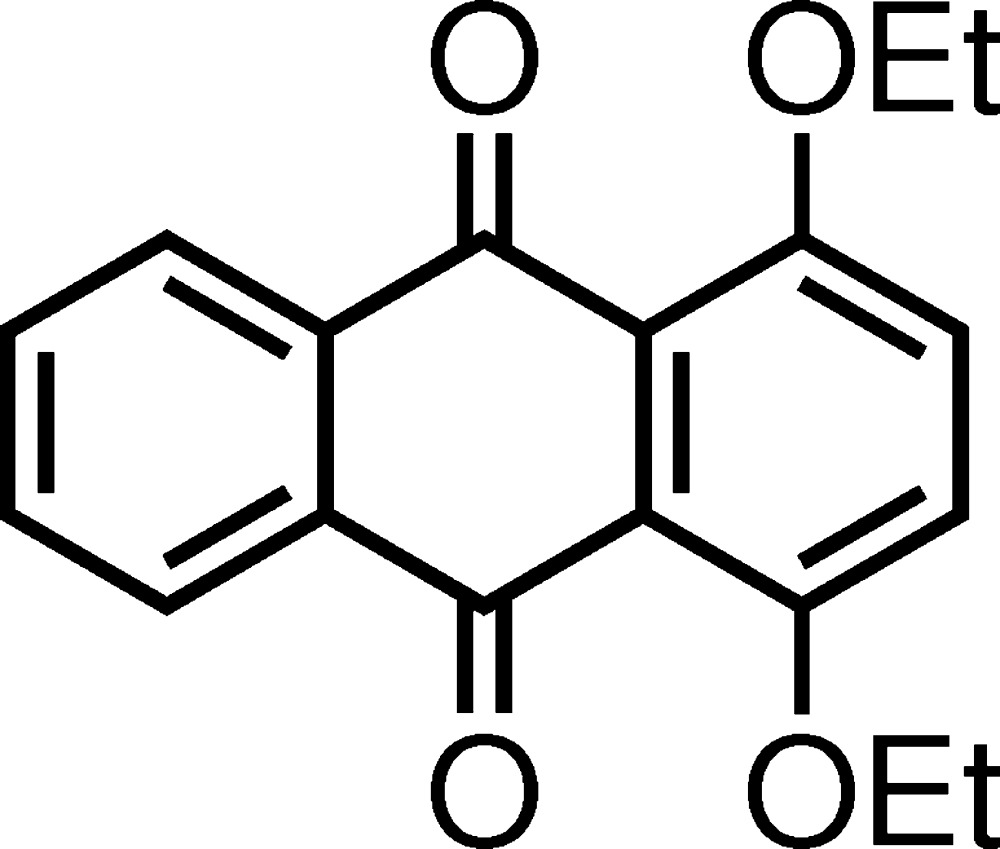
Experimental
Crystal data
C18H16O4
M r = 296.31
Monoclinic,

a = 13.5514 (11) Å
b = 14.7204 (11) Å
c = 14.5905 (10) Å
β = 90.604 (3)°
V = 2910.4 (4) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 223 K
0.56 × 0.40 × 0.36 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
27699 measured reflections
6645 independent reflections
3129 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.076
wR(F 2) = 0.273
S = 0.93
6645 reflections
397 parameters
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.48 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 1998 ▸); cell refinement: PROCESS-AUTO; data reduction: PROCESS-AUTO; program(s) used to solve structure: SIR2004 (Burla et al., 2005 ▸); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2015 ▸); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▸) and Mercury (Macrae et al., 2008 ▸); software used to prepare material for publication: WinGX (Farrugia, 2012 ▸).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989015011901/is5404sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015011901/is5404Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015011901/is5404Isup3.cml
. DOI: 10.1107/S2056989015011901/is5404fig1.tif
The asymmetric unit of the title compound, showing the atomic numbering and 40% probability displacement ellipsoids.
a . DOI: 10.1107/S2056989015011901/is5404fig2.tif
A packing diagram of the title compound viewed down the a axis, showing a dimer-herringbone pattern. Hydrogen atoms are omitted for clarity.
. DOI: 10.1107/S2056989015011901/is5404fig3.tif
A packing diagram of the title compound, showing C—H⋯O interactions (blue lines).
CCDC reference: 1008606
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C8AH8AO3B | 0.94 | 2.48 | 3.234(3) | 137 |
| C8BH8BO3A | 0.94 | 2.55 | 3.304(4) | 137 |
| C11AH11AO4B i | 0.94 | 2.60 | 3.325(3) | 135 |
| C11BH11BO4A ii | 0.94 | 2.46 | 3.199(4) | 135 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (C) (No. 15 K05482) from the JSPS.
supplementary crystallographic information
S1. Comment
9,10-Anthraquinone is an important framework as a dye. Various kinds of hydroxy-substituted anthraquinone dyes have been manufactured. However, there were little reports on alkoxy-substituted anthraquinone. In recent years, we presented the effects of the alkoxy substitution on the optical properties of 2,6-dialkoxy and 2,3,6,7-tetraalkoxy derivatives in solution as well as in the solid state (Ohta et al., 2012). Very recently, we have reported crystal structures of two polymorphs of 1,4-dipropoxy-9,10-anthraquinone, which contained red and yellow solids (Kitamura et al., 2015). The red crystal exhibited an anti-parallel arrangement along the stacking direction. On the other hand, the yellow crystal showed a slipped-parallel arrangement. To search the effect of alkyl chain length on molecular packing, we prepared the title compound, 1,4-diethoxy-9,10-anthraquinone, (I). In this paper, we present the crystal structure of (I).
The molecular structure of (I) is shown in Fig. 1. Two crystallographically independent molecules were found in the asymmetric unit, although the two molecules had almost the same molecular structure. There was a difference in planarity between the two molecules. Thus, the anthraquinone framework was slightly bent at the central quinone ring. For example, the dihedral angle between the two terminal benzene rings in the anthraquinone was 2.33 (8)° for one molecule and 13.31 (9)° for the other. The packing structure displays a dimer-herringbone pattern (Fig. 2), which is completely different from those of 1,4-dipropoxy-9,10-anthraquinone polymorphs (Kitamura et al., 2015). In the dimer part, the two molecules adopt slipped-parallel π-stack with an average interplanar distance of 3.45 Å, which would result in a yellow color in the solid state. The crystal structure is also stabilized by C—H···O interactions along the lateral direction of molecules (Fig. 3).
S2. Experimental
The title compound was prepared according to our previously reported method (Kitamura et al., 2004). A mixture of 1,4-hydrooxy-9,10-anthraquinone (2.20 g, 9.16 mmol), K2CO3 (2.51 g, 18.1 mmol), ethyl p-toluenesulfonate (5.02 g, 25.1 mmol) in o-dichlorobenzene (15 ml) was heated at reflux for 3 h under N2 gas. After cooling to room temperature, water (65 ml) was added to the reaction mixture. Then, the resulting solid was filtered off and washed with hexane to give the title compound (2.37 g, 87% yield) as a yellow solid. Single crystals suitable for X-ray analysis were obtained by slow evaporation from a CH2Cl2 solution (m.p. 172–175 °C). Elemental analysis for C18H16O4: C 72.96, H 5.44. Found: C 72.75, H 5.51. TOF-MS(EI): m/z Calcd C18H16O4: 296.1049. Found: 296.1074.
S3. Refinement
All the H atoms were positioned geometrically and refined using a riding model with C—H bonds of 0.94 Å, 0.98 Å, and 0.97 Å for aromatic, methylene and methyl groups, respectively, and Uiso(H) = 1.2Ueq(C) [Uiso(H) = 1.5Ueq(C) for methyl H atoms].
Figures
Fig. 1.

The asymmetric unit of the title compound, showing the atomic numbering and 40% probability displacement ellipsoids.
Fig. 2.

A packing diagram of the title compound viewed down the a axis, showing a dimer-herringbone pattern. Hydrogen atoms are omitted for clarity.
Fig. 3.

A packing diagram of the title compound, showing C—H···O interactions (blue lines).
Crystal data
| C18H16O4 | F(000) = 1248 |
| Mr = 296.31 | Dx = 1.352 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 11158 reflections |
| a = 13.5514 (11) Å | θ = 3–27.5° |
| b = 14.7204 (11) Å | µ = 0.10 mm−1 |
| c = 14.5905 (10) Å | T = 223 K |
| β = 90.604 (3)° | Prism, orange |
| V = 2910.4 (4) Å3 | 0.56 × 0.40 × 0.36 mm |
| Z = 8 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 3129 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed x-ray tube | Rint = 0.045 |
| Graphite monochromator | θmax = 27.5°, θmin = 3.0° |
| Detector resolution: 10 pixels mm-1 | h = −17→17 |
| ω scans | k = −19→19 |
| 27699 measured reflections | l = −16→18 |
| 6645 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | 0 constraints |
| R[F2 > 2σ(F2)] = 0.076 | H-atom parameters constrained |
| wR(F2) = 0.273 | w = 1/[σ2(Fo2) + (0.1824P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.93 | (Δ/σ)max < 0.001 |
| 6645 reflections | Δρmax = 0.27 e Å−3 |
| 397 parameters | Δρmin = −0.48 e Å−3 |
Special details
| Experimental. 1H-NMR: δ 1.56 (t, J = 7.0 Hz, 6H), 4.20 (q, J = 7.0 Hz, 4H), 7.32 (s, 2H), 7.69–7.72 (m, 2H), 8.17–8.19 (m, 2H); 13C-NMR: δ 14.9, 66.0, 122.1, 123.4, 126.4, 133.2, 134.2, 153.6, 183.3. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1A | 0.3813 (2) | 0.68438 (17) | 0.35528 (16) | 0.0548 (6) | |
| C2A | 0.3271 (2) | 0.71337 (18) | 0.27977 (17) | 0.0611 (7) | |
| H2A | 0.3605 | 0.7327 | 0.2271 | 0.073* | |
| C3A | 0.2261 (2) | 0.71464 (18) | 0.27990 (16) | 0.0596 (7) | |
| H3A | 0.1918 | 0.7347 | 0.2274 | 0.072* | |
| C4A | 0.1731 (2) | 0.68680 (17) | 0.35617 (16) | 0.0546 (6) | |
| C5A | 0.22559 (19) | 0.65664 (16) | 0.43427 (16) | 0.0518 (6) | |
| C6A | 0.1713 (2) | 0.63214 (19) | 0.51927 (18) | 0.0611 (7) | |
| C7A | 0.22803 (19) | 0.59444 (17) | 0.59796 (15) | 0.0527 (6) | |
| C8A | 0.1770 (2) | 0.56507 (18) | 0.67540 (17) | 0.0632 (7) | |
| H8A | 0.1078 | 0.5678 | 0.6766 | 0.076* | |
| C9A | 0.2293 (2) | 0.5320 (2) | 0.75003 (17) | 0.0707 (8) | |
| H9A | 0.1952 | 0.5121 | 0.8021 | 0.085* | |
| C10A | 0.3301 (2) | 0.5279 (2) | 0.74903 (18) | 0.0726 (8) | |
| H10A | 0.3647 | 0.5057 | 0.8005 | 0.087* | |
| C11A | 0.3814 (2) | 0.5564 (2) | 0.67253 (18) | 0.0681 (8) | |
| H11A | 0.4507 | 0.553 | 0.6717 | 0.082* | |
| C12A | 0.3296 (2) | 0.59013 (18) | 0.59665 (17) | 0.0572 (6) | |
| C13A | 0.3855 (2) | 0.6237 (2) | 0.5163 (2) | 0.0734 (9) | |
| C14A | 0.32994 (19) | 0.65421 (16) | 0.43355 (16) | 0.0530 (6) | |
| C15A | 0.5325 (2) | 0.7171 (2) | 0.2792 (2) | 0.0782 (9) | |
| H15A | 0.5119 | 0.7793 | 0.265 | 0.094* | |
| H15B | 0.5174 | 0.6787 | 0.226 | 0.094* | |
| C16A | 0.6399 (2) | 0.7146 (2) | 0.2998 (2) | 0.0814 (9) | |
| H16A | 0.676 | 0.7365 | 0.2471 | 0.122* | |
| H16B | 0.6542 | 0.7531 | 0.3523 | 0.122* | |
| H16C | 0.6598 | 0.6527 | 0.3134 | 0.122* | |
| C17A | 0.0200 (2) | 0.7234 (2) | 0.28108 (18) | 0.0672 (7) | |
| H17A | 0.0363 | 0.6897 | 0.2253 | 0.081* | |
| H17B | 0.0373 | 0.7874 | 0.2719 | 0.081* | |
| C18A | −0.0867 (2) | 0.7146 (2) | 0.3010 (2) | 0.0787 (9) | |
| H18A | −0.1251 | 0.7386 | 0.25 | 0.118* | |
| H18B | −0.1029 | 0.651 | 0.3099 | 0.118* | |
| H18C | −0.1019 | 0.7483 | 0.3562 | 0.118* | |
| O1A | 0.48108 (14) | 0.68464 (14) | 0.35759 (12) | 0.0673 (5) | |
| O2A | 0.07320 (14) | 0.68735 (14) | 0.35765 (12) | 0.0670 (5) | |
| O3A | 0.08375 (17) | 0.6462 (2) | 0.52801 (15) | 0.1020 (9) | |
| O4A | 0.47401 (18) | 0.6270 (3) | 0.5216 (2) | 0.1549 (16) | |
| C1B | −0.31908 (18) | 0.42295 (17) | 0.93060 (15) | 0.0515 (6) | |
| C2B | −0.2634 (2) | 0.3807 (2) | 0.99876 (16) | 0.0600 (7) | |
| H2B | −0.296 | 0.3526 | 1.0478 | 0.072* | |
| C3B | −0.1627 (2) | 0.37894 (19) | 0.99648 (16) | 0.0582 (7) | |
| H3B | −0.1276 | 0.3486 | 1.0432 | 0.07* | |
| C4B | −0.11091 (18) | 0.42118 (16) | 0.92621 (15) | 0.0507 (6) | |
| C5B | −0.16423 (18) | 0.46877 (16) | 0.85830 (14) | 0.0477 (6) | |
| C6B | −0.11215 (18) | 0.51742 (17) | 0.78342 (15) | 0.0517 (6) | |
| C7B | −0.17014 (18) | 0.54479 (16) | 0.70136 (15) | 0.0495 (6) | |
| C8B | −0.1216 (2) | 0.57178 (19) | 0.62221 (17) | 0.0636 (7) | |
| H8B | −0.0523 | 0.5707 | 0.6203 | 0.076* | |
| C9B | −0.1754 (2) | 0.6001 (2) | 0.54668 (17) | 0.0683 (8) | |
| H9B | −0.1426 | 0.6169 | 0.4928 | 0.082* | |
| C10B | −0.2760 (2) | 0.6040 (2) | 0.54962 (17) | 0.0680 (8) | |
| H10B | −0.312 | 0.6241 | 0.498 | 0.082* | |
| C11B | −0.3252 (2) | 0.57867 (18) | 0.62810 (17) | 0.0634 (7) | |
| H11B | −0.3944 | 0.5823 | 0.63 | 0.076* | |
| C12B | −0.27238 (18) | 0.54778 (16) | 0.70427 (15) | 0.0499 (6) | |
| C13B | −0.32523 (18) | 0.52046 (17) | 0.78847 (15) | 0.0520 (6) | |
| C14B | −0.26902 (17) | 0.47017 (16) | 0.86019 (14) | 0.0474 (5) | |
| C15B | −0.4706 (2) | 0.3711 (2) | 0.99707 (18) | 0.0672 (8) | |
| H15C | −0.462 | 0.4013 | 1.0565 | 0.081* | |
| H15D | −0.4444 | 0.3092 | 1.0022 | 0.081* | |
| C16B | −0.5768 (2) | 0.3682 (2) | 0.97143 (19) | 0.0714 (8) | |
| H16D | −0.6129 | 0.3351 | 1.0178 | 0.107* | |
| H16E | −0.6022 | 0.4296 | 0.9669 | 0.107* | |
| H16F | −0.5847 | 0.3378 | 0.9128 | 0.107* | |
| C17B | 0.04137 (19) | 0.36059 (19) | 0.98395 (17) | 0.0605 (7) | |
| H17C | 0.0159 | 0.2983 | 0.9816 | 0.073* | |
| H17D | 0.0335 | 0.3839 | 1.0464 | 0.073* | |
| C18B | 0.1478 (2) | 0.3621 (2) | 0.95790 (19) | 0.0681 (7) | |
| H18D | 0.1855 | 0.3244 | 1.0001 | 0.102* | |
| H18E | 0.1546 | 0.3388 | 0.8961 | 0.102* | |
| H18F | 0.1722 | 0.424 | 0.9606 | 0.102* | |
| O1B | −0.41883 (13) | 0.42021 (13) | 0.92745 (11) | 0.0609 (5) | |
| O2B | −0.01133 (12) | 0.41697 (12) | 0.91984 (11) | 0.0586 (5) | |
| O3B | −0.02445 (14) | 0.53591 (16) | 0.78861 (12) | 0.0763 (6) | |
| O4B | −0.41212 (14) | 0.54036 (16) | 0.79662 (13) | 0.0792 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1A | 0.0605 (17) | 0.0546 (14) | 0.0495 (13) | −0.0036 (11) | 0.0078 (11) | −0.0019 (11) |
| C2A | 0.0725 (19) | 0.0649 (16) | 0.0460 (13) | −0.0059 (13) | 0.0096 (12) | 0.0024 (11) |
| C3A | 0.0730 (19) | 0.0623 (15) | 0.0436 (13) | −0.0014 (13) | −0.0008 (12) | 0.0022 (11) |
| C4A | 0.0588 (17) | 0.0563 (14) | 0.0488 (13) | −0.0021 (11) | −0.0015 (11) | 0.0009 (11) |
| C5A | 0.0555 (15) | 0.0521 (13) | 0.0478 (13) | 0.0009 (10) | 0.0027 (11) | 0.0039 (10) |
| C6A | 0.0500 (16) | 0.0738 (18) | 0.0595 (15) | 0.0006 (12) | 0.0044 (12) | 0.0125 (13) |
| C7A | 0.0571 (16) | 0.0554 (14) | 0.0456 (13) | −0.0020 (11) | 0.0035 (11) | 0.0040 (10) |
| C8A | 0.0642 (18) | 0.0737 (17) | 0.0519 (14) | −0.0018 (13) | 0.0090 (12) | 0.0057 (12) |
| C9A | 0.085 (2) | 0.0814 (19) | 0.0452 (14) | −0.0047 (16) | 0.0060 (13) | 0.0117 (13) |
| C10A | 0.080 (2) | 0.087 (2) | 0.0507 (15) | −0.0024 (16) | −0.0112 (13) | 0.0135 (14) |
| C11A | 0.0623 (18) | 0.0812 (19) | 0.0605 (16) | −0.0011 (13) | −0.0049 (13) | 0.0164 (14) |
| C12A | 0.0600 (17) | 0.0620 (15) | 0.0497 (13) | −0.0015 (12) | −0.0007 (11) | 0.0084 (11) |
| C13A | 0.0499 (17) | 0.102 (2) | 0.0680 (17) | −0.0035 (15) | 0.0006 (13) | 0.0333 (16) |
| C14A | 0.0550 (15) | 0.0549 (14) | 0.0490 (13) | −0.0013 (11) | 0.0026 (11) | 0.0048 (10) |
| C15A | 0.072 (2) | 0.100 (2) | 0.0625 (17) | −0.0060 (17) | 0.0221 (15) | 0.0049 (16) |
| C16A | 0.068 (2) | 0.099 (2) | 0.078 (2) | −0.0059 (17) | 0.0214 (16) | 0.0005 (17) |
| C17A | 0.0692 (19) | 0.0800 (19) | 0.0522 (14) | 0.0096 (14) | −0.0100 (12) | 0.0001 (13) |
| C18A | 0.068 (2) | 0.102 (2) | 0.0660 (17) | 0.0023 (16) | −0.0154 (14) | −0.0043 (16) |
| O1A | 0.0570 (12) | 0.0860 (13) | 0.0593 (11) | −0.0029 (9) | 0.0144 (9) | 0.0075 (9) |
| O2A | 0.0574 (12) | 0.0872 (13) | 0.0563 (10) | −0.0004 (9) | −0.0067 (8) | 0.0116 (9) |
| O3A | 0.0555 (14) | 0.165 (3) | 0.0855 (15) | 0.0170 (14) | 0.0145 (11) | 0.0563 (15) |
| O4A | 0.0496 (16) | 0.289 (4) | 0.126 (2) | −0.0097 (19) | −0.0050 (14) | 0.127 (3) |
| C1B | 0.0491 (14) | 0.0616 (14) | 0.0440 (12) | −0.0026 (11) | 0.0030 (10) | 0.0047 (10) |
| C2B | 0.0609 (17) | 0.0720 (17) | 0.0473 (13) | −0.0048 (13) | 0.0066 (12) | 0.0168 (12) |
| C3B | 0.0557 (16) | 0.0688 (16) | 0.0500 (13) | 0.0000 (12) | −0.0007 (11) | 0.0154 (12) |
| C4B | 0.0514 (15) | 0.0590 (14) | 0.0416 (12) | 0.0001 (11) | −0.0002 (10) | 0.0020 (10) |
| C5B | 0.0534 (14) | 0.0556 (13) | 0.0342 (11) | −0.0005 (10) | 0.0029 (9) | 0.0021 (9) |
| C6B | 0.0455 (14) | 0.0653 (15) | 0.0442 (12) | −0.0025 (11) | 0.0026 (10) | 0.0057 (11) |
| C7B | 0.0543 (15) | 0.0537 (13) | 0.0407 (11) | −0.0022 (10) | 0.0020 (10) | 0.0059 (10) |
| C8B | 0.0624 (17) | 0.0802 (18) | 0.0483 (13) | −0.0060 (14) | 0.0066 (12) | 0.0134 (12) |
| C9B | 0.075 (2) | 0.085 (2) | 0.0451 (13) | −0.0091 (15) | 0.0059 (13) | 0.0173 (13) |
| C10B | 0.074 (2) | 0.0826 (19) | 0.0474 (14) | −0.0010 (15) | −0.0059 (13) | 0.0200 (13) |
| C11B | 0.0597 (17) | 0.0770 (18) | 0.0534 (14) | 0.0034 (13) | −0.0030 (12) | 0.0164 (13) |
| C12B | 0.0546 (15) | 0.0539 (13) | 0.0413 (12) | 0.0013 (10) | 0.0028 (10) | 0.0056 (10) |
| C13B | 0.0475 (14) | 0.0630 (15) | 0.0455 (12) | 0.0023 (11) | 0.0027 (10) | 0.0063 (11) |
| C14B | 0.0511 (14) | 0.0551 (13) | 0.0359 (11) | 0.0002 (10) | 0.0036 (9) | 0.0007 (9) |
| C15B | 0.0576 (18) | 0.0847 (19) | 0.0596 (16) | −0.0086 (14) | 0.0113 (13) | 0.0203 (14) |
| C16B | 0.0570 (18) | 0.095 (2) | 0.0620 (16) | −0.0103 (15) | 0.0097 (13) | 0.0086 (15) |
| C17B | 0.0567 (17) | 0.0672 (16) | 0.0572 (14) | −0.0002 (12) | −0.0105 (12) | 0.0086 (12) |
| C18B | 0.0550 (17) | 0.0804 (19) | 0.0688 (17) | 0.0069 (14) | −0.0070 (13) | 0.0045 (14) |
| O1B | 0.0471 (11) | 0.0822 (13) | 0.0535 (10) | −0.0018 (8) | 0.0076 (8) | 0.0159 (8) |
| O2B | 0.0478 (11) | 0.0773 (12) | 0.0508 (9) | 0.0029 (8) | −0.0036 (7) | 0.0139 (8) |
| O3B | 0.0512 (12) | 0.1171 (17) | 0.0605 (11) | −0.0142 (11) | −0.0020 (8) | 0.0288 (11) |
| O4B | 0.0525 (12) | 0.1193 (17) | 0.0659 (12) | 0.0183 (11) | 0.0086 (9) | 0.0321 (11) |
Geometric parameters (Å, º)
| C1A—O1A | 1.352 (3) | C1B—O1B | 1.353 (3) |
| C1A—C2A | 1.385 (4) | C1B—C2B | 1.388 (3) |
| C1A—C14A | 1.415 (3) | C1B—C14B | 1.419 (3) |
| C2A—C3A | 1.368 (4) | C2B—C3B | 1.366 (3) |
| C2A—H2A | 0.94 | C2B—H2B | 0.94 |
| C3A—C4A | 1.392 (3) | C3B—C4B | 1.395 (3) |
| C3A—H3A | 0.94 | C3B—H3B | 0.94 |
| C4A—O2A | 1.354 (3) | C4B—O2B | 1.355 (3) |
| C4A—C5A | 1.409 (3) | C4B—C5B | 1.407 (3) |
| C5A—C14A | 1.415 (3) | C5B—C14B | 1.421 (3) |
| C5A—C6A | 1.493 (3) | C5B—C6B | 1.490 (3) |
| C6A—O3A | 1.212 (3) | C6B—O3B | 1.221 (3) |
| C6A—C7A | 1.483 (3) | C6B—C7B | 1.481 (3) |
| C7A—C12A | 1.378 (4) | C7B—C12B | 1.387 (3) |
| C7A—C8A | 1.399 (3) | C7B—C8B | 1.393 (3) |
| C8A—C9A | 1.381 (4) | C8B—C9B | 1.379 (4) |
| C8A—H8A | 0.94 | C8B—H8B | 0.94 |
| C9A—C10A | 1.367 (4) | C9B—C10B | 1.366 (4) |
| C9A—H9A | 0.94 | C9B—H9B | 0.94 |
| C10A—C11A | 1.386 (4) | C10B—C11B | 1.383 (4) |
| C10A—H10A | 0.94 | C10B—H10B | 0.94 |
| C11A—C12A | 1.396 (4) | C11B—C12B | 1.392 (3) |
| C11A—H11A | 0.94 | C11B—H11B | 0.94 |
| C12A—C13A | 1.488 (4) | C12B—C13B | 1.484 (3) |
| C13A—O4A | 1.202 (3) | C13B—O4B | 1.220 (3) |
| C13A—C14A | 1.485 (4) | C13B—C14B | 1.485 (3) |
| C15A—O1A | 1.428 (3) | C15B—O1B | 1.436 (3) |
| C15A—C16A | 1.484 (4) | C15B—C16B | 1.484 (4) |
| C15A—H15A | 0.98 | C15B—H15C | 0.98 |
| C15A—H15B | 0.98 | C15B—H15D | 0.98 |
| C16A—H16A | 0.97 | C16B—H16D | 0.97 |
| C16A—H16B | 0.97 | C16B—H16E | 0.97 |
| C16A—H16C | 0.97 | C16B—H16F | 0.97 |
| C17A—O2A | 1.425 (3) | C17B—O2B | 1.435 (3) |
| C17A—C18A | 1.485 (4) | C17B—C18B | 1.495 (4) |
| C17A—H17A | 0.98 | C17B—H17C | 0.98 |
| C17A—H17B | 0.98 | C17B—H17D | 0.98 |
| C18A—H18A | 0.97 | C18B—H18D | 0.97 |
| C18A—H18B | 0.97 | C18B—H18E | 0.97 |
| C18A—H18C | 0.97 | C18B—H18F | 0.97 |
| O1A—C1A—C2A | 122.8 (2) | O1B—C1B—C2B | 123.1 (2) |
| O1A—C1A—C14A | 118.8 (2) | O1B—C1B—C14B | 118.4 (2) |
| C2A—C1A—C14A | 118.5 (3) | C2B—C1B—C14B | 118.5 (2) |
| C3A—C2A—C1A | 121.7 (2) | C3B—C2B—C1B | 121.8 (2) |
| C3A—C2A—H2A | 119.2 | C3B—C2B—H2B | 119.1 |
| C1A—C2A—H2A | 119.2 | C1B—C2B—H2B | 119.1 |
| C2A—C3A—C4A | 121.4 (2) | C2B—C3B—C4B | 121.4 (2) |
| C2A—C3A—H3A | 119.3 | C2B—C3B—H3B | 119.3 |
| C4A—C3A—H3A | 119.3 | C4B—C3B—H3B | 119.3 |
| O2A—C4A—C3A | 122.3 (2) | O2B—C4B—C3B | 122.6 (2) |
| O2A—C4A—C5A | 119.0 (2) | O2B—C4B—C5B | 118.6 (2) |
| C3A—C4A—C5A | 118.6 (3) | C3B—C4B—C5B | 118.7 (2) |
| C4A—C5A—C14A | 119.9 (2) | C4B—C5B—C14B | 120.0 (2) |
| C4A—C5A—C6A | 119.9 (2) | C4B—C5B—C6B | 120.8 (2) |
| C14A—C5A—C6A | 120.1 (2) | C14B—C5B—C6B | 119.24 (19) |
| O3A—C6A—C7A | 118.9 (2) | O3B—C6B—C7B | 119.8 (2) |
| O3A—C6A—C5A | 122.5 (2) | O3B—C6B—C5B | 122.0 (2) |
| C7A—C6A—C5A | 118.5 (2) | C7B—C6B—C5B | 118.2 (2) |
| C12A—C7A—C8A | 119.9 (2) | C12B—C7B—C8B | 119.8 (2) |
| C12A—C7A—C6A | 121.1 (2) | C12B—C7B—C6B | 120.4 (2) |
| C8A—C7A—C6A | 119.0 (2) | C8B—C7B—C6B | 119.8 (2) |
| C9A—C8A—C7A | 119.5 (3) | C9B—C8B—C7B | 119.9 (3) |
| C9A—C8A—H8A | 120.3 | C9B—C8B—H8B | 120 |
| C7A—C8A—H8A | 120.3 | C7B—C8B—H8B | 120 |
| C10A—C9A—C8A | 120.7 (3) | C10B—C9B—C8B | 120.4 (2) |
| C10A—C9A—H9A | 119.6 | C10B—C9B—H9B | 119.8 |
| C8A—C9A—H9A | 119.6 | C8B—C9B—H9B | 119.8 |
| C9A—C10A—C11A | 120.3 (3) | C9B—C10B—C11B | 120.3 (2) |
| C9A—C10A—H10A | 119.8 | C9B—C10B—H10B | 119.8 |
| C11A—C10A—H10A | 119.8 | C11B—C10B—H10B | 119.8 |
| C10A—C11A—C12A | 119.6 (3) | C10B—C11B—C12B | 120.0 (3) |
| C10A—C11A—H11A | 120.2 | C10B—C11B—H11B | 120 |
| C12A—C11A—H11A | 120.2 | C12B—C11B—H11B | 120 |
| C7A—C12A—C11A | 120.0 (2) | C7B—C12B—C11B | 119.4 (2) |
| C7A—C12A—C13A | 120.8 (2) | C7B—C12B—C13B | 120.5 (2) |
| C11A—C12A—C13A | 119.2 (3) | C11B—C12B—C13B | 120.0 (2) |
| O4A—C13A—C14A | 122.5 (3) | O4B—C13B—C12B | 119.3 (2) |
| O4A—C13A—C12A | 118.6 (3) | O4B—C13B—C14B | 122.6 (2) |
| C14A—C13A—C12A | 118.9 (2) | C12B—C13B—C14B | 118.0 (2) |
| C5A—C14A—C1A | 119.9 (2) | C1B—C14B—C5B | 119.5 (2) |
| C5A—C14A—C13A | 120.0 (2) | C1B—C14B—C13B | 120.6 (2) |
| C1A—C14A—C13A | 120.0 (2) | C5B—C14B—C13B | 119.9 (2) |
| O1A—C15A—C16A | 108.4 (3) | O1B—C15B—C16B | 108.4 (2) |
| O1A—C15A—H15A | 110 | O1B—C15B—H15C | 110 |
| C16A—C15A—H15A | 110 | C16B—C15B—H15C | 110 |
| O1A—C15A—H15B | 110 | O1B—C15B—H15D | 110 |
| C16A—C15A—H15B | 110 | C16B—C15B—H15D | 110 |
| H15A—C15A—H15B | 108.4 | H15C—C15B—H15D | 108.4 |
| C15A—C16A—H16A | 109.5 | C15B—C16B—H16D | 109.5 |
| C15A—C16A—H16B | 109.5 | C15B—C16B—H16E | 109.5 |
| H16A—C16A—H16B | 109.5 | H16D—C16B—H16E | 109.5 |
| C15A—C16A—H16C | 109.5 | C15B—C16B—H16F | 109.5 |
| H16A—C16A—H16C | 109.5 | H16D—C16B—H16F | 109.5 |
| H16B—C16A—H16C | 109.5 | H16E—C16B—H16F | 109.5 |
| O2A—C17A—C18A | 107.4 (2) | O2B—C17B—C18B | 107.5 (2) |
| O2A—C17A—H17A | 110.2 | O2B—C17B—H17C | 110.2 |
| C18A—C17A—H17A | 110.2 | C18B—C17B—H17C | 110.2 |
| O2A—C17A—H17B | 110.2 | O2B—C17B—H17D | 110.2 |
| C18A—C17A—H17B | 110.2 | C18B—C17B—H17D | 110.2 |
| H17A—C17A—H17B | 108.5 | H17C—C17B—H17D | 108.5 |
| C17A—C18A—H18A | 109.5 | C17B—C18B—H18D | 109.5 |
| C17A—C18A—H18B | 109.5 | C17B—C18B—H18E | 109.5 |
| H18A—C18A—H18B | 109.5 | H18D—C18B—H18E | 109.5 |
| C17A—C18A—H18C | 109.5 | C17B—C18B—H18F | 109.5 |
| H18A—C18A—H18C | 109.5 | H18D—C18B—H18F | 109.5 |
| H18B—C18A—H18C | 109.5 | H18E—C18B—H18F | 109.5 |
| C1A—O1A—C15A | 118.5 (2) | C1B—O1B—C15B | 119.11 (19) |
| C4A—O2A—C17A | 119.1 (2) | C4B—O2B—C17B | 118.09 (19) |
| O1A—C1A—C2A—C3A | −178.3 (2) | O1B—C1B—C2B—C3B | 175.1 (2) |
| C14A—C1A—C2A—C3A | 1.0 (4) | C14B—C1B—C2B—C3B | −4.0 (4) |
| C1A—C2A—C3A—C4A | 0.0 (4) | C1B—C2B—C3B—C4B | 1.3 (4) |
| C2A—C3A—C4A—O2A | −179.9 (2) | C2B—C3B—C4B—O2B | −176.9 (2) |
| C2A—C3A—C4A—C5A | −0.2 (4) | C2B—C3B—C4B—C5B | 2.0 (4) |
| O2A—C4A—C5A—C14A | 179.0 (2) | O2B—C4B—C5B—C14B | 176.5 (2) |
| C3A—C4A—C5A—C14A | −0.7 (4) | C3B—C4B—C5B—C14B | −2.4 (3) |
| O2A—C4A—C5A—C6A | −4.3 (4) | O2B—C4B—C5B—C6B | −3.0 (3) |
| C3A—C4A—C5A—C6A | 176.0 (2) | C3B—C4B—C5B—C6B | 178.1 (2) |
| C4A—C5A—C6A—O3A | −8.2 (4) | C4B—C5B—C6B—O3B | −17.1 (4) |
| C14A—C5A—C6A—O3A | 168.5 (3) | C14B—C5B—C6B—O3B | 163.4 (2) |
| C4A—C5A—C6A—C7A | 175.7 (2) | C4B—C5B—C6B—C7B | 163.8 (2) |
| C14A—C5A—C6A—C7A | −7.6 (4) | C14B—C5B—C6B—C7B | −15.7 (3) |
| O3A—C6A—C7A—C12A | −170.0 (3) | O3B—C6B—C7B—C12B | −161.9 (2) |
| C5A—C6A—C7A—C12A | 6.3 (4) | C5B—C6B—C7B—C12B | 17.2 (3) |
| O3A—C6A—C7A—C8A | 8.4 (4) | O3B—C6B—C7B—C8B | 15.1 (4) |
| C5A—C6A—C7A—C8A | −175.4 (2) | C5B—C6B—C7B—C8B | −165.8 (2) |
| C12A—C7A—C8A—C9A | 0.1 (4) | C12B—C7B—C8B—C9B | −0.9 (4) |
| C6A—C7A—C8A—C9A | −178.3 (2) | C6B—C7B—C8B—C9B | −177.9 (2) |
| C7A—C8A—C9A—C10A | 0.1 (4) | C7B—C8B—C9B—C10B | 1.6 (4) |
| C8A—C9A—C10A—C11A | −0.5 (5) | C8B—C9B—C10B—C11B | −0.7 (5) |
| C9A—C10A—C11A—C12A | 0.6 (5) | C9B—C10B—C11B—C12B | −0.9 (4) |
| C8A—C7A—C12A—C11A | 0.1 (4) | C8B—C7B—C12B—C11B | −0.6 (4) |
| C6A—C7A—C12A—C11A | 178.4 (3) | C6B—C7B—C12B—C11B | 176.3 (2) |
| C8A—C7A—C12A—C13A | −178.0 (3) | C8B—C7B—C12B—C13B | −179.2 (2) |
| C6A—C7A—C12A—C13A | 0.3 (4) | C6B—C7B—C12B—C13B | −2.2 (3) |
| C10A—C11A—C12A—C7A | −0.4 (4) | C10B—C11B—C12B—C7B | 1.5 (4) |
| C10A—C11A—C12A—C13A | 177.7 (3) | C10B—C11B—C12B—C13B | −179.9 (3) |
| C7A—C12A—C13A—O4A | 172.8 (4) | C7B—C12B—C13B—O4B | 165.4 (2) |
| C11A—C12A—C13A—O4A | −5.4 (5) | C11B—C12B—C13B—O4B | −13.1 (4) |
| C7A—C12A—C13A—C14A | −5.6 (4) | C7B—C12B—C13B—C14B | −14.1 (3) |
| C11A—C12A—C13A—C14A | 176.2 (3) | C11B—C12B—C13B—C14B | 167.4 (2) |
| C4A—C5A—C14A—C1A | 1.7 (4) | O1B—C1B—C14B—C5B | −175.6 (2) |
| C6A—C5A—C14A—C1A | −175.0 (2) | C2B—C1B—C14B—C5B | 3.5 (3) |
| C4A—C5A—C14A—C13A | 179.1 (3) | O1B—C1B—C14B—C13B | 4.0 (3) |
| C6A—C5A—C14A—C13A | 2.4 (4) | C2B—C1B—C14B—C13B | −176.8 (2) |
| O1A—C1A—C14A—C5A | 177.5 (2) | C4B—C5B—C14B—C1B | −0.3 (3) |
| C2A—C1A—C14A—C5A | −1.8 (4) | C6B—C5B—C14B—C1B | 179.2 (2) |
| O1A—C1A—C14A—C13A | 0.1 (4) | C4B—C5B—C14B—C13B | 180.0 (2) |
| C2A—C1A—C14A—C13A | −179.2 (3) | C6B—C5B—C14B—C13B | −0.5 (3) |
| O4A—C13A—C14A—C5A | −174.2 (4) | O4B—C13B—C14B—C1B | 16.2 (4) |
| C12A—C13A—C14A—C5A | 4.1 (4) | C12B—C13B—C14B—C1B | −164.3 (2) |
| O4A—C13A—C14A—C1A | 3.2 (5) | O4B—C13B—C14B—C5B | −164.1 (2) |
| C12A—C13A—C14A—C1A | −178.4 (2) | C12B—C13B—C14B—C5B | 15.4 (3) |
| C2A—C1A—O1A—C15A | 0.7 (4) | C2B—C1B—O1B—C15B | −0.4 (4) |
| C14A—C1A—O1A—C15A | −178.6 (2) | C14B—C1B—O1B—C15B | 178.7 (2) |
| C16A—C15A—O1A—C1A | 177.4 (2) | C16B—C15B—O1B—C1B | −172.6 (2) |
| C3A—C4A—O2A—C17A | −4.4 (4) | C3B—C4B—O2B—C17B | 5.2 (3) |
| C5A—C4A—O2A—C17A | 175.9 (2) | C5B—C4B—O2B—C17B | −173.7 (2) |
| C18A—C17A—O2A—C4A | 179.5 (2) | C18B—C17B—O2B—C4B | 175.7 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C8A—H8A···O3B | 0.94 | 2.48 | 3.234 (3) | 137 |
| C8B—H8B···O3A | 0.94 | 2.55 | 3.304 (4) | 137 |
| C11A—H11A···O4Bi | 0.94 | 2.60 | 3.325 (3) | 135 |
| C11B—H11B···O4Aii | 0.94 | 2.46 | 3.199 (4) | 135 |
Symmetry codes: (i) x+1, y, z; (ii) x−1, y, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: IS5404).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G. & Spagna, R. (2005). J. Appl. Cryst. 38, 381–388.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Kitamura, C., Hasegawa, M., Ishikawa, H., Fujimoto, J., Ouchi, M. & Yoneda, A. (2004). Bull. Chem. Soc. Jpn, 77, 1385–1393.
- Kitamura, C., Li, S., Takehara, M., Inoue, Y., Ono, K., Kawase, T. & Fujimoto, K. J. (2015). Bull. Chem. Soc. Jpn, 88, 713–715.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Ohta, A., Hattori, K., Kusumoto, Y., Kawase, T., Kobayashi, T., Naito, H. & Kitamura, C. (2012). Chem. Lett. 41, 674–676.
- Rigaku (1998). PROCESS-AUTO. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989015011901/is5404sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015011901/is5404Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015011901/is5404Isup3.cml
. DOI: 10.1107/S2056989015011901/is5404fig1.tif
The asymmetric unit of the title compound, showing the atomic numbering and 40% probability displacement ellipsoids.
a . DOI: 10.1107/S2056989015011901/is5404fig2.tif
A packing diagram of the title compound viewed down the a axis, showing a dimer-herringbone pattern. Hydrogen atoms are omitted for clarity.
. DOI: 10.1107/S2056989015011901/is5404fig3.tif
A packing diagram of the title compound, showing C—H⋯O interactions (blue lines).
CCDC reference: 1008606
Additional supporting information: crystallographic information; 3D view; checkCIF report


