Abstract
Microbial hosts engineered for the biosynthesis of plant natural products offer enormous potential as powerful discovery and production platforms. However, the reconstruction of these complex biosynthetic schemes faces numerous challenges due to the number of enzymatic steps and challenging enzyme classes associated with these pathways, which can lead to issues in metabolic load, pathway specificity, and maintaining flux to desired products. Cytochrome P450 enzymes are prevalent in plant specialized metabolism and are particularly difficult to express heterologously. Here, we describe the reconstruction of the sanguinarine branch of the benzylisoquinoline alkaloid pathway in Saccharomyces cerevisiae, resulting in microbial biosynthesis of protoberberine, protopine, and benzophenanthridine alkaloids through to the end-product sanguinarine, which we demonstrate can be efficiently produced in yeast in the absence of the associated biosynthetic enzyme. We achieved titers of 676 µg/L stylopine, 548 µg/L cis-N-methylstylopine, 252 µg/L protopine, and 80 µg/L sanguinarine from the engineered yeast strains. Through our optimization efforts, we describe genetic and culture strategies supporting the functional expression of multiple plant cytochrome P450 enzymes in the context of a large multi-step pathway. Our results also provided insight into relationships between cytochrome P450 activity and yeast ER physiology. We were able to improve the production of critical intermediates by 32-fold through genetic techniques and an additional 45-fold through culture optimization.
Keywords: cytochrome P450, synthetic biology, plant natural products, yeast
1 Introduction
Microbial biosynthesis of plant specialized metabolites is an important and growing field. Plants are a significant source of bioactive compounds; however, production and screening of plant specialized metabolites in their native plant hosts has faced several challenges. For example, many compounds exist only in trace amounts and the methods for isolating and purifying these compounds from the native plant hosts are generally inefficient and environmentally unsustainable. Microbial biosynthesis of plant specialized metabolites is an alternative production platform that provides several advantages over the native plant hosts, including well-developed genetic tools for pathway expression and manipulation, fast growth rates, and established large-scale culture methods. Recently, several drug-producing plant pathways have been successfully engineered in Saccharomyces cerevisiae including the anti-malarial drug artemisinin (Paddon et al., 2013; Ro et al., 2006; Westfall et al., 2012), the cancer therapeutic taxol (Dejong et al., 2006; Engels et al., 2008), and the analgesic morphine (Thodey et al., 2014).
Plant natural product pathways remain a significant challenge in the field of metabolic engineering, highlighted by the fact that few pathways have been reconstituted in microbial hosts compared with microbial natural product pathways. Plant specialized metabolite pathways tend to have numerous enzymatic steps and a branching architecture leading to many different structural classes of compounds, creating challenges in heterologous expression such as managing metabolic load, pathway stability, specificity toward desired branches and structures, and efficiency of flux through the pathway (Chemler and Koffas, 2008). Many pathways and enzymes have not been fully characterized or identified, greatly limiting the ability to access certain compounds of interest with synthetic systems.
In addition to the challenge of engineering large pathways, individual plant enzymes can be difficult to functionally express in microbial hosts. In particular, plant cytochrome P450s are notoriously difficult to functionally express in the context of multi-step heterologous biosynthetic pathways (Duan and Schuler, 2006; Renault et al., 2014). These enzymes have an N-terminal tag that directs their localization to the endoplasmic reticulum (ER) and can induce ER stress responses in a yeast host (Sandig et al., 1999; Zimmer et al., 1997). They require a partner enzyme, cytochrome P450 NADPH reductase (CPR), to interact with cofactors and shuttle electrons to the active site of the P450. In some cases P450s require another enzyme, cytochrome b5, to provide additional redox support to achieve full activity (Paddon et al., 2013). These unique properties and requirements for the functional expression of P450s, and their significant representation in plant specialized metabolite pathways (Mizutani and Ohta, 2010) make them a key challenge in constructing plant pathways in heterologous hosts.
To enable the engineering of microbial production platforms for these complex pathways, we sought to develop generalizable pathway optimization strategies with a focus on plant cytochrome P450 expression. To develop these methods, we engineered the biosynthesis of benzylisoquinoline alkaloids (BIAs) in S. cerevisiae. The BIAs are a diverse family of plant specialized metabolites naturally found in Ranunculales order (Hagel and Facchini, 2013). These compounds display a broad range of pharmacological properties, including anti-cancer (Kemény-Beke et al., 2006), anti-HIV (Kashiwada et al., 2005), and antimicrobial activities (Orhana et al., 2007). Several BIAs make up large drug markets, such as the opiates and their semi-synthetic derivatives (Hagel and Facchini, 2013). Yeast strains have previously been engineered to produce several BIA compounds including reticuline (Hawkins and Smolke, 2008; Minami et al., 2008) and morphine (Thodey et al., 2014). We focused on the sanguinarine pathway, which encompasses many of the challenges associated with engineering plant pathways including many enzymatic steps, multiple P450 enzymes, and multiple compound structural classes. The sanguinarine pathway branch was previously reconstituted in a yeast host (Fossati et al., 2014). However, the engineered strains and pathway were not optimized and relied on whole cell buffered assays, where cells were incubated in buffer with a small amount of substrate. We have applied metabolic engineering optimization techniques to improve enzymatic steps exhibiting low conversion efficiencies and develop strains that perform in conditions more relevant to industrial production.
In this work, we have engineered microbial production hosts for protoberberine, protopine, and benzophenanthridine alkaloids in the sanguinarine branch of the BIA biosynthetic pathway. Specifically, we report the highest titers to date in a microbial fermentation for the compounds cheilanthifoline, stylopine, cis-N-methylstylopine, protopine, and sanguinarine. These pathways represent some of the most complex plant natural products pathways reconstructed in a microbial host to date, based on number and type of heterologous enzymes. In particular, we describe strategies supporting the functional expression of multiple plant cytochrome P450 enzymes in the context of a large multi-step pathway in a microbial host, including expression of multiple enzyme variants, CPR pairing, expression stabilization, enzyme localization, and culture condition optimization. These general design strategies can be applied more broadly to support the engineering of diverse plant natural product pathways in yeast.
2 Materials and Methods
2.1 Plasmid and yeast strain construction
The S. cerevisiae strains described in this work are all derived from W303α (MATα leu2–3,112 trp1-1 can1–100 ura3-1 ade2-1 his3–11,15). A standard lithium acetate protocol was used for yeast transformations (Gietz and Woods, 2006). Oligonucleotides were synthesized by either the Stanford Protein and Nucleic Acid Facility (Stanford, CA) or Integrated DNA Technologies (Coralville, IA). Cloning was performed with chemically competent Escherichia coli (TOP10, LifeTech, F- mcrA Δ(mrr-hsdRMS-mcrBC) φp80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ−). E. coli were cultured in Luria-Bertani media (EMD Chemicals) with appropriate antibiotic: 100 µg/mL ampicillin (EMD Chemicals) or 50 µg/mL kanamycin (EMD Chemicals). Spin columns were used to purify plasmids from E. coli cultures according to the manufacturer’s instructions (Epoch Life Science). Sequencing was performed by Elim Biopharmaceuticals (Hayward, CA). DNA polymerases used were either PfuUltraII Fusion HS DNA Polymerase (Life Technologies) or Expand High Fidelity Polymerase (Roche), and PCR products were cleaned up using QIAquick PCR purification kit (Qiagen).
Gene sequences for EcCFS (BAG75113), EcCPR (AAC05022), EcSTS (BAD98250), and AmSTS (ABR14721) were yeast codon-optimized and assembled from oligonucleotides designed with DNAWorks (Hoover and Lubkowski, 2002). AmCFS (ABR14722), PsCFS (ADB89213), PsSTS (ADB89214), PsMSH (AGC92398), EcP6H (BAK20464), were yeast codon-optimized and synthesized by GeneArt (Life Technologies). PsCPR (AAC05021) and PsP6H (AGC92397) was codon-optimized by GeneArt, synthesized as two gBlock gene fragments by IDT, and assembled by PCR.
Most yeast expression vectors described in this work were constructed with Gateway Cloning Technology (Life Technologies). Enzyme coding regions were PCR amplified and cloned into the pENTR vector through either TOPO cloning or a BP recombination reaction using BP clonase II and the pDONR221 vector (Life Technologies). All genes were subsequently recombined into selected pAG expression vectors from the Lindquist laboratory (available through Addgene) (Alberti et al., 2007) using LR Clonase II (Life Technologies). For chromosomal integrations, the gene of interest was recombined into pCS2643 or pCS2644 (Thodey et al., 2014) using LR clonase II (Life Technologies), and the complete integration cassette (gene expression cassette and selection marker) was PCR amplified with the appropriate integration primers The integration selection markers were rescued using CRE recombinase (Güldener et al., 1996).
2.2 Growth conditions for assays
Overnight yeast cultures were grown at 30 °C, 260 rpm or in 500 µL cultures in deep 96 well plates covered with AeraSeal film (Excel Scientific) and grown at 30 °C, 480 rpm, 80% humidity in a Kuhner Lab-Therm LX-T plate shaker. Overnight cultures were back-diluted 75–100X into appropriate dropout media supplemented with 2 mM norlaudanosoline (Santa Cruz Biotech). More specifically, some assays were conducted with 500 µL cultures in deep 96 well plates covered with AeraSeal film (Excel Scientific) and grown at 30 °C, 480 rpm, 80% humidity in a Kuhner Lab-Therm LX-T plate shaker. Other assays were conducted using 2 mL cultures grown at 25 °C, 260 rpm in a New Brunswick Scientific I24 shaking incubator. Unless otherwise indicated, cultures were sampled 96 h following back-dilution.
2.3 Optimized growth conditions for assays
Overnight cultures were grown in 1X YNB synthetic complete dropout media then back-diluted 150X-200X into 3 mL 5X YNB synthetic complete dropout media and grown at 25 °C for 48 h. 2 mL of each culture was centrifuged for 5 minutes at 8000 rpm to harvest the cells, and the media discarded. The pellets were resuspended in 2 mL production phase media (1X YNB dropout media with 2 mM norlaudanosoline), grown at 25 °C for 96 h. For the production phase carbon source assays, 20% stocks of each carbon source were made in water and diluted to 2% in the assay conditions.
2.4 Analysis of metabolite production
Aliquots of yeast cultures were centrifuged at 13000 rpm for 10 min and growth media samples were taken for analysis by LC-MS/MS on either Agilent 6300 Series Ion Trap LC-MS or an Agilent 6420 triple quad LC-MS. Samples for the ion trap LC-MS were separated on an Agilent ZORBAX SB-Aq 4.6×50 mm, 5 µm column with 0.1% acetic acid as solvent A and methanol with 0.1% acetic acid as solvent B with the following method at a constant flow rate of 0.5 mL/min: 0–1 min, 0% to 27.5% B; 1–2 min, 27.5% B; 2–8 min, 27.5% to 60% B; 8–8.5, 35 to 100% B; 8.5–14 min, 100% B; followed by a 6 minute equilibration at 0% solvent B. After separation by HPLC, metabolites were injected into an Agilent 6320 ion trap mass spectrometer for detection and identification. Samples for the triple quad LC-MS were separated on an Agilent EclipsePlus C18, 2.1×50 mm, 1.8 µm column with 0.1% formic acid as solvent A and acetonitrile with 0.1% formic acid as solvent B with the following method at a constant flow rate of 0.4 mL/min: 0–0.1 min, 10% B; 0.1–5 min, 10 to 45% B; 5–5.5 min, 45 to 90% B; 5.5–7 min, 90% B; 7–7.01 min 10% B; followed by a 3 minute equilibration at 10% solvent B. Quantification of metabolites was based on integrated peak area of the extracted ion chromatogram peaks calculated using DataAnalysis for 6300 Series Ion Trap LC/MS version 3.4 (Bruker Daltonik GmbH) or Agilent MassHunter Workstation and reported as the mean ± s.d. We generated standard curves for stylopine (ChromaDex), protopine (Sigma), and sanguinarine (Sigma) and used the most similar standard chemical structure to estimate concentration of intermediates for which no standard was available. Percent conversion of an enzymatic step was calculated as a ratio of product concentration (µM) in the presence of the enzyme to substrate concentration (µM) in the absence of the enzyme.
2.5 Imaging P450 localization in live cells with confocal microscopy
Yeast cell cultures were inoculated into 3 mL of dropout media and grown for 8–12 h, to an approximate OD600 of 0.1. 1 mL of culture was pelleted at 6000 rpm for 30 seconds, the supernatant discarded and the cells resuspended in 25–50 µL dropout media. 1–3 µL of culture were mounted on 2% agarose pads made with appropriate dropout media to facilitate live cell imaging. Live yeast cells were imaged with a Leica SP5 multiphoton/confocal microscope at the Cell Sciences Imaging Facility (Stanford, CA) equipped with a 63.0X glycerine immersion objective.
2.6 Plasmid stability assay
Yeast strains harboring either a low-copy or high-copy plasmid expressing EcCFS were grown overnight in selective media (−TRP), and then the cultures were diluted so that approximately 200 cells were plated on selective (−TRP) and non-selective (YPD) media. The colony count on the selective plate was divided by the colony count on the non-selective plate to determine the percent of plasmid maintained.
3 Results
3.1 Engineering an optimized protoberberine microbial scaffold-producing strain
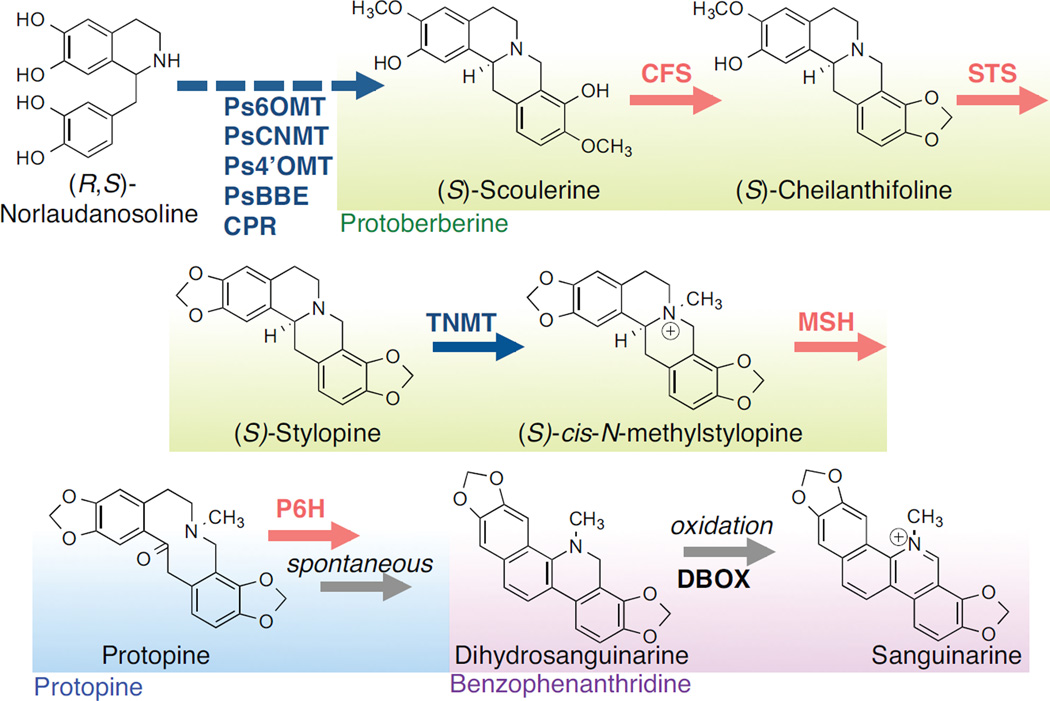
We targeted the reconstruction of the sanguinarine branch of the BIA biosynthetic pathway, which encompasses three structural subclasses of BIAs - the protoberberine, protopine, and benzophenanthridine alkaloids - in a yeast production host. The sanguinarine branch is comprised of ten enzymatic steps, four of which are catalyzed by plant cytochrome P450s, to reach sanguinarine (the end-product of the pathway) from norlaudanosoline (a commercially-available, early BIA precursor) (Fig. 1). Several downstream enzymes in this pathway have recently been cloned and characterized (Beaudoin and Facchini, 2013; Hagel et al., 2012; Takemura et al., 2013) enabling the engineering of a microbial production strain for intermediate metabolites along the sanguinarine branch.
Figure 1.
Engineered pathway for sanguinarine biosynthesis in yeast from (R, S)-norlaudanosoline. Red arrows indicate reactions catalyzed by a plant cytochrome P450 enzyme. Structural class of the metabolites are indicated as green, protoberberine; blue, protopine; purple, benzophenanthridine. Ps6OMT, P. somniferum 6-O-methyltransferase; PsCNMT, P. somniferum coclaurine N-methyltransferase; Ps4’OMT, P. somniferum 4’-O-methyltransferase; PsBBE, P. somniferum berberine bridge enzyme; CPR, cytochrome P450 reductase; CFS, cheilanthifoline synthase; STS, stylopine synthase; TNMT, tetrahydroprotoberberine N-methyltransferase; MSH, cis-N-methylstylopine 14-hydroxylase; P6H, protopine 6-hydroxylase; DBOX, dihydrobenzophenanthridine oxidase.
Our starting point was a yeast strain that had been previously engineered to produce the protoberberine scaffold scoulerine from the fed substrate norlaudanosoline (Hawkins and Smolke, 2008). This strain expressed four plant enzymes: three methyltransferases (Papaver somniferum norcoclaurine 6-O-methyltransferase, Ps6OMT; P. somniferum coclaurine-N-methyltransferase, PsCNMT; P. somniferum 3’hydroxy-N-methylcoclaurine 4’-O-methyltransferase, Ps4’OMT) and berberine bridge enzyme (P. somniferum berberine bridge enzyme, PsBBE), the enzyme that converts the key branch point metabolite (S)-reticuline to (S)-scoulerine. In the original strain the methyltransferases were integrated into the chromosome and PsBBE was expressed from a high-copy plasmid. We integrated the PsBBE expression cassette into the yeast chromosome (CS Y953), and examined the effect of altering PsBBE expression on scoulerine production. Strains were fed 2 mM norlaudanosoline and grown for 96 h at 30 °C. Growth media was analyzed for metabolite accumulation using high-performance liquid chromatography coupled to tandem mass spectrometry (LC-ESI-MS/MS), and production of scoulerine was confirmed by comparison of the fragmentation pattern of the extracted ion (MS-MS) to reported spectra (Schmidt et al., 2007). By altering expression of PsBBE from a high-copy plasmid to the chromosome, we increased the production of scoulerine from 419 µg/L to 717 µg/L and improved the conversion efficiency of reticuline to scoulerine from 23% to 67% (Fig. S1).
3.2 Plant cytochrome P450 expression strategies for cheilanthifoline production
The first dedicated step in the sanguinarine branch is the conversion of scoulerine to cheilanthifoline by the enzyme cheilanthifoline synthase (CFS), the first cytochrome P450 in the sanguinarine pathway. As plant cytochrome P450s are often challenging to functionally express in microbial hosts, we examined various strategies to support functional heterologous expression of this P450 enzyme and optimize its activity in yeast (Fig. 2A).
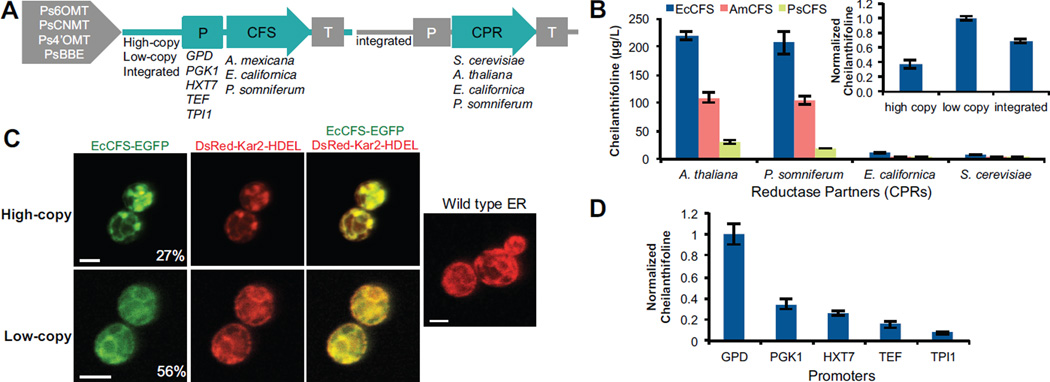
Figure 2.
Optimization of (S)-cheilanthifoline production through genetic strategies. (A) Schematic representing genetic strategies used to optimize cheilanthifoline production, including expression method (plasmids or integrated), promoter, species variants, and CPR variants. All strains contain Ps6OMT, PsCNMT, Ps4’OMT, and PsBBE chromosomally integrated. (B) Cheilanthifoline production in yeast strains as a function of CFS and CPR variants. Variants of CFS from E. californica (EcCFS), A. mexicana (AmCFS), and P. somniferum (PsCFS) were expressed from low-copy plasmids in yeast strains with an integrated cytochrome P450 reductase enzyme (CPR) from either the native yeast (CSY953) or various plant sources (A. thaliana, CSY844; E. californica, CSY850; P. somniferum, CSY985). Inset, cheilanthifoline production with EcCFS and ATR1 as a function of expression method, normalized to production from the low-copy plasmid. (C) Confocal microscopy analysis of EcCFS C-terminally tagged with GFP on high-copy (top) or low-copy (bottom) plasmids coexpressed with ER marker Kar2-DsRed-HDEL in CSY844. Wild-type ER (no heterologous P450 expressed) is shown for comparison (right). Percentages indicate portion of the yeast population that are GFP positive under the indicated expression condition. Scale bars are 4 µm. Images are representative of at least 3 independent experiments. (D) Cheilanthifoline production in yeast strains engineered to express EcCFS from a low-copy plasmid under the control of different promoters in CSY844. Data are normalized to production from PGPD. (B, D) Metabolite production is determined by LC-MS analysis of culture media after indicated strains were grown for 96 h. Data is reported as the mean ± s.d. of at least 3 independent experiments.
We assayed yeast codon-optimized versions of all available species variants of CFS - Eschscholzia californica (EcCFS) (Ikezawa et al., 2009), Argemone mexicana (AmCFS) (Díaz Chávez et al., 2011), and P. somniferum (PsCFS) (Fossati et al., 2014). The CFS variants were expressed on either a high- or low-copy plasmid from a strong yeast promoter (PGPD), and we examined the pairing of various plant cytochrome P450 reductases (CPRs) with CFS variants. Three plant CPR expression cassettes, from A. thaliana (ATR1) (Urban et al., 1997), E. californica (EcCPR) (Rosco et al., 1997), and P. somniferum (PsCPR) (Rosco et al., 1997), were individually integrated into the yeast chromosome at the TRP1 locus under the control of the PTEF promoter (CSY844, CSY850, and CSY985, respectively) and tested with each of the CFS variants. Cheilanthifoline production was confirmed through comparison of the fragmentation pattern to reported patterns (Schmidt et al., 2007) (Fig. S2A, B).
The data demonstrate that the specific CPR pairing and P450 expression level substantially impact the functional activity of the plant P450 in a heterologous microbial host. Specifically, ATR1 and PsCPR supported similar levels of activity with all CFS variants and resulted in up to 219 µg/L cheilanthifoline production, while the native yeast CPR and EcCPR were able to support a low level of activity from EcCFS (Fig. 2B). All CFS variants resulted in cheilanthifoline production when paired with an appropriate CPR; however, EcCFS and AmCFS exhibited substantially higher activity than PsCFS (Fig. 2B). The difference in activity observed between the EcCFS and AmCFS variants in yeast is supported by the reported Km values for these enzymes (EcCFS, 900 nM; AmCFS, 1.9 µM; PsCFS, no reported Km) (Bauer and Zenk, 1991; Díaz Chávez et al., 2011). Our data also demonstrate that cheilanthifoline production levels were up to 2.6-fold higher when the CFS variants were expressed from a low-copy plasmid or the chromosome (CSY903) than from a high-copy plasmid (Fig. 2B, inset and Fig. S2C).
We further explored the mechanisms underlying the observed differences in P450 activity between high- and low-copy plasmid expression. Plant P450s naturally localize to the endoplasmic reticulum (ER), and previous studies have shown that overexpression of eukaryotic P450s can cause a stress response in which the ER membranes proliferate in yeast (Sandig et al., 1999). To examine the connection between ER morphology and P450 activity, we C-terminally tagged the EcCFS with EGFP, expressed the fusion from a high- or low-copy plasmid, and examined the resulting enzyme expression and subcellular localization across the cell population. Live cells harboring these constructs were imaged by confocal microscopy (Fig. 2C), and ER localization of the tagged enzymes was confirmed by co-localization with the ER marker Kar2-DsRed-HDEL expressed from a high-copy plasmid with the PTEF promoter (Bevis et al., 2002).
We observed two major differences between the cells expressing the tagged CFS variant from high- or low-copy plasmids. First, a significant reduction in the number of GFP positive cells was observed in cells harboring a P450-GFP fusion construct compared to a GFP-only construct, and the difference was more significant when the fusion was expressed from a high-copy plasmid (27%) than from a low-copy plasmid (56%). To confirm a difference in plasmid retention, we performed a plasmid stability assay on high-copy and low-copy plasmids expressing EcCFS to test for the presence of the selection marker instead of relying on GFP as the readout. The low-copy plasmid was 90% maintained while the high-copy plasmid was only 10.7% maintained, which supports the observations through microscopy. Second, the morphology of the ER was distinct between cells harboring the P450-GFP expression cassettes in high- and low-copy plasmids. In cells harboring the high-copy plasmids, the P450-GFP fusion proteins were generally observed in highly concentrated bright patches. In contrast, in cells harboring the low-copy plasmids, the ER membranes remained distributed throughout the cell and GFP fluorescence levels were dim, indicating a lower concentration of P450s. Since expression from a low-copy plasmid resulted in high P450 activity, we concluded that a more wild-type ER morphology was favorable for enzyme activity. We observed similar ER localization patterns for AmCFS and PsCFS (Fig. S2D).
Finally, we examined the impact of promoter strength and regulation on plant P450 expression level and cheilanthifoline production in the yeast host. Cheilanthifoline production was examined with EcCFS expressed from a low-copy plasmid under the control of five different promoters: PGPD, PTEF1, PPGK1, PTPI1, and PHXT7. PGPD, PTEF1, PPGK1, and PTPI1 are generally considered to be constitutive promoters, while pHXT7 is a strong promoter during glucose-depletion, during late stages in the growth cycle. However, PGPD and PPGK1 tend be very strong during the glucose consumption phase and drop off during later stages of growth (Partow et al., 2010). The GPD promoter provided up to a 10-fold improvement in cheilanthifoline production compared to the other tested promoters (Fig. 2D). The data indicate that both expression level and regulation pattern of the promoters play important roles in optimizing the activity of plant P450s in a heterologous yeast host. For example, the TEF promoter exhibits similar strength as PGPD, and PPGK1 exhibits a similar regulatory profile as PGPD with a different strength; however, each of these promoters resulted in different cheilanthifoline production levels. Taken together, the optimized cheilanthifoline-producing yeast strain was engineered to express the plant P450 EcCFS from either a low-copy plasmid or the chromosome, under the control of a GPD promoter, and paired with the ATR1 CPR. This optimized strain produced up to 219 µg/L cheilanthifoline with a scoulerine to cheilanthifoline conversion efficiency of 31%.
3.3 Optimizing multiple P450 enzyme expression and culture methods for stylopine production
The next step in the sanguinarine branch is the conversion of (S)-cheilanthifoline to (S)-stylopine by stylopine synthase (STS), a plant cytochrome P450 closely related to CFS. As a starting point to engineering stylopine-producing strains we utilized the P450 optimization strategies elucidated with CFS; specifically, (i) characterizing yeast codon-optimized versions of enzyme variants from multiple species (E. californica, EcSTS (Ikezawa et al., 2007); A. mexicana, AmSTS (Díaz Chávez et al., 2011); P. somniferum, PsSTS (Fossati et al., 2014)), (ii) expressing the plant P450 from a low-copy plasmid, (iii) controlling the expression of the P450 from the GPD promoter, and (iv) pairing the P450 with the ATR1 CPR. Since the biosynthetic pathway to stylopine includes two plant P450s, we examined additional strategies for further optimizing the activities of these enzymes within the context of the multi-step pathway.
We first examined the pairings of plant variants of STS and CFS to explore any synergistic effects between particular pairings. Yeast strains with the ATR1 CPR expression cassette integrated into the chromosome (CSY844) and harboring different combinations of CFS and STS variants expressed from low-copy plasmids were assayed. Stylopine production was confirmed through comparison of the retention time and fragmentation pattern to a stylopine standard and reported patterns (Ikezawa et al., 2007) (Fig. S3A). The data demonstrate that heterologous expression of EcSTS and PsSTS result in stylopine production from the engineered yeast strains, whereas AmSTS was not active under these conditions. The most productive P450 enzyme pairing was EcCFS and EcSTS, which produced 15 µg/L stylopine, up to a 5.5-fold improvement over the other enzyme combinations (Fig. 3A). The difference in activity observed between the STS variants in yeast is supported by the reported Km values for these enzymes (EcSTS, 400 nM; AmSTS, 5.2 µM; PsSTS, no reported Km) (Bauer and Zenk, 1991; Díaz Chávez et al., 2011). However, even with the most productive P450 pairings, the conversion efficiency of cheilanthifoline to stylopine was only 6.7%.
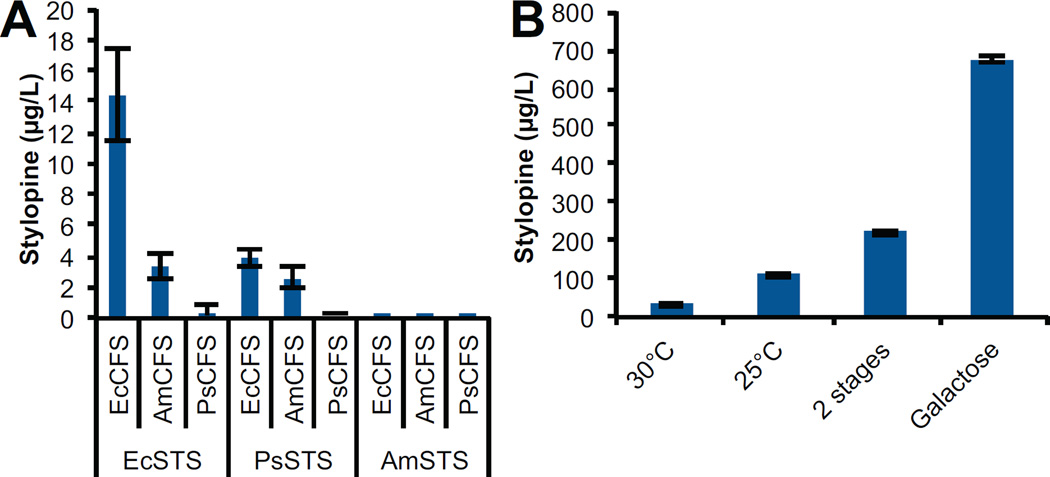
Figure 3.
Optimization of stylopine production through genetic and culture strategies. (A) Stylopine production in yeast strains as a function of the combination of the species variants of CFS and STS. CFS and STS variants were expressed from separate low-copy plasmids in CSY844. (B) Stylopine production in CSY904 grown under various culture conditions. 30 °C: 30 °C growth temperature; 25 °C: 25 °C growth temperature. 2 Stages: cultured at 25 °C with an initial growth phase followed by a production phase; Galactose: grown as described in 2 stages with 2% galactose used as the carbon source during the production phase. Metabolite production is determined by LC-MS analysis of culture media after indicated strains were grown for 96 h. Data is reported as the mean ± s.d. of at least 3 independent experiments.
We investigated culture optimization as a way to improve stylopine production. First, we integrated EcSTS into the chromosome (CSY904) to circumvent the need for selective media and potential plasmid stability issues. We examined the effect of growth temperature on plant P450 activity, as growth at lower temperatures has been shown to improve cytochrome P450 activity in yeast (Zimmer et al., 1997). Stylopine production increased by 3.4-fold from a titer of 43 µg/L to 112 µg/L when the strain was grown at 25 °C compared to 30 °C (Fig. 3B). The data indicate the strains are more productive on a per OD basis (Fig. S3C), such that the improved production of stylopine is due to an increase in the bulk activity of the enzymes and not due to higher cell densities.
We next developed a culture method to support higher cell culture densities while maintaining the per cell productivity by separating the culture into two-stage: an initial growth stage followed by a BIA production stage. During the growth phase we cultured the stylopine-producing strain in 5X YNB dropout media with 2% dextrose for 48 h at 25 °C. We harvested the cells by centrifugation and resuspended the cell pellet in 1X YNB dropout media supplemented with 2% dextrose and 2 mM norlaudanosoline and grew for 96 h at 25 °C. Using this culture strategy we improved stylopine titer by an additional 2-fold to 222 ug/L in the densely grown culture, while maintaining the per OD productivity of the cells (Fig. S3D).
We further optimized the two-stage growth system by varying the carbon source during the production phase. After a 48 hour growth phase in 5X YNB dropout media with 2% dextrose, we harvested the cells by centrifugation and resuspended the cell pellets in 1X YNB dropout media with 2 mM norlaudanosoline and either 2% dextrose, 2% galactose, 2% sucrose, 4% glycerol or no carbon source (Fig. S3E). The strains were grown at 25 °C for 96 h (Fig. 3B). Stylopine titer was improved by a further 3-fold to 676 µg/L when the cultures were grown with galactose in the production phase. Production in galactose increased the overall BIA titer by 2.2-fold (Fig. S3F). The total increase in stylopine production through all optimization methods was 45-fold.
3.4 Engineering a microbial production host for (S)-cis-N-methylstylopine and protopine
The next step in the pathway is the conversion of (S)-stylopine to (S)-cis-N-methylstylopine by the enzyme tetrahydroprotoberberine-N-methyltransferase (TNMT). We first examined the activity of two TNMT variants (E. californica, EcTNMT (Liscombe et al., 2009); P. somniferum, PsTNMT (Liscombe and Facchini, 2007)) by expressing each from a low-copy plasmid in CSY904. We found that PsTNMT was more active than EcTNMT (Fig. S4D), and we integrated PsTNMT into CSY904 to create CSY968. We tested CSY968 in culture conditions optimized for stylopine production and cis-N-methylstylopine production was confirmed through comparison of the retention time and reported fragmentation patterns (Schmidt et al., 2007). We found that CSY968 was very efficient and produced 548 ug/L cis-N-methylstylopine with 77% conversion efficiency from stylopine (Fig. 4A). Due to the high efficiency of PsTNMT, no additional optimization was performed.
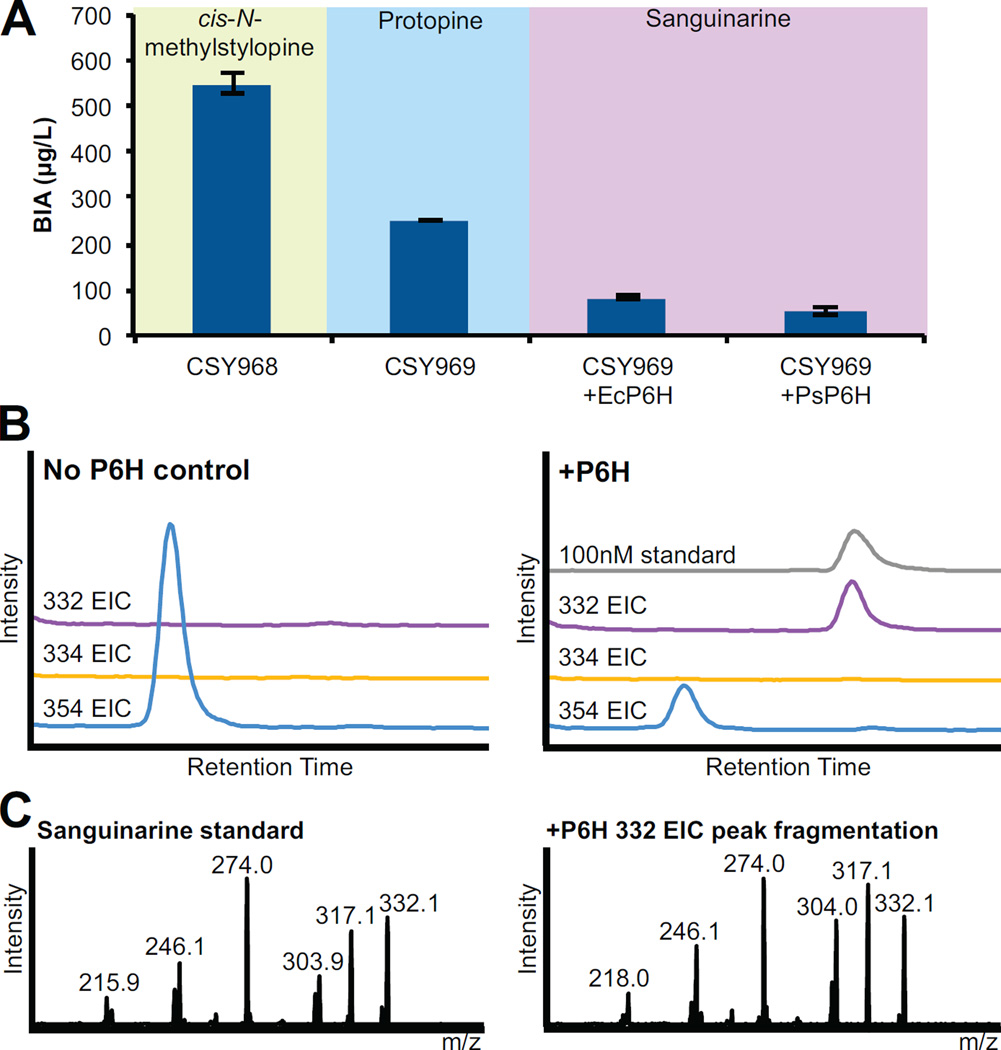
Figure 4.
Microbial production of cis-N-methylstylopine, protopine, and sanguinarine in optimized culture conditions. (A) Production of cis-N-methylstylopine, protopine, and sanguinarine yeast strains under optimized growth conditions. CSY968 contains PsTNMT integrated and CSY969 contains both PsTNMT and PsMSH integrated. P6H variants were expressed from a low-copy plasmid in CSY969. Metabolite production is determined by LC-MS analysis of culture media after indicated strains were grown for 96 h (CSY968) or 10 d (CSY969, CSY969 with P6H). Data is reported as the mean ± s.d. of at least 3 independent experiments. (B) LC-MS analysis of growth media of CSY969 with EcP6H. Traces are shown for a no EcP6H enzyme control strain (left) and an engineered strain expressing EcP6H (right). EICs for compounds corresponding to protopine (354 m/z), dihydrosanguinarine (334 m/z), and sanguinarine (332 m/z) are shown. A sanguinarine standard is included for comparison. (C) MS2 spectra for the sanguinarine standard (left) and the 332 EIC peak produced from the engineered yeast strain (right). LC-MS traces and fragmentation patterns are representative of at least 3 independent experiments.
Protopine is produced through the hydroxylation of (S)-cis-N-methylstylopine by the cytochrome P450 (S)-cis-N-methylstylopine 14-hydroxylase (MSH), the third P450 enzyme in the sanguinarine branch and converts the protoberberine scaffold to a protopine scaffold. Only one species variant of MSH (P. somniferum MSH, PsMSH) has been reported (Beaudoin and Facchini, 2013). We utilized our plant P450 optimization strategies and expressed a yeast codon-optimized version of this variant from the GPD promoter. The expression construct was integrated into the chromosome of CSY968 to build the protopine-producing strain CSY969. Protopine production was confirmed by comparison of the retention time and fragmentation pattern to a protopine standard and reported patterns (Schmidt et al., 2007) (Fig. S4A–C). When we characterized CSY969 in optimized conditions, we found that the strain grew more slowly, likely due to the overexpression of multiple P450s, so production was examined on day 4, 7, and 10 (Fig. S4D). On day 10, 252 µg/L of protopine was produced with 61% conversion efficiency from cis-N-methylstylopine (Fig. 4A).
3.5 Microbial production of sanguinarine from protopine through expression of a single plant P450 enzyme
The next step in the sanguinarine pathway changes the protopine scaffold to the benzophenanthridine scaffold by oxidizing protopine to dihydrosanguinarine via protopine 6-hydroxylase (P6H), the fourth cytochrome P450 in the sanguinarine branch. We applied our P450 optimization strategies and expressed two yeast codon optimized variants of P6H (E. californica, EcP6H (Takemura et al., 2013); P. somniferum, PsP6H (Beaudoin and Facchini, 2013)) each from a low-copy plasmid with a GPD promoter in the protopine-producing strain CSY969. When we assayed the strains in the optimized culture conditions after 4, 7 and 10 d growth, we were unable to detect dihydrosanguinarine (334 m/z) accumulation. However, after 7 d of growth, we detected a peak with m/z = 332, which corresponds to the m/z value of sanguinarine, the end-product of the pathway (Fig S4E). We compared the retention time and fragmentation spectra of this peak to a sanguinarine standard (Fig. 4B, C) and confirmed the identity of this compound as sanguinarine. We observed that EcP6H in our yeast strains produced 80 ug/L sanguinarine with a conversion efficiency from protopine of 24% while PsP6H produced 52 µg/L sanguinarine (Fig. 4A). Our results indicate that the oxidation of dihydrosanguinarine to sanguinarine can occur in the context of our engineered yeast strain without the addition of the final enzyme dihydrobenzophenanthridine oxidase (DBOX). However, the exact mechanism of the oxidation of dihydrosanguinarine is unclear.
4 Conclusions
We engineered a set of yeast strains as production hosts for the protoberberine, protopine, and benzophenanthridine scaffolds, three subclasses of BIAs with promising pharmaceutical properties. We demonstrate production of sanguinarine in an optimized yeast strain from the fed substrate norlaudanosoline, which required the heterologous expression of 10 enzymes, including four plant cytochrome P450 enzymes (EcCFS, EcSTS, PsMSH, EcP6H), a class of enzymes notoriously difficult to functionally express in heterologous microbial hosts. We report the highest titers achieved to date in a microbial fermentation for the compounds cheilanthifoline, stylopine, cis-N-methylstylopine, protopine, and sanguinarine. Our approach emphasized the application of classic metabolic engineering techniques with industrially relevant culture and assay strategies as well as identifying generalizable genetic and culture strategies to optimize the functional expression of multiple plant cytochrome P450s in the context of a complex biosynthetic pathway.
The greatest gains in BIA production at bottleneck steps along the pathway were achieved by testing all available enzyme species variants. A similar approach was used to screen four species variants of CYP71D for the conversion of (+)-valencene to nootkatone in yeast (Gavira et al., 2013). These examples demonstrate the importance of exploring the functional space defined by the natural sequence diversity in the heterologous yeast host in the context of a heterologous biosynthetic pathway to inform pathway engineering strategies. For example, previous work in the dihydrosanguinarine pathway in yeast only examined CFS and STS enzymes from P. somniferum (Fossati et al., 2014). However, we found that the E. californica CFS exhibited 7.4-fold higher activity compared to P. somniferum CFS when paired with ATR1, and the combination of EcCFS and EcSTS resulted in 38-fold higher stylopine production in our yeast strains than the pairing of PsCFS and PsSTS variants as previously reported (Fossati et al., 2014). By examining multiple species variants of CPRs, we found that the CPR from A. thaliana is a versatile CPR able to pair effectively with all four P450s in this pathway. Our results highlight that it is not necessary to match P450s with the same species CPR and that ATR1 can be used as a generalizable CPR in plant pathway reconstruction in yeast. Further optimization of the pairing between the plant P450s and the CPR may be achieved through altering the expression level of the CPR relative to the P450s (Paddon et al., 2013).
We also found that the regulatory expression profile of plant cytochrome P450s can substantially impact their activity in a yeast host. Expression from a low-copy plasmid or the chromosome improved P450 activity compared to expression from a high-copy plasmid. We linked the observed differences in activity to differences in population-level expression and ER morphology. It is likely that high-level expression of endomembrane-localized P450s is stressful to the yeast host and creates a selection pressure to lose the plasmid. The development of bright aggregates within subcompartments of the ER suggests accumulation of unstable or otherwise non-functional enzymes. This differential subcellular localization has been previously observed for heterologous P450 expression in S. cerevisiae (Sandig et al., 1999; Zimmer et al., 1997). Our observations are the first to examine these physiological impacts on P450 expression in the context of an engineered metabolic pathway. Our promoter studies further suggest that the optimization of P450 expression is likely a balance between stability of expression and high transcriptional strength. Promoters that exhibit strong expression profiles earlier in the growth cycle, such as PGPD and PPGK1, support the highest activity levels from plant P450s in the yeast host.
We developed culture conditions that improve the activity of plant P450s and increase the overall flux through the pathway in yeast. Previous work on the dihydrosanguinarine pathway relied on whole-cell buffered assays, which preclude culture optimization approaches. Through our culture optimization techniques, we were able to achieve a conversion efficiency of 11% between stylopine and sanguinarine, whereas previous work reached only 2% conversion (Fossati et al., 2014). We found that growing the cells at reduced temperature increases some P450 activities, particularly stylopine synthase. The mechanism of this improvement is most likely related to improved protein folding. We also found that separating the culture into a growth stage and production stage, allowed us to reach higher ODs while maintaining the per cell productivity. Two-stage culture methods have been described with an initial growth stage followed by expression of the pathway components through activation by an inducer molecule or a nutrient, such as for the optimized production of amorphadiene (Westfall et al., 2012) and β-nootkatol (Gavira et al., 2013) in yeast and reticuline in E. coli (Nakagawa et al., 2012). In our system, the pathway is expressed during both stages and production of BIAs is controlled by the addition of the fed precursor molecule in the production stage. We also found that the carbon source used during the production phase can substantially alter the culture’s productivity, with galactose resulting in the highest levels of overall BIA production. Galactose induces a variety of metabolic and transcriptional responses in yeast; future studies may focus on identifying which of these responses are related to the observed improvements in BIA titers.
Finally, we found that sanguinarine can be synthesized by yeast cultures expressing the sanguinarine pathway through P6H and lacking the DBOX enzyme. Characterization of P6H was previously performed in yeast microsomes (Takemura et al., 2013), where dihydrosanguinarine was reported as the major product under these conditions. In whole-cell buffered assays of dihydrosanguinarine production in yeast, the detection of sanguinarine was reported as a trace product (Fossati et al., 2014). Our data indicate that actively growing yeast cultures promote the efficient oxidation of dihydrosanguinarine to sanguinarine, possibly through non-enzymatic mechanisms (e.g., media components, aeration, low pH), endogenous enzyme activities, and/or uncharacterized activities of heterologously expressed enzymes. Future studies may focus on elucidating the mechanism of the efficient formation of sanguinarine in this system.
This work provides a set of strategies for engineering complex plant natural product pathways in yeast, particularly with respect to the expression of pathways incorporating multiple plant P450 enzymes. In combination with recent advances in opioid (Thodey et al., 2014), sesquiterpene (Gavira et al., 2013; Paddon et al., 2013; Westfall et al., 2012), triterpene (Dai et al., 2013; Moses et al., 2014), and glucosinate (Mikkelsen et al., 2012) production in yeast, the work presented here demonstrates that S. cerevisiae is a robust platform for the production of plant natural products. Microbial biosynthesis is a promising technology to address manufacturing needs for pharmaceutically important plant-derived compounds and also as tool for the discovery and screening of new compounds and associated enzymes.
Supplementary Material
Highlights.
Yeast strains produce three structural subclasses of benzylisoquinoline alkaloids.
Strategies to increase plant P450 activities in multi-step pathways are described.
Culture methods significantly increase flux through plant pathways in yeast.
The end-product sanguinarine can be made without the associated enzyme.
Acknowledgements
We thank Stanford Cell Sciences Imaging Facility for providing fluorescence microscopy access (Leica SP5, NIH grant SIG number: 1S10RR02557401) and training; S. Galanie, Y. Li, M. McKeague, C. Schmidt, M. Siddiqui, K. Thodey for discussions and feedback in the preparation of this manuscript. This work was supported by the National Institutes of Health (grant to C.D.S., AT007886), National Science Foundation (grant to C.D.S., CBET-1066100; fellowship to I.J.T.), Bill and Melinda Gates Foundation (grant to C.D.S., OPP1058690), and ARCS (fellowship to I.J.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
IJT and CDS conceived of the project, designed the experiments, analyzed the results, and wrote the manuscript. IJT performed the experiments.
Competing interests statement The authors declare competing financial interests in the form of a pending patent application.
References
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. 2007:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W, Zenk M. Two methylenedioxy bridge forming cytochrome P-450 dependent enzymes are involved in (S)-stylopine biosynthesis. Phytochemistry. 1991:30. [Google Scholar]
- Beaudoin GW, Facchini PJ. Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem. Biophys. Res. Commun. 2013;431:597–603. doi: 10.1016/j.bbrc.2012.12.129. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- Chemler JA, Koffas MG. Metabolic engineering for plant natural product biosynthesis in microbes. Curr. Opin. Biotechnol. 2008;19:597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Dai Z, Liu Y, Zhang X, Shi M, Wang B, Wang D, Huang L, Zhang X. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab. Eng. 2013;20:146–156. doi: 10.1016/j.ymben.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2006;93:212–224. doi: 10.1002/bit.20694. [DOI] [PubMed] [Google Scholar]
- Díaz Chávez ML, Rolf M, Gesell A, Kutchan TM. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 2011;507:186–193. doi: 10.1016/j.abb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Duan H, Schuler MA. Heterologous expression and strategies for encapsulation of membrane-localized plant P450s. Phytochem. Rev. 2006;5:507–523. [Google Scholar]
- Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Fossati E, Ekins A, Narcross L, Zhu Y, Falgueyret J-P, Beaudoin GAW, Facchini PJ, Martin VJJ. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 2014;5:3283. doi: 10.1038/ncomms4283. [DOI] [PubMed] [Google Scholar]
- Gavira C, Höfer R, Lesot A, Lambert F, Zucca J, Werck-Reichhart D. Challenges and pitfalls of P450-dependent (+)-valencene bioconversion by Saccharomyces cerevisiae. Metab. Eng. 2013;18:25–35. doi: 10.1016/j.ymben.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Yeast Transformation by the LiAc/SS Carrier DNA/PEG Method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Beaudoin G, a W, Fossati E, Ekins A, Martin VJJ, Facchini PJ. Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J. Biol. Chem. 2012;287:42972–42983. doi: 10.1074/jbc.M112.420414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 2013;54:647–672. doi: 10.1093/pcp/pct020. [DOI] [PubMed] [Google Scholar]
- Hawkins K, Smolke C. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa N, Iwasa K, Sato F. Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 2007;274:1019–1035. doi: 10.1111/j.1742-4658.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Ikezawa N, Iwasa K, Sato F. CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 2009;28:123–133. doi: 10.1007/s00299-008-0624-8. [DOI] [PubMed] [Google Scholar]
- Kashiwada Y, Aoshima A, Ikeshiro Y, Chen Y-P, Furukawa H, Itoigawa M, Fujioka T, Mihashi K, Cosentino LM, Morris-Natschke SL, Lee K-H. Anti-HIV benzylisoquinoline alkaloids and flavonoids from the leaves of Nelumbo nucifera, and structure-activity correlations with related alkaloids. Bioorg. Med. Chem. 2005;13:443–448. doi: 10.1016/j.bmc.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Kemény-Beke A, Aradi J, Damjanovich J, Beck Z, Facskó A, Berta A, Bodnár A. Apoptotic response of uveal melanoma cells upon treatment with chelidonine, sanguinarine and chelerythrine. Cancer Lett. 2006;237:67–75. doi: 10.1016/j.canlet.2005.05.037. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ. Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 2007;282:14741–14751. doi: 10.1074/jbc.M611908200. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Ziegler J, Schmidt J, Ammer C, Facchini PJ. Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 2009;60:729–743. doi: 10.1111/j.1365-313X.2009.03980.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Buron LD, Salomonsen B, Olsen CE, Hansen BG, Mortensen UH, Halkier BA. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Minami H, Kim J-S, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- Moses T, Pollier J, Almagro L, Buyst D, Van Montagu M, Pedreño MA, Martins JC, Thevelein JM, Goossens A. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1634–1639. doi: 10.1073/pnas.1323369111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Minami H, Kim J-S, Koyanagi T, Katayama T, Sato F, Kumagai H. Bench-top fermentative production of plant benzylisoquinoline alkaloids using a bacterial platform. Bioeng. Bugs. 2012;3:49–53. doi: 10.4161/bbug.3.1.18446. [DOI] [PubMed] [Google Scholar]
- Orhana I, Ozçelik B, Karaoglu T, Sener B. Antiviral and antimicrobial profiles of selected isoquinoline alkaloids from Fumaria and Corydalis species. Z. Naturforsch. C. 2007;62:19–26. doi: 10.1515/znc-2007-1-204. [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai a, Main a, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjørn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- Renault H, Bassard J-E, Hamberger B, Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr. Opin. Plant Biol. 2014;19:27–34. doi: 10.1016/j.pbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho Ka, Eachus Ra, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rosco A, Pauli HH, Priesner W, Kutchan TM. Cloning and heterologous expression of NADPH-cytochrome P450 reductases from the Papaveraceae. Arch. Biochem. Biophys. 1997;348:369–377. doi: 10.1006/abbi.1997.0374. [DOI] [PubMed] [Google Scholar]
- Sandig G, Kärgel E, Menzel R, Vogel F, Zimmer T, Schunck WH. Regulation of endoplasmic reticulum biogenesis in response to cytochrome P450 overproduction. Drug Metab. Rev. 1999;31:393–410. doi: 10.1081/dmr-100101926. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Boettcher C, Kuhnt C, Kutchan TM, Zenk MH. Poppy alkaloid profiling by electrospray tandem mass spectrometry and electrospray FT-ICR mass spectrometry after [ring-13C6]-tyramine feeding. Phytochemistry. 2007;68:189–202. doi: 10.1016/j.phytochem.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Takemura T, Ikezawa N, Iwasa K, Sato F. Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry. 2013;91:100–108. doi: 10.1016/j.phytochem.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer T, Vogel F, Ohta A, Takagi M, Schunck WH. Protein quality--a determinant of the intracellular fate of membrane-bound cytochromes P450 in yeast. DNA Cell Biol. 1997;16:501–514. doi: 10.1089/dna.1997.16.501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.