Abstract
The CAS (Crk-associated substrate) adaptor protein family consists of four members: CASS1/BCAR1/p130Cas, CASS2/NEDD9/HEF1/Cas-L, CASS3/EFS/Sin and CASS4/HEPL. While CAS proteins lack enzymatic activity, they contain specific recognition and binding sites for assembly of larger signaling complexes that are essential for cell proliferation, survival, migration, and other processes. All family members are intermediates in integrin-dependent signaling pathways mediated at focal adhesions, and associate with FAK and SRC family kinases to activate downstream effectors regulating the actin cytoskeleton. Most studies of CAS proteins to date have been focused on the first two members, BCAR1 and NEDD9, with altered expression of these proteins now appreciated as influencing disease development and prognosis for cancer and other serious pathological conditions. For these family members, additional mechanisms of action have been defined in receptor tyrosine kinase (RTK) signaling, estrogen receptor signaling or cell cycle progression, involving discrete partner proteins such as SHC, NSP proteins, or AURKA. By contrast, EFS and CASS4 have been less studied, although structure-function analyses indicate they conserve many elements with the better-known family members. Intriguingly, a number of recent studies have implicated these proteins in immune system function, and the pathogenesis of developmental disorders, autoimmune disorders including Crohn’s Disease, Alzheimer’s Disease, cancer, and other diseases. In this review, we summarize the current understanding of EFS and CASS4 protein function in the context of the larger CAS family group.
Keywords: EFS, CASS3, SIN, CASS4, HEPL, CAS family protein, inflammation, signal transduction, prognosis
Introduction
The CAS (CRK-associated substrate) family of adaptor proteins consists of four members: BCAR1 (also known as p130Cas and CASS1) [1], NEDD9 (also known as HEF1, Cas-L, and CASS2) [2, 3], EFS/SIN/CASS3 [4] and HEPL/CASS4 [5]. Each of these proteins contains specific binding and recognition sites allowing them to serve as scaffolds to assemble larger signaling complexes, and lacks known enzymatic activity. Intensive study predominantly of BCAR1 and NEDD9 in the 1990s led to the early development of a paradigm for the function of this protein group (reviewed in [6]). In brief, engagement of integrins at the cell surface causes activation of focal adhesion kinase (FAK) or its paralog PTK2B (also known as RAFTK or Pyk2), which binds and phosphorylates a CAS protein. Subsequently, a SRC family kinase binds a site created by the FAK-phosphorylation event and hyperphosphorylates CAS, creating binding sites for CRK and associated proteins that reorganized the actin cytoskeleton in a manner that influences integrin-mediated attachment, cell spreading, and migration. Disruption of the function of these complexes led to anoikis (detachment-induced cell death), while overexpression or hyperphosphorylation of CAS proteins accompanies and supports cell transformation.
In this context, a literature addressing the paralogous EFS and CASS4 family members has been slower to emerge. In general, an advantage of studies in lower eukaryotes is the opportunity to dissect protein function in a signaling environment in which few paralogues are present. In mammals, it is often challenging to distinguish redundant versus unique signaling features, and therefore establish the biological importance of each gene within expanded protein families. Given the near-ubiquitous expression of BCAR1 in particular [7], and the abundant expression of NEDD9 in many tissues [3, 8], identifying discrete roles for other CAS family members is not trivial. However, there are a growing number of cases where genes that were thought to have predominantly redundant function were subsequently shown to take part in distinct feedback loops and synergistic signaling, or to have partially opposing function that can have important biological consequences and influence clinical outcomes in the case of disease. As one example, clinical effectiveness of RAF inhibitors in the context of V600E mutant (constitutively active) versus wild type BRAF is influenced strongly by the functionality of the paralogous CRAF protein, based on heterodimerization and transactivation processes specific to each of the two proteins [9]. Such examples reinforce the importance of understanding the role of individual paralogues.
In the CAS family, as discussed below, all family members share common structural features and have at least some similar functions in cell signaling, related to the paradigm noted above. However, individual roles have emerged for the better-studied BCAR1 and NEDD9 proteins that differentiate part of their function from the remainder of the CAS group. For example, BCAR1 expression regulates tamoxifen resistance in breast cancer [10], and plays a crucial role in cardiovascular system and liver development [11] and osteoclast-dependent bone resorption [12]. BCAR1 supports myelin formation by oligodendrocytes in the central nervous system [13], acts as a downstream effector of serotonin (5-HT) signaling in the process of vasoconstriction [14], and serves as an important mediator for several infectious diseases including infection by Salmonella typhimurium [15], Plasmodium falciparum-associated malaria [16], pathogenic Yersinia [17, 18], and Kaposi’s sarcoma-associated herpesvirus [19]. NEDD9 known regulate neural crest cells migration during embryogenesis, which is crucial for proper neural system development [20], functions at centrosomes to activate the Aurora-A (AURKA) mitotic kinase [21], and at the basal body to activate AURKA in causing disassembly of the primary cilia [22]. Based on these non-canonical roles, NEDD9 function has recently been shown to modulate pathogenesis of the ciliopathy autosomal dominant polycystic kidney disease (ADPKD) [23].
In contrast, the literature on EFS and CASS4 has been slower to develop. Nevertheless, over the past several years, a growing number of studies have begun to define similarities and differences between these and other CAS proteins, and to implicate EFS and CASS4 as causative factors or effectors for clinically important human diseases, including developmental disorders, neurodegenerative syndromes, autoimmune disorders, and cancer. In this review, we summarize the current status of the literature on these two proteins, their structure, functions and roles in signaling transduction of signaling pathways.
Identification of EFS and CASS4
EFS (Embryonal Fyn-associated Substrate), also known as SIN (Src INteracting or Signal INtegrating protein) was discovered in two independent studies, conducted by Ishino et al. [4] in 1995 and Alexandropoulos et al. [24] in 1996. Ishino and colleagues performed a cDNA library screening of a mouse embryonal library for proteins containing SH3-interacting domains to identify EFS, while Alexandropoulos screened a mouse embryonal library looked for proteins interacting with the SRC SH3 domain, leading to the designation SIN.
CASS4, the last described member of the CAS family, was detected by Singh et al. [5] in 2008 over a decade after the description of the other family members. CASS4 was identified following in silico screening of databases describing expressed sequence tags from an evolutionarily diverse group of organisms, using the mRNAs for the three previously defined CAS proteins as templates. Subsequently, Singh et al cloned and characterized the CASS4 gene, originally assigning the name HEPL (HEF1-EFS-p130Cas-like) for similarity to the other three defined CAS genes.
Gene and mRNA expression for EFS and CASS4
The EFS gene is localized to chromosome 14q11.2, with genomics coordinates 14: 23356400–23365633 on the reverse strand in GRChB38p2 [25] The chromosomal location of the CASS4 gene is 20q13.31, with genomic coordinates of 20: 56411548–56459340 on the forward strand in GRChB38p2 [26].
Relatively little work has been done to directly study the transcriptional regulation of EFS and CASS4. Initial studies profiling EFS mRNA indicated broad expression, with maximal levels in the placenta, the embryonal central nervous system, heart, testes and lungs [27]. EFS expression in the thymus and lymphocytes is functionally important for T cell maturation and prevention of autoimmunity, discussed below [28–30]. A screen for implantation-related genes regulated by progesterone, 17β-estradiol and progesterone found this regimen downregulated EFS mRNA in explants of the late proliferative phase endometrium, suggesting hormone responsiveness [31]. The initial publication describing CASS4 used RT-PCR to demonstrate that CASS4 mRNA is most highly expressed in spleen and lung among normal tissues, and is highly expressed in ovarian and leukemia cell lines [5].
Some further hints as to the transcriptional control of these genes can be gleaned from SABiosciences’ DECODE database, which is based on UCSC Bioinformatics Genome Browser [32]. This resource proposes several transcriptional regulators for EFS based on consensus binding sites in its promoter region for ATF (Activating transcription factor), NF-κβ, NF-κβ1, GATA-3, C/EBPα (CCAAT/enhancer-binding protein alpha), glucocorticoid receptors α and β, and p53. Transcription factor binding motifs in the CASS4 promoter are similar to those for EFS. They include NF-κβ, p53, LCR-F1 (NFE2-L1, nuclear factor, erythroid 2-like1), MAX1, C/EBPα, CHOP-10 (C/EBP homologous protein 10), POU3F1 (POU domain, class 3, transcription factor 1, aka Oct-6), AREB6 (ZEB1, Zinc finger E-box binding homeobox 1).
These are compatible with regulated expression in lymphocytes in relation to differentiation and inflammation [33, 34], hormonal regulation [35], and deregulation in cancer [36, 37]. There are three transcript variants for EFS in humans. Isoform 1 contains 6 exons end encodes the full-length protein (561 amino acids); isoform 2 contains 5 exons and encodes a shorter protein (468 amino acids); and isoform 3 contains 6 exons and encodes the shortest protein (392 amino acids). In humans, four transcript variants of CASS4 are known. The first and second variants 7 exons each and encode the same full-length protein isoform a (786 amino acids, considered the major isoform). The third variant contains 6 exons and encodes a shorter isoform b (732 amino acids), and the fourth variant contains 5 exons and encodes the shortest isoform c (349 amino acids).
Protein structure
The evolutionary divergence of the CAS proteins family members is discussed by Singh et al in detail [5]. There are no CAS homologues in single cell eukaryotes and nematodes such as C. elegans. A single ancestral member, DCAS, is found in Drosophila melanogaster that has common structural characteristics and functions with other proteins within the group [38]. The characteristic domains and motifs are summarized in Table 1 and graphically represented in Figure 1.
Table 1.
Interactive partners defined for association with specific domains of EFS or CASS4.
| Domain | Protein | Amino acids | Length | Interactors | Refs |
|---|---|---|---|---|---|
| SH3 | EFS | 5–68 | 64 | FAK, PTK2B, C3G, PTP-PEST | [45, 50, 99] |
| CASS4 | 11–73 | 63 | FAK | [5] | |
| SH2-binding | EFS | 69–350 | 282 | Crk1/2, CrkL | [24, 53] |
| CASS4 | 72–372 | 301 | CrkL, PKA/PKG | [51], [85] | |
| Serine rich | EFS | 351–488 | 138 | ||
| CASS4 | 373–429 | 57 | |||
| C-terminal | EFS | 489–561 | 73 | SRC, Fyn | [24, 28, 53] |
| CASS4 | 430–786 | 357 | SRC | [5] |
Figure 1. Schematic domain structure and phosphorylation sites comparison of the CAS family proteins.
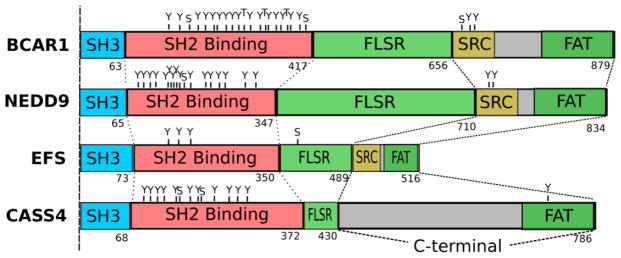
Shown are the longest isoforms for the members of CAS protein family. SH3 = SRC homology 3 binding domain; SH2 Binding = tyrosine phosphosite-enriched domain containing binding sites for partners with SH2 domains; FAT = focal adhesion targeting domain (the central serine rich region (FLSR) of the CAS proteins although has the structure of FAT domain); SRC = SRC family kinase-binding region, contains binding sites for both SRC SH2 and SH3 domains in BCAR1 and EFS, and only SRC SH2 domain in NEDD9). Domain amino acid lengths were obtained from uniprot.org database, indicated in numbering below each image. Phosphorylation sites are designated as follows: Y=tyrosine; S=serine; T=threonine; and were obtained from phosphosite.org.
All CAS proteins maintain a single N-terminal and highly conserved SH3-domain, which allows binding to proline-rich motif containing proteins [4]. This encompasses amino acids 5–68 and 11–73 for EFS and CASS4 respectively. This SH3 domain is highly conserved, with 81% of overall similarity between the CAS family proteins. For EFS and CASS4, the SH3 domains are the most highly conserved domains: for example, the murine and human EFS SH3 domains are 100% identical, while the remainder of the coding sequences are only 78% identical [27], which indicates an important functional role. Similarly, for CASS4, the murine and human SH3 domains are 87.3% identical, while the remainder of coding sequence has 59.8% identity. Important binding partners for CAS family members for this region include FAK [3], PTK2B [39], C3G [40], PTP-PEST [41], PTP1B [42], CIZ [43] and FRNK [44]. Singh et al. used a yeast two-hybrid approach to directly demonstrate an interaction between the CASS4 SH3 domain and FAK. EFS has also been shown to interact with FAK, with an affinity comparable to p130Cas/BCAR1 and NEDD9 [45]. PTP-PEST, a soluble protein tyrosine phosphatase that is ubiquitously expressed in mice both during embryonic development and in adult tissues, opposes FAK and PTK2B activity, as it dephosphorylates PTK2B, FAK and CAS family members, among other proteins [46]. The PTP-PEST proline-rich sequence 332PPKPPR337 has been shown to interact directly with the SH3 domain of members of EFS [47]. This set of binding partners for a highly conserved domain reflects the fundamental role(s) of this SH3 domain in mediating integrin-dependent cell adhesion.
Carboxy-terminal to the SH3 domain, CAS proteins contain a “substrate domain” (amino acids 69–350 for human EFS and 74–372 for human CASS4) with multiple repeats of specific sequences (YxxP) which are phosphorylated on tyrosines by SRC or SRC-related kinases, creating binding motifs for SH2 domains-containing signaling proteins [48]. Both EFS and CASS4 have a limited number of potential SH2 binding sites (estimated at 9 and 10, respectively), in contrast to BCAR1 and NEDD9, which have 20 and 18 of these motifs [5]. Important interactive partners binding this region include Crk1/2 and Crk-L, SH2- and SH3 domain-containing adaptor proteins that are important for regulation of cellular motility and migration [2, 5, 27, 49]; and C3G, a guanine nucleotide exchange factor (GEF) for RAP1 [50]. Interactions with CRK and C3G subsequently promote reorganization of the actin cytoskeleton [38]. Direct binding has been identified between CRKL and CASS4 [51], [52], while EFS has been shown to bind Crk1/2 [24] and CrkL [53]. Studies of BCAR1 have shown that YxxP sequences in this region become more available to phosphorylation by SRC upon mechanical stretch of the protein [54] which implies a role of CAS proteins as direct intracellular mechanoreceptors mediating cellular response to mechanical forces, especially considering that all four CAS proteins localize at focal adhesions [2, 5, 50, 55].
Carboxy-terminal to the substrate domain is a serine-rich domain encompassing a 4 α-helix bundle (amino acids 351–488 for human EFS and 373–429 for CASS4). Although primary amino acid sequence shows considerable divergence versus other CAS family members in this region, structural analysis predicts that this bundle has a highly conserved fold and provides a docking site for family members. Direct interacting partners for this region have not yet been identified for CASS4 and EFS.
The C-terminal domain (around 120 amino acids in length) is second most strongly conserved between family members (after the SH3 domain) at both the level of primary amino acid sequence and predicted fold [5]. The overall similarity for this sequence is estimated at 51%. This domain has been shown to mediate to provide homo-dimerization for NEDD9 [56], and is involved in interactions with some important binding partners including SRC for CASS4 [5] and EFS [24] and Fyn kinase for EFS [28]. For BCAR1 and NEDD9, important interactors in this region also include the NSP proteins, which provide connections to receptor tyrosine kinase signaling [37]; interactions of EFS and CASS4 in this region have been minimally probed. In study of mechanosensation by BCAR1 discussed above [54], anchoring of the protein based on interactions between FAK at the SH3 domain and a complex involving SRC-family kinases at the C-terminal domain provides the two poles between which the substrate domain is stretched to become active.
In terms of short motifs, all CAS proteins except CASS4 contain a flanking YDYVHL sequence. Typically, phosphorylation of this motif by FAK or PTK2B creates a binding site for the SH2 domain of a SRC-family protein [57], which then hyper-phosphorylates the substrate domain. In contrast to other family members, EFS and BCAR1 have also been shown to contain two and one proline-rich motifs, respectively [24], which may enhance the binding and phosphorylation by SRC family kinases, based on interactions with the SRC SH3 domain [58].
Despite the reduced overall sequence similarity of CASS4 to other family members, molecular modeling analysis performed by Singh and colleagues [5] using p130CAS/BCAR1 structures as templates suggested an almost identical fold between CASS4 and p130CAS/BCAR1 within their SH3 domains, and substantial similarity within 432–591 residues of CASS4 and 449–610 residues of p130Cas/BCAR1 at the level of secondary and tertiary structures. Also, the similar periodicity of α-helices and β-sheets in both CASS4 and p130Cas/BCAR1 provides another confirmation for the idea of well-conserved structures within the family members.
EFS, CASS4, and canonical CAS protein functions
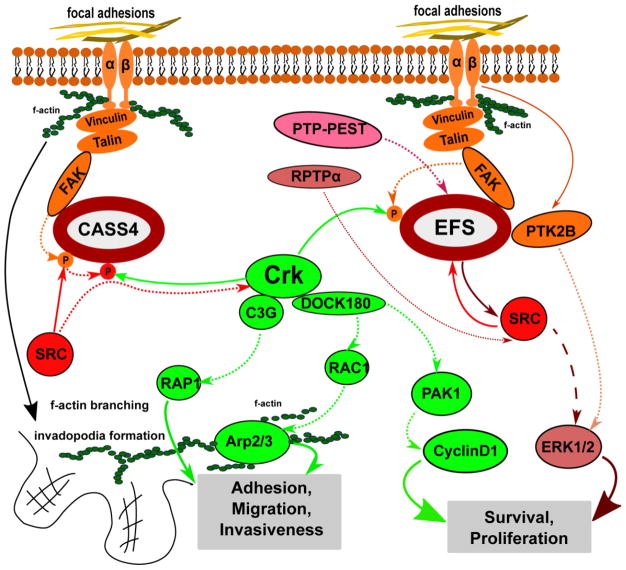
The canonical signaling activities of EFS and CASS4 proteins are outlined in Figure 2. These effects are based on the ability of CAS proteins to act as scaffolds for other catalytically active or adaptor proteins.
Figure 2. Scheme of signaling pathways involving EFS and CASS4 proteins.
Integrin (αβ) heterodimer activation at focal adhesions triggers phosphorylation of CAS proteins by FAK and SRC, allowing them to act as scaffolds for CRK and downstream factors that reorganize the actin cytoskeleton, and influence proliferation and survival signaling. Dotted lines stand for phosphorylation events, solid lines stand for direct interactions between proteins.
In their initial report of EFS, Ishino et al. [4] showed the protein bound directly to the SRC-family kinase Fyn in vitro and in vivo, and showed that Fyn directly phosphorylated EFS. Alexandropoulos and colleagues showed that SRC directly phosphorylates residues Y576 and Y577 tyrosine sites on the EFS, thus enhancing FAK targeting, and eventually the solubility and/or stability of the complex. Reciprocally, EFS activates SRC signaling through c-CRK and RAP1 [50], a process associated with cell migration for multiple family members [8, 59]. EFS contains two high-affinity class I Src-SH3-binding sites [60] it might be involved in the regulation of c-Src. Through a series of in vitro and in vivo assays with truncated and full length EFS constructs, this group carefully dissected the mechanism of c-SRC-EFS complex formation and further activation of c-SRC by EFS [24], as necessary for allowing a direct interaction between EFS and CRK. This work defined the DVP motifs carboxy-terminal to the phosphorylated tyrosines in the EFS substrate domain as contributing to a consensus recognition site for the CRK SH2 domain [48].
Activation of EFS to reorganize the actin cytoskeleton downstream of CRK has been shown to play a role in neurite outgrowth. Yang et al. identified EFS as an essential component of an additional signaling pathway involved in neurite processes formation, connected to the canonical ERK activation pathway [61, 62]. RPTPα (receptor-protein-tyrosine phosphatase α) is abundant in neural tissue and associates physically with SRC and Fyn [63, 64], and its overexpression activates these kinases [65] in response to EGF stimulation. In this study, the authors showed that stimulation by RPTPα caused SRC phosphorylation of EFS, and increased recruitment of CRK and NCK to EFS [66]. Alexandropoulos et al. have also investigated EFS expression in T lymphocytes and in immune responses [33], discussed further below.
Very few papers address the mechanistic action of CASS4, with these beginning with the hypothesis that it has biological activities similar to those of other family members in influencing cell attachment and movement. Singh et al. [5] as stated above, confirmed direct interaction between CASS4 SH3 domain and FAK, also authors revealed colocalization of CASS4 with paxillin within focal adhesions in HOP-62 cells. In direct comparison to NEDD9, overexpression of CASS4 was shown to act similarly in activating FAK phosphorylation and promoting cell spreading, but to a lesser degree than for NEDD9, and only in a subset of cells overexpressing the protein. Depletion of CASS4 reduced the spreading and FAK activation in a subset of cells. Despite the fact that CASS4 lacks the YDYVHL site that coordinates docking with the Src SH2 domain for other family members [57], attachment-induced Src phosphorylation of CASS4 was observed, suggesting that additional interactions between CASS4, FAK, and Src are sufficient to drive this modification. Unusually, CASS4 depletion had a bimodal effect on cell migration, causing some cells to have lower velocity and others to have higher velocity than control cells. This study concluded that CASS4 was a weaker paralogue of BCAR1, and also suggested the function of CASS4 may be cell-type specific and dependent upon the presence or absence of expression of other CAS family members because of the structural differences noted above.
EFS and immune system function
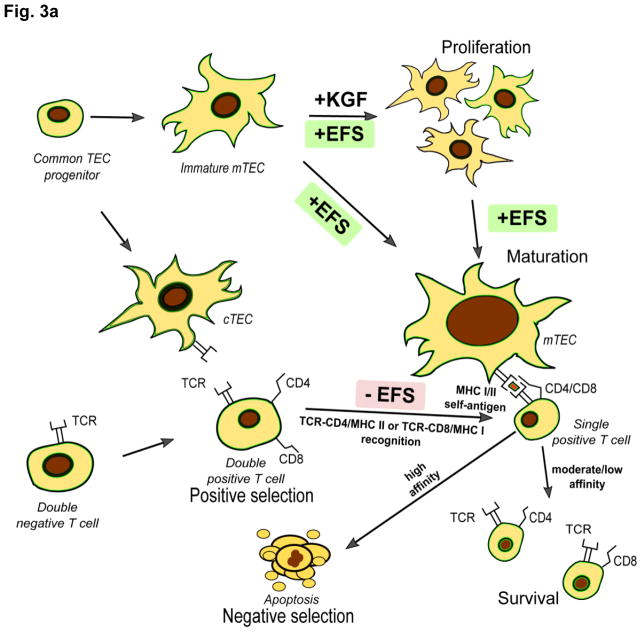
Several studies have addressed the role of EFS in T-cell function and maturation, indicating important roles in maintaining immune homeostasis and preventing development of autoimmunity (Figure 3A). Medullary thymus epithelial cells (mTECs) are important for proper T-cell maturation and negative selection of autoreactive clones, required for development of central immunological self-tolerance. Two studies have reported that EFS expression by (mTECs) is important for negative selection of T-cells during their development, while cortical thymus epithelial cells, required for positive T cell selection, were shown not to express EFS at the protein level [29, 30]. EFS expression in mTECs was shown to be mechanistically crucial for their functional maturation and keratinocyte growth-factor (KGF)-induced expansion. In these studies, mice with genetic reduction of EFS levels developed normally during embryogenesis but after 7–14 months of life developed massive inflammatory lesions with T-cells infiltration in multiple tissues including the small intestine, liver, kidneys and lungs that bore a striking histological resemblance to inflammatory bowel disorders resembling Crohn’s disease [29]. In this context, it is interesting that He et al. [67] linked EFS single nucleotide polymorphisms (SNPs) to Crohn’s disease using a novel Sherlock algorithm on a study of 3,230 cases versus 4,829 controls despite the fact that initial genome-wide association studies (GWAS) of Crohn’s disease did not identify EFS [68]. Then they performed a replication analysis on an independent sample from another GWAS [69] of total 6,333 patients with Crohn’s disease versus 15,056 controls and confirmed this association (p-value 0.039). The 16 SNPs linked to EFS in this work were trans-acting, potentially affecting the level of EFS expression but not its coding sequence.
Figure 3. EFS and immune system activity.
A. A common thymic epithelial cell (TEC) progenitor gives rise to cortical TECs (cTECs) and medullary TECs (mTECs). Immature mTECs are distinguished by lack of expression of the Aire (autoimmune regulator) transcription factor. Mature TECs have high expression of the Major Histocompatibility Complexes (MHCI and MHCII), which are required for representation of antigens, including self-antigens. During their differentiation, T lymphocytes progress from a double negative (CD4−CD8−) to a double positive (CD4+CD8+) stage, at which point positive selection by cTECs occurs. Only T cells moderately binding to the MHCI (for CD8+) or MHCII (for CD4+) molecules expressed by cTECs are provided with a pro-survival co-stimulatory signal. EFS dephosphorylation and downregulation of its effectors in T lymphocytes is crucial for them to move from the double positive to the single positive (CD4+CD8− or CD4−CD8+) stage. This process is accompanied by T cell migration to the thymic medulla, where negative selection by mTECs to remove auto-reactive clones occurs. EFS signaling is required for KGF (keratinocyte growth factor)-mediated expansion and functional maturation of mTECs. T cells binding MHC-self-antigen complexes with high affinity undergo apoptosis.
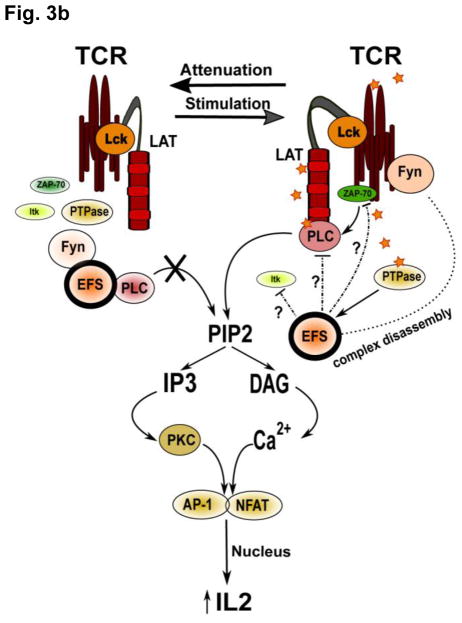
B. A dual role of EFS in mature T cells was identified in a Jurkat T cell model [71]. Both overexpression and siRNA knockdown of EFS led to decreased transcriptional activation of IL-2-dependent promoters following TCR stimulation. Phosphorylated EFS bound to Fyn and PLC-γ sequesters signaling molecules away from the TCR complex, acting as a negative regulator in resting cells. TCR activation leads to dephosphorylation of EFS, most likely through the action of a protein tyrosine phosphatase (PTPase), resulting in release of binding partners and activating downstream signaling: for instance, activated PLC-γ cleaves phosphoinositide PIP2 into the second messengers IP3 and DAG. IP3 induces intracellular calcium release, causing activation of NFAT, and DAG activates PKC, resulting in increased AP-1 transcriptional activity: NFAT and AP-1 bind and contribute to activation of the IL2 promoter. Attenuation of TCR signaling induces inhibitory reassociation of the EFS-Fyn-PLCγ complex.
Besides a role in mTECs, there is also evidence that EFS-mediated signaling in lymphocytes is crucial during their maturation. Donlin and colleagues [28] have shown that transgenic expression of a truncated form of EFS (SinDeltaC) from the human CD2 promoter, which causes transgene expression in both thymocytes and mature T cells, resulted in a constitutively activated protein that was bound more effectively and was hyperphosphorylated by the SRC-family FYN kinase. Lymphocyte maturation was impaired beyond the double positive stage (CD4+CD8+), with the transgenic phenotype associated with 2-fold increased lymphocyte apoptosis and decreased levels of single positive (CD4+CD8− and CD4−CD8+) T cells. Crossing these mice to Fyn heterozygous or null mice alleviated the apoptotic phenotype, but mice still were deficient in development of single positive cells, suggesting additional SRC kinases or other kinases may be involved in this process. SinDeltaC expression did not affect T cell receptor (TCR)-proximal signaling or JNK activity, but did inhibit the ERK activation that is required during lymphocytes positive selection [70]. These results indicated that EFS inactivation (dephosphorylation) was necessary for appropriate T cell differentiation.
In other work, EFS was shown to play repressive role on processes associated with the activation of mature T-cells, including IL-2 pro-inflammatory cytokine secretion and IL-2-dependent clonal expansion of T cells [29, 71]. In contrast to many other signal transducers which are phosphorylated upon TCR stimulation, EFS was found to be phosphorylated and bound to FYN kinase and phospholipase C-γ in resting T-cells. EFS dephosphorylation after T-cell receptor (TCR) stimulation and release of the SRC family kinase FYN and phospholipase C-γ normally lead to self-limitation of the immune response. EFS overexpression in murine wild type T cells decreased IL-2 concentration in supernatants versus controls in response to TCR stimulation [71]. Conversely, in Jurkat T cells with EFS expression reduced by siRNA, NFAT mediated transcriptional activation was increased [29]. The authors of one study [71] have proposed a complex dualistic role of EFS in mature T cells functions, because both overexpression and siRNA knockdown of this protein in cell models resulted in decreased transcriptional activation of IL-2 dependent promoters following TCR stimulation (Figure 3B).
Another study has raised the possibility that elevated EFS expression might contribute to susceptibility to acute rheumatoid fever [72]. In this work, peripheral blood mononuclear cells (PBMCs) from patients with rheumatoid heart disease (RHD) and control subjects that had never experienced acute rheumatoid fever were stimulated with rheumatogenic and non-rheumatogenic group A streptococci (GAS) strains. EFS is one of only four genes with significantly increased expression associated with rheumatogenic GAS exposure. In possibly related work, Tchernev et al. [73] have connected EFS function to Chediak-Higashi syndrome (CHS) is rare and severe autosomal recessive disorder associated with partial albinism, peripheral neuropathy, mild coagulation defects and propensity to recurrent bacterial and fungal infections, caused by incomplete phagocytosis due to failure in phagolysosome formation. In this study, the authors identified a direct physical interaction in vitro and in vivo between EFS and LYST (lysosomal trafficking regulator, aka CHS1 (Chediak-Higashi syndrome 1), a large protein that is mutated in CHS and regulates intracellular trafficking of proteins through endosomes. These results may imply the role of EFS as a disease progression modifier, although further testing and establishment of mechanism is necessary. The connections to CHS and to GAS infection is also of interest because of the well-documented role of BCAR1 in regulating infection by microbial pathogens including Salmonella, Plasmodium, herpesvirus, and Yersinia noted above [17, 18], and recent implication of NEDD9 as a regulator of infection by Candida albicans [74]: it is possible that EFS has similar function. This in turn may be relevant to a role in Crohn’s disease, which is linked to recurrent infection with pathogenic microorganisms [75]. In sum, these data are consistent with the idea that EFS makes multiple contributions to T lymphocyte development and immune system maturation and that it acts both directly in lymphocytes and indirectly through mTECs.
By contrast, almost no work has addressed possible roles of CASS4 in immune system function. However, changes in CASS4 expression have been linked to atopic asthma in a study performed by Esnault et al. [76]. In this work, CASS4 was reported to be an eosinophil-associated gene, with expression in sputum cells significantly increased after whole lung allergen challenge. The CASS4 mRNA was also upregulated in cells collected by bronchoalveolar lavage after segmental broncho-provocation with an allergen. When this procedure was performed following administration of mepolizumab (a humanized monoclonal anti-IL-5 antibodies which reduces excessive eosinophilia) CASS4 mRNA was reciprocally downregulated, therefore suggesting relation of CASS4 activity with immune response during atopic asthma development. Direct functional testing is warranted.
Implication of EFS and CASS4 in other disorders
A number of studies not directly focused on EFS or CASS4 have provided initial suggestions that the activity of these proteins is relevant to other diseases (summarized in Table 2).
Table 2.
Identification of EFS or CASS4 in studies employing high throughput or GWAS screening methods.
| Disease associations | |||
|---|---|---|---|
| Screen purpose | Protein | Observation | Reference |
| GWAS | |||
| Alzheimer’s disease | CASS4 | SNP rs7274581 T/C linked to risk. Odds ratio 0.72; p-value 0.011 | [79] |
| SNP rs7274581 T/C linked to risk. Odds ratio 0.88; p-value 2.5*10−8 | [77] | ||
| SNP rs7274581 T/C linked to risk. Odds ratio 0.8888; p-value 1.75 ×10−7 | [78] | ||
| SNP rs6024870, RegulomeDB score 2b, meaning that this SNP is likely to affect transcription factor binding. | [80] | ||
| SNP rs16979934 T/G linked to risk. Odds ratio 0.5956; p-value 0.03. | [81] | ||
| Cystic fibrosis | CASS4 | Possible correlation with severity of the lung manifestation of the disease | [84] |
| Crohn’s disease | EFS | EFS gene linkage to Crohn’s disease confirmation (p-value 0.039) in humans. | [67] |
| Expression or gene dosage change association | |||
| Atopic asthma | CASS4 | Significantly increased expression in eosinophils after antigen exposure | [76] |
| Non small cell lung cancer | CASS4 | Association of high CASS4 expression in tumors with disease severity and poor prognosis. | [97] |
| Colorectal cancer | CASS4 | Increased CASS4 expression is associated with higher rate of chromosomal instabilities and worse prognosis in colorectal cancer. | [98] |
| Rheumatic fever | EFS | Significant increase in expression after peripheral blood mononuclear cells stimulation with rheumatogenic group A streptococci from patients with rheumatoid heart disease. | [72] |
| Prostate cancer | EFS | EFS gene CpG sites hypermethylation and decreased EFS expression is associated with prostate cancer recurrence and metastatic potential. | [89] |
| Uveal melanoma | EFS | EFS hypermethylation is associated with high metastatic rate. | [92] |
| Breast cancer | EFS | EFS expression may play role in trastuzumab resistance development in HER2+ breast cancer. | [93] |
| Prolactinoma | EFS | EFS association to stem cell regulation, tumor cell invasion, tumor recurrence, and drug resistance. | [100] |
| Gestational choriacarcinoma | EFS | EFS is one of more than 100 genes located in frequently amplified chromosomal region. | [95] |
| Glioblastoma multiforne | EFS | Differential expression of EFS between glioblastoma multiforme subtypes. | [96] |
| Endometrium expression profiling | EFS | 17β-estradiol or progesterone stimulation decreases EFS expression in explants of late proliferative phase endometrium. | [31] |
| Other | |||
| Phosphoproteome of resting human platelets | CASS4 | Phosphorylation on S305 PKA/PKG consensus site. | [85] |
| Platelet activation by oxidized phospholipids | CASS4 | KODA-PC induces S249 phosphorylation in platelets | [86] |
| Chediak-Higashi syndrome | EFS | Direct interaction with LYST. | [73] |
Alzheimer’s disease has been shown to have strong pathological association with CASS4 by the IGAP (International Genomics of Alzheimer’s Disease Project), the largest genetic epidemiological investigation of AD risk to date, involving assessment of 74,046. In this work, Lambert et al. [77] found a locus associated with lower susceptibility to AD on chromosome 20 at the location of the CASS4 gene, and identified a corresponding SNP - rs7274581 T/C (OR = 0.87) - which reached genome-wide significance in both the first stage (p-value 1.6*10−6 in the analysis of 4 combined samples totaling 17,008 cases versus 37,154 controls) and the second stage (p-value 4.1*10−3 in the replication study by genotyping SNPs showed moderate significance in stage 1 in an independent sample of 8,572 cases versus 11,312 controls) analysis. Several additional studies have investigated this association. Ruiz et al. conducted a follow-up study comparing 1,808 patients with AD symptoms versus 2,564 unrelated healthy individuals [78], with results showing that this SNP was not predictive. However, in the GWAS performed by Beecham et al. [79] CASS4 showed a significant correlation with clinical pathological features of AD such as neurofibrillary tangles and neuritic plaques. Two additional CASS4 SNPs were reported to be associated with AD susceptibility: rs6024870 (RegulomeDB score 2b) by Rosenthal et al. [80] and rs16979934 T/G (OR=0.5956, p-value 0.03) by Wang et al. [81]. Given the likely conserved CAS family cytoskeletal function of CASS4, summarized above, it has been speculated that it may have a role in axonal transport and influence the expression of the amyloid precursor (APP) and tau proteins, which are pathologically affected in AD [82]. Possible mechanisms for CASS4 action in AD are discussed by Beck et al. [83].
Cystic fibrosis severity, progression and comorbid conditions have been reported to be associated with altered expression of CASS4. A GWAS study performed by Wright et al. [84] involved 3,467 individuals in three independent cohorts with different recruitment parameters. The Gene Modifier Study (GMS) included unrelated affected individuals homozygous for CTFR allele p.Phe508del, common in cystic fibrosis; the Canadian Consortium for Genetic Studies (CGS) was a population-based study of patients with cystic fibrosis; and the Twins & Sibs Study (TSS) assessed families with two or more surviving children with cystic fibrosis. Evidence of CASS4 association was first identified in patients in the GMS and CGS groups, and then linkage was analyzed in 486 sibling pairs in family-based TSS. A number of modifier loci for lung disease severity in cystic fibrosis were identified during this analysis, including the 20q13.2 locus (log10 odds = 5.03) that contains five genes (CBLN4, MC3R, CASS4, CSTF1, and AURKA), expressed in fetal or adult lung, or bronchial epithelia. Taking in consideration that the CAS family member NEDD9 has been shown to interact directly with AURKA (encoding Aurora-A kinase) in cell cycle regulation [21] and ciliary resorption [22], it is possible that CASS4 may similarly interact with Aurora-A kinase. Further investigation is necessary.
Thrombosis
CASS4 signaling may contribute to platelet activation and aggregation. Zahedi et al [85] performed a phosphoproteome analysis of resting human platelets and identified a PKA/PKG phosphorylation site in CASS4 on residue S305 in the substrate domain; the functional significance of this phosphorylation is currently unknown. Zimman et al. [86] showed significantly increased phosphorylation on S249 of CASS4, also in the substrate domain, after platelet stimulation with the oxidized phospholipid KODA-PC (9-keto-12-oxo-10-dodecenoic acid ester of 2-lyso-phosphocholine, a CD36 receptor agonist) versus thrombin treatment, which may implicate CASS4-mediated signaling in platelet hyperreactivity. Direct evaluation of these ideas is required.
Cancer
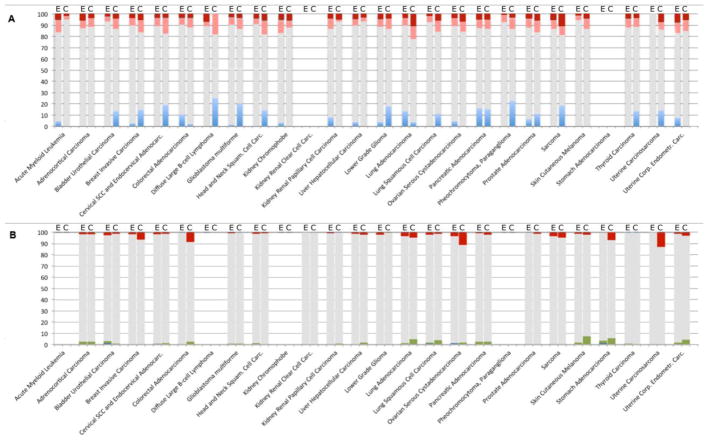
Many CAS family proteins have altered activity and functional roles in cancer initiation, progression, metastasis, and drug resistance [35, 36]. Based on information in the genomic databases available through the cBioportal site (The Cancer Genome Atlas (TCGA) and others), the CASS4 and EFS genes show copy number, gene expression changes, or mutation in a subset of tumors for most tumor types for which information is available (Figure 4). CASS4 is more frequently amplified than EFS, particularly in tumor types that often have genomic instability and a higher mutational burden: this may reflect a specific preference for elevated CASS4, or alternatively may reflect the fact that CASS4 is part of a common amplicon with the AURKA oncogene, which is often elevated in cancer (and see below discussion). Gene expression changes generally indicate the CAS genes, and particularly EFS, are more commonly overexpressed than downregulated in tumors. More thorough analysis may assign changes in CAS expression to tumor stage and grade that provide insights into roles of these proteins at specific points in tumor progression.
Figure 4. Genomic alterations and expression changes in CASS4 and EFS in cancer.
Data available from the TCGA was extracted using the cBioPortal site (http://www.cbioportal.org). For some tumor sites, data was unavailable for both genomic and mRNA profiles, reflected by empty columns. A. mRNA expression profiles for EFS (E) and CASS4 (C). Dark red: percent of tumor samples with mRNA upregulation with Z-score >2; pink: upregulation with Z-score >1; light blue: percent of tumor samples with mRNA downregulation Z-score <−1; gray: no significant change (−1 < z <1). B. Copy number variations and somatic mutations for EFS (E) and CASS4 (C). Red: percent of tumor samples with gene amplifications; blue: homozygous deletions; green: mutations. Gray represents no change. The fraction of samples with both CNV and mutations in the genes of interest was not significant and is not shown.
In focused analyses, a number of studies connect EFS to prostate cancer. In one study, the local and systemic recurrence of prostate cancer was associated with CpG site hypermethylation of number of genes, including EFS, FLNC, ECRG4, PITX2, PDLIM4, and KCNMA1. Further investigation indicated that methylation of CpG sites that is specifically predicted to result in reduction of gene expression was restricted to the FLNC and EFS genes (p ≤ .03), both involved in cell attachment [87]. In a study conducted by Marques et al., EFS expression was strongly downregulated in hormonal therapy-resistant versus therapy-responsive PC346C prostate cancer cells [88], in general agreement with a second study, in which low EFS expression correlated with malignant behavior of the PC-3 and LNCaP prostate cancer cells [89]. Additional work showed decreased EFS mRNA expression levels in prostate cancer samples with higher Gleason scores [90]. Finally, in a 2013 study of castration-resistant prostate cancer, EFS was identified as having significantly increased gross phosphorylation levels in samples from short or long term androgen-deprived (AD), or castration-resistant prostate carcinoma xenografts, versus in AD-naıve xenograft samples [91]. This paints a complex picture of reduced expression, but increased protein activity, in the case of aggressive disease, and requires further study.
Methylation of the EFS CpG island was observed in 69% of cases of uveal melanoma (UM) [92], with methylation only observed in cases of metastatic disease. As anticipated, RT-PCR expression analysis revealed a significant inverse relationship between EFS mRNA expression with EFS methylation in UM. EFS methylation was tissue-specific with full methylation in peripheral blood cells, but no methylation in other tissues such as fetal muscle, kidney and brain was detected.
EFS and other proteins involved in SRC kinase signaling (CDCP1/Trask and Paxillin) were showed to have increased expression in a study of breast cancer using trastuzumab (Herceptin)-resistant versus -sensitive BT474 cell line derivatives [93]. Importantly, EFS knockdown with siRNA restored trastuzumab sensitivity [93]. Reflecting the importance of post-translational modification of CAS proteins, in a study of cell lines and tumor tissue in malignant melanoma, EFS phosphorylation and activity significantly decreased (p<0.05) in response to vemurafenib treatment in BRAF wild-type melanoma tumors compared to tumors with a BRAF V600E mutation with additional resistance to vemurfenib [94].
In more tenuous connections to cancer, gestational choriocarcinoma is characterized by overexpression of the centromeric 10.21 Mb “minimal critical region” on Chromosome 14, containing more than 100 genes, including EFS [95]. The EFS mRNA was also identified as differentially expressed in two of the three groups of glioblastoma multiforme as identified by gene expression profiles (GEPs) [96]. EFS was differentially expressed in GEP1 and GEP3 groups, which were associated with worse prognosis, and increased cytogenetic abnormalities.
CASS4 has been much less studied in the context of human malignancies. Miao et al. correlated elevated CASS4 expression with lymph node metastasis and high TNM stage for non-small cell lung cancer (NSCLC) [97]. In addition, this study detected a significant difference in cytoplasmic accumulation of CASS4 protein between high (H1299 and BE1) and low (LTE and A549) metastatic potential lung cancer cell lines. These may suggest CASS4 as a possible prognostic marker in clinical management of NSCLC. CASS4 was highlighted in a study by Bond et al [98], which performed a stratification of the BRAF (V600E) microsatellite stable (MSS) and microsatellite-unstable (MSI) mutations in colorectal cancers. MSS mutations confer a poor patient prognosis, whereas MSI mutant colorectal cancers have an excellent prognosis. CASS4 was among the genes located on chromosome 20q (along with AURKA, BCAS1 and ZBP1 genes) that displayed statistically significant (p=0.01) increase in presence of copy number aberrations that are indicative of chromosomal instability in BRAF mutant/MSS cancers (n = 33) compared to BRAF mutant/MSI cancers (n = 30). This study suggested CASS4 and associated genes possible prognostic markers in human colorectal pathologies.
Conclusions
Twenty years after its initial description, EFS is now emerging as an important determinant of multiple immune cell processes relevant to auto-immunity and control of infection. EFS deserves more attention as a potential biomarker for these conditions, and study of EFS is likely to inform understanding of disease mechanisms for conditions including Crohn’s Disease, CHS, and other inflammatory syndromes. For the more recently identified CASS4, the most intriguing direct linkage to a clinical condition is to Alzheimer’s disease, where it may prove of value as a predictive biomarker, and study of CASS4 in this setting may help elucidate underlying pathogenic processes. For both proteins, expression or phosphorylation may be useful as a marker of disease progression and prognosis in some types of cancer. There are currently no therapeutic approaches targeting EFS and CASS4, and given the proteins lack a catalytic domain and extracellular moieties, it may be challenging to generate such agents. However, given increasingly detailed knowledge of conserved versus unique domain-specific interactions of EFS, CASS4, and their partner proteins, it is possible that protein-interaction disrupting tools can be devised. Clearly, greater investigation of CASS4 and EFS in their own right, rather than as less commonly expressed CAS paralogues, is now entirely merited.
Acknowledgments
We thank Dr. Ilya Serebriiskii for guidance in analyzing data in cBioPortal. The authors were supported by NCI Core Grant P30 CA006927 (to Fox Chase Cancer Center) and NCI CA181287 (to EAG); by a project of the Russian Government to support the Program for Competitive Growth of Kazan Federal University (to AD); and by the Russian Ministry of Science and Education, as task #17.1891.2014/K (to VK). This review and two corresponding Gene Wiki articles were written as part of the Gene Wiki Review series--a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM083924). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The corresponding Gene Wiki entries for this review can be found here: http://en.wikipedia.org/wiki/Embryonal_fyn-associated_substrate and http://en.wikipedia.org/wiki/CASS4
Abbreviations list
- 5-HT
5-hydroxytryptamine, serotonin
- A549
non-small cell lung cancer cell line
- AD
Alzheimer’s Disease
- ADPKD
autosomal dominant polycystic kidney disease
- Aire
autoimmune regulator
- AP-1
Activator protein 1
- APP
amyloid precursor protein
- AREB6
ZEB1
- ATF
Activating transcription factor
- AURKA
Aurora A kinase
- BCAR1
Breast cancer anti-estrogen resistance 1
- BE1
non-small cell lung cancer cell line
- BRAF
v-Raf murine sarcoma viral oncogene homolog B
- c-CRK
cellular CRK
- c-RAF
cellular RAF
- c-SRC
cellular SRC kinase
- C/EBPα
CCAAT/enhancer-binding protein alpha
- C20orf32
Chromosome 20 open read frame 32
- C3G
RAPGEF1, CRK SH3-binding GNRP
- CAKb
PTK2B, cell adhesion kinase
- CAS
Crk-associated substrate
- Cas-L
NEDD9, Crk-associated substrate, lymphocyte type
- CASS3
Crk-associated substrate 3
- CASS4
Crk-associated substrate 4
- CBLN4
Cerebellin-4
- CCAAT
DNA sequence
- CD4/8
cluster of differentiation 4/8
- CDCP1
CUB domain-containing protein 1
- CGS
Canadian Consortium for Genetic Studies
- CHOP-10
C/EBP homologous protein 10
- CHS
Chediak-Higashi syndrome
- CHS1
Chediak-Higashi syndrome protein 1
- CIZ
Cas-interacting zinc finger protein
- CRK
v-Crk avian sarcoma virus CT10 oncogene homolog
- Crk-L
Crk-like protein
- CSTF1
Cleavage stimulation factor subunit 1
- DAG
diacylglycerol
- DCAS
Drosophila Crk-associated substrate
- DECODE
DECipherment Of DNA Elements
- ECRG4
Esophageal cancer-related gene 4 protein, Augurin
- EFS
Embryonal fyn-associated substrate
- EFS1
Enbryonal fyn-associated substrate 1
- EFS2
Enbryonal fyn-associated substrate 2
- ERK
Extracellular-signal-regulated kinase
- FAK
Focal adhesion kinase
- FLNC
Filamin-C
- FRNK
FAK-related non-kinase
- GAS
group A streptococci
- GATA-3
transcription factor binding GATA DNA sequence
- GEF
guanine nucleotide exchange factor
- GEP
gene expression profile
- GMS
Gene Modifier Study
- GRChB38p2
Genome Reference Consortium Human Build 38 patch release 2
- GWAS
genome-wide association study
- H1299
non-small cell lung cancer cell line
- HEF1
NEDD9, Human enhancer of filamentation 1
- HEFL
HEF like protein
- HEFS
Human EFS
- HEPL
HEF1-EFS-p130Cas-like protein
- HOP-62
lung adenocarcinoma cell line
- IGAP
International Genomics of Alzheimer’s Disease Project
- IL-2
interleukin 2
- IP3
inositol three phosphate
- KCNMA1
Potassium large conductance calcium-activated channel, subfamily M, alpha member 1
- KGF
keratinocyte growth factor
- KODA-PC
9-keto-12-oxo-10-dodecenoic acid ester of 2-lyso-phosphocholine
- LCR-F1
Locus Control Region-Factor 1
- LNCaP
prostate cancer cell line
- LTE
non-small cell lung cancer cell line
- LYST
lysosomal trafficking regulator
- MAX1
Myc-associated factor X
- MC3R
Melanocortin receptor 3
- MHC I/II
major histocompatibility complexes I/II
- MSI
microsatellite instability
- MSS
microsatellite stable
- mTEC
medullar thymus epithelial cells
- Nck
Non-catalytic region of tyrosine kinase adaptor protein
- NEDD9
Neural precursor cell expressed, developmentally down-regulated 9
- NF-κβ
nuclear factor kappa-light-chain-enhancer of activated B cells
- NF-κβ1
Nuclear factor NF-kappa-B p105 subunit
- NFAT
Nuclear factor of activated T-cells
- NFE2-L1
nuclear factor, erythroid 2-like1
- NSCLC
non-small cell lung cancer
- NSP
Novel SH2 containing protein
- Oct-6
Octamer transcription factor 6
- OR
odds ratio
- PBMC
peripheral blood mononuclear cell
- PC
prostate cancer
- PDLIM4
PDZ and LIM domain 4
- PIP2
phosphoinositide diphosphate
- PITX2
Pituitary homeobox 2
- PLC-γ
Phosphoinositide phospholipase C gamma
- POU
Pit-Oct-Unc,
- POU3F1
POU domain, class 3, transcription factor 1, aka Oct-6
- PTK2B
Protein tyrosine kinase 2B
- PTP-PEST
Protein tyrosine phosphatase containing C-terminal PEST motif
- PTP1B
Protein tyrosine phosphatase 1B
- PTPase
Protein tyrosine phosphatase
- Pyk2
PTK2B, protein tyrosine (Y) kinase
- RAFTK
PTK2B, related adhesion focal tyrosine kinase
- RAP1
Ras-proximate-1 or Ras-related protein 1
- RAPGEF1
Rap guanine nucleotide exchange factor 1
- RHD
rheumatoid heart disease
- RPTPα
receptor-protein-tyrosine phosphatase alfa
- RTK
receptor tyrosine kinase
- SH2
Src Homology 2 domain
- SH3
Src homology 3 domain
- SHC
Src homology 2 domain containing
- Sin
Src interacting or signal integrating protein
- SNP
Single nucleotide polymorphism
- TCR
T-cell receptor
- TEC
thymus epithelial cell
- TNM
tumor, nodules, metastases scale
- Trask
CDCP1, Transmembrane and associated with src kinases
- TSS
Twins & Sibs Study
- UCSC
University of California Santa Cruz
- UM
uveal melanoma
- v-RAF
Virus-induced Rapidly Accelerated Fibrosarcoma
- ZEB1
Zinc finger E-box binding homeobox 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sakai R, et al. Characterization, partial purification, and peptide sequencing of p130, the main phosphoprotein associated with v-Crk oncoprotein. J Biol Chem. 1994;269(52):32740–6. [PubMed] [Google Scholar]
- 2.Law SF, et al. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(7):3327–37. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minegishi M, et al. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996;184(4):1365–75. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishino M, et al. Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which contains a Src homology 3 domain and associates with Fyn. Oncogene. 1995;11(11):2331–8. [PubMed] [Google Scholar]
- 5.Singh MK, et al. A novel Cas family member, HEPL, regulates FAK and cell spreading. Mol Biol Cell. 2008;19(4):1627–36. doi: 10.1091/mbc.E07-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Neill GM, Fashena SJ, Golemis EA. Integrin signalling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10(3):111–9. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- 7.Barrett A, et al. p130Cas: a key signalling node in health and disease. Cell Signal. 2013;25(4):766–77. doi: 10.1016/j.cellsig.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Fashena SJ, et al. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115(Pt 1):99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 9.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Agthoven T, et al. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27(4):542–9. doi: 10.1200/JCO.2008.17.1462. [DOI] [PubMed] [Google Scholar]
- 11.Tazaki T, et al. p130Cas, Crk-associated substrate plays essential roles in liver development by regulating sinusoidal endothelial cell fenestration. Hepatology. 2010;52(3):1089–99. doi: 10.1002/hep.23767. [DOI] [PubMed] [Google Scholar]
- 12.Nagai Y, et al. p130Cas, Crk-associated substrate, plays important roles in osteoclastic bone resorption. J Bone Miner Res. 2013;28(12):2449–62. doi: 10.1002/jbmr.1936. [DOI] [PubMed] [Google Scholar]
- 13.Gonsior C, et al. Oligodendroglial p130Cas is a target of Fyn kinase involved in process formation, cell migration and survival. PLoS One. 2014;9(2):e89423. doi: 10.1371/journal.pone.0089423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogden K, et al. A new signaling paradigm for serotonin: use of Crk-associated substrate in arterial contraction. Am J Physiol Heart Circ Physiol. 2006;291(6):H2857–63. doi: 10.1152/ajpheart.00229.2006. [DOI] [PubMed] [Google Scholar]
- 15.Shi J, Casanova JE. Invasion of host cells by Salmonella typhimurium requires focal adhesion kinase and p130Cas. Mol Biol Cell. 2006;17(11):4698–708. doi: 10.1091/mbc.E06-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis SP, et al. Plasmodium falciparum-induced CD36 clustering rapidly strengthens cytoadherence via p130CAS-mediated actin cytoskeletal rearrangement. FASEB J. 2012;26(3):1119–30. doi: 10.1096/fj.11-196923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce-Staskal PJ, et al. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J Cell Sci. 2002;115(Pt 13):2689–700. doi: 10.1242/jcs.115.13.2689. [DOI] [PubMed] [Google Scholar]
- 18.Weidow CL, et al. CAS/Crk signalling mediates uptake of Yersinia into human epithelial cells. Cell Microbiol. 2000;2(6):549–60. doi: 10.1046/j.1462-5822.2000.00079.x. [DOI] [PubMed] [Google Scholar]
- 19.Bandyopadhyay C, et al. p130Cas scaffolds the signalosome to direct adaptor-effector cross talk during Kaposi’s sarcoma-associated herpesvirus trafficking in human microvascular dermal endothelial cells. J Virol. 2014;88(23):13858–78. doi: 10.1128/JVI.01674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aquino JB, et al. The retinoic acid inducible Cas-family signaling protein Nedd9 regulates neural crest cell migration by modulating adhesion and actin dynamics. Neuroscience. 2009;162(4):1106–19. doi: 10.1016/j.neuroscience.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7(10):937–46. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugacheva EN, et al. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129(7):1351–63. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikonova AS, et al. Nedd9 restrains renal cystogenesis in Pkd1−/− mice. Proc Natl Acad Sci U S A. 2014;111(35):12859–64. doi: 10.1073/pnas.1405362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel p130Cas-related protein, Sin. Genes Dev. 1996;10(11):1341–55. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 25.EntrezGene. EFS gene. 2015 http://www.ncbi.nlm.nih.gov/gene/10278.
- 26.EntrezGene. CASS4 gene. 2015 http://www.ncbi.nlm.nih.gov/gene/57091.
- 27.Ishino M, et al. Identification of an Efs isoform that lacks the SH3 domain and chromosomal mapping of human Efs. Oncogene. 1997;15(14):1741–5. doi: 10.1038/sj.onc.1201346. [DOI] [PubMed] [Google Scholar]
- 28.Donlin LT, et al. Defective thymocyte maturation by transgenic expression of a truncated form of the T lymphocyte adapter molecule and Fyn substrate, Sin. J Immunol. 2002;169(12):6900–9. doi: 10.4049/jimmunol.169.12.6900. [DOI] [PubMed] [Google Scholar]
- 29.Donlin LT, et al. Deficiency in expression of the signaling protein Sin/Efs leads to T-lymphocyte activation and mucosal inflammation. Mol Cell Biol. 2005;25(24):11035–46. doi: 10.1128/MCB.25.24.11035-11046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danzl NM, Donlin LT, Alexandropoulos K. Regulation of medullary thymic epithelial cell differentiation and function by the signaling protein Sin. J Exp Med. 2010;207(5):999–1013. doi: 10.1084/jem.20092384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dassen H, et al. Progesterone regulation of implantation-related genes: new insights into the role of oestrogen. Cell Mol Life Sci. 2007;64(7–8):1009–32. doi: 10.1007/s00018-007-6553-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UCSC. Bioinformatics Genome Browser. http://www.sabiosciences.com/chipqpcrsearch.php.
- 33.Alexandropoulos K, et al. Sin: good or bad? A T lymphocyte perspective. Immunol Rev. 2003;192:181–95. doi: 10.1034/j.1600-065x.2003.00021.x. [DOI] [PubMed] [Google Scholar]
- 34.Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2010;67(7):1025–48. doi: 10.1007/s00018-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikonova AS, et al. CAS proteins in health and disease: an update. IUBMB Life. 2014;66(6):387–95. doi: 10.1002/iub.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tornillo G, Defilippi P, Cabodi S. Cas proteins: dodgy scaffolding in breast cancer. Breast Cancer Res. 2014;16(5):443. doi: 10.1186/s13058-014-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallez Y, et al. NSP-CAS Protein Complexes: Emerging Signaling Modules in Cancer. Genes Cancer. 2012;3(5–6):382–93. doi: 10.1177/1947601912460050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tikhmyanova N, et al. Dcas supports cell polarization and cell-cell adhesion complexes in development. PLoS One. 2010;5(8):e12369. doi: 10.1371/journal.pone.0012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Astier A, et al. The related adhesion focal tyrosine kinase differentially phosphorylates p130Cas and the Cas-like protein, p105HEF1. J Biol Chem. 1997;272(32):19719–24. doi: 10.1074/jbc.272.32.19719. [DOI] [PubMed] [Google Scholar]
- 40.Kirsch KH, Georgescu MM, Hanafusa H. Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G. J Biol Chem. 1998;273(40):25673–9. doi: 10.1074/jbc.273.40.25673. [DOI] [PubMed] [Google Scholar]
- 41.Garton AJ, et al. Association of PTP-PEST with the SH3 domain of p130cas; a novel mechanism of protein tyrosine phosphatase substrate recognition. Oncogene. 1997;15(8):877–85. doi: 10.1038/sj.onc.1201279. [DOI] [PubMed] [Google Scholar]
- 42.Liu F, Sells MA, Chernoff J. Protein tyrosine phosphatase 1B negatively regulates integrin signaling. Curr Biol. 1998;8(3):173–6. doi: 10.1016/s0960-9822(98)70066-1. [DOI] [PubMed] [Google Scholar]
- 43.Nakamoto T, et al. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol Cell Biol. 2000;20(5):1649–58. doi: 10.1128/mcb.20.5.1649-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harte MT, et al. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J Biol Chem. 1996;271(23):13649–55. doi: 10.1074/jbc.271.23.13649. [DOI] [PubMed] [Google Scholar]
- 45.Ohba T, et al. Dot far-western blot analysis of relative binding affinities of the Src homology 3 domains of Efs and its related proteins. Anal Biochem. 1998;262(2):185–92. doi: 10.1006/abio.1998.2772. [DOI] [PubMed] [Google Scholar]
- 46.Davidson D, Veillette A. PTP-PEST, a scaffold protein tyrosine phosphatase, negatively regulates lymphocyte activation by targeting a unique set of substrates. Embo j. 2001;20(13):3414–26. doi: 10.1093/emboj/20.13.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cote JF, et al. Combination of gene targeting and substrate trapping to identify substrates of protein tyrosine phosphatases using PTP-PEST as a model. Biochemistry. 1998;37(38):13128–37. doi: 10.1021/bi981259l. [DOI] [PubMed] [Google Scholar]
- 48.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–78. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 49.Sakai R, et al. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13(16):3748–56. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing L, et al. c-Src signaling induced by the adapters Sin and Cas is mediated by Rap1 GTPase. Mol Cell Biol. 2000;20(19):7363–77. doi: 10.1128/mcb.20.19.7363-7377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brehme M, et al. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci U S A. 2009;106(18):7414–9. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsia DA, et al. Differential regulation of cell motility and invasion by FAK. J Cell Biol. 2003;160(5):753–67. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu C, et al. Systematic identification of SH3 domain-mediated human protein-protein interactions by peptide array target screening. Proteomics. 2007;7(11):1775–85. doi: 10.1002/pmic.200601006. [DOI] [PubMed] [Google Scholar]
- 54.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–26. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petch LA, et al. Adhesion-induced tyrosine phosphorylation of the p130 src substrate. J Cell Sci. 1995;108( Pt 4):1371–9. doi: 10.1242/jcs.108.4.1371. [DOI] [PubMed] [Google Scholar]
- 56.Law SF, et al. Dimerization of the docking/adaptor protein HEF1 via a carboxy-terminal helix-loop-helix domain. Exp Cell Res. 1999;252(1):224–35. doi: 10.1006/excr.1999.4609. [DOI] [PubMed] [Google Scholar]
- 57.Tachibana K, et al. Tyrosine phosphorylation of Crk-associated substrates by focal adhesion kinase. A putative mechanism for the integrin-mediated tyrosine phosphorylation of Crk-associated substrates. J Biol Chem. 1997;272(46):29083–90. doi: 10.1074/jbc.272.46.29083. [DOI] [PubMed] [Google Scholar]
- 58.Pellicena P, Miller WT. Processive phosphorylation of p130Cas by Src depends on SH3-polyproline interactions. J Biol Chem. 2001;276(30):28190–6. doi: 10.1074/jbc.M100055200. [DOI] [PubMed] [Google Scholar]
- 59.Cary LA, et al. Identification of p130Cas as a mediator of focal adhesion kinase-promoted cell migration. J Cell Biol. 1998;140(1):211–21. doi: 10.1083/jcb.140.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexandropoulos K, Cheng G, Baltimore D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci U S A. 1995;92(8):3110–4. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stofega MR, et al. Activation of extracellular signal-regulated kinase (ERK) by mitogenic stimuli is repressed in v-Src-transformed cells. Cell Growth Differ. 1997;8(1):113–9. [PubMed] [Google Scholar]
- 62.Hakak Y, Martin GS. Cas mediates transcriptional activation of the serum response element by Src. Mol Cell Biol. 1999;19(10):6953–62. doi: 10.1128/mcb.19.10.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng XM, Resnick RJ, Shalloway D. A phosphotyrosine displacement mechanism for activation of Src by PTPalpha. EMBO J. 2000;19(5):964–78. doi: 10.1093/emboj/19.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhandari V, Lim KL, Pallen CJ. Physical and functional interactions between receptor-like protein-tyrosine phosphatase alpha and p59fyn. J Biol Chem. 1998;273(15):8691–8. doi: 10.1074/jbc.273.15.8691. [DOI] [PubMed] [Google Scholar]
- 65.den Hertog J, et al. Receptor protein tyrosine phosphatase alpha activates pp60c-src and is involved in neuronal differentiation. EMBO J. 1993;12(10):3789–98. doi: 10.1002/j.1460-2075.1993.tb06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang LT, Alexandropoulos K, Sap J. c-SRC mediates neurite outgrowth through recruitment of Crk to the scaffolding protein Sin/Efs without altering the kinetics of ERK activation. J Biol Chem. 2002;277(20):17406–14. doi: 10.1074/jbc.M111902200. [DOI] [PubMed] [Google Scholar]
- 67.He X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92(5):667–80. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alberola-Ila J, et al. Positive and negative selection invoke distinct signaling pathways. J Exp Med. 1996;184(1):9–18. doi: 10.1084/jem.184.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing L, et al. The adapter molecule Sin regulates T-cell-receptor-mediated signal transduction by modulating signaling substrate availability. Mol Cell Biol. 2004;24(10):4581–92. doi: 10.1128/MCB.24.10.4581-4592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bryant PA, et al. Susceptibility to acute rheumatic fever based on differential expression of genes involved in cytotoxicity, chemotaxis, and apoptosis. Infect Immun. 2014;82(2):753–61. doi: 10.1128/IAI.01152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tchernev VT, et al. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med. 2002;8(1):56–64. [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, et al. New signaling pathways govern the host response to C. albicans infection in various niches. Genome Res. 2015;25(5):679–89. doi: 10.1101/gr.187427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stappenbeck TS, et al. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy. 2011;7(4):355–74. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esnault S, et al. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One. 2013;8(7):e67560. doi: 10.1371/journal.pone.0067560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruiz A, et al. Follow-up of loci from the International Genomics of Alzheimer’s Disease Project identifies TRIP4 as a novel susceptibility gene. Transl Psychiatry. 2014;4:e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beecham GW, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenthal SL, et al. Connecting the dots: potential of data integration to identify regulatory SNPs in late-onset Alzheimer’s disease GWAS findings. PLoS One. 2014;9(4):e95152. doi: 10.1371/journal.pone.0095152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, et al. Genetic determinants of disease progression in Alzheimer’s disease. J Alzheimers Dis. 2015;43(2):649–55. doi: 10.3233/JAD-140729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beck TN, et al. Adaptors for disorders of the brain? The cancer signaling proteins NEDD9, CASS4, and PTK2B in Alzheimer’s disease. Oncoscience. 2014;1(7):486–503. doi: 10.18632/oncoscience.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–46. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zahedi RP, et al. Phosphoproteome of resting human platelets. J Proteome Res. 2008;7(2):526–34. doi: 10.1021/pr0704130. [DOI] [PubMed] [Google Scholar]
- 86.Zimman A, et al. Phosphoproteomic analysis of platelets activated by pro-thrombotic oxidized phospholipids and thrombin. PLoS One. 2014;9(1):e84488. doi: 10.1371/journal.pone.0084488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanaja DK, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27(5):549–60. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marques RB, et al. Bypass mechanisms of the androgen receptor pathway in therapy-resistant prostate cancer cell models. PLoS One. 2010;5(10):e13500. doi: 10.1371/journal.pone.0013500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sertkaya S, et al. Decreased expression of EFS is correlated with the advanced prostate cancer. Tumour Biol. 2014 doi: 10.1007/s13277-014-2703-5. [DOI] [PubMed] [Google Scholar]
- 90.Nakagawa T, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3(5):e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roe K, et al. Hypoxic tumor kinase signaling mediated by STAT5A in development of castration-resistant prostate cancer. PLoS One. 2013;8(5):e63723. doi: 10.1371/journal.pone.0063723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neumann LC, et al. EFS shows biallelic methylation in uveal melanoma with poor prognosis as well as tissue-specific methylation. BMC Cancer. 2011;11:380. doi: 10.1186/1471-2407-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boyer AP, et al. Quantitative proteomics with siRNA screening identifies novel mechanisms of trastuzumab resistance in HER2 amplified breast cancers. Mol Cell Proteomics. 2013;12(1):180–93. doi: 10.1074/mcp.M112.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tahiri A, et al. Differential inhibition of ex-vivo tumor kinase activity by vemurafenib in BRAF(V600E) and BRAF wild-type metastatic malignant melanoma. PLoS One. 2013;8(8):e72692. doi: 10.1371/journal.pone.0072692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Poaty H, et al. Genome-wide high-resolution aCGH analysis of gestational choriocarcinomas. PLoS One. 2012;7(1):e29426. doi: 10.1371/journal.pone.0029426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vital AL, et al. Gene expression profiles of human glioblastomas are associated with both tumor cytogenetics and histopathology. Neuro Oncol. 2010;12(9):991–1003. doi: 10.1093/neuonc/noq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miao Y, et al. Overexpression and cytoplasmic accumulation of Hepl is associated with clinicopathological parameters and poor prognosis in non-small cell lung cancer. Tumour Biol. 2013;34(1):107–14. doi: 10.1007/s13277-012-0517-x. [DOI] [PubMed] [Google Scholar]
- 98.Bond CE, et al. Microsatellite Stable Colorectal Cancers Stratified by the BRAF V600E Mutation Show Distinct Patterns of Chromosomal Instability. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.JFC, et al. Combination of gene targeting and substrate trapping to identify substrates of protein tyrosine phosphatases using PTP-PEST as a model. Biochemistry. 1998;37(38):13128–37. doi: 10.1021/bi981259l. [DOI] [PubMed] [Google Scholar]
- 100.Tong Y, et al. Genomic characterization of human and rat prolactinomas. Endocrinology. 2012;153(8):3679–91. doi: 10.1210/en.2012-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]