Abstract
A novel gyrovirus genome found in the feces of an adult with diarrhea is described. The genome shows the three expected main ORFs encoding a structural protein (VP1), nonstructural protein (VP2), and Apoptin protein (VP3), which shared identities of 41, 42, and 38 % with those of the most closely related gyrovirus proteins, respectively. Given the high divergence in its genome, this gyrovirus may be considered the prototype for a new viral species (GyV9) in the Gyrovirus genus. Because the closest relatives of this gyrovirus infect chicken, a possible dietary origin for the presence of this virus in human feces is discussed.
Keywords: Gyrovirus, Diarrhea, Feces, Human, Metagenomics
The study
Gyroviruses are non-enveloped, icosahedral viruses with small circular single-stranded DNA genomes of ~2-kb in length and comprise the Gyrovirus genus currently classified together with circoviruses within the Circoviridae family [1]. At least eight gyrovirus genomes have been described which all include three overlapping ORFs encoding structural protein VP1, non-structural protein VP2, and VP3/Apoptin protein. Gyroviruses can be divided phylogenetically into three clades A (CAV, HGyV/AGV2, GyV3, −6 and −7), B (GyV4, −5), and C (GyV8) [2, 3]. Chicken anemia virus (CAV), the prototype of the Gyrovirus genus, is responsible for severe anemia and immunosuppression in young chickens [4]. HGyV/AGV2 (human gyrovirus/avian gyrovirus 2) has been detected using PCR in tissues and sera of diseased chicken from Brazil [5], skins and plasma of healthy French adults [6], blood from solid organ transplant patients [7], and healthy blood donors [8]. The third gyrovirus species (GyV3) was found in feces of human diarrhea cases from Chile [9] and Hong Kong [10]. A second genotype of GyV3 was recently reported in feces of ferrets [11]. GyV4 was detected in the feces from unexplained cases of human diarrhea and in chicken meats and skin specimens in Hong Kong [10]. Two additional species, GyV5 and GyV6, were identified in Tunisian diarrheal feces [3]. Recently, GyV7 was reported to infect chicken [12] and GyV8 was found in tissues of a fulmar (sea bird) [2], respectively. Here we describe the genome of a proposed new gyrovirus species (GyV9) found in a human diarrhea sample.
Viral metagenomics was first used to analyze a fecal specimen collected from a French adult with unexplained diarrhea and fever. Because the sample was pre-existing and provided anonymized, it is considered non-human subject research under UCSF CHR guidelines (http://www.research.ucsf.edu/chr/Guide/chrExemptApp.asp). The sample had previously tested negative for adenovirus, astrovirus, group A rotavirus, norovirus, and sapovirus using the assays described in this Ref. [13]. The fecal suspension was first vortexed and clarified by 15,000×g centrifugation for 10 min. The supernatant was filtered through a 0.45-μm filter (Millipore) to remove bacterium-sized and larger particles. The filtrate was treated with a mixture of nuclease enzymes to digest unprotected nucleic acids, and then viral nucleic acids were extracted [14]. Random RT-PCR was used to amplify RNA and DNA, and a library was constructed for Illumina sequencing (MiSeq 2 × 250 bases) using Nextera™ XT Sample Preparation Kit. The average length in nt of the reads obtained was 228. The reads were denovo assembled using EnsembleAssembler [15]. Translated sequence reads showing similarity to viral sequences with E score <10−5 were identified using BLASTx.
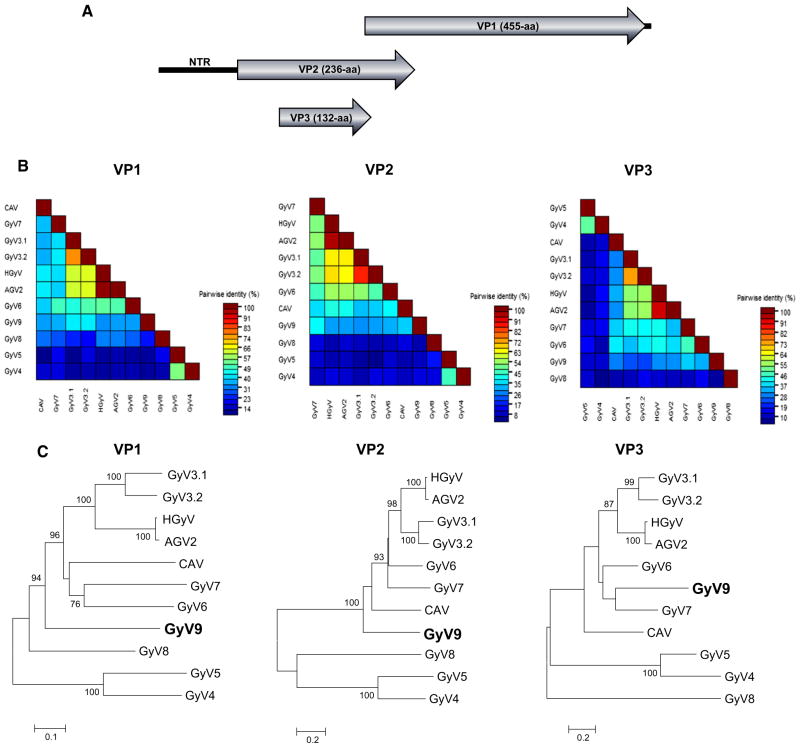
Out of 225,421 sequence reads, eight anellovirus reads, six circovirus-related reads, and six reads encoding gyrovirus-related proteins (BLASTx E score of 3 × 10−7 to 1 × 10−42). The nearly complete genome of GyV9 (GenBank KP742975) was then amplified using inverse PCR and the amplicon directly Sanger sequenced by primer walking. Two pairs of primers for inverse PCR were designed from the initial gyrovirus sequence reads. Primers GyV9-F1 (5′-ACA GAA ATG GAT GAC CCT AGA CCC T-3′) and GyV9-R1 (5′-CTG ATC CCT GTG CTC TTT GAG T-3′) were used for the first round of PCR. Primers GyV9-F2 (5′-TGA AAC ACT TAT TGG AGA ACT GTA CCT-3′) and GyV9-R2 (5′-TTG TCC TCT CTT TGA GCC TCT GTC-3′) were used for the second round of PCR, producing ~1.9-kb product (1841 bases actually acquired by Sanger sequencing). Putative ORFs in the circular genomes were predicted by NCBI ORF finder. The 2242-base-long genome contained two major ORFs encoding a 455-aa structural protein (VP1) and a 236-aa nonstructural protein (VP2) (Fig. 1a). The non-translated region (NTR) could not be entirely sequenced due to a region of high GC content. The NTR was 353-bp in length and contained a polyadenylation signal (AATAAA). The alignment of the partial NTR sequences of GyV9 and the closest relatives revealed that 61 nucleotides were likely missing. The NTR possessed three tandem repeats of a CAV promoter (TGTACAGGGGGGGTACGTCA) containing a putative estrogen-response element (in boldface) that can up-regulate transcription [16]. Sequence identity was measured using BioEdit and SDT [17]. VP1 and VP2 shared the best aa identities of 41 and 42 %, respectively, with other members of the Gyrovirus genus (Fig. 1b). GyV9 showed conserved VP1 motif ELX2AQ for rolling-circle replication (RCR) function [18], and phosphatase motif CX5R and WX7HX3-CXCX5H in VP2 [19, 20]. A smaller ORF (132-aa) was also identified in GyV9 that showed highest identity of 38 % to the similarly positioned VP3/Apoptin protein in GyV7. Phylogenetic analyses were based on translated amino acid sequences and were performed using CLUSTAL X with the default settings [21]. Neighbor-joining phylogenetic trees were generated using MEGA version 5 with the Poisson Indel Process. [22]. Bootstrap values for each node are shown if>70 %. Phylogenetic analyses of the VP1, VP2, and VP3 proteins confirmed that GyV9 was distinct from other known gyroviruses (Fig. 1c). Using the maximum likelihood [22] and Bayesian [23] phylogenetic methods produced trees with identical topologies (data not shown). SimPlot analysis was also used to test for recombination [24]. No recombination was detected between GyV9 and GyV1–GyV8.
Fig. 1.
New gyrovirus genome and phylogeny. a Organization of the circular DNA gyrovirus 9 genome shown here in a linear form. b Pairwise comparison of VP1, VP2, and VP3 proteins of the new gyrovirus and other species in the Gyrovirus genus. c Phylogenetic trees generated with VP1, VP2, and VP3 proteins of the new gyrovirus and other species in the Gyrovirus genus. The scale indicated amino acid substitutions per position. Bootstrap values (based on 100 replicates) for each node are given if >70. CLUSTAL X with the default settings included gap opening penalty (10), gap extension penalty (0.2), protein weight matrix (gonnet), residue-specific penalities (on), hydrophilic penalties (on), gap separation distance (4), and end gap separation (off)
A total of three eukaryotic viruses were detected in this fecal sample. Anelloviruses are nearly universal in human blood and acquired very early in infancy and have been shown to be shed over long period in feces of infants [25]. Anellovirus genome detection in feces might indicate the presence of blood in this diarrhea sample or replication in the digestive track. Sequences were also detected that were closely related to a group of small circular DNA viral genomes previously reported in the feces of wild chimpanzees and animals including pigs, cows, a turkey, and urban rats [26–32]. The potential role of these three viruses in this patient’s diarrhea, if any, is not known. Because pathogenic bacteria and parasites were not tested for their presence and possible role in this patient’s diarrhea remains a possibility.
The numerous gyroviruses described to date have been detected in chickens and/or on human skin, blood, or fecal samples. Gyrovirus CAV is a very frequent chicken infection, and CAV DNA is commonly seen in human feces [9]. Gyrovirus DNA has also been reported in the feces of carnivore, possibly from the consumption of chicken or other birds [33, 34]. The detection of diverse gyrovirus DNA in human feces may reflect recent consumption of infected chicken, a frequent source of diarrhea-causing bacterial infections [35, 36], rather than active gyrovirus replication in human cells. A PCR study for HGyV DNA in human CD34 + hematopoietic cells and CD34-cells from ten hematological patients did not detect gyrovirus DNA [37]. Gyrovirus DNA may therefore survive transit through the human gut without playing any role in this patient’s diarrhea [38, 39]. Despite the detection of gyrovirus DNA detection in human blood, on human skin, and in feces of human and carnivorous mammals [6–8, 33, 34], gyrovirus replication in non-avian species such as mammals remains to be conclusively demonstrated.
Acknowledgments
We acknowledge NHLBI Grant R01 HL105770 to E.L.D and the Blood Systems Research Institute for support. A.C.C. was supported by Sao Paulo Research Foundation (FAPESP), Grant Numbers 2014/05211-2 and 2012/03417-7. We thank Sylvie Pillet and Philippe Berthelot for collection of clinical information and samples.
Footnotes
Nucleotide sequence accession number
The nearly complete genome sequence of GyV9 is available in GenBank under accession number KP742975.
References
- 1.Hino S, Prasetyo AA. Curr Top Microbiol Immunol. 2009;331:117–130. doi: 10.1007/978-3-540-70972-5_8. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Pesavento AP, Gaynor MA, Duerr SR, Phan GT, Zhan W, Deng X, Delwart E. Arch Virol. 2015 doi: 10.1007/s00705-015-2468-1. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gia Phan T, Phung Vo N, Sdiri-Loulizi K, Aouni M, Pothier P, Ambert-Balay K, Deng X, Delwart E. Virology. 2013;446:346–348. doi: 10.1016/j.virol.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schat KA. Curr Top Microbiol Immunol. 2009;331:151–183. doi: 10.1007/978-3-540-70972-5_10. [DOI] [PubMed] [Google Scholar]
- 5.Rijsewijk FA, Dos Santos HF, Teixeira TF, Cibulski SP, Varela AP, Dezen D, Franco AC, Roehe PM. Arch Virol. 2011;156:1097–1100. doi: 10.1007/s00705-011-0971-6. [DOI] [PubMed] [Google Scholar]
- 6.Sauvage V, Cheval J, Foulongne V, Gouilh MA, Pariente K, Manuguerra JC, Richardson J, Dereure O, Lecuit M, Burguiere A, Caro V, Eloit M. J Virol. 2011;85:7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maggi F, Macera L, Focosi D, Vatteroni ML, Boggi U, Antonelli G, Eloit M, Pistello M. Emerg Infect Dis. 2012;18:956–959. doi: 10.3201/eid1806.120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biagini P, Bedarida S, Touinssi M, Galicher V, de Micco P. Emerg Infect Dis. 2013;19:1014–1015. doi: 10.3201/eid1906.130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan TG, Li L, O’Ryan MG, Cortes H, Mamani N, Bonkoungou IJ, Wang C, Leutenegger CM, Delwart E. J Gen Virol. 2012;93:1356–1361. doi: 10.1099/vir.0.041731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu DK, Poon LL, Chiu SS, Chan KH, Ng EM, Bauer I, Cheung TK, Ng IH, Guan Y, Wang D, Peiris JS. J Clin Virol. 2012;55:209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feher E, Pazar P, Lengyel G, Phan TG, Banyai K. Virus Genes. 2015;50:137–141. doi: 10.1007/s11262-014-1128-y. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Li L, Deng X, Kapusinszky B, Delwart E. Virology. 2014;468–470:303–310. doi: 10.1016/j.virol.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sdiri-Loulizi K, Gharbi-Khelifi H, de Rougemont A, Chouchane S, Sakly N, Ambert-Balay K, Hassine M, Guediche MN, Aouni M, Pothier P. J Clin Microbiol. 2008;46:1349–1355. doi: 10.1128/JCM.02438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. Nucleic Acids Res. 2015;43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MM, Jarosinski KW, Schat KA. J Virol. 2005;79:2859–2868. doi: 10.1128/JVI.79.5.2859-2868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhire BM, Varsani A, Martin DP. PLoS One. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilyina TV, Koonin EV. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters MA, Jackson DC, Crabb BS, Browning GF. J Biol Chem. 2002;277:39566–39573. doi: 10.1074/jbc.M201752200. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Iwasa Y, Hijikata M, Mishiro S. Arch Virol. 2000;145:979–993. doi: 10.1007/s007050050689. [DOI] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 24.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapusinszky B, Minor P, Delwart E. J Clin Microbiol. 2012;50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikorski A, Massaro M, Kraberger S, Young LM, Smalley D, Martin DP, Varsani A. Virus Res. 2013;177:209–216. doi: 10.1016/j.virusres.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Sachsenroder J, Twardziok S, Hammerl JA, Janczyk P, Wrede P, Hertwig S, Johne R. PLoS One. 2012;7:e34631. doi: 10.1371/journal.pone.0034631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachsenroder J, Braun A, Machnowska P, Ng TF, Deng X, Guenther S, Bernstein S, Ulrich RG, Delwart E, Johne R. J Gen Virol. 2014;95:2734–2747. doi: 10.1099/vir.0.070029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuter G, Boros A, Delwart E, Pankovics P. Arch Virol. 2014;159:2161–2164. doi: 10.1007/s00705-014-2025-3. [DOI] [PubMed] [Google Scholar]
- 30.Kim HK, Park SJ, Nguyen VG, Song DS, Moon HJ, Kang BK, Park BK. J Gen Virol. 2012;93:635–639. doi: 10.1099/vir.0.037838-0. [DOI] [PubMed] [Google Scholar]
- 31.Cheung AK, Ng TF, Lager KM, Bayles DO, Alt DP, Delwart EL, Pogranichniy RM, Kehrli ME., Jr Arch Virol. 2013;158:2157–2162. doi: 10.1007/s00705-013-1701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blinkova O, Victoria J, Li Y, Keele BF, Sanz C, Ndjango JB, Peeters M, Travis D, Lonsdorf EV, Wilson ML, Pusey AE, Hahn BH, Delwart EL. J Gen Virol. 2010;91:74–86. doi: 10.1099/vir.0.015446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Liu Y, Ji J, Chen F, Sun B, Xue C, Ma J, Bi Y, Xie Q. BioMed Res Int. 2014;2014:313252. doi: 10.1155/2014/313252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feher E, Pazar P, Kovacs E, Farkas SL, Lengyel G, Jakab F, Martella V, Banyai K. Arch Virol. 2014;159:3401–3406. doi: 10.1007/s00705-014-2203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodhidatta L, Srijan A, Serichantalergs O, Bangtrakulnonth A, Wongstitwilairung B, McDaniel P, Mason CJ. Southeast Asian J Trop Med Public Health. 2013;44:259–272. [PubMed] [Google Scholar]
- 36.Bless PJ, Schmutz C, Suter K, Jost M, Hattendorf J, Mausezahl-Feuz M, Mausezahl D. Eur J Epidemiol. 2014;29:527–537. doi: 10.1007/s10654-014-9917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macera L, Focosi D, Giannelli R, Bulleri M, Zucca A, Scatena F, Pistello M, Ceccherini Nelli L, Maggi F. J Clin Virol. 2013;57:182–183. doi: 10.1016/j.jcv.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Oude Munnink BB, Canuti M, Deijs M, de Vries M, Jebbink MF, Rebers S, Molenkamp R, van Hemert FJ, Chung K, Cotten M, Snijders F, Sol CJ, van der Hoek L. BMC Infect Dis. 2014;14:22. doi: 10.1186/1471-2334-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smuts HE. Adv Virol. 2014:321284. doi: 10.1155/2014/321284. [DOI] [PMC free article] [PubMed] [Google Scholar]