Abstract
Background and Purpose
Intracerebral hemorrhage (ICH) has a substantial genetic component. We performed a preliminary search for rare coding variants associated with ICH.
Methods
757 cases and 795 controls were genotyped using the Illumina HumanExome Beadchip (Illumina, Inc. San Diego, CA, USA). Meta-analyses of single-variant and gene-based association were computed.
Results
No rare coding variants were associated with ICH. Three common variants on chromosome 19q13 at an established susceptibility locus, encompassing TOMM40, APOE, and APOC1 met genome-wide significance (p<5e-08). After adjusting for the APOE epsilon alleles, this locus was no longer convincingly associated with ICH. No gene reached genome-wide significance level in gene-based association testing.
Conclusions
While no coding variants of large effect were detected, this study further underscores a major challenge for the study of genetic susceptibility loci – large sample sizes are required for sufficient power except for loci with large effects.
Keywords: intracerebral hemorrhage, exome array, rare variant association study
Introduction
Genetic variation plays a substantial role in the risk of intracerebral hemorrhage (ICH).1 Genome-wide association studies (GWAS) have identified common variants associated with risk of ICH, both lobar and non-lobar subtypes.2 The degree to which rare genetic variants, those with minor allele frequencies far smaller than those of variants typically discovered through GWAS, contribute to this risk is unknown. Preliminary targeted sequencing studies have supported a possible role for rare variants in sporadic ICH.3 Recently, the exome array has emerged as an efficient, cost-effective tool to bridge array-based common variant association studies and whole-exome or whole-genome sequencing in order to identify coding variation underlying common conditions. The goal of this study was to explore the role of exonic variants in risk of ICH, using exome array.
Methods
Study subjects, genotyping and quality control procedures are described in the Supplemental Methods.
Scores and minor allele frequency (MAF) for individual variants, and a covariance matrix for each gene were computed, including age, gender, and the first two principal components as covariates in the model. Inverse variance-weighted meta-analysis of score tests was computed for both common and rare variants.
As MAF decreases, single-variant analysis loses the power to reach genome-wide significance, even in the presence of a true association. Therefore, variants within each gene or region of interest are aggregated in order to increase the power to detect variants with small effects. We applied SKAT, SKAT-O, and T1 count tests for gene-based analysis.4 In analysis using SKAT, each SNP was weighted by the inverse of its standard error and its MAF, where variants with lower MAF are relatively up-weighted. In the T1 count test each variant was weighted equally irrespective of their MAF. The association models were adjusted for age, gender and the first two principal components.
We performed association analysis in all subjects, as well as separately for lobar ICH. Analysis of non-lobar ICH was not performed due to small sample size. Quality control was performed using PLINK v1.07. All other analyses were carried out using seqMeta package in R version 3.0.2.5
Results
After excluding subjects for quality (n=31) and genetic outliers (n=56), there were 1553 subjects for analysis (Supplemental Table I).
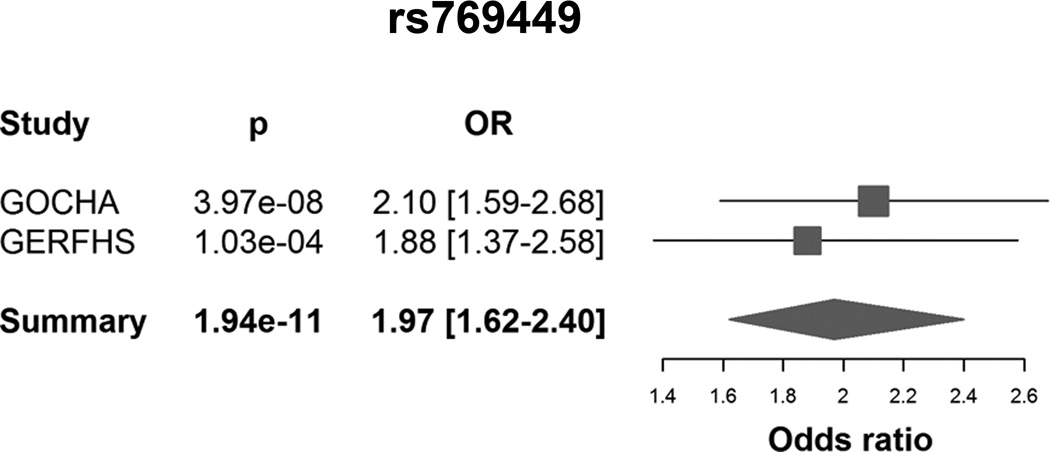
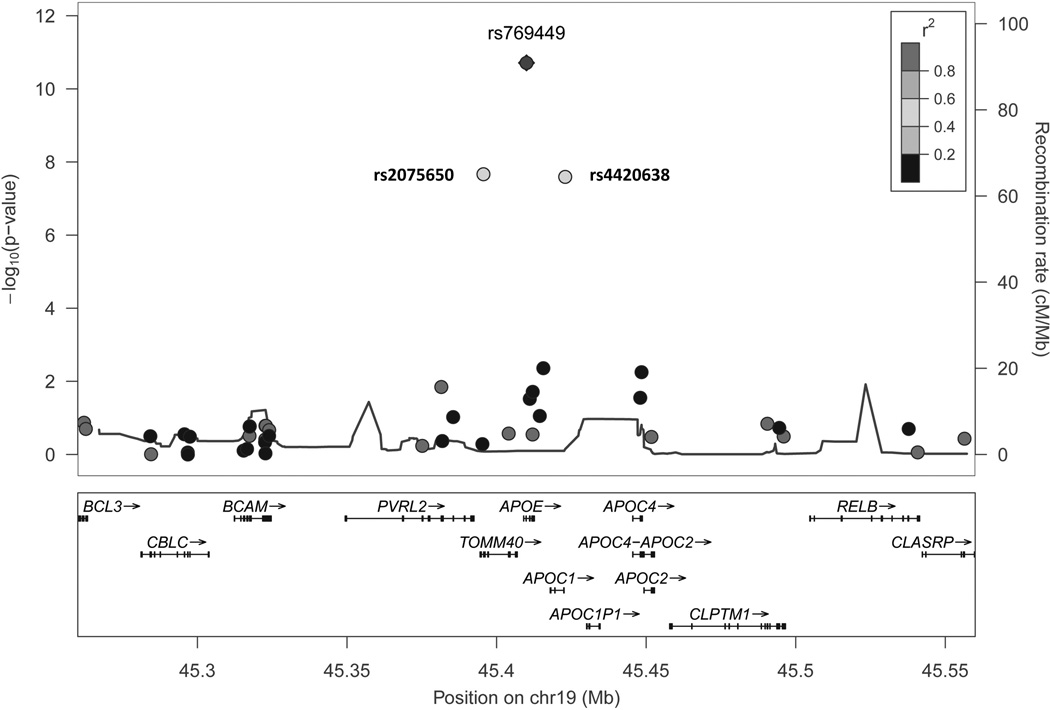
In single-variant analysis, we identified a susceptibility locus at chr19q13 (p<5e-08), including 3 common variants with MAF ranging 13%-19% (Table 1) (Supplemental Figure I). The top variant at this locus was rs769449, which is an intronic SNP on APOE (p=1.94e-11; OR=1.97 [95%CI: 1.62–2.40]). There was no evidence of heterogeneity across two studies (Figure 1). These variants are in moderate linkage disequilibrium, with r2 estimates ranging 0.4–0.6 (Figure 2).
Table 1.
Variants associated with ICH at p<5e-08 in meta-analysis
| CHR | SNP | Gene | BPP | Coded Allele |
MAF | Unadjusted for ε alleles | Adjusted for ε alleles | ||
|---|---|---|---|---|---|---|---|---|---|
| OR [95%CI] | p | OR [95%CI] | p | ||||||
| 19 | rs769449 | APOE | 45410002 | A | 0.13 | 1.97 [1.62–2.40] | 1.94e-11 | 1.99 [1.23–3.22] | 0.006 |
| 19 | rs2075650 | TOMM40 | 45395619 | G | 0.15 | 1.70 [1.42–2.03] | 2.14e-08 | 1.26 [0.91–1.74] | 0.15 |
| 19 | rs4420638 | APOC1 | 45422946 | G | 0.19 | 1.62 [1.38–1.89] | 2.54e-08 | 1.11 [0.76–1.61] | 0.58 |
BPP: Base pair position
CHR: Chromosome
ε: APOE epsilon alleles 2 and 4
MAF: Minor allele frequency
Figure 1.
Forest plot depicting effect estimates for rs769449
Figure 2.
Regional association plot for 19q13 locus
The 19q13 locus encompasses TOMM40, APOE, and APOC1. Common variants in this locus have been associated with several traits, including lipid levels, Alzheimer’s disease, cerebral amyloid angiopathy, and ICH.6,7 Given the association of APOE ε2 and ε4 alleles with ICH, we adjusted for these alleles, which had been previously genotyped in the majority of study subjects.8 (Supplemental Table II) This adjustment resulted in loss of the observed signal, suggesting that these associations arose from the effect of ε2 and ε4 alleles (Table 1).
No low frequency variant or gene emerged as associated with ICH or the lobar subtype using SKAT, SKAT-O, or burden tests before and after adjustment for the ε2 and ε4 alleles. The strongest association for the gene-based analyses was observed for GADL1 in the T1 count test after adjustment for the epsilon alleles (p=6.37e-05; cumulative MAF=3.3%).
Discussion
Common genetic variation appears to play a substantial role in ICH risk and key clinical features, including clinical outcome.1 Ongoing GWAS are designed to detect common variants, but they may miss rare variation. The contribution from rare variation is less substantiated.
The present analysis did not identify any rare coding variants for ICH. Our effort to identify coding variation with modest effect sizes was limited by inadequate statistical power. The gene-based tests can partially compensate for this limitation, but still lack of sufficient number of observations of rare variants in such small sample sizes prohibits taking full advantage of this approach. We estimated our power was approximately 7% for detection of a significant association at p<1e-06 (corrected for multiple tests in the gene-based analysis) at maximum OR=5 when MAF=0.0001.9 Our data therefore suggest that most genetic risk for ICH resides within common and rare variants with modest effect size. Accurate estimation of the extent to which rare variants contribute to risk of ICH will require larger scale sequencing studies with coverage of both common and rare variants.
Development of international consortia has facilitated recruitment of hundreds of thousands of subjects with common diseases like ischemic stroke and accelerated the rate of genetic discoveries for complex traits. With decreasing costs of sequencing studies and further expansion of consortia, genetic characterization of less common conditions like ICH, will become more feasible.
Supplementary Material
Acknowledgments
Sources of Funding
This study was supported by grants R01NS073344, R01NS059727, and 5K23NS059774 from the NIH–National Institute of Neurological Disorders and Stroke (NIH-NINDS) and, 0755984T from American Heart Association. Computation support provided, in part, by the Wake Forest School of Medicine Center for Public Health Genomics.
Disclosures
C.D.A received research grants from NIH-NINDS, American Brain Foundation, and Massachusetts General Hospital Institute for Heart, Vascular and Stroke Care. B.B.W. received a research grant from the NIH, and is the Associate Editor for the journal Neurology. J.R. received research grants from the NIH, and is a consultant to Boehringer Ingelheim.
References
- 1.Devan WJ, Falcone GJ, Anderson CD, Jagiella JM, Schmidt H, Hansen BM, et al. Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013;44:1578–1583. doi: 10.1161/STROKEAHA.111.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeanne M, Labelle-Dumais C, Jorgensen J, Kauffman WB, Mancini GM, Favor J, et al. Col4a2 mutations impair col4a1 and col4a2 secretion and cause hemorrhagic stroke. American journal of human genetics. 2012;90:91–101. doi: 10.1016/j.ajhg.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.R: A Language and Environment for Statistical Computing. Version 3.0.2. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 6.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, et al. Variants at apoe influence risk of deep and lobar intracerebral hemorrhage. Annals of neurology. 2010;68:934–943. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly LA, Palmer CN, Whitley AL, Lang CC, Doney AS, Morris AD, et al. Apolipoprotein e genotypes are associated with lipid-lowering responses to statin treatment in diabetes: A go-darts study. Pharmacogenetics and genomics. 2008;18:279–287. doi: 10.1097/FPC.0b013e3282f60aad. [DOI] [PubMed] [Google Scholar]
- 8.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, et al. Variants at apoe influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol. 2010;68:934–943. doi: 10.1002/ana.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.