Abstract
The nature of the complexes of histones H1 and H5 and their globular domains (GH1 and GH5) with DNA suggested two DNA-binding sites which are likely to be the basis of the preference of H1 and H5 for the nucleosome, compared with free DNA. More recently the X-ray and NMR structures of GH5 and GH1, respectively, have identified two basic clusters on opposite sides of the domains as candidates for these sites. Removal of the positive charge at either location by mutagenesis impairs or abolishes the ability of GH5 to assemble cooperatively in 'tramline' complexes containing two DNA duplexes, suggesting impairment or loss of its ability to bind two DNA duplexes. The mutant forms of GH5 also fail to protect the additional 20 bp of nucleosomal DNA that are characteristically protected by H1, H5 and wild-type recombinant GH5. They still bind to H1/H5-depleted chromatin, but evidently inappropriately. These results confirm the existence of, and identify the major components of, two DNA-binding sites on the globular domain of histone H5, and they strongly suggest that both binding sites are required to position the globular domain correctly on the nucleosome.
Full text
PDF
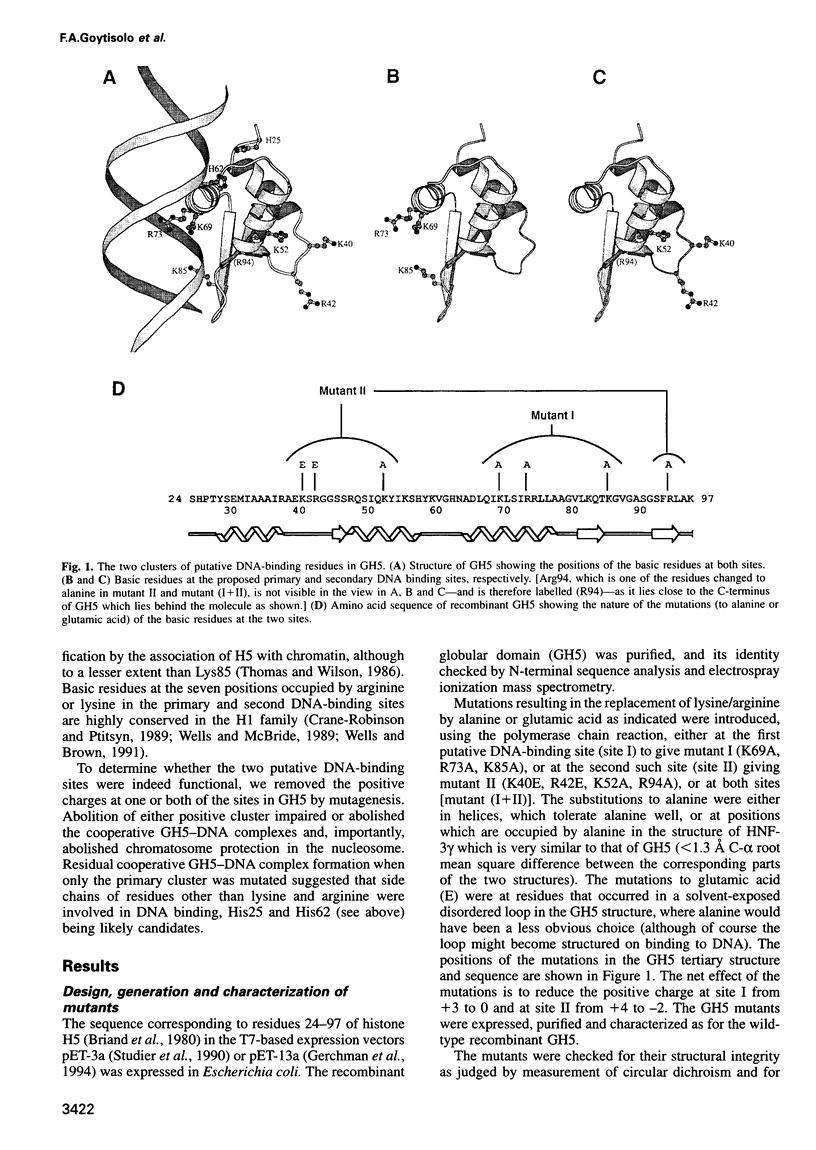
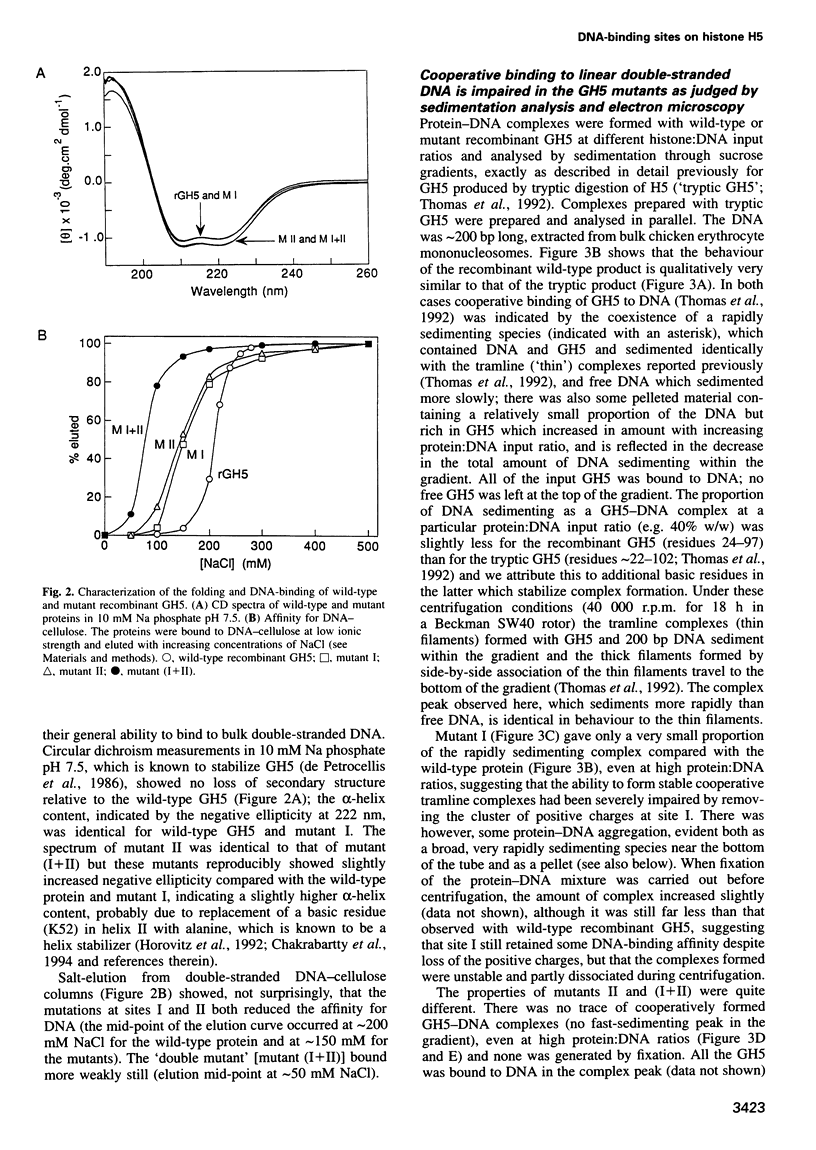
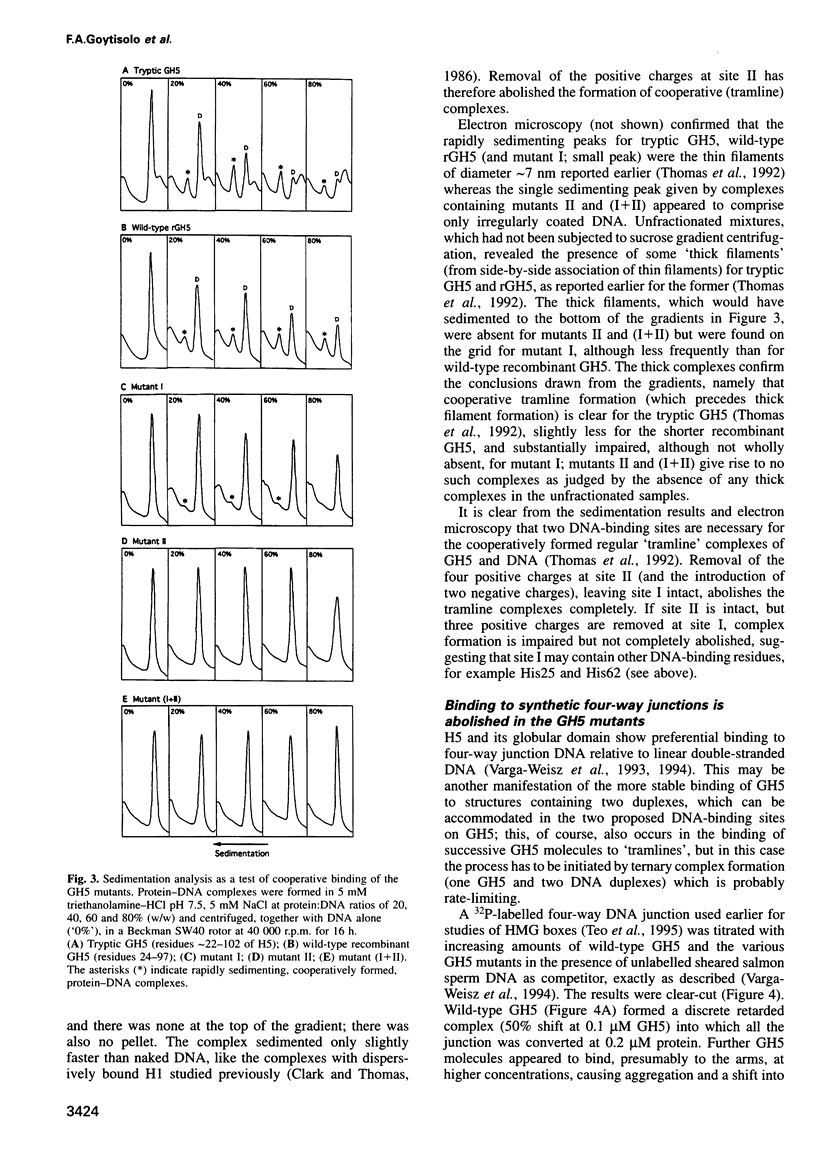
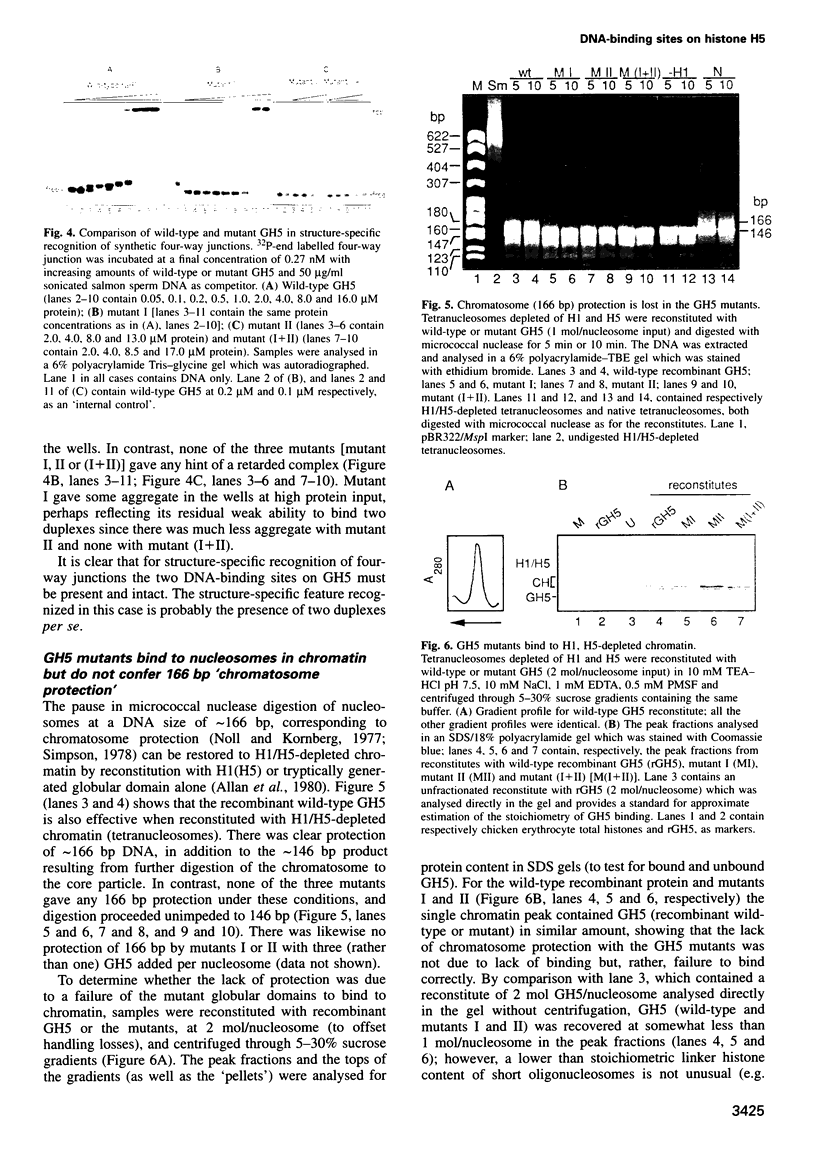




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Belyavsky A. V., Bavykin S. G., Goguadze E. G., Mirzabekov A. D. Primary organization of nucleosomes containing all five histones and DNA 175 and 165 base-pairs long. J Mol Biol. 1980 May 25;139(3):519–536. doi: 10.1016/0022-2836(80)90144-8. [DOI] [PubMed] [Google Scholar]
- Briand G., Kmiecik D., Sautiere P., Wouters D., Borie-Loy O., Biserte G., Mazen A., Champagne M. Chicken erythrocyte histone H5. IV. Sequence of the carboxy-termined half of the molecule (96 residues) and complete sequence. FEBS Lett. 1980 Apr 7;112(2):147–151. doi: 10.1016/0014-5793(80)80167-0. [DOI] [PubMed] [Google Scholar]
- Buckle R. S., Maman J. D., Allan J. Site-directed mutagenesis studies on the binding of the globular domain of linker histone H5 to the nucleosome. J Mol Biol. 1992 Feb 5;223(3):651–659. doi: 10.1016/0022-2836(92)90981-o. [DOI] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Cerf C., Lippens G., Ramakrishnan V., Muyldermans S., Segers A., Wyns L., Wodak S. J., Hallenga K. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry. 1994 Sep 20;33(37):11079–11086. doi: 10.1021/bi00203a004. [DOI] [PubMed] [Google Scholar]
- Chakrabartty A., Kortemme T., Baldwin R. L. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 1994 May;3(5):843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Differences in the binding of H1 variants to DNA. Cooperativity and linker-length related distribution. Eur J Biochem. 1988 Dec 1;178(1):225–233. doi: 10.1111/j.1432-1033.1988.tb14447.x. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Crane-Robinson C., Ptitsyn O. B. Binding of the globular domain of linker histones H5/H1 to the nucleosome: a hypothesis. Protein Eng. 1989 Aug;2(8):577–582. doi: 10.1093/protein/2.8.577. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L., Quagliarotti G., Tomei L., Geraci G. Structuring of H1 histone. Evidence of high-affinity binding sites for phosphate ions. Eur J Biochem. 1986 Apr 1;156(1):143–148. doi: 10.1111/j.1432-1033.1986.tb09559.x. [DOI] [PubMed] [Google Scholar]
- Dong F., Hansen J. C., van Holde K. E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draves P. H., Lowary P. T., Widom J. Co-operative binding of the globular domain of histone H5 to DNA. J Mol Biol. 1992 Jun 20;225(4):1105–1121. doi: 10.1016/0022-2836(92)90108-v. [DOI] [PubMed] [Google Scholar]
- Gerchman S. E., Graziano V., Ramakrishnan V. Expression of chicken linker histones in E. coli: sources of problems and methods for overcoming some of the difficulties. Protein Expr Purif. 1994 Jun;5(3):242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
- Graziano V., Gerchman S. E., Schneider D. K., Ramakrishnan V. Histone H1 is located in the interior of the chromatin 30-nm filament. Nature. 1994 Mar 24;368(6469):351–354. doi: 10.1038/368351a0. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Pruss D., Wolffe A. P. Contacts of the globular domain of histone H5 and core histones with DNA in a "chromatosome". Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Wolffe A. P. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Rimmer J. M., Green B. N., Finch J. T., Thomas J. O. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 1991 Jul;10(7):1939–1948. doi: 10.1002/j.1460-2075.1991.tb07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Horovitz A., Matthews J. M., Fersht A. R. Alpha-helix stability in proteins. II. Factors that influence stability at an internal position. J Mol Biol. 1992 Sep 20;227(2):560–568. doi: 10.1016/0022-2836(92)90907-2. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 1983 Feb 11;11(3):619–627. doi: 10.1093/nar/11.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990 Nov 30;96(1):125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Lindsey G. G., Orgeig S., Thompson P., Davies N., Maeder D. L. Extended C-terminal tail of wheat histone H2A interacts with DNA of the "linker" region. J Mol Biol. 1991 Apr 20;218(4):805–813. doi: 10.1016/0022-2836(91)90268-b. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Pruss D. V., Ebralidse K. K. Chromatin superstructure-dependent crosslinking with DNA of the histone H5 residues Thr1, His25 and His62. J Mol Biol. 1990 Jan 20;211(2):479–491. doi: 10.1016/0022-2836(90)90366-T. [DOI] [PubMed] [Google Scholar]
- Muyldermans S., Travers A. A. DNA sequence organization in chromatosomes. J Mol Biol. 1994 Jan 21;235(3):855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Pehrson J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9149–9153. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings S., Meersseman G., Bradbury E. M. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D., Hayes J. J., Wolffe A. P. Nucleosomal anatomy--where are the histones? Bioessays. 1995 Feb;17(2):161–170. doi: 10.1002/bies.950170211. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993 Mar 18;362(6417):219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z., Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988 Dec 1;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Teo S. H., Grasser K. D., Hardman C. H., Broadhurst R. W., Laue E. D., Thomas J. O. Two mutations in the HMG-box with very different structural consequences provide insights into the nature of binding to four-way junction DNA. EMBO J. 1995 Aug 1;14(15):3844–3853. doi: 10.1002/j.1460-2075.1995.tb00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Finch J. T. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992 Jan 25;20(2):187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Wilson C. M. Selective radiolabelling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J. 1986 Dec 20;5(13):3531–3537. doi: 10.1002/j.1460-2075.1986.tb04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Muyldermans S. V. A DNA sequence for positioning chromatosomes. J Mol Biol. 1996 Apr 5;257(3):486–491. doi: 10.1006/jmbi.1996.0178. [DOI] [PubMed] [Google Scholar]
- Ura K., Hayes J. J., Wolffe A. P. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 1995 Aug 1;14(15):3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P., Zlatanova J., Leuba S. H., Schroth G. P., van Holde K. Binding of histones H1 and H5 and their globular domains to four-way junction DNA. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3525–3529. doi: 10.1073/pnas.91.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P., van Holde K., Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J Biol Chem. 1993 Oct 5;268(28):20699–20700. [PubMed] [Google Scholar]
- Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., Brown D. Histone and histone gene compilation and alignment update. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2173–2188. doi: 10.1093/nar/19.suppl.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]






