Abstract
The aggregation of human amylin has been strongly implicated in the progression of Type II diabetes. This 37-residue peptide forms a variety of secondary structures, including random coils, α-helices, and β-hairpins. The balance between these structures depends on the chemical environment, making amylin an ideal candidate to examine inherent biases in force fields. Rat amylin differs from human amylin by only 6 residues; however, it does not form fibrils. Therefore it provides a useful complement to human amylin in studies of the key events along the aggregation pathway. In this work, the free energy of rat and human amylin was determined as a function of α-helix and β-hairpin content for the Gromos96 53a6, OPLS-AA/L, CHARMM22/CMAP, CHARMM22*, Amberff99sb*-ILDN, and Amberff03w force fields using advanced sampling techniques, specifically bias exchange metadynamics. This work represents a first systematic attempt to evaluate the conformations and the corresponding free energy of a large, clinically relevant disordered peptide in solution across force fields. The NMR chemical shifts of rIAPP were calculated for each of the force fields using their respective free energy maps, allowing us to quantitatively assess their predictions. We show that the predicted distribution of secondary structures is sensitive to the choice of force-field: Gromos53a6 is biased towards β-hairpins, while CHARMM22/CMAP predicts structures that are overly α-helical. OPLS-AA/L favors disordered structures. Amberff99sb*-ILDN, AmberFF03w and CHARMM22* provide the balance between secondary structures that is most consistent with available experimental data. In contrast to previous reports, our findings suggest that the equilibrium conformations of human and rat amylin are remarkably similar, but that subtle differences arise in transient alpha-helical and beta-strand containing structures that the human peptide can more readily adopt. We hypothesize that these transient states enable dynamic pathways that facilitate the formation of aggregates and, eventually, amyloid fibrils.
Introduction
Molecular dynamics (MD) and Monte Carlo (MC) simulations [1] are widely used to study the structure of polypeptides. A number of advanced sampling techniques have been proposed to improve the efficiency of simulations. Examples include thermodynamic integration [2], umbrella sampling [3], parallel tempering [4], metadynamics [5,6], bias exchange metadynamics [7], and flux-tempered metadynamics [8].
Recently, these techniques have been applied to the study of protein folding for systems such as insulin [9], amylin [10–20], and amyloid fibrils [21]. The force field used to describe intermolecular interactions determines peptide structure. However, available force fields often display inherent biases in the secondary structures, as summarized in Table 1.
Table 1. Reported biases for selected force fields.
| Name | Water Model | Year | α-helix Bias? | β Bias? | Other Notes |
|---|---|---|---|---|---|
| Amberff99 [22] | TIP3P [23] | 2000 | High [24,25] | Low [26] | |
| Amberff99sb [27] | TIP3P | 2006 | Low [28,29] | Low [30] | Models ILDN residues poorly [31] |
| Amberff99sb-ILDN [31] | TIP3P | 2010 | Low [29] | ||
| Amberff03 [32] | TIP3P | 2003 | High [28,29,33,34] | Low [35] | |
| Amberff03* [29] | TIP3P | 2009 | High [28] | Low [35] | |
| Amberff03w [36] | TIP4P/2005 [37] | 2010 | Struggled with Ubiquitin [38] | ||
| CHARMM22 [39] | TIPS3P [23] | 1998 | High [28,34] | Low [26] | |
| CHARMM27 [40,41] | TIPS3P, TIP4P | 2004 | High [28,42,43] | Low [30] | |
| Gromos53a6 [44] | SPC [45] | 2004 | Low [33] | ||
| OPLS-AA/L [46] | TIP4P | 2001 | Low [26,33,43] | High [43]/Low [30] | Favors Disordered states [30,43] |
A value of low for α-helix bias indicates that the force field failed to form the experimentally expected α-helically structure or failed to remain stable when experiments suggested that an α-helix was stable. A high value for α-helix bias indicates that the force field formed an α-helix when experimental evidence suggests that an α-helix is disfavored. Similarly, β-Bias indicates biases related to forming parallel and anti-parallel β-sheets.
These biases can cause a secondary structure to misfold, despite experimental evidence for its stability [26,28,30,33,43,47,48]. Likewise, biases may prevent simulations from sampling experimentally relevant secondary structures with the correct probability.
Several recent studies have investigated force field biases, usually by comparison with experimental NMR data [35,38,49]. Such studies have been largely limited to folded proteins, and have not attempted to calculate the free energy of the molecule. Studies of disordered polypeptides have been scarce. Long molecular dynamics or parallel tempering simulations are often used to identify metastable states. However, both techniques suffer from significant drawbacks. Large free energy barriers can prevent sampling of metastable states and, for molecular dynamics simulations and umbrella sampling, the resulting free energy estimates can exhibit large errors. Molecular dynamics simulations only explore those regions of low free energy that are accessible on the time scale of a simulation, making difficult a comparison of free energies corresponding to secondary structures from different regions.
Bias exchange simulations facilitate quantitative comparison of free energy minima corresponding to different secondary structures [7]. Large free energy barriers can be overcome, thereby enabling sampling of favorable and unfavorable states. It is based on the metadynamics method [5], which, like most density-of-states based sampling techniques, relies on history-dependent biases to overcome barriers [50]. The choice of order parameter is essential; if slow degrees of freedom are left out, systematic biases in the free energy can arise [51]. Due to the complex dynamics of proteins, choosing a small set of order parameters to study secondary structure is difficult [15]. Each order parameter can be sampled in a different simulation box and, for metadynamics simulations, the time to explore the free energy surface increases exponentially with the number of order parameters that are incorporated in a calculation. Bias exchange simulations allow the use of many simultaneous order parameters. These help each other overcome barriers in free energy along order parameters that are orthogonal to the one being sampled in a distinct simulation box or window. For non-periodic order parameters, such as the α-helical content of a peptide, errors can accumulate near the boundaries [52]; such errors, however, can be avoided by modifying the shape of the history dependent potential according to methods that we have recently developed [53].
Amylin is an important test case for the evaluation of force fields, as it is known to adopt random coils, β-hairpins, and α-helices [54]. It has been strongly implicated in the development of Type II Diabetes Mellitus (T2DM) [55]. In over 90% of patients with type II diabetes [56–59], amyloid fibrils have been found in the β-cells of the Islets of Langerhans [60–63]. These fibrils are composed primarily of islet amyloid polypeptide [61,62] (known also as amylin or IAPP) in the form of primarily β-sheets [64–67]. The formation of these fibrils in the β-cells of the Islets of Langerhans cause membrane disruption [68,69] and cell death [70,71]. Furthermore, the presence of negatively charged membranes has been shown to accelerate the aggregation and membrane disruption processes [72,73]. The disruption of the membrane occurs prior to the formation of the mature fibrils [74]. Indeed, mature fibrils are of limited cytotoxicity compared to the early oligomers [69], suggesting that the early aggregates are the cytotoxic species.
Several sources suggest a causal relation between IAPP and Type II Diabetes. While humans, cats, and dogs readily develop the disease and form aggregates, species such as mice and rats do not typically form aggregates or display diabetes like symptoms [75]. However, when human IAPP (referred to as hIAPP) is transgenically expressed in mice and rats, fibrils are formed and symptoms typical of T2DM become manifest [76–78]. Furthermore, the addition of solutions of hIAPP oligomers and fibrils to healthy Pancreatic β-cells quickly results in cell death [69,71]. Finally, a mutation in the hIAPP gene has been linked to the early onset of T2DM and quicker rates of fibrillization [79,80].
Because of the rapid rate of aggregation, the detailed structures of hIAPP aggregates in their early stages are not fully understood. The structure of the monomer in solution has been studied by circular dichroism (CD) [54,64–66,81,82], molecular simulations [10–20], 2D-IR [83], and NMR [84]. It is believed to be mostly a random coil, with all residues above the cysteine bridge transiently adopting α-helical structures. Small sections adopt a β-hairpin structure. Residues 2–5 are held in a β-loop due to a cysteine bond between residues 2 and 7. CD [73], NMR [85,86] and EPR [87] studies show that hIAPP adopts an α-helical conformation as it binds to the phospholipid micelles as a monomer [88] in the head group region [89]. The spectra of this conformation decreases inversely with the increase of fluorescently labeled fibrillar structures and the formation of β-sheet structures [66]. However, the detailed structures of the oligomers and toxic intermediates are presently unknown.
Rat islet amyloid polypeptide (or rat amylin or rIAPP) provides a useful counterpart to human amylin. Despite differing by only six residues (Table 2), rat amylin does not form fibrils and, as mentioned earlier, rats do not display symptoms of T2DM [90]. Like hIAPP, rIAPP is predominantly random coil in solution [54,82,91], and transiently adopts an α-helical structure between residues 5–23. Upon binding to a negatively charged lipid micelle or bilayer, it forms an α-helix whose structure has been solved via NMR [92], Unlike human amylin, rIAPP does not form β-hairpins. This is thought to be due to the presence of three prolines, which disrupt the secondary structure in the C-terminus [93].
Table 2. Sequence and charges used for the simulation of rat and human amylin.
| Residue Number | 1 5 10 15 20 25 30 35 |
| hIAPP Sequence | KCNTATCATQRLANFLVHSSNNFGAILSSTNVGSNTY-NH2 |
| Charge Used | ++ + |
| rIAPP Sequence | KCNTATCATQRLANFLVRSSNNLGPVLPPTNVGSNTY-NH2 |
| Charge Used | ++ + + |
Differences in sequence are bolded.
Extensive experimental information is available for rat and human amylin in solution, including IR spectra, NMR chemical shifts, and CD data. Results from simulations can be compared quantitatively to experiments, thereby providing a stringent test of available force fields. While amylin is predominantly random coil in solution, it also transiently samples α-helical states and small β-turns and hairpins. Therefore, in contrast to larger proteins where these three structural elements are stable, the free energy differences between distinct states are small and differ from the typical funnel view of the free energy of protein secondary structures. Bias exchange simulations allow us to sample a variety of random coils, α-helices, and β-hairpins. Furthermore, we can determine the free energy differences between clusters of these structures. In this work, we evaluate the ability of Amberff99sb*-ILDN [29,31,34,35], Amberff03w [36], CHARMM22/CMAP [40,41], CHARMM22* [34], Gromos96 53a6 [44], and OPLS-AA/L [46] to describe the structure of rat and human amylin in solution.
Materials and Methods
Simulation Parameters
Molecular dynamics simulations were run using the Gromacs 4.6.2 package [94]. The structures predicted from NMR by the Ramamaoorthy group for human [86] and rat [92] amylin on a micelle were used as the starting points for bias exchange simulations, which can quickly explore other, different configurations. In both human and rat amylin, the C-terminus is amidated. Only the +3 charge version of human amylin was simulated here. A disulfide bond is present between residues 2 and 7. The peptide was placed in the center of a 8.5 nm dodecahedral box and solvated with water. The force field and water model combinations considered for rat and human amylin are shown in Table 3. The water model used to parameterize each force field is chosen for each force field and water combination. In addition, three additional combinations were chosen to evaluate the effect nonstandard force field and water pairings can have: Amberff99sb*-ILDN with TIP4P, Amberff03w with TIP4P, and CHARMM22* with TIP4P. For hIAPP, Amberff03w with TIP4P was omitted because Amberff03w was optimized with TIP4P2005, not TIP4P. CHARMM22* with TIPS3P was also omitted since the TIP4P water model better predicted the NMR shifts for rIAPP.
Table 3. Force field and water combinations used to simulate human and rat amylin.
| Name | Year of Publication | Water Models Used |
|---|---|---|
| Amberff99sb*-ILDN | 2011 | TIP3P, TIP4P |
| Amberff03w | 2010 | TIP4P, TIP4P2005 |
| CHARMM22/CMAP | 2004 | TIPS3P |
| CHARMM22* | 2011 | TIPS3P, TIP4P |
| Gromos96 53a6 | 2004 | SPC |
| OPLS-AA/L | 2001 | TIP4P |
Four Cl- were used to neutralize rIAPP, and three Cl- were used to neutralize hIAPP. The energy was minimized first by using the steepest descent algorithm for 500 steps and a step size of 0.01 nm without constraints, and then for 10000 steps with all bonds constrained. The system was then equilibrated for 100 ps at 310 K at constant volume, and then for 10 ns at 310 K and a constant pressure of 1.0 bar. The velocity-rescaling temperature coupling [95] and Parrinello-Rahman [96,97] barostat were used for the equilibration simulations. The temperature was coupled separately for the solvent and protein. A 2 fs time step was used. Center of mass motion was removed every 10 ns. Electrostatic interactions were handled using a particle mesh Ewald method [98,99] (PME), with a rcoloumb value of 0.9 nm, an order of 4, 0.12 nm spacing, a tolerance of 1e-5, and 3D geometry. A long range dispersion correction was applied for both energy and pressure. Bonds involving hydrogen were constrained during the simulation using the LINCS [100] algorithm.
Bias Exchange Simulations
Bias exchange metadynamics simulations [7] were run for 600 ns for all force fields except Amberff99sb*-ILDN/TIP3P for rat, which was run for 1000 ns in order to confirm convergence. A Fourier spacing of 0.33 was used for PME. The Nose-Hoover [101,102] algorithm was used for temperature control, with unit chain length. Protein and non-protein atoms were coupled separately. LINCS [100] was used to constrain the bonds involving hydrogen. The system was run in the canonical ensemble from the configuration obtained at the end of the NPT equilibration runs described above. The αRMSD and anti-parallel βRMSD [103] order parameters have been shown to allow efficient sampling of α-helical and β-sheet structures. A bias exchange simulation was run with four replicas: one unbiased box, a box biasing along the αRMSD order parameter, a box biasing along βRMSD, and one box biasing on both αRMSD and βRMSD. A Gaussian hill of height 0.1 kJ/mol and standard deviation 2.0 was deposited every 1 ps. The parameters used for the switching function in the RMSD collective variables were: r0 = 0.8, n = 8, and m = 12. The proteins were aligned as a single molecule at every step and periodic boundary conditions were then disabled for the subsequent RMSD calculations. Preliminary simulations showed that, without a boundary correction, significant artifacts accumulated near values of 0 RMSD. Here we emphasize that a standard invert method [52] is unable to correct for errors around 0 αRMSD and 0 βRMSD, and such errors can be significant. To circumvent this problem, we used the boundary correction modification of McGovern et al. [53] to account for errors near the boundaries. A lower and upper boundary of 0 and 32 were used for both αRMSD and βRMSD for this method. After 200ns, a grid was implemented between collective variable values of -5 and 42 and with 2351 bins. A wall on the maximum distance between the backbone nitrogen of the third residue and the backbone carboxyl C on the 35th residue was placed at 7.0 nm, with a kappa value of 100.0 kJ/mol. The backbone atoms of the protein used for RMSD were aligned. The boundary correction version [53] of PLUMED 1.3 [104] was used for simulations without a grid. This was further modified to support the use of a grid.
In order to calculate the free energy versus αRMSD and βRMSD, the bias applied to the box sampling both αRMSD and βRMSD was averaged from the beginning of the bias exchange simulations using custom scripts. The free energy was calculated in a 160 by 160 grid of αRMSD and βRMSD points ranging from 0 to 32 in each dimension.
Molecular Dynamics Simulations
For comparison with the bias exchange results, molecular dynamics simulations with the Amberff03w and Gromos96 53a6 force fields were run with rat amylin for 500 ns. The starting points were configurations found during the bias exchange simulation, including local free energy minima for the α-helix, β-hairpin, random coil, as well as other, unstable structures. Five starting configurations were used for Gromos96 53a6/SPC, and four for Amberff03w/TIP4P2005. The βRMSD was then plotted as a function of αRMSD and compared to the figures of the free energy vs. αRMSD and βRMSD.
NMR Calculations
Structures were taken every 10 ps from each of the bias exchange simulations for rat amylin. The NMR chemical shifts were calculated for the Hα, CO, Cα, HN, and Cβ atoms using the SPARTA+ program [105]. The default random coil values from the SPARTA+ program were used. The average free energy of each bin in the free energy surface was calculated by averaging the NMR shifts of all structures belonging to that bin. The predicted secondary shift for each atom was then found by calculating the weighted average of the average secondary shift for each bin multiplied by the probability corresponding to its free energy. These results are compared to those from Williamson and Miranker [106] for amylin at 278 K, with the secondary shifts calculated by subtracting the random coil shifts from SPARTA+ from the shifts from in the Biological Magnetic Resonance Database under accession number 7311. The secondary shifts for HN were calculated at 310 K using the experimental temperature coefficients reported by these authors; other temperature coefficients, however, are not available and we therefore can only compare our results to data at 278 K. In the case of HN, we were able to compare the errors between the predicted shifts and the experimental measurements at 310 K and 278 K, respectively. We found that the same force fields that performed well with the NMR values uncorrected for temperature perform well with the corrected values. Therefore, the experimental shifts at 278 K are used for Cα, Cβ, Hα, and N to compare with simulation predictions.
To ascertain the effect of temperature on our calculations, we conducted additional simulations with Amberff99sb*-ILDN with TIP3P, Amberff03w with TIP4P/2005, and CHARMM22* with TIP4P at T = 280, 290, 300, 310, and 320 K. The purpose of these additional calculations was to determine the average change in Cα shifts that one might expect in the range of temperatures encompassing the experimental measurements and at a physiological temperature. Five independent simulations were performed at each of these temperatures, starting from configurations belonging to local minima identified using bias exchange metadynamics. These simulations were conducted in the NPT ensemble with the same parameters used above in the equilibration procedure. After generating a 5 ns trajectory for each configuration, the average Cα shift over a 1 ns period was calculated for every residue at each of these temperatures. The mean differences between the average Cα shifts (averaged over all 37 residues and over five independent runs) at 310 K and at each of the other four temperatures T mentioned above are denoted by μ(T), and are shown in S1 Table.
A two-sided t-test was performed on the resulting data for all residues and all runs at a given temperature, with the null-hypothesis that the difference was 0. For all t-tests, we failed to find a statistically significant difference at a confidence level of 0.05. These results are shown in S1 Table. In summary, the results of our additional calculations indicate that there is no statistically significant difference between the shifts computed at 310 K and those observed at 280K. These results are consistent with the experimental observations of Soong et al., whose measurements of the circular dichroism signal, which depends on the secondary structure of rIAPP, are relatively unchanged between 273–313 K [107].
Secondary Structure Calculations
The secondary structure of each frame was calculated using DSSP 2.04 [108,109]. For each element of secondary structure calculated by DSSP (for example, α-helix, β-sheet, β-bridge, etc.), the average fraction of structures corresponding to a given αRMSD and βRMSD bin was calculated. For each residue, the average fraction of structures containing a secondary structure element was calculated by weighting the average secondary structure fraction of that bin by the probability corresponding to the free energy of that bin. The average fraction of peptide in a secondary structure was found by averaging the average fraction of a residue in a secondary structure over all residues. These were then summed into two groups: helix (α-helix, 310 Helix or π Helix) or strand (β-sheet or β-Bridge). This was performed because π and 310 helices are often misclassified as α-helices, and because in DSSP β-Bridges correspond to the same hydrogen bonding pattern as β-Sheets, but are shorter in length.
Results and Discussion
Free Energy Maps
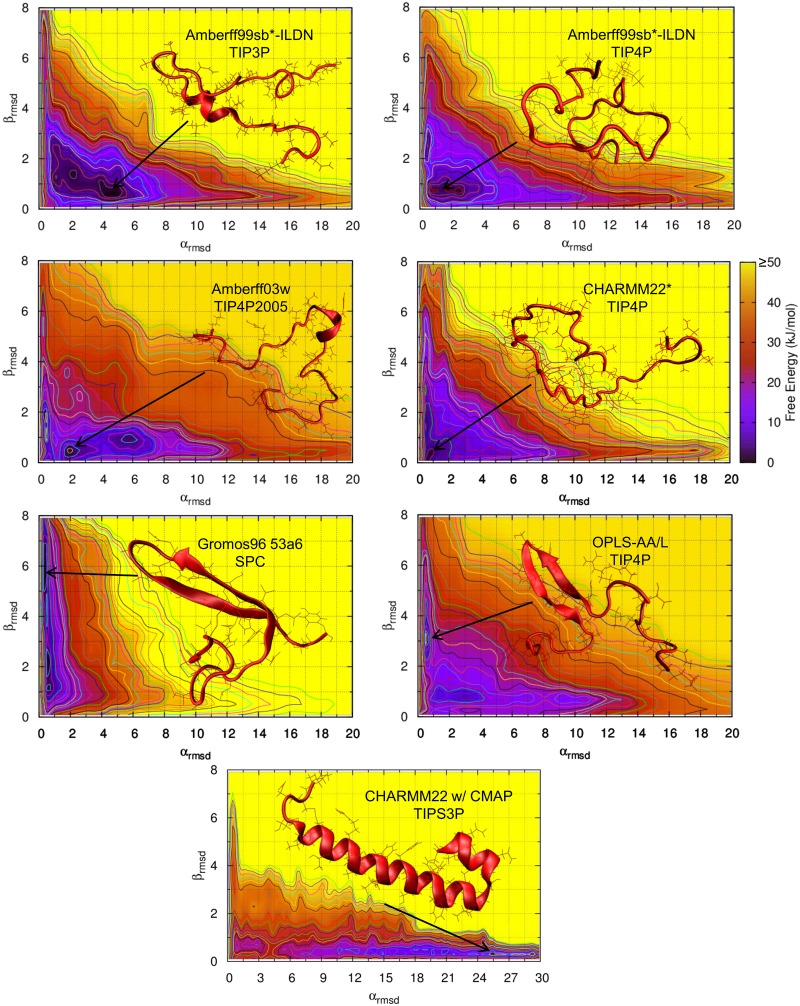
The free energy of rIAPP as a function of αRMSD and βRMSD is shown in Fig 1 for various combinations of force fields and water models. Amberff03w with TIP4P is shown in S1 Fig. Note that αRMSD and βRMSD are proportional to the number of residues in the respective secondary structure; a larger value of αRMSD indicates that a larger fraction of rIAPP is in an α-helical state. Once the αRMSD increases beyond approximately 3 units, α-helices exist in a majority of structures. Likewise, β-sheets exist in most structures with a βRMSD greater than approximately 2 units. The color of the plots provides the magnitude of the free energy in kJ/mol. Significant qualitative differences between many of the force fields considered here are immediately apparent. Most force fields favor a predominantly random coil, with the deepest local minima below 3 αRMSD and 2 βRMSD. However, CHARMM22/CMAP heavily favors α-helical structures, with a deep minimum around 16 RMSD. In contrast, Gromos96 53a6 favors β-hairpin structures: its deepest minimum occurs around 0.5 αRMSD and 2.5 βRMSD. The Amberff99sb*-ILDN force fields predict predominantly random coil structures. However, the α-helical and β-hairpin structures are not completely disfavored, and significant fractions of amylin exist in both states. Amberff03w with TIP4P2005 is almost completely disordered. There are no local minima in the α-helical or β-hairpin states. CHARMM22* with TIPS3P exhibits a deep minimum at 1 αRMSD and 0.5 βRMSD, well within the random coil region. The OPLS-AA/L force field exhibits a qualitatively different free energy surface from those of the other force fields, with long α-helical and 2 β-hairpin structures being highly disfavored. It is, however, less α-helical and more heavily β-hairpin than Gromos96 53a6 with SPC. Importantly, these free energy differences are substantial and lead to qualitatively different conclusions regarding the pathways for amyloid formation.
Fig 1. Free energy of rat amylin as a function of αRMSD and βRMSD for various force fields.
The darker regions indicate regions of lower free energy.
The free energy surfaces of human amylin are shown in Fig 2. The Gromos96 53a6 force field predicts much more β-sheet character than the other force fields—much greater than that predicted for rat amylin. The other force fields follow trends similar to those seen above for rIAPP: the Amber and CHARMM22* force fields predict mostly random coil structures with a small amount of α-helical and β-hairpin structures. OPLS-AA/L is also mostly random coil and predicts more β-sheet structures compared with rIAPP. The free energy of the β-sheet structures is also lower than that observed in all other force fields except Gromos96 53a6.
Fig 2. Free energy of human amylin as a function of αRMSD and βRMSD for various force fields.
The darker regions indicate regions of lower free energy.
Molecular Dynamics Simulations
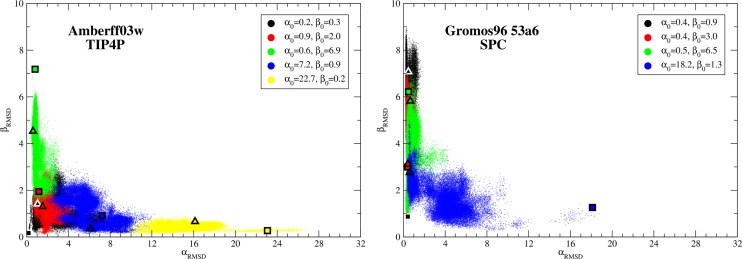
Molecular dynamics simulations were run for 500ns starting from several selected structures for Amberff03w with TIP4P and Gromos96 53a6 with SPC. The αRMSD and βRMSD are shown in Fig 3 every 10 ps. The starting configurations are shown as squares and the ending configurations as triangles. As can be seen in the figure, the two force fields behave qualitatively different. The simulations for Amberff03w retain greater amounts of α-helical character than Gromos96 53a6. For example, structures starting in the high α-helical state retain large amounts of α-helical character throughout the 500 ns for Amberff03w. In Gromos96 53a6, the α-helices quickly unravel. In contrast, the large β-hairpins are unstable in Amberff03w while in Gromos96 53a6 a large β-hairpin forms from an initially low βRMSD state.
Fig 3. αRMSD vs. βRMSD every 10 ps of a molecular dynamics simulation of rIAPP starting at different configurations.
The starting points are shown as squares while the ending points are shown as triangles. Each run was 500 ns long. The results for Amberff03w with TIP4P are shown on the left, while those for Gromos 96 53a6 are shown on the right.
These simulations are consistent with the free energy surfaces from bias exchange simulations discussed above. Amberff03w predicted a balance between α-helical and β-hairpin structures and indeed the molecular dynamics simulations drifted towards these free energy minima. In contrast, the Gromos96 53a6 simulations disfavor α-helical structures and prefer β-hairpin structures.
While the highly α-helical structure for Amberff03w remains in a high αRMSD state predicted to be relatively unstable, this is most likely due to the slow conformational dynamics and not an inherent preference of the protein under this force field. This simulation provides evidence that caution is needed when analyzing MD simulations of proteins, even when half-microsecond simulations are considered. In this example, the underlying free energy landscape allows the peptide to remain trapped in relatively unfavorable conformations for at least 500 ns.
Secondary Structures Predicted
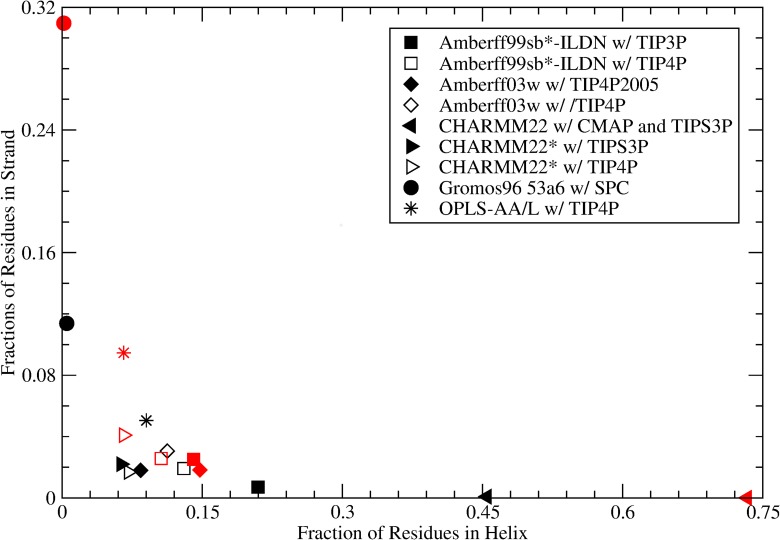
The free energy surfaces were used to calculate the fraction of amino acids in a helix (α-helix, 310 helix, or π-helix) or strand (β-sheet or β-bridge) as described above in the Methods section. The results at 310 K are shown in Fig 4 and span a range of helix and strand fractions. Depending on the force field, amylin can be found to adopt a helical structure with no β-sheet character, or a mostly random coil state with varying amounts of β-sheet structures. The force field which predicts the largest helical content for amylin is CHARMM22/CMAP with TIPS3P. Approximately 47% of amylin’s residues are predicted to be in an α-helix, 310 helix, or π-helix structure. This is not surprising, as this force field has been previously noted to exhibit a strong α-helical bias [28,42,43]. The force field predicting the second highest helical content is Amberff99sb*-ILDN with TIP3P (21%). Gromos96 53a6 predicts less than 0.5% helical content and the highest strand content, namely 11%. The next highest prediction of β-sheet content is by the OPLS-AA/L force field with TIP4P, which yields 9% helical content and 5% strand content.
Fig 4. Fraction of structures with a helix (α-helix, 310 Helix or π Helix) or a strand (β-sheet or β-Bridge) for rIAPP (black symbols) and hIAPP (red symbols) as predicted with various force fields.
Force field and solvent model combinations where the force field was optimized with that solvent model are shown with a filled symbol; otherwise an unfilled symbol is shown.
The difference in structural propensities between human and rat amylin predicted depends on the force field chosen. For Gromos96 53a6, for example, the fraction of residues in a β-sheet or β-bridge conformation is 0.31 for human versus 0.11 for rat. OPLS-AA/L predicts a change of this fraction of residues in a strand conformation from 0.095 in hIAPP to 0.051 in rIAPP, while CHARMM22* with TIP4P predicts the fraction changing from 0.041 to 0.017 for human and rat respectively. In contrast, Amberff03w’s strand content is 0.018 for both hIAPP and rIAPP, but the helical fraction is 0.15 in hIAPP compared to 0.084 in rIAPP. The amount of helical content is almost identical to that observed by Miller et al. [12] in replica exchange simulations with the same force field and water model (0.15 for hIAPP, and 0.086 for rIAPP). However, Miller et al. predicts more strand content in rIAPP and hIAPP (0.053 for rIAPP and 0.038 for hIAPP). The small differences in secondary structure content may be the result of differences in temperature, the amidation state of the C-terminus, or salt concentration. For most other force fields, the amount of helical character in human amylin is smaller than the amount of strand character, and the difference in the predicted number of residues in an α-helical or β-hairpin character between hIAPP and rIAPP depends on force field.
The predicted strand and helix fractions of rat amylin for different water models are also shown in Fig 4. Three non-standard water models were investigated: Amberff99sb*-ILDN w/ TIP3P and TIP4P, Amberff03w with TIP4P and TIP4P2005, and CHARMM22* with TIPS3P and TIP4P. The corresponding free energies relative to the coil state are shown in Table 4. For Amberff99sb*-ILDN, the use of TIP4P instead of the recommended TIP3P results in an increase in the free energy of the helix state for the average residue of 0.6 kT relative to the coil structure. The free energy of the strand fraction decreased by 0.9 kT. These differences correspond to a change in fraction of residues in a helix and strand of -0.08 and 0.01 respectively. For Amberff03w, the free energy of the helix states and strand states decreased by 0.4 and 0.6 kT, corresponding to increases in fraction of residues of 0.03 and 0.01, respectively. In CHARMM22*, the use of TIP4P instead of TIP3P changed the free energies by -0.1 and 0.2 kT, altering the fractions of helix and strand by +0.01 and -0.005, respectively. Amberff99sb*-ILDN exhibits the most dramatic free energy change, where the fraction of helix decreases by almost half, bringing its predictions for helix into rough agreement with those of Ambeff03w, CHARMM22*, and OPLS-AA/L. Its strand content also increases to 0.02, in agreement with these force fields. In contrast CHARMM22*’s secondary structure composition remains roughly unaltered. This suggests that different water models may be used with relatively little ill effect on CHARMM22*, as already suggested by Bjelkmar et al. [41]. Amberff03w also exhibits a slight decrease in strand and helix content, making it very similar to CHARMM22*’s predictions.
Table 4. Free energy of secondary structure appearing in a residue in rIAPP relative to the coil secondary structure in units of kT at 310 K.
| Force Field | Water Model | Helix | Strand | Turn | Bend |
|---|---|---|---|---|---|
| Amberff99sb*-ILDN | TIP3P | 0.52 | 3.91 | 0.47 | 0.53 |
| Amberff99sb*-ILDN | TIP4P | 1.12 | 3.03 | 0.51 | 0.63 |
| Amberff03w | TIP4P2005 | 1.72 | 3.26 | 1.24 | 0.47 |
| Amberff03w | TIP4P | 1.36 | 2.67 | 1.00 | 0.54 |
| CHARMM22/CMAP | TIPS3P | -0.47 | 5.84 | 1.08 | 0.56 |
| CHARMM22* | TIPS3P | 2.12 | 3.17 | 1.39 | 0.70 |
| CHARMM22* | TIP4P | 2.00 | 3.43 | 1.38 | 0.71 |
| Gromos53a6 | SPC | 4.66 | 1.53 | 1.91 | 0.64 |
| OPLS-AA/L | TIP4P | 1.57 | 2.15 | 1.09 | 0.42 |
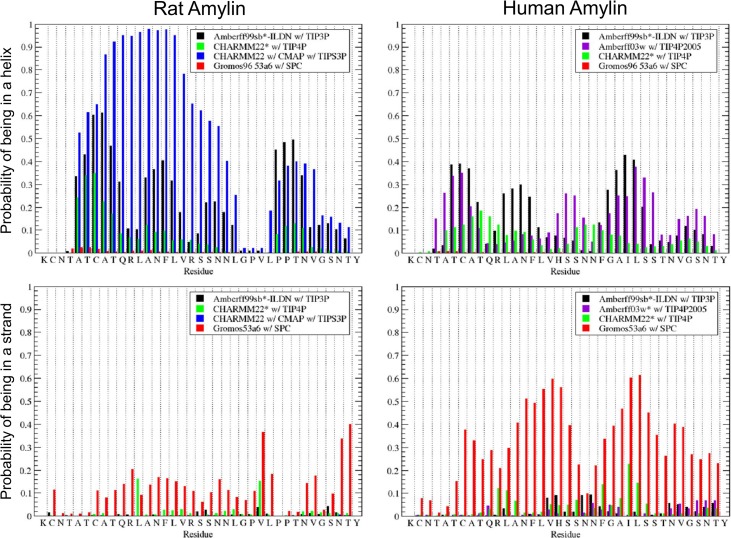
These overall helix and strand propensities are broken down by residue in Fig 5. CHARMM22/CMAP exhibits the most helical behavior. This is concentrated between residues 5–23 as observed in experiments and secondary structure prediction algorithms. For this force field, these residues are usually in an α-helix. This α-helix is broken by glycine at residue 24. Another helix occasionally forms in the tail between residues 27 and 36. This is predicted to be present roughly 30% of the time by CHARMM22/CMAP. Amberff99sb*-ILDN is the next most helical force field. It follows the same broad trends with intermittent helices between residues 5–23 and 27–36. These are followed by Amberff03w with TIP4P2005 and CHARMM22* with TIPS3P. CHARMM22* predicts a helix more frequently between residues 27–36 than between 7–23.
Fig 5. Predicted fraction of time each residue is in a helix (α-helix, 310 Helix or π Helix) or strand (β-sheet or β-Bridge).
The predicted fractions for rat are shown on the left for the Amberff99sb*-ILDN with TIP3P, CHARMM22* with TIP4P, CHARMM22/CMAP with TIPS3P, and Gromos96 53a6 with SPC force fields. On the right, the predicted fractions of human amylin are shown for Amberff99sb*-ILDN with TIP3P, Amberff03w with TIP4P, CHARMM22* with TIP4P, and Gromos96 53a6 with SPC.
Gromos96 53a6 with SPC has the largest strand propensities by residue. These are relatively evenly distributed between residues 7–36, with a break at the double prolines at residues 28 and 29. In general, the strand is predicted to be much less frequent than the helix. Amberff99sb*-ILDN with TIP3P has very little strand propensity throughout. Amberff03w and CHARMM22* have roughly equal strand propensities.
Convergence
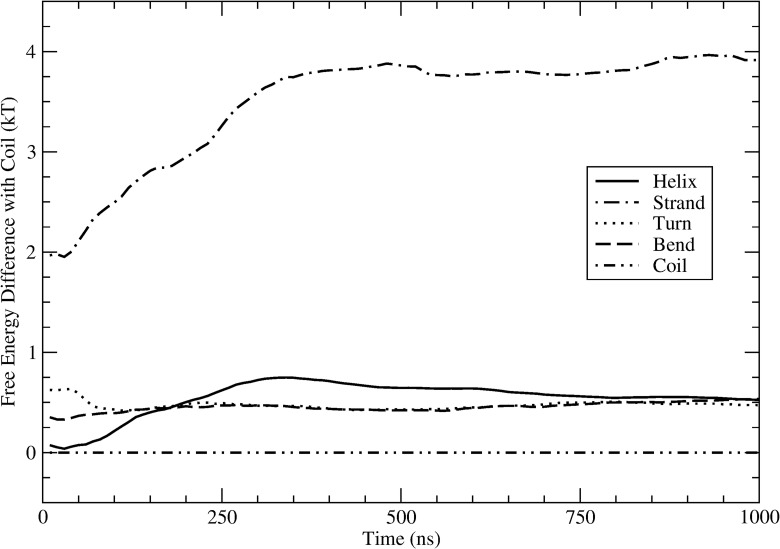
Fig 6 shows the free energy of rIAPP in a helix, strand, turn, and bend (relative to the coil) versus time for Amberff99sb*-ILDN with TIP3P. The free energy of the bend and turn states converges within approximately 100 ns, reaching a value that is 0.5 kT greater than the coil state. The helix and strand states take longer to converge. The difference in free energy between the helix and coil states starts small but grows to a maximum of 0.75 kT before slowly decreasing. At the end of the simulation the difference is 0.53 kT. The free energy difference between strand and coil increases rapidly until about 350 ns when it reaches a value of 3.8 kT. It then fluctuates around this value for the rest of the simulation, finishing at 3.9 kT. While the fraction of helix and possibly strand may not have completely converged even after 1000 ns, as evidenced by the small increase in fraction of helix, the differences are relatively unchanged after 350 ns. When these free energy differences are converted to fractions of the structure that are in a helix or strand versus time, they become even smaller and remain within 0.05 of the value at 100 ns. These fractions are the important weights when determining properties of the molecule, such as NMR shifts, hydrogen bonding, and chemical reaction rates. Small changes of less than 0.05 fraction of these structures will not greatly influence the results. Furthermore, these changes are small compared to the large differences between the force fields. The convergence plots for the other force fields are shown in S2 and S3 Figs for rIAPP and hIAPP respectively.
Fig 6. Free energy difference with coil for fraction of residues in a secondary structure predicted for rIAPP vs. time for Amberff99sb*-ILDN with TIP3P.
The secondary structure of each residue was determined using DSSP.
NMR Secondary Shifts
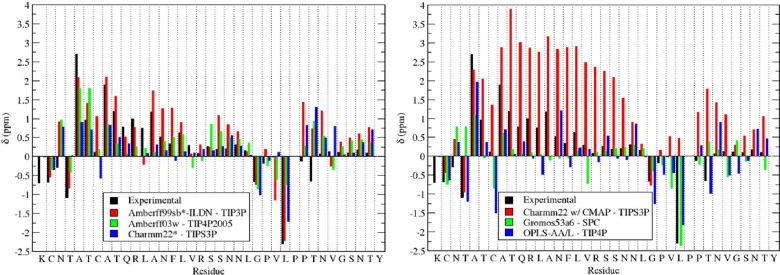
The NMR secondary shifts were computed for amylin and compared with the experimental results from Williamson and Miranker [106]. These were calculated for the Cα, Cβ, Hα, HN, and N atoms. The random coil shifts from SPARTA+ [110] were used for these calculations. These random coil shifts were subtracted from the chemical shifts obtained by Williamson and Miranker in order to compare secondary shifts calculated using the same random coil shifts. The Cα secondary shifts provide a measure of the secondary structure propensity of the residue; a value greater than 0 suggests a propensity towards helical structures; a value of 0 indicates a random coil structure, and values less than 0 correspond to β-sheet structures. The Cα secondary shifts are shown in Fig 7 for several force field and water combinations. Amberff99sb*-ILDN with TIP3P qualitatively reproduces the major trends in the chemical shift pattern. Residues 5–25 are up-shifted, corresponding to the increased α-helical propensity. It also captures the down field secondary shifts around the glycine and first proline. The agreement with the experimental results is the poorest at the C-terminus. The values predicted are higher than the experimental chemical shifts, corresponding to a slight propensity in the simulations to form a small α-helix at the C-terminus. Most experimental evidence suggests that this region is predominantly unstructured. Because random coil regions involve a large number of low energy states their sampling is challenging and prone to statistical uncertainties, which may explain the differences between the predicted and experimental results. Fig 7 shows a similar result for Amberff03w with TIP4P2005 and CHARMM22* with TIPS3P: the discrepancy between the predicted and experimental NMR results is greatest in the disordered C-terminus of rIAPP.
Fig 7. Predicted Cα secondary chemical shifts for Amberff99sb*-ILDN with TIP3P, Amberff03w with TIP4P2005, CHARMM22* with TIPS3P, CHARMM22/CMAP with TIPS3P, Gromos96 53a6 with SPC, and OPLS-AA/L with TIP4P.
The predictions were made using SPARTA+.
The second half of the figure shows the shifts for CHARMM22/CMAP with TIPS3P, Gromos53a6 with SPC, and OPLS-AA/L with TIP4P. CHARMM22/CMAP with TIPS3P is drastically upshifted of the experimental results as a result of its high helix propensity. In contrast, Gromos96 53a6, which does not predict significant amounts of α-helical character, consistently underestimates the Cα coefficients at the N-terminus. OPLS-AA/L with TIP4P is the most downfield shifted and fails to reproduce the NMR shifts at both the N-terminus and C-terminus.
The average error for the predicted NMR shifts is shown in Table 5 in units of ppm / residue. They were calculated using the following formula:
where n is the number of residues, and δi,pred and δi,ex are the predicted and experimental shifts for the ith residue respectively. The best performing force fields are Ambeff99sb*-ILDN, Amberff03w, and CHARMM22*. For Amberff99sb*-ILDN, the secondary shifts with TIP4P are better than with the recommended water model TIP3P. This may correspond to the lower overall α-helical content of this model. With Amberff03w, the C secondary shifts are much lower with TIP4P2005 than with TIP4P, suggesting that the distinction between the two water models may be important. CHARMM22* with TIP4P produces the lowest errors for the C and H chemical shifts, even compared to the recommended water model TIPS3P. This agrees with the suggestion by the authors of the Gromacs implementation of the CHARMM22/CMAP force field that TIP4P may be a preferable alternative to TIPS3P [41]. Surprisingly, Gromos96 53a6 has the smallest error for the backbone nitrogen atoms, while also showing large errors for the carbon and hydrogen secondary shifts.
Table 5. Average error in the NMR secondary shifts (ppm/residue) predicted for rIAPP for different force field and water models at 310 K.
| Force Field | Water Model | Cα | Cβ | Hα | HN | N |
|---|---|---|---|---|---|---|
| Amberff99sb*-ILDN | TIP3P | 0.52 | 0.42 | 0.10 | 0.12 | 2.64 |
| Amberff99sb*-ILDN | TIP4P | 0.47 | 0.39 | 0.10 | 0.09 | 2.34 |
| Amberff03w | TIP4P | 0.62 | 0.44 | 0.11 | 0.12 | 1.48 |
| Amberff03w | TIP4P2005 | 0.51 | 0.38 | 0.11 | 0.14 | 1.44 |
| CHARMM22/CMAP | TIPS3P | 1.24 | 0.49 | 0.09 | 0.10 | 2.43 |
| CHARMM22* | TIPS3P | 0.51 | 0.40 | 0.12 | 0.13 | 1.20 |
| CHARMM22* | TIP4P | 0.38 | 0.36 | 0.10 | 0.09 | 1.38 |
| Gromos53a6 | SPC | 0.55 | 0.50 | 0.16 | 0.24 | 0.99 |
| OPLS-AA/L | TIP4P | 0.54 | 0.45 | 0.14 | 0.10 | 2.09 |
The smallest percent error is in bold for each secondary shift.
Comparison of Human and Rat Amylin
As summarized in Fig 4, the fraction of amylin predicted to adopt a helical or strand conformation varies appreciably across force fields. Most force fields predict that both human and rat adopt a predominantly random coil structure in solution, consistent with experiments. CHARMM22* with TIP4P, the force field that most accurately predicted the NMR shifts, predicts that the strand content is twice as large in human versus rat amylin, though it remains below 5%. For Amberff03w with TIP4P2005, rat and human amylin exhibit approximately equal fractions of strand character, consistent with the results of Miller et al. [12]. All other force fields predict modest increases in the amount of strand character. However, except for Gromos96 53a6, the strand content remains a small fraction of the overall secondary structure of the peptide.
For several key residues, however, the fraction of time spent in a helical or strand state differs between rat and human amylin, as seen in Fig 5. In rat amylin, little helical content is predicted between residues 24–26. Despite varying estimates of the overall amount of α-helical character between force fields, this decrease in helical propensity is consistently observed across all force fields examined here, even for CHARMM22/CMAP with TIP3SP. This is consistent with the tendency of proline to cap α-helices [111], and with NMR data indicating that the N-terminus α-helix ends with Leucine-23 [92]. In contrast, the helical propensity does not decrease sharply at these residues in human amylin. The aggregation of α-helices has been proposed to be an important intermediate in the formation of oligomers [112]. The ability of human amylin to form α-helices throughout the peptide could increase its propensity to form oligomers composed of helices. Larger helices could also be involved in oligomeric states along the pathway to the formation of amyloid fibrils in hIAPP; rIAPP may not be able to form these intermediate states composed of helices past residue 23.
Similarly, as shown in Fig 5, the double proline at residues 28 and 29 disrupts the β-sheet and β-bridge character for rat amylin, consistent with studies showing that prolines disrupt β-sheets [111]. While the strand content of rIAPP is generally low, no β-sheets are ever observed involving these prolines. However, for hIAPP, strand character is observed across these residues. Gromos96 53a6, with the largest prediction of β-sheet character, provides the most clear observation of this trend. In Amberff99sb*-ILDN with TIP3P or CHARMM22* with TIP4P, hIAPP occasionally forms β-sheets involving these residues, in contrast to rIAPP where no strand is ever observed involving residues 28 and 29. The ability to form β-sheets in this region agrees with experimental [75] and computational [20] evidence indicating that fibril formation is greatly inhibited when the peptide contains a proline at residue 25, 28, and/or 29.
The absence of this decrease in strand propensity is reflected in the formation of larger β-hairpins in hIAPP compared to rIAPP. Fig 8 shows the free energy versus βRMSD of rIAPP and hIAPP for Amberff99sb*-ILDN with TIP3P, Amberff03w with TIP4P2005, and CHARMM22* with TIP4P. When the free energy along the αRMSD coordinate is integrated out, a noticeable trend appears. In all three cases, when the βRMSD is large, as in the case of a large hairpin, the free energy of hIAPP is less than that of rIAPP. We hypothesize that the ability of human amylin to form α-helices and β-hairpins across residues 24–29 allows the formation of a much more diverse set of potential transition structures, which could be responsible for the propensity of hIAPP to form fibrils. Indeed, for both the 2-fold [113] and 3-fold [93] models for amylin, large β-sheets are present. Furthermore, residues 28 and 29, which had no observed strand content in rIAPP but occasionally formed β-sheets for hIAPP, are predicted to be part of the β-strands experimentally observed in hIAPP. These residues are in the β-sheet region of conformations identified as intermediates in the formation of amyloid fibrils [114]. The formation of a β-hairpin involving these atoms has also been identified as part of a transition from the α-helical to β-hairpin conformations of hIAPP [115].
Fig 8. Helmholtz free energy in kT versus βRMSD for rat and human amylin.
The βRMSD is correlated with the number of residues in a β-hairpin. The results for rIAPP are shown using solid lines, while the results for hIAPP are given in dashed lines. The Helmholtz free energy is shown for Amberff99sb*-ILDN with TIP3P (black), Amberff03w with TIP4P2005 (red), and CHARMM22* with TIP4P (blue).
Conclusions
The free energy of amylin was calculated as a function of αRMSD and βRMSD and used to determine the fraction of rat and human amylin in a helix or strand for various force field and water combinations. These results show that the choice of force field greatly influences the fraction of helix and strand, even for recent force fields such as Amberff99sb*-ILDN and CHARMM22*. Older force fields such as CHARMM22/CMAP and Gromos96 53a6 have strong biases towards α-helices and β-hairpins respectively. More elaborate calculations concerning the formation of dimers and oligomers are likely to be affected as well. Depending on the biases of the force field being used, the favorability of certain fibril precursors is likely influenced by the force field choice. CHARMM22* with TIP4P predicted the experimental rIAPP NMR secondary shifts most accurately. There was no general trend in the change of overall fraction of helix and strand between rat and human amylin across the different force fields. While Gromos96 53a6 predicts a large increase in β-hairpin character, CHARMM22* with TIP4P predicts only a small increase. Amberff03w with TIP4P2005, however, predicts a slight increase in helical character. Despite these differences, all force fields agree that the prolines in rIAPP cap helices in the C-terminus and disrupt the β-hairpins.
Taken together, the results of simulations of amylin across force fields present a picture in which the human and rat versions of the peptide are intrinsically disordered in solution, with only subtle structural differences between them. The human peptide, however, can more readily form transient α helices and β strands, which in the rat version are disrupted by the prolines at residues 25, 28 and 29. We hypothesize that these transient states confer to the human peptide a greater propensity to adopt β sheet structures that are stabilized by the presence of other peptides in aggregates or fibrils.
More generally, the study presented here represents a first systematic attempt to examine the structure and free energy of large disordered peptides across multiple force fields. Our results demonstrate that, for amylin, the preference towards α-helices, β-hairpins, or random coils is sensitive to the force field chosen; because of its intrinsically disordered nature, accurate force fields are essential in order to make predictions of secondary structure. In light of these findings, past simulations involving amylin (including our own) should be re-examined in the context of the force field chosen, especially when the resulting predictions align with given force field’s biases. Furthermore, the same issues could arise for other amyloid forming disordered peptides. A second note of caution is that the secondary structure of amylin was observed to change slowly. For example, in Gromos96 53a6, helices predicted to be unfavorable, remained relatively stable even for 500 ns. In light of this and other evidence that protein folding can take on the order of microseconds [116], the stability of a structure over a few hundred nanoseconds does not necessarily indicate that it corresponds to equilibrium.
Supporting Information
The corresponding p-values are shown. Using the standard deviation of the means, and a power of 0.95, the predicted difference in ppm between the two temperatures that could be resolved is shown as Δ.
(DOCX)
The darker regions indicate regions of lower free energy.
(TIFF)
The secondary structure of each residue was determined using DSSP.
(TIFF)
The secondary structure of each residue was determined using DSSP.
(TIFF)
Acknowledgments
We thank Sadanand Singh for many helpful discussions concerning the preparation of these simulations.
Data Availability
The data are available at: doi:10.7910/DVN/29368.
Funding Statement
This work is supported by the National Science Foundation, Division of Engineering, Chemical, Bioengineering, Environmental and Thermal Systems, CBET 1264021. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Frenkel D, Smit B. Understanding Molecular Simulation, Second Edition: From Algorithms to Applications. 2nd ed Academic Press; 2001. [Google Scholar]
- 2. Carter EA, Ciccotti G, Hynes JT, Kapral R. Constrained reaction coordinate dynamics for the simulation of rare events. Chemical Physics Letters. 1989;156: 472–477. 10.1016/S0009-2614(89)87314-2 [DOI] [Google Scholar]
- 3. Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. Journal of Computational Physics. 1977;23: 187–199. 10.1016/0021-9991(77)90121-8 [DOI] [Google Scholar]
- 4. Earl DJ, Deem MW. Parallel tempering: theory, applications, and new perspectives. Phys Chem Chem Phys. 2005;7: 3910–3916. [DOI] [PubMed] [Google Scholar]
- 5. Laio A, Parrinello M. Escaping free-energy minima. PNAS. 2002;99: 12562–12566. 10.1073/pnas.202427399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barducci A, Bussi G, Parrinello M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys Rev Lett. 2008;100: 020603 10.1103/PhysRevLett.100.020603 [DOI] [PubMed] [Google Scholar]
- 7. Piana S, Laio A. A Bias-Exchange Approach to Protein Folding. The Journal of Physical Chemistry B. 2007;111: 4553–4559. 10.1021/jp067873l [DOI] [PubMed] [Google Scholar]
- 8. Singh S, Chiu C, Pablo JJ de. Flux Tempered Metadynamics. J Stat Phys. 2011;145: 932–945. 10.1007/s10955-011-0301-0 [DOI] [Google Scholar]
- 9. Todorova N, Marinelli F, Piana S, Yarovsky I. Exploring the Folding Free Energy Landscape of Insulin Using Bias Exchange Metadynamics. J Phys Chem B. 2009;113: 3556–3564. 10.1021/jp809776v [DOI] [PubMed] [Google Scholar]
- 10. Reddy AS, Wang L, Lin Y-S, Ling Y, Chopra M, Zanni MT, et al. Solution Structures of Rat Amylin Peptide: Simulation, Theory, and Experiment. Biophysical Journal. 2010;98: 443–451. 10.1016/j.bpj.2009.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reddy AS, Wang L, Singh S, Ling YL, Buchanan L, Zanni MT, et al. Stable and Metastable States of Human Amylin in Solution. Biophysical Journal. 2010;99: 2208–2216. 10.1016/j.bpj.2010.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller C, Zerze GH, Mittal J. Molecular Simulations Indicate Marked Differences in the Structure of Amylin Mutants, Correlated with Known Aggregation Propensity. J Phys Chem B. 2013;117: 16066–16075. 10.1021/jp409755y [DOI] [PubMed] [Google Scholar]
- 13. Qi R, Luo Y, Ma B, Nussinov R, Wei G. Conformational Distribution and α-Helix to β-Sheet Transition of Human Amylin Fragment Dimer. Biomacromolecules. 2014;15: 122–131. 10.1021/bm401406e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andrews MN, Winter R. Comparing the structural properties of human and rat islet amyloid polypeptide by MD computer simulations. Biophysical Chemistry. 2011;156: 43–50. 10.1016/j.bpc.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 15. Murphy RD, Conlon J, Mansoor T, Luca S, Vaiana SM, Buchete N-V. Conformational dynamics of human IAPP monomers. Biophysical Chemistry. 2012;167: 1–7. 10.1016/j.bpc.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 16. Dupuis NF, Wu C, Shea J-E, Bowers MT. Human Islet Amyloid Polypeptide Monomers Form Ordered β-hairpins: A Possible Direct Amyloidogenic Precursor. Journal of the American Chemical Society. 2009;131: 18283–18292. 10.1021/ja903814q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dupuis NF, Wu C, Shea J-E, Bowers MT. The Amyloid Formation Mechanism in Human IAPP: Dimers Have β-Strand Monomer−Monomer Interfaces. J Am Chem Soc. 2011;133: 7240–7243. 10.1021/ja1081537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang G, Zhao J, Yu X, Zheng J. Comparative Molecular Dynamics Study of Human Islet Amyloid Polypeptide (IAPP) and Rat IAPP Oligomers. Biochemistry. 2013;52: 1089–1100. 10.1021/bi301525e [DOI] [PubMed] [Google Scholar]
- 19. Wu C, Shea J-E. Structural Similarities and Differences between Amyloidogenic and Non-Amyloidogenic Islet Amyloid Polypeptide (IAPP) Sequences and Implications for the Dual Physiological and Pathological Activities of These Peptides. PLoS Comput Biol. 2013;9: e1003211 10.1371/journal.pcbi.1003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiu C, Singh S, de Pablo JJ. Effect of Proline Mutations on the Monomer Conformations of Amylin. Biophysical Journal. 2013;105: 1227–1235. 10.1016/j.bpj.2013.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baftizadeh F, Biarnes X, Pietrucci F, Affinito F, Laio A. Multidimensional View of Amyloid Fibril Nucleation in Atomistic Detail. J Am Chem Soc. 2012;134: 3886–3894. 10.1021/ja210826a [DOI] [PubMed] [Google Scholar]
- 22. Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? Journal of Computational Chemistry. 2000;21: 1049–1074. [DOI] [Google Scholar]
- 23. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79: 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- 24. Okur A, Strockbine B, Hornak V, Simmerling C. Using PC clusters to evaluate the transferability of molecular mechanics force fields for proteins. Journal of Computational Chemistry. 2003;24: 21–31. 10.1002/jcc.10184 [DOI] [PubMed] [Google Scholar]
- 25. García AE, Sanbonmatsu KY. α-Helical stabilization by side chain shielding of backbone hydrogen bonds. PNAS. 2002;99: 2782–2787. 10.1073/pnas.042496899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoda T, Sugita Y, Okamoto Y. Comparisons of force fields for proteins by generalized-ensemble simulations. Chemical Physics Letters. 2004;386: 460–467. 10.1016/j.cplett.2004.01.078 [DOI] [Google Scholar]
- 27. Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics. 2006;65: 712–725. 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Best RB, Buchete N-V, Hummer G. Are Current Molecular Dynamics Force Fields too Helical? Biophysical Journal. 2008;95: L07–L09. 10.1529/biophysj.108.132696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Best RB, Hummer G. Optimized Molecular Dynamics Force Fields Applied to the Helix−Coil Transition of Polypeptides. J Phys Chem B. 2009;113: 9004–9015. 10.1021/jp901540t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cino EA, Choy W-Y, Karttunen M. Comparison of Secondary Structure Formation Using 10 Different Force Fields in Microsecond Molecular Dynamics Simulations. J Chem Theory Comput. 2012;8: 2725–2740. 10.1021/ct300323g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics. 2010;78: 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry. 2003;24: 1999–2012. 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- 33. Todorova N, Legge FS, Treutlein H, Yarovsky I. Systematic Comparison of Empirical Forcefields for Molecular Dynamic Simulation of Insulin. J Phys Chem B. 2008;112: 11137–11146. 10.1021/jp076825d [DOI] [PubMed] [Google Scholar]
- 34. Piana S, Lindorff-Larsen K, Shaw DE. How Robust Are Protein Folding Simulations with Respect to Force Field Parameterization? Biophysical Journal. 2011;100: L47–L49. 10.1016/j.bpj.2011.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. Systematic Validation of Protein Force Fields against Experimental Data. PLoS ONE. 2012;7: e32131 10.1371/journal.pone.0032131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Best RB, Mittal J. Protein Simulations with an Optimized Water Model: Cooperative Helix Formation and Temperature-Induced Unfolded State Collapse. J Phys Chem B. 2010;114: 14916–14923. 10.1021/jp108618d [DOI] [PubMed] [Google Scholar]
- 37. Abascal JLF, Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. The Journal of Chemical Physics. 2005;123: 234505–12. 10.1063/1.2121687 [DOI] [PubMed] [Google Scholar]
- 38. Beauchamp KA, Lin Y-S, Das R, Pande VS. Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements. J Chem Theory Comput. 2012;8: 1409–1414. 10.1021/ct2007814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacKerell, Bashford D, Bellott, Dunbrack, Evanseck JD, Field MJ, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins†. J Phys Chem B. 1998;102: 3586–3616. 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 40. Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of Computational Chemistry. 2004;25: 1400–1415. 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- 41. Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J Chem Theory Comput. 2010;6: 459–466. 10.1021/ct900549r [DOI] [PubMed] [Google Scholar]
- 42. Freddolino PL, Park S, Roux B, Schulten K. Force Field Bias in Protein Folding Simulations. Biophys J. 2009;96: 3772–3780. 10.1016/j.bpj.2009.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Patapati KK, Glykos NM. Three Force Fields’ Views of the 310 Helix. Biophysical Journal. 2011;101: 1766–1771. 10.1016/j.bpj.2011.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oostenbrink C, Villa A, Mark A, Van Gunsteren W. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6 RID A-1554-2010 RID A-8799-2011. J Comput Chem. 2004;25: 1656–1676. 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- 45. Berendsen HJC, Postma JPM, Van Gunsteren WF, Hermans J. Interaction models for Water in Relation to Protein Hydration, Intermolecular Forces. Intermolecular Forces. 1981;11: 331–338. [Google Scholar]
- 46. Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides†. J Phys Chem B. 2001;105: 6474–6487. 10.1021/jp003919d [DOI] [Google Scholar]
- 47. Lange OF, van der Spoel D, de Groot BL. Scrutinizing Molecular Mechanics Force Fields on the Submicrosecond Timescale with NMR Data. Biophysical Journal. 2010;99: 647–655. 10.1016/j.bpj.2010.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mittal J, Best RB. Tackling Force-Field Bias in Protein Folding Simulations: Folding of Villin HP35 and Pin WW Domains in Explicit Water. Biophysical Journal. 2010;99: L26–L28. 10.1016/j.bpj.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li D-W, Brüschweiler R. Certification of Molecular Dynamics Trajectories with NMR Chemical Shifts. J Phys Chem Lett. 2010;1: 246–248. 10.1021/jz9001345 [DOI] [Google Scholar]
- 50. Singh S, Chopra M, de Pablo JJ. Density of States—Based Molecular Simulations. Annual Review of Chemical and Biomolecular Engineering. 2012;3: 369–394. 10.1146/annurev-chembioeng-062011-081032 [DOI] [PubMed] [Google Scholar]
- 51. Branduardi D, Gervasio FL, Cavalli A, Recanatini M, Parrinello M. The Role of the Peripheral Anionic Site and Cation−π Interactions in the Ligand Penetration of the Human AChE Gorge. J Am Chem Soc. 2005;127: 9147–9155. 10.1021/ja0512780 [DOI] [PubMed] [Google Scholar]
- 52. Crespo Y, Marinelli F, Pietrucci F, Laio A. Metadynamics convergence law in a multidimensional system. Phys Rev E. 2010;81: 055701 10.1103/PhysRevE.81.055701 [DOI] [PubMed] [Google Scholar]
- 53. McGovern M, de Pablo J. A boundary correction algorithm for metadynamics in multiple dimensions. The Journal of Chemical Physics. 2013;139: 084102 10.1063/1.4818153 [DOI] [PubMed] [Google Scholar]
- 54. Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Müller SA, et al. Amyloid Fibril Formation from Full-Length and Fragments of Amylin. Journal of Structural Biology. 2000;130: 352–362. 10.1006/jsbi.2000.4268 [DOI] [PubMed] [Google Scholar]
- 55. Hull RL, Westermark GT, Westermark P, Kahn SE. Islet Amyloid: A Critical Entity in the Pathogenesis of Type 2 Diabetes. JCEM. 2004;89: 3629–3643. 10.1210/jc.2004-0405 [DOI] [PubMed] [Google Scholar]
- 56. Höppener J, Ahrén B, Lips C. Islet Amyloid and Type 2 Diabetes Mellitus. N Engl J Med. 343: 411–419. [DOI] [PubMed] [Google Scholar]
- 57. Westermark P. Quantitative studies on amyloid in the islets of Langerhans. Ups J Med Sci. 1972;77: 91–94. [DOI] [PubMed] [Google Scholar]
- 58. Clark A, Saad MF, Nezzer T, Uren C, Knowler WC, Bennett PH, et al. Islet amyloid polypeptide in diabetic and non-diabetic Pima Indians. Diabetologia. 1990;33: 285–289. 10.1007/BF00403322 [DOI] [PubMed] [Google Scholar]
- 59. Clark A, Lewis C., Willis A., Cooper GJ., Morris J., Reid KB., et al. Islet Amyloid Formed from Diabetes-Associated Peptide May Be Pathogenic in Type-2 Diabetes. The Lancet. 1987;330: 231–234. 10.1016/S0140-6736(87)90825-7 [DOI] [PubMed] [Google Scholar]
- 60. Opie EL. The Relation Oe Diabetes Mellitus to Lesions of the Pancreas. Hyaline Degeneration of the Islands Oe Langerhans. J Exp Med. 1901;5: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Westermark P, Wernstedt C, O’Brien TD, Hayden DW, Johnson KH. Islet amyloid in type 2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am J Pathol. 1987;127: 414–417. [PMC free article] [PubMed] [Google Scholar]
- 62. Cooper GJ, Willis AC, Clark A, Turner RC, Sim RB, Reid KB. Purification and Characterization of a Peptide from Amyloid-Rich Pancreases of Type 2 Diabetic Patients. PNAS. 1987;84: 8628–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Westermark P, Wilander E. The influence of amyloid deposits on the islet volume in maturity onset diabetes mellitus. Diabetologia. 1978;15: 417–421. 10.1007/BF01219652 [DOI] [PubMed] [Google Scholar]
- 64. Jaikaran ETA., Higham CE, Serpell LC, Zurdo J, Gross M, Clark A, et al. Identification of a novel human islet amyloid polypeptide β-sheet domain and factors influencing fibrillogenesis. Journal of Molecular Biology. 2001;308: 515–525. 10.1006/jmbi.2001.4593 [DOI] [PubMed] [Google Scholar]
- 65. Higham CE, Jaikaran ETAS, Fraser PE, Gross M, Clark A. Preparation of synthetic human islet amyloid polypeptide (IAPP) in a stable conformation to enable study of conversion to amyloid-like fibrils. FEBS Letters. 2000;470: 55–60. 10.1016/S0014-5793(00)01287-4 [DOI] [PubMed] [Google Scholar]
- 66. Kayed R, Bernhagen J, Greenfield N, Sweimeh K, Brunner H, Voelter W, et al. Conformational transitions of islet amyloid polypeptide (IAPP) in amyloid formation in Vitro. Journal of Molecular Biology. 1999;287: 781–796. 10.1006/jmbi.1999.2646 [DOI] [PubMed] [Google Scholar]
- 67. Sumner Makin O, Serpell LC. Structural Characterisation of Islet Amyloid Polypeptide Fibrils. Journal of Molecular Biology. 2004;335: 1279–1288. 10.1016/j.jmb.2003.11.048 [DOI] [PubMed] [Google Scholar]
- 68. Mirzabekov TA, Lin M, Kagan BL. Pore Formation by the Cytotoxic Islet Amyloid Peptide Amylin. Journal of Biological Chemistry. 1996;271: 1988–1992. 10.1074/jbc.271.4.1988 [DOI] [PubMed] [Google Scholar]
- 69. Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48: 491–498. 10.2337/diabetes.48.3.491 [DOI] [PubMed] [Google Scholar]
- 70. Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66: S74 –S78. 10.1212/01.wnl.0000192103.24796.42 [DOI] [PubMed] [Google Scholar]
- 71. Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus., Published online: 21 April 1994;. 1994;368: 756–760. 10.1038/368756a0 [DOI] [PubMed] [Google Scholar]
- 72. Knight JD, Miranker AD. Phospholipid Catalysis of Diabetic Amyloid Assembly. Journal of Molecular Biology. 2004;341: 1175–1187. 10.1016/j.jmb.2004.06.086 [DOI] [PubMed] [Google Scholar]
- 73. Jayasinghe SA, Langen R. Lipid Membranes Modulate the Structure of Islet Amyloid Polypeptide†. Biochemistry. 2005;44: 12113–12119. 10.1021/bi050840w [DOI] [PubMed] [Google Scholar]
- 74. Scalisi S, Sciacca MFM, Zhavnerko G, Grasso DM, Marletta G, La Rosa C. Self-Assembling Pathway of HiApp Fibrils within Lipid Bilayers. Chem Eur J of Chem Bio. 2010;11: 1856–1859. 10.1002/cbic.201000090 [DOI] [PubMed] [Google Scholar]
- 75. Westermark P, Engström U, Johnson KH, Westermark GT, Betsholtz C. Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87: 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Janson J, Soeller WC, Roche PC, Nelson RT, Torchia AJ, Kreutter DK, et al. Spontaneous Diabetes Mellitus in Transgenic Mice Expressing Human Islet Amyloid Polypeptide. PNAS. 1996;93: 7283–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Butler AE, Jang J, Gurlo T, Carty MD, Soeller WC, Butler PC. Diabetes Due to a Progressive Defect in B-Cell Mass in Rats Transgenic for Human Islet Amyloid Polypeptide (HIP Rat) A New Model for Type 2 Diabetes. Diabetes. 2004;53: 1509–1516. 10.2337/diabetes.53.6.1509 [DOI] [PubMed] [Google Scholar]
- 78. Matveyenko AV, Butler PC. Islet amyloid polypeptide (IAPP) transgenic rodents as models for type 2 diabetes. ILAR J. 2006;47: 225–233. [DOI] [PubMed] [Google Scholar]
- 79. Sakagashira S, Sanke T, Hanabusa T, Shimomura H, Ohagi S, Kumagaye KY, et al. Missense Mutation of Amylin Gene (S20G) in Japanese NIDDM Patients. Diabetes. 1996;45: 1279–1281. [DOI] [PubMed] [Google Scholar]
- 80. Sakagashira S, Hiddinga HJ, Tateishi K, Sanke T, Hanabusa T, Nanjo K, et al. S20G Mutant Amylin Exhibits Increased in Vitro Amyloidogenicity and Increased Intracellular Cytotoxicity Compared to Wild-Type Amylin. The American Journal of Pathology. 2000;157: 2101–2109. 10.1016/S0002-9440(10)64848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jha S, Sellin D, Seidel R, Winter R. Amyloidogenic Propensities and Conformational Properties of ProIAPP and IAPP in the Presence of Lipid Bilayer Membranes. Journal of Molecular Biology. 2009;389: 907–920. 10.1016/j.jmb.2009.04.077 [DOI] [PubMed] [Google Scholar]
- 82. Knight JD, Hebda JA, Miranker AD. Conserved and Cooperative Assembly of Membrane-Bound α-Helical States of Islet Amyloid Polypeptide†. Biochemistry. 2006;45: 9496–9508. 10.1021/bi060579z [DOI] [PubMed] [Google Scholar]
- 83. Shim S-H, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. PNAS. 2009;106: 6614–6619. 10.1073/pnas.0805957106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yonemoto IT, Kroon GJA, Dyson HJ, Balch WE, Kelly JW. Amylin Proprotein Processing Generates Progressively More Amyloidogenic Peptides that Initially Sample the Helical State†. Biochemistry. 2008;47: 9900–9910. 10.1021/bi800828u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Patil SM, Xu S, Sheftic SR, Alexandrescu AT. Dynamic α-Helix Structure of Micelle-bound Human Amylin. J Biol Chem. 2009;284: 11982–11991. 10.1074/jbc.M809085200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nanga RPR, Brender JR, Vivekanandan S, Ramamoorthy A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2011;1808: 2337–2342. 10.1016/j.bbamem.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Apostolidou M, Jayasinghe SA, Langen R. Structure of α-Helical Membrane-bound Human Islet Amyloid Polypeptide and Its Implications for Membrane-mediated Misfolding. J Biol Chem. 2008;283: 17205–17210. 10.1074/jbc.M801383200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Engel MFM, Yigittop H, Elgersma RC, Rijkers DTS, Liskamp RMJ, de Kruijff B, et al. Islet Amyloid Polypeptide Inserts into Phospholipid Monolayers as Monomer. Journal of Molecular Biology. 2006;356: 783–789. 10.1016/j.jmb.2005.12.020 [DOI] [PubMed] [Google Scholar]
- 89. Lee C-C, Sun Y, Huang HW. How Type II Diabetes-Related Islet Amyloid Polypeptide Damages Lipid Bilayers. Biophysical Journal. 2012;102: 1059–1068. 10.1016/j.bpj.2012.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huang C-J, Haataja L, Gurlo T, Butler AE, Wu X, Soeller WC, et al. Induction of Endoplasmic Reticulum Stress-Induced B-Cell Apoptosis and Accumulation of Polyubiquitinated Proteins by Human Islet Amyloid Polypeptide. Am J Physiol Endocrinol Metab. 2007;293: E1656–E1662. 10.1152/ajpendo.00318.2007 [DOI] [PubMed] [Google Scholar]
- 91. Balasubramaniam A, Renugopalakrishnan V, Stein M, Fischer JE, Chance WT. Syntheses, structures and anorectic effects of human and rat amylin. Peptides. 1991;12: 919–924. 10.1016/0196-9781(91)90038-Q [DOI] [PubMed] [Google Scholar]
- 92. Nanga RPR, Brender JR, Xu J, Hartman K, Subramanian V, Ramamoorthy A. Three-Dimensional Structure and Orientation of Rat Islet Amyloid Polypeptide Protein in a Membrane Environment by Solution NMR Spectroscopy. J Am Chem Soc. 2009;131: 8252–8261. 10.1021/ja9010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Middleton CT, Marek P, Cao P, Chiu C, Singh S, Woys AM, et al. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat Chem. 2012;4: 355–360. 10.1038/nchem.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4: 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- 95. Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling RID A-1776-2009 RID C-6971-2008. J Chem Phys. 2007;126 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 96. Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 1981;52: 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- 97. Nosé S, Klein ML. Constant pressure molecular dynamics for molecular systems. Molecular Physics. 1983;50: 1055–1076. 10.1080/00268978300102851 [DOI] [Google Scholar]
- 98. Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98: 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- 99. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103: 8577 10.1063/1.470117 [DOI] [Google Scholar]
- 100. Hess B, Bekker H, Berendsen H, Fraaije J. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18: 1463–1472. [DOI] [Google Scholar]
- 101. Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Molecular Physics. 1984;52: 255–268. 10.1080/00268978400101201 [DOI] [Google Scholar]
- 102. Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31: 1695–1697. 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- 103. Pietrucci F, Laio A. A Collective Variable for the Efficient Exploration of Protein Beta-Sheet Structures: Application to SH3 and GB1. Journal of Chemical Theory and Computation. 2009;5: 2197–2201. 10.1021/ct900202f [DOI] [PubMed] [Google Scholar]
- 104. Bonomi M, Branduardi D, Bussi G, Camilloni C, Provasi D, Raiteri P, et al. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Computer Physics Communications. 2009;180: 1961–1972. 10.1016/j.cpc.2009.05.011 [DOI] [Google Scholar]
- 105. Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR. 2011;50: 43–57. 10.1007/s10858-011-9478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Williamson JA, Miranker AD. Direct detection of transient α-helical states in islet amyloid polypeptide. Protein Science. 2007;16: 110–117. 10.1110/ps.062486907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Soong R, Brender JR, Macdonald PM, Ramamoorthy A. Association of Highly Compact Type II Diabetes Related Islet Amyloid Polypeptide Intermediate Species at Physiological Temperature Revealed by Diffusion NMR Spectroscopy. J Am Chem Soc. 2009;131: 7079–7085. 10.1021/ja900285z [DOI] [PubMed] [Google Scholar]
- 108. Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22: 2577–2637. 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- 109. Joosten RP, Beek TAH te, Krieger E, Hekkelman ML, Hooft RWW, Schneider R, et al. A series of PDB related databases for everyday needs. Nucl Acids Res. 2011;39: D411–D419. 10.1093/nar/gkq1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48: 13–22. 10.1007/s10858-010-9433-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47: 251–276. 10.1146/annurev.bi.47.070178.001343 [DOI] [PubMed] [Google Scholar]
- 112. Abedini A, Raleigh DP. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys Biol. 2009;6: 015005 10.1088/1478-3975/6/1/015005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Luca S, Yau W-M, Leapman R, Tycko R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid-State NMR†. Biochemistry. 2007;46: 13505–13522. 10.1021/bi701427q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Buchanan LE, Dunkelberger EB, Tran HQ, Cheng P-N, Chiu C-C, Cao P, et al. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. PNAS. 2013;110: 19285–19290. 10.1073/pnas.1314481110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Singh S, Chiu C, Reddy AS, Pablo JJ de. α-helix to β-hairpin transition of human amylin monomer. The Journal of Chemical Physics. 2013;138: 155101 10.1063/1.4798460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lindorff-Larsen K, Trbovic N, Maragakis P, Piana S, Shaw DE. Structure and Dynamics of an Unfolded Protein Examined by Molecular Dynamics Simulation. J Am Chem Soc. 2012;134: 3787–3791. 10.1021/ja209931w [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The corresponding p-values are shown. Using the standard deviation of the means, and a power of 0.95, the predicted difference in ppm between the two temperatures that could be resolved is shown as Δ.
(DOCX)
The darker regions indicate regions of lower free energy.
(TIFF)
The secondary structure of each residue was determined using DSSP.
(TIFF)
The secondary structure of each residue was determined using DSSP.
(TIFF)
Data Availability Statement
The data are available at: doi:10.7910/DVN/29368.