Abstract
Without a doubt, our current antimicrobials are losing the battle in the fight against newly-emerged multidrug-resistant pathogens. There is a pressing, unmet need for novel antimicrobials and novel approaches to develop them; however, it is becoming increasingly difficult and costly to develop new antimicrobials. One strategy to reduce the time and cost associated with antimicrobial innovation is drug repurposing, which is to find new applications outside the scope of the original medical indication of the drug. Ebselen, an organoselenium clinical molecule, possesses potent antimicrobial activity against clinical multidrug-resistant Gram-positive pathogens, including Staphylococcus, Streptococcus, and Enterococcus, but not against Gram-negative pathogens. Moreover, the activity of ebselen against Gram-positive pathogens exceeded those activities determined for vancomycin and linezolid, drugs of choice for treatment of Enterococcus and Staphylococcus infections. The minimum inhibitory concentrations of ebselen at which 90% of clinical isolates of Enterococcus and Staphylococcus were inhibited (MIC90) were found to be 0.5 and 0.25 mg/L, respectively. Ebselen showed significant clearance of intracellular methicillin-resistant S. aureus (MRSA) in comparison to vancomycin and linezolid. We demonstrated that ebselen inhibits the bacterial translation process without affecting mitochondrial biogenesis. Additionally, ebselen was found to exhibit excellent activity in vivo in a Caenorhabditis elegans MRSA-infected whole animal model. Finally, ebselen showed synergistic activities with conventional antimicrobials against MRSA. Taken together, our results demonstrate that ebselen, with its potent antimicrobial activity and safety profiles, can be potentially used to treat multidrug resistant Gram-positive bacterial infections alone or in combination with other antibiotics and should be further clinically evaluated.
Introduction
Infections caused by Gram-positive drug-resistant pathogens are a leading cause of mortality. Three species—methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pneumoniae and vancomycin-resistant enterococcus (VRE)—are responsible annually for at least 84% of the antibiotic-resistant bacteria mortality in the United States alone. Further exacerbating the issue of bacterial resistance is the slow rate of the development and approval of new antimicrobials. For almost 80 years, antimicrobials have been crucial allies in the treatment of bacterial infections caused by these pathogens. However, multidrug resistant strains have recently emerged that are resistant to almost all antimicrobials once deemed effective, including fluoroquinolones, macrolides, and β-lactams [1]. Collectively, this points to an urgent need for the discovery of new antimicrobials and novel strategies to develop them. One novel strategy that warrants more attention as a unique method for development of new antimicrobials is drug repurposing [2]. Our recent attempt to identify non-antibiotic drugs with potent antimicrobial activity, within an applicable clinical range, identified organoselenium compound ebselen (EB) as having potent antibacterial activities against Gram-positive pathogens [3, 4]. EB is considered a clinically safe molecule but without proven use yet [5]. It has anti-oxidative, anti-inflammatory, and anti-atherosclerotic properties [6]. Additionally, EB has been shown to exhibit antimicrobial activity in vitro and in vivo [3, 4, 7–9]. EB exhibited antimicrobial activity by inhibition of thioredoxin reducatse (TrxR) enzyme of Escherichia coli and H+-ATPase function and proton-translocation function in yeast [7, 8, 10]. However, the antibacterial mechanism of action of EB against Gram-positive bacteria remains unidentified [8].
The potent antimicrobial activity of EB against Gram-positive pathogens motivated us to further investigate the therapeutic applications of EB. The aims of the present study are to investigate the antibacterial activity of EB against Gram-positive clinical pathogens, including MRSA and VRE in vitro, to identify antibacterial mechanism of action, to analyze the ability of EB to clear MRSA intracellular infection, to evaluate antibacterial efficacy in MRSA-infected Caenorhabditis elegans whole animal models, to evaluate the effect on mitochondrial biogenesis and toxicity in C. elegans, and to assess whether EB is capable of working synergistically with conventional antibiotics against MRSA in in vitro and in infected cell cultures. This study provided valuable insights into potential therapeutic applications of EB for use as antimicrobial agents for the treatment of multidrug-resistant Gram-positive infections.
Materials and Methods
Bacterial strains and reagents
Bacterial strains employed in this study are presented in Table 1. Mannitol salt agar (MSA) was purchased from Hardy Diagnostics (Santa Maria, CA). Muller-Hinton broth (MHB) was purchased from Sigma-Aldrich (St. Louis, MO). Trypticase soy broth (TSB) and Trypticase soy agar (TSA) were purchased from Becton, Dickinson (Cockeysville, MD). EB was purchased from (Adipogen corp, San Diego), vancomycin hydrochloride (Gold Biotechnology, St. Louis, MO), linezolid (Selleck Chemicals, Houston, TX), clindamycin (TCI chemicals, Portland, OR), erythromycin, rifampicin, ampicillin, gentamicin, chloramphenicol and fetal bovine serum (FBS) were purchased from Sigma-Aldrich (St. Louis, MO). DMEM media were purchased from Life technologies and MTS reagent (Promega, Madison, WI, USA).
Table 1. The MIC and MBC of EB against Gram-positive and Gram-negative bacteria.
| Bacterial Strains | Strain ID | Source | Phenotypic Characteristics | MIC/MBC (μg/ml) |
|---|---|---|---|---|
| Enterococcus spp | E. faecalis ATCC49533 | Blood, Wisconsin | Resistant to streptomycin | 0.25/8 |
| E. faecalis ATCC7080 | Meat involved in food poisoning, New York | - | 0.25/8 | |
| E. faecalis ATCC49532 | Blood, Wisconsin | Resistant to gentamicin | 0.25/8 | |
| E. faecalis ATCC14506 | Quality control strain | - | 0.5/8 | |
| E. faecalis ATCC 51229 (VRE) | Peritoneal fluid, St. Louis, MO | Resistant to vancomycin. Sensitive to teichoplanin | 0.5/0.5 | |
| E. faecalis SF24397 | Urine, Michigan | Resistance to erythromycin (ermB+) and gentamicin | 0.125/4 | |
| E. faecalis SF24413 (VRE) | Urine, Michigan | Resistant to erythromycin, gentamicin and vancomycin | 0.125/4 | |
| E. faecalis SF28073 (VRE) | Urine, Michigan | Resistant to erythromycin, gentamicin and vancomycin | 0.0625/8 | |
| E. faecalis HH22 | Urine, Texas | Resistance to penicillin, erythromycin, tetracycline and high levels of aminoglycosides | 0.125/4 | |
| E. faecalis MMH594 | Blood, Wisconsin | Resistance to erythromycin and gentamicin | 0.125/4 | |
| E. faecalis SV587 (VRE) | Urine Missouri | Resistance to vancomycin | 0.125/8 | |
| E. faecium E1162 | Blood, France | Resistance to ampicillin. | 0.25/16 | |
| E. faecium E0120 (VRE) | Ascites fluid, Netherlands | Resistant to gentamicin and vancomycin | 0.5/32 | |
| E. faecium ERV102 (VRE) | Oral sputum, Colombia | Resistant to ampicillin and vancomycin, and displays high levels of resistance to streptomycin | 0.5/16 | |
| E. faecium ATCC6569 | Human feces | - | 1/32 | |
| E. faecium ATCC 700221 (VRE) | Human feces, Connecticut | Resistant to vancomycin and teicoplanin | 0.5/1 | |
| Staphylococcus spp | MSSA (NRS 72) | United Kingdom | Resistant to penicillin | 0.25/0.5 |
| MRSA (NRS 384) | United States (Mississippi) | Resistant to erythromycin, methicillin, and tetracycline | 0.125/0.125 | |
| MRSA (NRS119) | United States (Massachusetts) | Resistant to linezolid | 0.125/0.25 | |
| MRSA (NRS 123) | United States | Resistant to methicillin; susceptible to nonbeta-lactam antibiotics | 0.25/0.5 | |
| MRSA (NRS194) | United States (North Dakota) | Resistant to methicillin | 0.25/1 | |
| MRSA (NRS108) | France | Resistant to gentamicin | 0.25/0.25 | |
| MRSA (NRS70) | Japan | Resistant to clindamycin, erythromycin and spectinomycin | 0.25/0.25 | |
| VISA (NRS 1) | Japan | Resistant to aminoglycosides and tetracycline (minocycline) | 0.125/0.125 | |
| VISA (NRS 19) | United States (Illinois) | Glycopeptide-intermediate S. aureus | 0.25/0.025 | |
| VRSA11a | United States | Resistant to erythromycin and spectinomycin | 0.125/0.25 | |
| VRSA11b | United States | Resistant to erythromycin and spectinomycin | 0.25/0.25 | |
| VRSA12 | United States | Resistant to vancomycin | 0.25/0. 5 | |
| VRSA13 | United States | Resistant to vancomycin | 0.25/0.25 | |
| Streptococcus spp | Streptococcus pyogenes ATCC 12344 | - | Quality control strain | 0.5/1 |
| Streptococcus agalactiae MNZ938 | Human blood | Beta-hemolytic, Serogroup: Group B | 0.5/0.5 | |
| Streptococcus agalactiae MNZ 933 | Human blood | Beta-hemolytic, Serogroup: Group B | 0.5/0.5 | |
| Streptococcus agalactiae MNZ 929 | Human blood | Beta-hemolytic, Serogroup: Group B | 0.5/0.5 | |
| Gram-negative bacteria | Acinetobacter baumannii ATCC BAA1605 | MDR strain isolated from the sputum of a Canadian soldier | Resistant to ceftazidime, gentamicin, ticarcillin, piperacillin, aztreonam, cefepime, ciprofloxacin, imipenem and meropemem | 16/ND |
| E. coli O157:H7 ATCC 700728 | Quality control strain | - | 32/ND | |
| Salmonella Typhimurium ATCC 700720 | Isolated from a natural source | - | 32/ND | |
| Klebsiella pneumonia ATCC BAA 2146 | Human urine | Clinical isolate New Delhi Metallo- β-Lactamase (NDM-1) positive | 64/ND | |
| Pseudomonas aeruginosa ATCC 9721 | Quality control strain | - | >256/ND |
VRE: vancomycin-resistant Enterococcus; MSSA: methicillin-sensitive S. aureus; MRSA: methicillin-resistant S. aureus; VISA: vancomycin-intermediate S. aureus; VRSA: vancomycin-resistant S. aureus; ND: not determined
In vitro antibacterial assays
Minimum inhibitory concentrations (MICs) were evaluated using micro dilution broth as per the standards of Clinical and Laboratory Standards Institute (CLSI) [11]. MICs of drugs were interpreted as the lowest concentration of the drug which inhibits the growth of bacteria after incubating for at least 16–24 h at 37°C. The minimum bactericidal concentration (MBC) was determined by sub-culturing 10 μl from the wells were no growth was observed onto TSA plates. The plates were incubated for 24 h before the MBCs were determined. The MBC was categorized as the concentration where ⩾99.9% reduction in bacterial cell count was observed [1].
Intracellular infection assay
J774A.1 murine macrophage-like cells were seeded at a density of 20,000 cells per well in 96-well tissue culture plates. Cells were infected with MRSA USA300 (NRS 384–0114; ST-8) for 30 min at a 1:100 multiplicity of infection (MOI). Then the cells were washed three times with DMEM medium containing 10 IU lysostaphin to kill the extracellular bacteria [12]. Drugs (vancomycin, linezolid and EB) were added at a concentration of 1 μg/ml to the DMEM medium supplemented with 10% FBS and 4 IU lysostaphin. After 24 h incubation, the cells were washed three times with phosphate buffered saline (PBS) and lysed with 0.1% Triton X-100. Lysates were diluted and plated on TSA plates and MRSA colony forming units (CFU) were counted.
Toxicity assay
The toxicity assays were performed in cell culture and C. elegans. (a) Cell culture: J774A.1 murine macrophage-like cells at a density of 20,000 cells per well were seeded and allowed to adhere in a 96-well tissue culture plate in DMEM media containing 10% FBS overnight. EB at various concentrations ranging from 0 to 256 μg/ml were added to the cells in DMEM media with FBS. After 24 h incubation with the drug, cells were washed with PBS and the MTS assay reagent,3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H tetrazolium) in DMEM medium was added and incubated for 4 h at 37°C. Absorbance was measured at 490 nm using ELISA microplate reader (Molecular Devices, Sunnyvale, CA, USA). Cell viability after treatment with EB was expressed as a percentage of the control, DMSO. (b) C. elegans: Temperature-sensitive C. elegans AU37 (sek-1; glp-4) strain (glp-4(bn2) was used for toxicity studies and the worms were synchronized as described before [13]. Synchronized L4-stage worms were re-suspended in buffer containing 50% M9 buffer and 50% TSB. Then 100 μl of the buffer containing approximately 15–20 worms were deposited in each well in 96-well plates and EB (4 and 8 μg/ml) and vancomycin (8 μg/ml) were added. Worms were counted daily for three days and the percent of live worms was calculated in each group. At least triplicate wells were used for each treatment
Cell-free bacterial and mammalian transcription/translation assay
The cell-free bacterial translation and mammalian translation assays were performed by the commercially available Escherichia coli S30 System and Rabbit Reticulocyte Lysate System (Promega), respectively. The assays were performed as described by the manufacturer, in conjunction with appropriate positive control (chloramphenicol) and negative control (ampicillin) antibiotics. In bacterial translation assay, the reaction mixtures were incubated at 37°C for 1 h. Mammalian translation assay reaction mixtures were incubated at 30°C for 1 h. Luciferase assay reagent was added to the reaction and the intensity of the luminescence was measured by luminescence microplate reader (FLx800 BioTek Instruments, Inc. Winooski, Vermont) according to the manufacturer’s instructions. Average luciferase readout of protein production from two replicates from two independent experiments was calculated.
Mitochondrial biogenesis assay
The mitobiogenesis assay was done using In-Cell ELISA Kit (MitoSciences Inc., Eugene, OR) as per the manufactures instruction [14]. Briefly, J774A.1 cells were seeded (40,000 cells per well) in 96-well plates for overnight. EB and control antimicrobials (chloramphenicol and ampicillin) were added to the cells and the cells were allowed to grow for approximately 3 days with the drugs. Media were removed and cells were washed with PBS, then fixed with 4% paraformaldehyde. After fixing, cells were washed with PBS and permeabilization and blocking processes were done according to the manufacturer’s instructions. Primary antibodies to detect the levels of two proteins (subunit I of Complex IV (COX-I), which is mitochondrial DNA (mtDNA)-encoded, and the 70 kDa subunit of Complex II (SDH-A), which is nuclear DNA (nDNA)-encoded were added and incubated for overnight at 4°C. After incubation, cells were washed with PBS and secondary antibodies were added and incubated at room temperature for 1 h. The expression of SDH-A and COX-1 were measured after washing and development at 405 nm and 600 nm wavelength, respectively. The ratio between COX-I and SDH-A was calculated and the percent of inhibition of mitochondrial biogenesis was measured.
Efficacy of EB in infected animal model (C. elegans)
L4-stage worms of C. elegans AU37 (sek-1; glp-4) strain (glp-4(bn2) were used to test the antimicrobial efficacy of EB as described before [13]. Briefly, worms were infected with MRSA USA300 (NRS 384–0114; ST-8) in nematode growth media agar plate for 8 h at room temperature. After 8 h of infection, worms were collected and washed with M9 buffer four times before incubation with the drugs. Worms were transferred to 96-well plates (20 worms per well) and the drugs (EB and vancomycin) were added to the wells in triplicates to achieve a final concentration of either 4 or 8 μg/ml. After 24 h incubation with the drugs, worms were transferred to 2-ml centrifuge tubes, washed four times with PBS and 100 mg 1.0-mm silicon carbide particles (Biospec Products, Bartlesville, OK) were added to each tube. The tubes were vortexed for one minute at maximum speed to disrupt the worms without affecting bacterial survival [13]. The resulting suspension was diluted and plated onto MSA plates to count the MRSA CFU. The total CFU obtained from each well was divided by the number of worms in respective wells and the results were expressed as percent of bacterial reduction per worm.
Synergistic activities of EB with conventional antibiotics in vitro and in cell culture
(a) In vitro synergistic assay: The synergistic activities of EB with conventional antibiotics were evaluated using the Bliss Independence Model as described before [4, 15]. Briefly, the optical density of the bacteria grown in the presence of antibiotics and EB (f AB), antibiotics alone (f A0), EB alone (f 0B) and in the absence of drugs (f 00) were measured and a degree of synergy (S) was calculated using the formula: S = (f A0 /f 00 )(f 0B /f 00 )-(f AB /f 00 ). Positive and negative values represent the degree of synergism and antagonism, respectively. (b) Intracellular synergistic assay in J774A.1 cells: J774A.1 cells were seeded and infected as described before under intracellular infection assay. EB at concentration of 0.5 μg/ml was added to infected cells alone or in in combination with control antibiotics such as linezolid (4 μg/ml), clindamycin (1 μg/ml), vancomycin (4 μg/ml), chloramphenicol (4 μg/ml), erythromycin (8 μg/ml), rifampicin (0.5 μg/ml) and gentamicin (1 μg/ml). Untreated cells, and cells treated with antibiotics alone were used as a control. After 24 h incubation, the cells were lysed and intracellular MRSA CFU were determined as described above. Percent bacterial reduction was calculated in relative to the untreated groups. Combination therapy was compared with single antibiotic therapy treatment groups.
Statistical analyses
Statistical analyses were done using Graph Pad Prism 6.0 (GraphPad Software, La Jolla, CA). P values were calculated by the one-tailed Student t test. P values of ˂ 0.05 were considered as significant.
Results and Discussion
In vitro antibacterial assays
In an attempt to repurpose approved drugs as antimicrobial agents, we investigated the antimicrobial activity of EB against various multidrug-resistant clinical isolates of Gram-positive and Gram-negative pathogens (Table 1). EB exhibited potent bactericidal activity, in a nanogram range, against all tested Gram-positive strains regardless of their resistance phenotype. EB showed potent activity against clinical isolates of Enterococcus faecalis and Enterococcus faecium with MIC90 of 0.5 μg/ml (see Table 1). EB also showed potent activity against vancomycin-resistant strains of Enterococcus (VRE). Next, we tested the activity of EB against the clinical isolates of multidrug-resistant S. aureus. EB showed more potent activity against methicillin-sensitive S. aureus, MRSA, vancomycin-intermediate S. aureus (VISA) and vancomycin-resistant S. aureus (VRSA) strains than VRE with MIC90 of 0.25 μg/ml (see Table 1). Finally, EB also showed potent activity against clinical isolates of Streptococcus pyogenes and Streptococcus agalactiae with MIC of 0.5 μg/ml (see Table 1). On the other hand, EB did not show potent antimicrobial activity (MIC ≥16 μg/ml) against Gram-negative pathogens, including Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumonia, Salmonella Typhimurium, and Acinetobacter baumannii. The lack of activity of EB against Gram-negative pathogens might be due to its reduced ability to enter the cells due to outer membrane barrier or the efflux pump rather than lack of target of EB inside Gram-negative bacteria [16–19].
Intracellular infection and cell toxicity
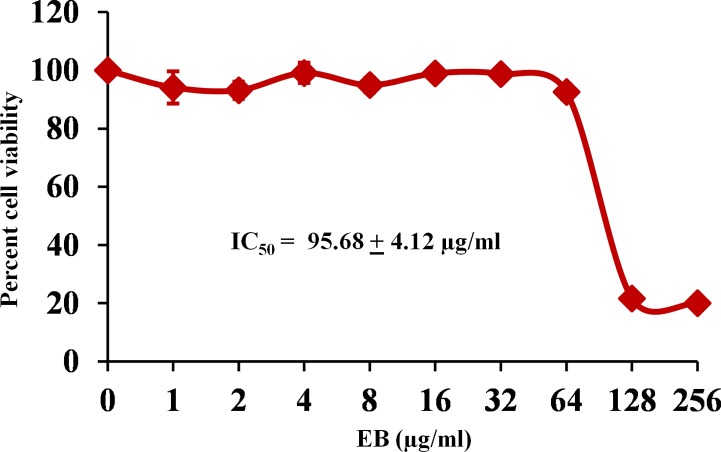
Some extracellular pathogens such as S. aureus are also capable of invading and surviving within the mammalian host cells, leading to persistent chronic infections. Moreover, during the S. aureus intracellular invasion phase, treatment with antimicrobials is very challenging because most antibiotics do not actively pass through cellular membranes [20–27]. Therefore, clinical failures of drug of choice, such as vancomycin, to cure S. aureus pneumonia have exceeded 40% and have been attributed mainly to poor intracellular penetration of the drug and consequently to the failure to kill intracellular MRSA in alveolar macrophages [28]. Hence, finding antimicrobials that possess both extra- and intracellular activity would be an optimum strategy to treat such invasive intracellular S. aureus infections. Therefore, we investigated if EB possesses intracellular anti-staphylococcal activity. As shown in Fig 1, EB at a concentration of 1 μg/ml significantly reduced the intracellular MRSA by 32%. In contrast, the conventional antimicrobials such as vancomycin and linezolid (drugs of choice for treatment of MRSA infections) at the same concentration reduced intracellular MRSA by only 16% and 21%, respectively. EB toxicity was assayed against J774A.1 cells at a concentration ranging from 0 to 256 μg/ml for 24 h. The results shown in Fig 2 indicate that EB does not show toxicity up to 64 μg/ml. The concentration of the EB that causes 50% toxicity (half inhibitory concentration: IC50) in J774A.1 cells is 95.68 ± 4.12 μg/ml. This value is more than 380-fold higher than the concentration required to inhibit MRSA. Collectively, these results suggest that EB has great potential for treatment of S. aureus infections where not only is eradication of extracellular bacteria important, but the killing of intracellular bacteria is also critical [29].
Fig 1. Activity of EB, vancomycin and linezolid against intracellular MRSA USA300 in J774A.1 cells.
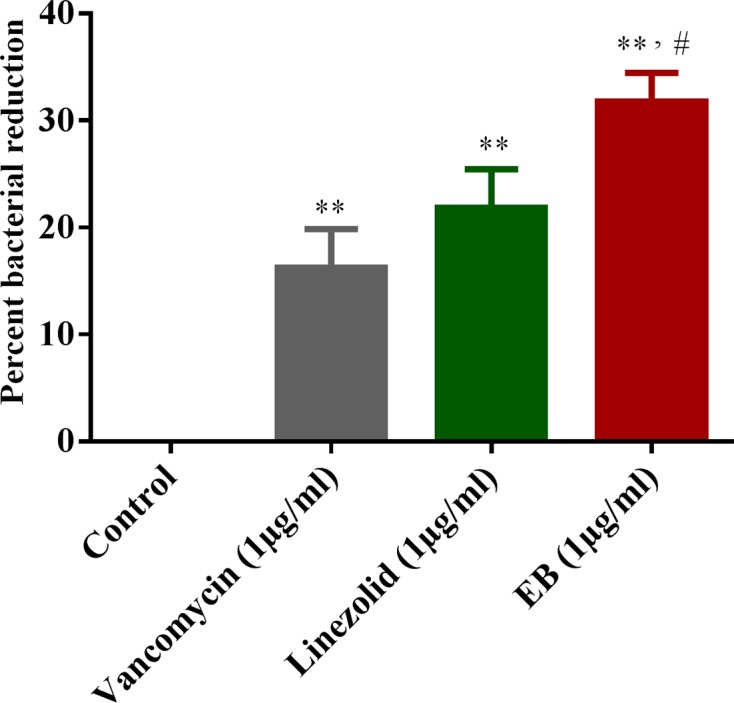
MRSA infected J774A.1 cells were treated with EB and control antibiotics (vancomycin and linezolid) for 24 h and the percent bacterial reduction was calculated compared to untreated control groups. The results are given as means ± SD (n = 3). P values of (**, # ≤ 0.05) are considered as significant. EB was compared to controls (**) and to antibiotics (#).
Fig 2. Cytotoxicity assay in murine macrophage-like cells (J774A.1) cells.
J774A.1 cells were treated with different concentration of EB ranging from 0 to 256 μg/ml. DMSO was used as a negative control. Cell viability was measured by MTS assay and IC50 of EB to cause cytotoxicity in J774A.1 cells was calculated.
Cell-free bacterial transcription/translation assay
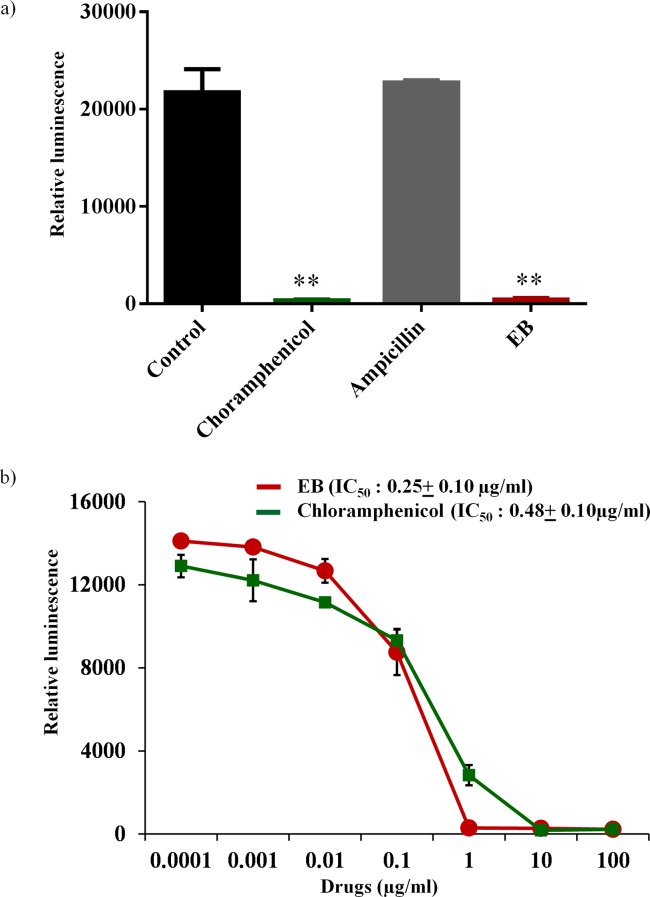
Antimicrobials that target microbial protein synthesis such as oxazolidinones and lincomycins are considered excellent choices for the treatment of toxin-mediated bacterial infections caused by S. aureus, such as toxic shock syndrome (TSS) and pneumonia [30–33]. In addition to the suppression of S. aureus toxins such as Panton-Valentine leucocidin (PVL), α-hemolysin (hla), and toxic shock syndrome toxin–1 (TSST-1), these antimicrobials also reduce excessive host-inflammatory responses associated with these toxins [34, 35]. Hence, protein synthesis inhibitors are often preferred in clinical practice for the treatment of toxin-associated staphylococcal infections [30–33]. We tested the effects of EB in our study on bacterial, mammalian and mitochondrial protein-synthesis. For bacterial protein-synthesis inhibition, we used E. coli cellular extracts in a transcription and translation assay that monitors protein production via luciferase readout. Unlike the antibiotic ampicillin that inhibits cell wall synthesis, EB strongly inhibited bacterial transcription/translation process similar to chloramphenicol antibiotic that inhibits protein synthesis (Fig 3A). EB inhibited bacterial protein synthesis in the cell-free transcription-translation, exhibiting IC50 of 0.25±0.10 μg/ml which is comparable to IC50 of chloramphenicol antibiotic 0.48 ± 0.10 μg/ml (Fig 3B). These results indicate that EB acts by a favorable mechanism of action and inhibits bacterial protein synthesis and, most likely, toxin production. However, inhibition of bacterial protein synthesis does not exclude other possible mechanism of action of EB.
Fig 3. Effects of EB on coupled transcription-translation (TT) in S30 extracts from E. coli.
(a) Average luciferin protein production in the presence of EB, ampicillin and chloramphenicol at the concentration of 2 μg/ml were shown. The results are given as means ± SD (n = 3). (b) Concentration dependent TT-inhibition of EB and chloramphenicol were shown. IC50 of the drugs required to inhibit 50% TT-activity were determined. P values of (** ≤ 0.05) are considered as significant.
Cell-free mammalian transcription/translation assay and mitochondrial biogenesis
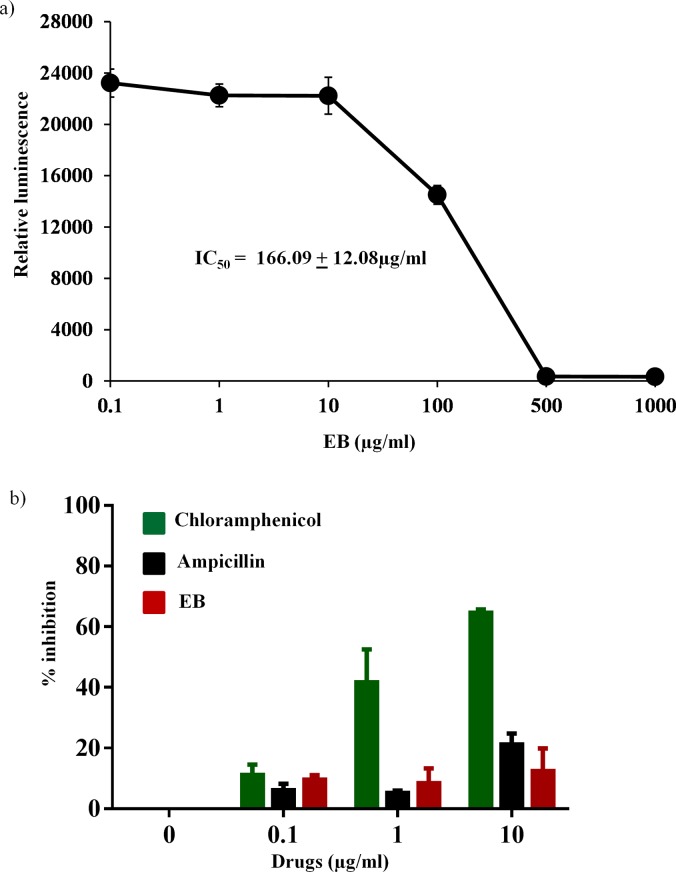
Due to concern about the possible mitochondrial toxicities associated with many antibacterial protein synthesis inhibitors such as linezolid and chloramphenicol [36–42], we tested the effect of EB on the inhibition of eukaryotic transcription/translation process using the rabbit reticulocyte lysate system with the cellular components necessary for mammalian protein synthesis [43, 44]. As shown in Fig 4A, EB showed high safety profile with IC50 of mammalian protein synthesis of 166.09 ± 12.08 μg/ml. This value is more than 660-fold higher than the concentration required to inhibit protein synthesis in bacteria. However, in order to test the effect of EB more specifically on mitochondrial biogenesis and to confirm the above in vitro results obtained from rabbit reticulocyte lysate system, we measured the effect of EB on mitochondrial protein synthesis directly within the mammalian cells. In-cell ELISA was performed in J774A.1 cells treated with EB and chloramphenicol for three days to detect the levels of mtDNA-encoded COX-I and nDNA-encoded SDH-A proteins. Results shown in Fig 4B indicate that EB had no significant inhibition (less than 10%) of mitobiogenesis, similar to the effect of ampicillin, which does not interfere with mitochondrial protein synthesis process. At the same time, chloramphenicol had more than 60% inhibition of mitochondrial protein synthesis. These results provide valuable information about EB’s safety profile and the lack of interference with mammalian protein synthesis and mitobiogenesis.
Fig 4. Effects of EB on mammalian protein synthesis.
(a) Concentration dependent inhibition of protein synthesis were determined using rabbit reticulocyte lysate extract system. IC50 of the EB required to inhibit 50% translational activity were determined. (b) Effect of EB, chloramphenicol and ampicillin on mitobiogenesis. J774A.1 cell In cell- ELISA was carried out in the presence and absence of these drugs, and the levels of mitochondrial (mt)-DNA encoded protein (COX-I) and nuclear-DNA encoded protein (SDH-A) were quantified. Ratio of COX-I and SDH-A were calculated and the results were shown as percent inhibition of mitochondrial biogenesis.
Efficacy of EB in infected animal model (C. elegans)
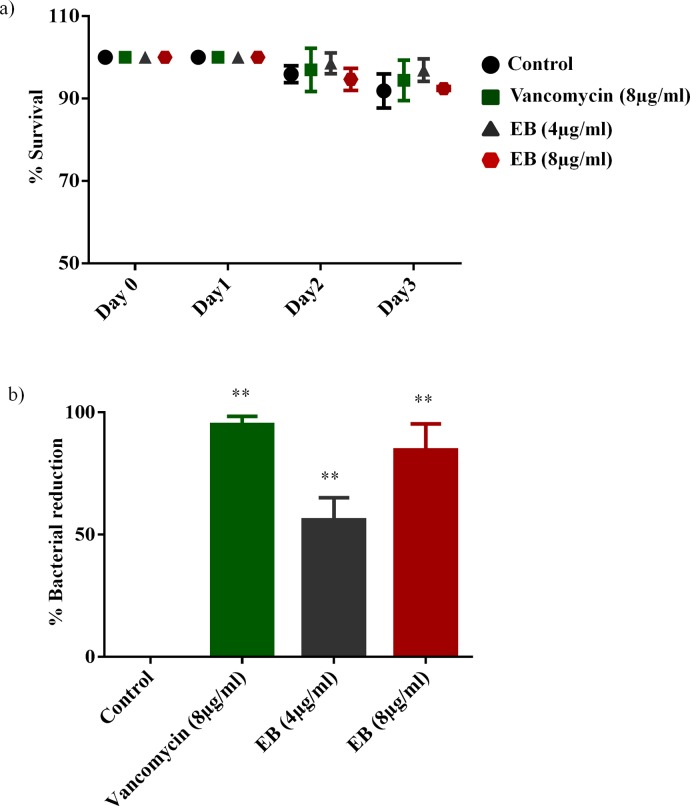
To investigate if the potent in vitro antimicrobial activity of EB translates to antimicrobial efficacy in vivo, we tested antimicrobial efficacy of EB in an infected C. elegans whole animal model. A whole animal model, such as C. elegans, represents a great platform for drug discovery and enables simultaneous assessment of efficacy and toxicity of the tested drugs. Additionally, using a C. elegans model reduces the associated cost of drug discovery and lowers the burden for extensive animal testing [13, 45]. Prior to testing the efficacy of treatment with EB in infected C. elegans, we tested toxicity of EB in non-infected C. elegans. As shown in Fig 5A, treatment of C. elegans with EB at 4 and 8 μg/ml for three days did not show any significant toxicity, similar to control groups. With no observable toxicity noticed in EB treated groups at a concentration of 4 and 8 μg/ml, we moved forward with an in vivo infection model using C. elegans infected with MRSA. As seen in Fig 5B, treatment with EB had a significant reduction in bacterial load when compared to untreated groups. EB at a concentration of 4 and 8 μg/ml significantly reduced the mean bacterial count by 56% and 85%, respectively. Moreover, treatment with EB at a concentration of 8 μg/ml showed comparable effect to treatment with the drug of choice vancomycin in reducing MRSA burden in infected C. elegans. Taken together, these results show that EB exhibits potent in vivo antistapylococcal efficacy in MRSA-infected C. elegans.
Fig 5. Evaluation of toxicity and antimicrobial efficacy of EB in C. elegans model.
(a) C. elegans strain glp-4; sek-1 (L4-stage) were grown for three days in the presence of EB (4μg and 8 μg/ml) and vancomycin (8 μg/ml). Live worms were counted and the results were expressed as percent live worms in relative to the untreated control groups. (b) MRSA USA300 infected L4-stage worms were treated with EB (4μg and 8 μg/ml) and vancomycin (8 μg/ml) for 24 h. Worms were lysed and the CFU were counted and the percent bacterial reduction per worm in treated groups were calculated in relative to the untreated control groups. P values of (** ≤ 0.05) are considered as significant.
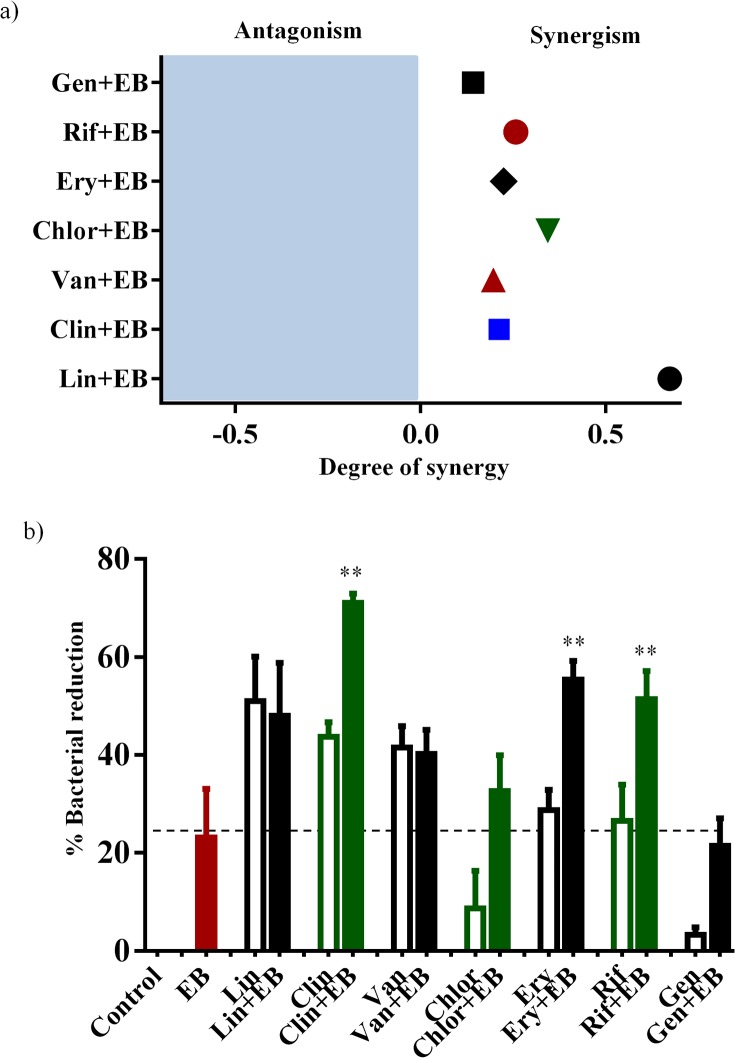
Synergistic activities of EB with conventional antibiotics in vitro and in cell culture
After confirming that EB has a potential use as an antibacterial agent for the treatment of infections caused by multidrug resistant pathogens, it was important to explore the synergistic relationship of EB with conventional antibiotics in vitro and in cell culture. With the rapid emergence of multidrug-resistant strains of S. aureus, monotherapy with single antibiotic has become less effective [46, 47]. Therefore, alternative strategies such as combinational therapy have been used in the healthcare setting to improve the morbidity associated with MRSA infections and to reduce the likelihood of emergence of resistant strains [1, 46, 48, 49]. To ascertain whether EB has the potential to be combined in vitro and in cell culture with conventional antimicrobials such as linezolid, clindamycin, vancomycin, chloramphenicol, erythromycin, rifampicin, and gentamicin against MRSA USA300, we used the in vitro Bliss independence model of synergism and infected cell culture assay [15]. In vitro results from the Bliss independence model of synergism are presented in Fig 6A. EB was found to exhibit a synergistic relationship with all tested conventional antimicrobials in vitro against MRSA USA300. Results of synergistic relationship of EB with conventional antimicrobials in infected cell culture against intracellular MRSA USA300 are presented in Fig 6B. Conventional antimicrobials (clindamycin, erythromycin, and rifampicin) showed synergistic activity when combined with EB and significantly reduced intracellular MRSA when compared to monotherapy. However, EB did not show synergistic activity with linezolid, vancomycin, chloramphenicol, or gentamicin in clearing intracellular MRSA. Identifying antibiotics that can be synergistically paired with EB can potentially prolong the clinical utility of these antibiotics and reduce the likelihood of emergence of resistant strains.
Fig 6. Synergistic activities of EB with conventional antibiotics in vitro and in cell culture.
(a) The Bliss Model for Synergy confirms the in vitro synergism with conventional antimicrobials (gentamicin, rifampicin, erythromycin, chloramphenicol, vancomycin, clindamycin and linezolid) against MRSA USA300. Degree of synergy was calculated in the presence of EB (0.0312 μg/ml) in combination with sub-inhibitory concentrations of conventional antimicrobials. (b) Synergistic activity of EB with conventional antimicrobials in infected cell culture. Efficacy of EB (0.5μg/ml) in combination with linezolid (4μg/ml), clindamycin (1μg/ml), vancomycin (4μg/ml), chloramphenicol (4μg/ml), erythromycin (8μg/ml), rifampicin (0.5μg/ml) and gentamicin (1μg/ml) in clearing intracellular MRSA USA300 was determined in J774A.1 cells. Percent bacterial reduction was calculated in relative to the untreated groups. The results are given as means ± SD (n = 3). Combination therapy was compared to monotherapy and the P values of (**, ≤ 0.05) are considered as significant.
In conclusion, we have successfully explored the potential applications of EB in vitro, in cell culture, and in vivo to combat multidrug-resistant Gram-positive pathogens, especially MRSA. We demonstrated that EB inhibits the bacterial translation process without affecting mitochondrial biogenesis. Additionally, we demonstrated the efficacy of EB in vivo in a C. elegans MRSA-infected model. Finally, we identified potential antibiotics that can be synergistically combined with EB to prolong the clinical utility of these antibiotics and reduce the likelihood of the emergence of resistant strains. Taken together, our study results demonstrate that EB, with its potent antimicrobial activity and safety profiles, might be a potential candidate drug for systemic and (or) topical applications to treat multidrug resistant Gram-positive bacterial infections alone or in combination with other antibiotics and should therefore be further clinically evaluated.
Acknowledgments
The authors would like to thank the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH Contract # HHSN272200700055C and BEI resources, NIAID, NIH for providing bacterial strains used in this study.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Mohammad H, Mayhoub AS, Cushman M, Seleem MN. Anti-biofilm activity and synergism of novel thiazole compounds with glycopeptide antibiotics against multidrug-resistant Staphylococci. The Journal of antibiotics. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thangamani S, Mohammad H, Younis W, Seleem MN. Drug Repurposing for the Treatment of Staphylococcal Infections. Current pharmaceutical design. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Younis W, Thangamani S, Seleem MN. Repurposing Non-antimicrobial Drugs and Clinical Molecules to Treat Bacterial Infections. Current pharmaceutical design. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thangamani S, Younis W, Seleem MN. Repurposing ebselen for treatment of multidrug-resistant staphylococcal infections. Scientific reports. 2015;5:11596 10.1038/srep11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306(5699):1138–9. [DOI] [PubMed] [Google Scholar]
- 6. Schewe T. Molecular actions of ebselen—an antiinflammatory antioxidant. General pharmacology. 1995;26(6):1153–69. [DOI] [PubMed] [Google Scholar]
- 7. Nozawa R, Yokota T, Fujimoto T. Susceptibility of methicillin-resistant Staphylococcus aureus to the selenium-containing compound 2-phenyl-1,2-benzoisoselenazol-3(2H)-one (PZ51). Antimicrobial agents and chemotherapy. 1989;33(8):1388–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J, Vlamis-Gardikas A, Kandasamy K, Zhao R, Gustafsson TN, Engstrand L, et al. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2013;27(4):1394–403. [DOI] [PubMed] [Google Scholar]
- 9. Favrot L, Grzegorzewicz AE, Lajiness DH, Marvin RK, Boucau J, Isailovic D, et al. Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nature communications. 2013;4:2748 10.1038/ncomms3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan G, Hardej D, Santoro M, Lau-Cam C, Billack B. Evaluation of the antimicrobial activity of ebselen: role of the yeast plasma membrane H+-ATPase. Journal of biochemical and molecular toxicology. 2007;21(5):252–64. [DOI] [PubMed] [Google Scholar]
- 11. CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A7 CLSI, Wayne, PA: 2007. [Google Scholar]
- 12. Seral C, Van Bambeke F, Tulkens PM. Quantitative analysis of gentamicin, azithromycin, telithromycin, ciprofloxacin, moxifloxacin, and oritavancin (LY333328) activities against intracellular Staphylococcus aureus in mouse J774 macrophages. Antimicrob Agents Chemother. 2003;47(7):2283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alajlouni RA, Seleem MN. Targeting listeria monocytogenes rpoA and rpoD genes using peptide nucleic acids. Nucleic acid therapeutics. 2013;23(5):363–7. 10.1089/nat.2013.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen DJ, Newman E, Iqbal S, Chang RY, Potmesil M, Ryan T, et al. Postoperative intraperitoneal 5-fluoro-2'-deoxyuridine added to chemoradiation in patients curatively resected (R0) for locally advanced gastric and gastroesophageal junction adenocarcinoma. Annals of surgical oncology. 2012;19(2):478–85. 10.1245/s10434-011-1940-8 [DOI] [PubMed] [Google Scholar]
- 15. Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. Silver enhances antibiotic activity against gram-negative bacteria. Science translational medicine. 2013;5(190):190ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochimica et biophysica acta. 2009;1794(5):808–16. 10.1016/j.bbapap.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakae T. The problems in the outer membrane permeability and the antibiotic resistance of Pseudomonas aeruginosa. The Kitasato archives of experimental medicine. 1991;64(2–3):115–21. [PubMed] [Google Scholar]
- 18. Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clinical microbiology reviews. 2015;28(2):337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aeschlimann JR. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy. 2003;23(7):916–24. [DOI] [PubMed] [Google Scholar]
- 20. Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. The Journal of antimicrobial chemotherapy. 2009;64(3):441–6. 10.1093/jac/dkp241 [DOI] [PubMed] [Google Scholar]
- 21. Garzoni C, Kelley WL. Staphylococcus aureus: new evidence for intracellular persistence. Trends in microbiology. 2009;17(2):59–65. 10.1016/j.tim.2008.11.005 [DOI] [PubMed] [Google Scholar]
- 22. Ellington JK, Harris M, Webb L, Smith B, Smith T, Tan K, et al. Intracellular Staphylococcus aureus. A mechanism for the indolence of osteomyelitis. The Journal of bone and joint surgery British volume. 2003;85(6):918–21. [PubMed] [Google Scholar]
- 23. Fowler VG Jr., Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, et al. Staphylococcus aureus endocarditis: a consequence of medical progress. Jama. 2005;293(24):3012–21. [DOI] [PubMed] [Google Scholar]
- 24. Ellington JK, Harris M, Hudson MC, Vishin S, Webb LX, Sherertz R. Intracellular Staphylococcus aureus and antibiotic resistance: implications for treatment of staphylococcal osteomyelitis. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2006;24(1):87–93. [DOI] [PubMed] [Google Scholar]
- 25. Seleem MN, Jain N, Pothayee N, Ranjan A, Riffle JS, Sriranganathan N. Targeting Brucella melitensis with polymeric nanoparticles containing streptomycin and doxycycline. FEMS microbiology letters. 2009;294(1):24–31. 10.1111/j.1574-6968.2009.01530.x [DOI] [PubMed] [Google Scholar]
- 26. Seleem MN, Munusamy P, Ranjan A, Alqublan H, Pickrell G, Sriranganathan N. Silica-antibiotic hybrid nanoparticles for targeting intracellular pathogens. Antimicrobial agents and chemotherapy. 2009;53(10):4270–4. 10.1128/AAC.00815-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soofi MA, Seleem MN. Targeting essential genes in Salmonella enterica serovar typhimurium with antisense peptide nucleic acid. Antimicrobial agents and chemotherapy. 2012;56(12):6407–9. 10.1128/AAC.01437-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cruciani M, Gatti G, Lazzarini L, Furlan G, Broccali G, Malena M, et al. Penetration of vancomycin into human lung tissue. J Antimicrob Chemoth. 1996;38(5):865–9. [DOI] [PubMed] [Google Scholar]
- 29. Peltola H, Paakkonen M, Kallio P, Kallio MJT. Bad Bugs, No Drugs: No ESCAPE Revisited Reply. Clinical Infectious Diseases. 2009;49(6):993–. [DOI] [PubMed] [Google Scholar]
- 30. Stevens DL, Ma YS, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195(2):202–11. [DOI] [PubMed] [Google Scholar]
- 31. Diep BA, Afasizheva A, Le HN, Kajikawa O, Matute-Bello G, Tkaczyk C, et al. Effects of Linezolid on Suppressing In Vivo Production of Staphylococcal Toxins and Improving Survival Outcomes in a Rabbit Model of Methicillin-Resistant Staphylococcus aureus Necrotizing Pneumonia. J Infect Dis. 2013;208(1):75–82. 10.1093/infdis/jit129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otto MP, Martin E, Badiou C, Lebrun S, Bes M, Vandenesch F, et al. Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J Antimicrob Chemoth. 2013;68(7):1524–32. [DOI] [PubMed] [Google Scholar]
- 33. Karau MJ, Tilahun AY, Schmidt SM, Clark CR, Patel R, Rajagopalan G. Linezolid Is Superior to Vancomycin in Experimental Pneumonia Caused by Superantigen-Producing Staphylococcus aureus in HLA Class II Transgenic Mice. Antimicrobial agents and chemotherapy. 2012;56(10):5401–5. 10.1128/AAC.01080-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nau R, Eiffert H. Modulation of release of proinflammatory bacterial compounds by antibacterials: potential impact on course of inflammation and outcome in sepsis and meningitis. Clinical microbiology reviews. 2002;15(1):95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diep BA, Equils O, Huang DB, Gladue R. Linezolid effects on bacterial toxin production and host immune response: review of the evidence. Current therapeutic research, clinical and experimental. 2012;73(3):86–102. 10.1016/j.curtheres.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrobial agents and chemotherapy. 2006;50(6):2042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gerson SL, Kaplan SL, Bruss JB, Le V, Arellano FM, Hafkin B, et al. Hematologic effects of linezolid: Summary of clinical experience. Antimicrobial agents and chemotherapy. 2002;46(8):2723–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuter DJ, Tillotson GS. Hematologic effects of antimicrobials: Focus on the oxazolidinone linezolid. Pharmacotherapy. 2001;21(8):1010–3. [DOI] [PubMed] [Google Scholar]
- 39. McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrobial agents and chemotherapy. 2006;50(6):2042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manyan DR, Arimura GK, Yunis AA. Chloramphenicol-induced erythroid suppression and bone marrow ferrochelatase activity in dogs. The Journal of laboratory and clinical medicine. 1972;79(1):137–44. [PubMed] [Google Scholar]
- 41. Parashar S, Rao R, Tikare SK, Tikare SS. Chloramphenicol induced reversible bone marrow suppression. A case report. Journal of postgraduate medicine. 1972;18(2):90–2. [PubMed] [Google Scholar]
- 42. Yunis AA. Chloramphenicol-induced bone marrow suppression. Seminars in hematology. 1973;10(3):225–34. [PubMed] [Google Scholar]
- 43. Skripkin E, McConnell TS, DeVito J, Lawrence L, Ippolito JA, Duffy EM, et al. R chi-01, a new family of oxazolidinones that overcome ribosome-based linezolid resistance. Antimicrobial agents and chemotherapy. 2008;52(10):3550–7. 10.1128/AAC.01193-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan K, Madden L, Choudhry AE, Voigt CS, Copeland RA, Gontarek RR. Biochemical characterization of the interactions of the novel pleuromutilin derivative retapamulin with bacterial ribosomes. Antimicrobial agents and chemotherapy. 2006;50(11):3875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajamuthiah R, Fuchs BB, Conery AL, Kim W, Jayamani E, Kwon B, et al. Repurposing Salicylanilide Anthelmintic Drugs to Combat Drug Resistant Staphylococcus aureus. PloS one. 2015;10(4):e0124595 10.1371/journal.pone.0124595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deresinski S. Vancomycin in combination with other antibiotics for the treatment of serious methicillin-resistant Staphylococcus aureus infections. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009;49(7):1072–9. [DOI] [PubMed] [Google Scholar]
- 47. Appelbaum PC. Microbiology of antibiotic resistance in Staphylococcus aureus. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2007;45 Suppl 3:S165–70. [DOI] [PubMed] [Google Scholar]
- 48. Huang V, Rybak MJ. Pharmacodynamics of cefepime alone and in combination with various antimicrobials against methicillin-resistant Staphylococcus aureus in an in vitro pharmacodynamic infection model. Antimicrobial agents and chemotherapy. 2005;49(1):302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drago L, De Vecchi E, Nicola L, Gismondo MR. In vitro evaluation of antibiotics' combinations for empirical therapy of suspected methicillin resistant Staphylococcus aureus severe respiratory infections. BMC infectious diseases. 2007;7:111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.