Abstract
Background
No opioid receptor, mu 1 (OPRM1) gene polymorphisms, including the functional single nucleotide polymorphism (SNP) rs1799971, have been conclusively associated with heroin/other opioid addiction, despite their biological plausibility. We used evidence of polymorphisms altering OPRM1 expression in normal human brain tissue to nominate and then test associations with heroin addiction.
Methods
We tested 103 OPRM1 SNPs for association with OPRM1 mRNA expression in prefrontal cortex from 224 European Americans and African Americans of the BrainCloud cohort. We then tested the 16 putative cis-quantitative trait loci (cis-eQTL) SNPs for association with heroin addiction in the Urban Health Study and two replication cohorts, totaling 16,729 European Americans, African Americans, and Australians of European ancestry.
Results
Four putative cis-eQTL SNPs were significantly associated with heroin addiction in the Urban Health Study (smallest P=8.9×10−5): rs9478495, rs3778150, rs9384169, and rs562859. Rs3778150, located in OPRM1 intron 1, was significantly replicated (P=6.3×10−5). Meta-analysis across all case-control cohorts resulted in P=4.3×10−8: the rs3778150-C allele (frequency=16%-19%) being associated with increased heroin addiction risk. Importantly, the functional SNP allele rs1799971-A was associated with heroin addiction only in the presence of rs3778150-C (P=1.48×10−6 for rs1799971-A/rs3778150-C and P=0.79 for rs1799971-A/rs3778150-T haplotypes). Lastly, replication was observed for six other intron 1 SNPs which had prior suggestive associations with heroin addiction (smallest P=2.7×10−8 for rs3823010).
Conclusions
Our findings show that common OPRM1 intron 1 SNPs have replicable associations with heroin addiction. The haplotype structure of rs3778150 and nearby SNPs may underlie the inconsistent associations between rs1799971 and heroin addiction.
Keywords: genetic association study, heroin, multiancestry, opioid, OPRM1, prefrontal cortex
Introduction
According to the 2010 Global Burden of Disease Health Measurement Survey, around 15.5 million people worldwide were dependent on heroin and other opioid drugs (1). Three regions had prevalence rates significantly higher than the global rate: Australasia, western Europe, and North America (1). In the United States, the 2013 National Survey on Drug Use and Health estimated that 669,000 people aged 12 or older abused heroin in the past year, representing a 78% increase since 2007 (2). To address this public health burden, a better understanding of the pathogenesis leading to heroin addiction is needed. Genetic vulnerability is recognized as a major risk factor contributing to heroin and other opioid addiction, as evidenced by twin studies showing that genetic factors account for 40% to 60% of the population variability (3-6). A few genome-wide association (7-9) and numerous candidate gene studies (10-19) in humans have implicated genes encoding opioid receptors (OPRM1, OPRD1, and OPRK1), potassium channels (KCNG1 and KCNG2), and others as contributing to heroin/opioid addiction phenotypes. In supporting the biological plausibility of the candidate genes, particularly genes in the opioid system (20), knockout mouse models have been used to study behavioral effects resulting from genetic perturbations. However, conclusively identified associations of specific genetic variants remain elusive.
The current study focuses on the opioid receptor, mu 1 (OPRM1) gene, the most widely studied candidate gene (17), which encodes a predominant target for heroin and other opioid molecules. The OPRM1 missense polymorphism rs1799971 has been widely studied for its functional consequences, including reduced signaling efficiency and reduced expression of the receptor (21-25). However, rs1799971 associations with heroin and other drug addictions (13-17) have been modest and often inconsistent. Additional OPRM1 single nucleotide polymorphisms (SNPs) have been tested for association with heroin/opioid addiction (10, 12, 13, 18, 19), but none of the findings have been independently replicated.
To enhance the detection of replicable OPRM1 SNP associations with heroin addiction, our study focused only on SNPs with evidence for altering OPRM1 mRNA expression in human brain, thereby reducing the multiple testing burden with a limited number of plausible regulatory SNPs carried forward for disease association testing. This approach was motivated by prior findings that psychiatric and other disease-associated SNPs tag expression quantitative trait loci (eQTL) more often than unassociated SNPs (26-28). Other studies have successfully used cis-eQTL mapping to nominate SNPs and consequently find associations with complex diseases, such as Crohn's disease (29), chronic obstructive pulmonary disease (30), and amyotrophic lateral sclerosis (31). We used cis-eQTL mapping to identify SNPs associated with OPRM1 mRNA expression in human prefrontal cortex from 224 BrainCloud cohort participants who had no evidence of drug use/abuse at the time of death and tested the putative cis-eQTL SNPs for association with heroin addiction across three independent cohorts totaling 16,729 (4,287 cases and 12,442 controls). Our findings revealed replicable and highly significant SNP associations with heroin addiction and provided strong support of OPRM1 as an important susceptibility gene for heroin addiction.
Methods and Materials
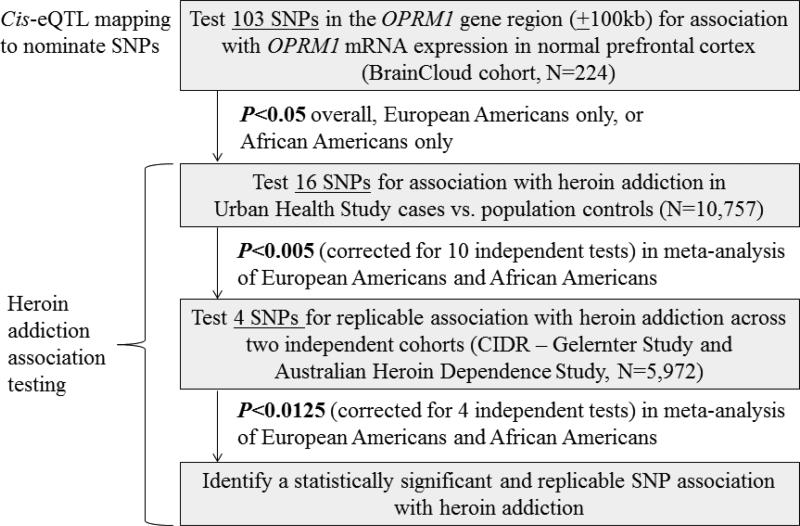
Figure 1 outlines our overall study design of conducting cis-eQTL mapping for OPRM1 in non-addicted BrainCloud participants and testing the nominated SNPs for association in heroin addiction case-control cohorts. All study protocols received Institutional Review Board approval at their respective sites, and all study participants or their legal next of kin provided informed consent.
Figure 1.
Overview of study design.
Cis-eQTL mapping using human prefrontal cortex in the BrainCloud cohort
To identify putative cis-eQTL SNPs for OPRM1, we utilized SNP genotype and gene expression data (see Supplement) available on post-mortem dorsolateral prefrontal cortex samples from 110 European Americans and 114 African Americans, ranging in age from 0 to 78 years old, who had no neuropathological or neuropsychiatric diagnoses, no drug or alcohol abuse, and no positive toxicology result (32-34).
Normalized ratios of gene expression levels from the single OPRM1 probe available in BrainCloud (Figure S1) were log-transformed and tested for association with additive three-level SNP genotypes using the BrainCloud software (32). Genotyped SNP association results were generated from the best fit general linear model with race, sex, age, life stage (infant, child, teen, and adult), and an age-by-life stage interaction included as the range of covariates. Inclusion of age, life stage, and the corresponding interaction was used to account for nonlinear trajectories of age-dependent gene expression over the lifespan (33). We evaluated associations overall and stratified by ancestry for the 103 SNPs genotyped across OPRM1 and its 100kb flanking regions. A P value threshold of 0.00125, which takes into account the correlations among the 103 OPRM1 SNPs (α=0.05/40 independent tests) (35, 36), was used to declare statistical significance. Given the hypothesis-generating nature of our cis-eQTL mapping, SNPs associated with OPRM1 expression at the nominal significance threshold of P<0.05 were carried forward for association testing with heroin addiction.
Discovery cohort for heroin addiction association testing: Urban Health Study (UHS) cases vs. population controls
European American and African American cases were drawn from the UHS, one of the largest studies of street-recruited injection drugs users in North America (37). See the Supplement for further details on the UHS design. Stored serum samples from 3,227 UHS participants were selected for genotyping on the Illumina Omni1-Quad BeadChip. Over 60% of the genotyped UHS participants met the Office of National Drug Control Policy definition of heroin abuse (injecting 10+ times in the past 30 days) (38, 39), which is highly correlated with clinical levels of dependence on the Severity of Dependence Scale (40, 41) and with DSM-IV (42) heroin abuse/dependence in analyses of the National Household Survey on Drug Use and Health data (87% positive predictive value; see Table S1 and Supplemental Methods). These UHS participants, who abused heroin an average of 80.9 times in the past month and were very likely dependent on it, are henceforth referred to as heroin addiction cases. The remaining genotyped UHS participants, who were addicted to cocaine or other substances but not addicted to heroin, were not included in the current study.
For comparison with the UHS heroin addiction cases, we used six study cohorts from the database of Genotypes and Phenotypes (dbGaP) as a source of controls. Several prior studies have been reliably conducted using a similar design with study cases and population controls (43-48). See Table S2, Figure S2, and the Supplemental Methods for an outline of our cohort selection and quality control processes. There were no apparent biases underlying our control dataset, as indicated by genomic control inflation factor (λgc) <1.05 for all pair-wise combinations of dbGaP cohorts (Tables S3 and S4). Our final analysis dataset included 7,095 European Americans (711 UHS heroin addiction cases and 6,384 population controls) and 3,662 African Americans (1,293 UHS heroin addiction cases and 2,369 population controls).
To capture all of the nominated cis-eQTL SNPs, genotype imputation was conducted using IMPUTE2 (49) with reference to all 1000 Genomes haplotype panels (50, 51). To avoid potential bias due to UHS cases and population controls being genotyped on different arrays, imputations were based on the intersection of SNPs available across all participants (52). Further details on quality control and imputation are provided in the Supplement.
Replication cohorts for heroin addiction association testing
Two independent cohorts, comprised of heroin and other opioid abuse/dependent participants (henceforth referred to as heroin addiction cases) and their own controls, were used to replicate SNP associations with heroin addiction. First, we used African Americans from the “Alcohol Dependence [genome-wide association study] GWAS in European- and African Americans (CIDR – Gelernter Study)” (dbGaP accession number phs000425.v1.p1) (7). Our final analysis dataset included 852 African Americans (307 DSM-IV-defined cases of heroin/other opioid abuse or dependence and 545 controls with no illicit drug abuse or dependence, see Table S2 and Supplemental Methods). European Americans from this cohort were not analyzed because there was an insufficient number of independent controls (<5) who met our inclusion criteria.
Second, we used DSM-IV defined cases of heroin dependence and general population controls gathered from Australian datasets, many of which are deposited in dbGaP (accession number phs000277.v1.p1). We henceforth refer to them as the Australian Heroin Dependence Study. A subset of this case-control cohort and descriptions of the study design have been previously described (10). Our final analysis dataset totaled 5,120 Australians of European ancestry (1,976 cases and 3,144 controls, see Table S2 and Supplemental Methods).
Statistical analyses: Association testing with heroin addiction
Putative cis-eQTL SNPs were tested for association with heroin addiction across both ancestry groups in the discovery cohort (UHS vs. population controls). Significantly associated SNPs were then tested for replication in the CIDR – Gelernter Study and the Australian Heroin Dependence Study. Observed SNP genotypes or imputed SNP genotype dosages were tested for association using logistic regression models, separately by cohort and by ancestry group, with adjustment for sex and principal component eigenvectors (see Supplement). SNP association results were compared using the Forest Plot Viewer (53) and combined via fixed-effects sample size-weighted meta-analyses, as used in prior multiancestry meta-analyses (54, 55). As shown in Figure 1, at each stage of the association testing, linkage disequilibrium among the tested SNPs was taken into account to compute the P value thresholds for declaring statistical significance (35, 36) (α=0.05 / number of independent tests): meta-analysis P<0.0050 based on 10 independent tests in the discovery stage and meta-analysis P<0.0125 based on 4 independent tests in the replication stage.
Bioinformatics analyses
Linkage disequilibrium patterns were discerned using LocusZoom (56) and Haploview (57) with reference to two 1000 Genomes panels: EUR (comprised of European Americans, Finns, Britons, Spaniards, and Italians) and AFR (comprised of African Americans, Kenyans, and Nigerians). SNPs were annotated according to their location in the longest principal isoform for OPRM1, according to the APPRIS database (58). Regulatory SNP annotations were taken from the HaploReg database containing information on chromatin states, conservation, and regulatory motif alterations from the Encyclopedia of DNA Elements and elsewhere (59).
Results
Cis-eQTL mapping in human prefrontal cortex
Sixteen of the 103 SNPs tested for association with OPRM1 expression in the BrainCloud cohort were nominated as cis-eQTL SNPs (Figure 1 and Table 1): 9 SNPs overall, 2 SNPs when analyzing only European Americans, and 5 SNPs when analyzing only African Americans. Three of the cis-eQTL SNPs were significantly associated with OPRM1 expression at P<0.00125, based on an overall α=0.05 corrected for 40 independent tests among the 103 OPRM1 SNPs (35, 36); the other 13 SNPs were nominally associated at P<0.05. Results of all tested SNPs are shown in Table S5.
Table 1.
Sixteen putative cis-eQTL SNPs for OPRM1 in the BrainCloud cohort and tested for association with heroin addiction in European American and African American cases from the Urban Health Study (UHS) vs. population controls. SNPs are sorted by the meta-analysis P value for association with heroin addiction, with statistically significant results shown in bold (meta-analysis P<0.005).
| SNP (Minor Allele) | Base Pair Position (NCBI build 36) | SNP Type (Distance to OPRM 1 for intergenic SNPs) | Association with OPRM1 expression in BrainCloud | Association with OPRM1 expression in BrainCloud | Association with heroin addiction | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African Americans (N=1,293 cases and 2,369 controls) | European Americans (N=711 cases and 6,384 controls) | Meta analysis P | ||||||||||
| P, over all | P, European Americans only (N=110) | P, Africa Americans only (N=114) | MAF | P | OR (95% CI) | MAF | P | OR (95% CI) | ||||
| rs9478495 (A) | 154,365,602 | intergenic (−36,533) | 0.22 | 0.89 | 0.015 | 0.22 | 0.019 | 1.16 (1.02-1.30) | 0.16 | 0.0017 | 1.27 (1.09-1.47) | 8.9×10−5 |
| rs3778150 (C) | 154,425,351 | intronic | 0.089 | 0.042 | 0.38 | 0.19 | 0.037 | 1.15 (1.01-1.30) | 0.16 | 0.0012 | 1.28 (1.10-1.47) | 1.2×10−4 |
| rs9384169 (C) | 154,354,864 | intergenic (−47,271) | 0.25 | 0.90 | 0.046 | 0.31 | 0.029 | 1.12 (1.01-1.25) | 0.32 | 0.025 | 1.15 (1.02-1.30) | 2.0×10−3 |
| rs562859 (C) | 154,456,266 | synonymous | 0.054 | 0.016 | 0.96 | 0.38 | 0.018 | 1.13 (1.02-1.25) | 0.33 | 0.070 | 1.12 (0.99-1.26) | 4.3×10−3 |
| rs613355 (C) | 154,491,542 | intronic | 0.010 | 0.012 | 0.72 | 0.39 | 0.071 | 1.10 (0.99-1.22) | 0.34 | 0.067 | 1.12 (0.99-1.26) | 0.011 |
| rs13203628 (G) | 154,502,815 | intronic | 0.037 | 0.010 | 0.82 | 0.34 | 0.068 | 1.11 (0.99-1.23) | 0.26 | 0.11 | 1.11 (0.97-1.28) | 0.019 |
| rs671531 (A) | 154,482,434 | intronic | 0.015 | 0.010 | 0.92 | 0.39 | 0.11 | 1.08 (0.98-1.20) | 0.33 | 0.079 | 1.11 (0.99-1.26) | 0.019 |
| rs9479779 (G) | 154,544,688 | intronic | 0.30 | 0.57 | 0.042 | 0.085 | 0.57 | 1.05 (0.88-1.25) | 0.059 | 0.20 | 0.84 (0.65-1.10) | 0.48 |
| rs558948 (T) | 154,483,556 | intronic | 0.0017 | 0.00094 | 0.71 | 0.16 | 0.76 | 1.02 (0.89-1.16) | 0.25 | 0.58 | 1.04 (0.91-1.19) | 0.53 |
| rs650825 (A) | 154,470,229 | intronic | 0.0034 | 0.0017 | 0.45 | 0.17 | 0.86 | 0.99 (0.87-1.12) | 0.25 | 0.57 | 1.04 (0.91-1.19) | 0.72 |
| rs10485060 (A) | 154,557,793 | intronic | 0.039 | 0.34 | 0.24 | 0.022 | 0.23 | 0.81 (0.57-1.15) | 0.047 | 0.66 | 1.06 (0.81-1.39) | 0.73 |
| rs647192 (G) | 154,464,437 | intronic | 0.0085 | 0.0018 | 0.63 | 0.085 | 0.80 | 0.98 (0.82-1.17) | 0.25 | 0.60 | 1.04 (0.91-1.18) | 0.78 |
| rs647303 (C) | 154,324,902 | intergenic (−77,233) | 0.79 | 0.28 | 0.0010 | 0.11 | 0.29 | 1.09 (0.93-1.27) | 0.26 | 0.60 | 0.97 (0.85-1.10) | 0.85 |
| rs6900805 (G) | 154,329,725 | intergenic (−72,410) | 0.79 | 0.28 | 0.0010 | 0.11 | 0.34 | 1.08 (0.92-1.26) | 0.26 | 0.63 | 0.97 (0.85-1.10) | 0.87 |
| rs538174 (C) | 154,494,229 | intronic | 0.012 | 0.0028 | 1.00 | 0.076 | 0.27 | 0.89 (0.74-1.09) | 0.24 | 0.35 | 1.06 (0.93-1.22) | 0.91 |
| rs1852629 (T) | 154,564,432 | intronic | 0.044 | 0.090 | 0.38 | 0.42 | 0.96 | 1.00 (0.90-1.10) | 0.43 | 0.87 | 1.01 (0.90-1.13) | 0.92 |
MAF, minor allele frequency; OR, odds ratio; CI, confidence interval
HaploReg (59) corroborated the regulatory potential of the putative cis-eQTL SNPs, showing that almost all of them are predicted to alter one or more of the regulatory motifs indicative of transcription factor binding sites (60, 61) (Table S6). Moreover, rs3778150 resides in a highly conserved region, and two of the SNPs (rs562859 and rs13203628) are located in DNase hypersensitivity and enhancer sites (Table S6).
Testing cis-eQTL SNPs for association with heroin addiction
The 16 putative cis-eQTL SNPs for OPRM1 were tested for association with heroin addiction in UHS cases vs. population controls (Figure 1 and Table 1). There was no evidence to suggest heterogeneity for any of the 16 SNPs (smallest P=0.15), and the imputation quality of all tested SNPs was high (info>0.9). Meta-analysis across the ancestry groups revealed 4 SNPs significantly associated with heroin addiction at P<0.005, based on an overall α=0.05 corrected for 10 independent tests among the 16 cis-eQTL SNPs (35, 36): rs9478495, rs3778150, rs9384169, and rs562859. Minor alleles of these four SNPs were common (frequencies ranging from 16% to 38%), and they had consistent directions of association: odds ratios (ORs) ranging from 1.12 to 1.28 across the ancestry groups. The two most significantly associated SNPs for heroin addiction (rs9478495 and rs3778150) were in moderate linkage disequilibrium in the 1000 Genomes EUR panel (r2=0.48, Figure S3A) but weak linkage disequilibrium in the AFR panel (r2=0.04, Figure S3B).
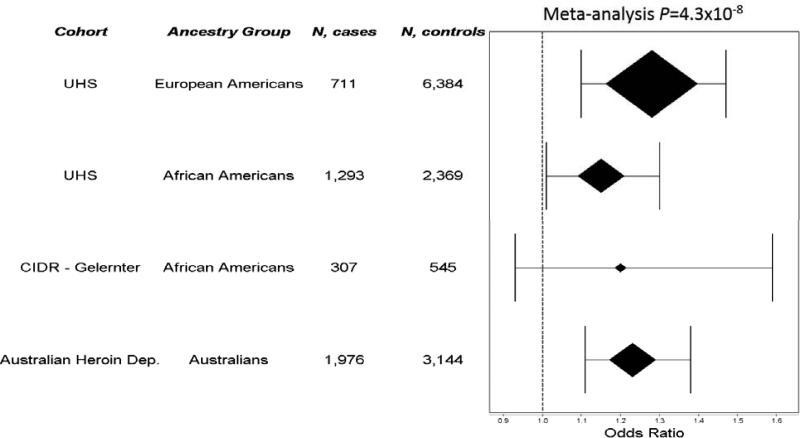
The four SNPs significantly associated with heroin addiction in the discovery cohort were then tested in African Americans from the CIDR – Gelernter Study and Australians of European ancestry from the Australian Heroin Dependence Study (Figure 1). Results are presented in Table S7. The rs3778150 association with heroin addiction was significantly replicated at P=6.8×10−5, well below the corrected replication threshold P<0.0125, based on an overall α=0.05 corrected for 4 independent tests (35, 36). Meta-analysis of rs3778150 combining discovery and replication cohorts resulted in P=4.3×10−8. As shown by the OR estimates in Figure 2, the rs3778150 minor allele (C) was consistently associated with an increased heroin addiction risk in participants of European ancestry (1.28 in the UHS and 1.23 in the Australian Heroin Dependence Study) and African Americans (1.15 in the UHS and 1.20 in the CIDR – Gelernter Study), and it was associated at P<0.05 in both UHS ancestry groups and the Australian Heroin Dependence Study. The direction of the rs3778150-C association was consistent in the CIDR – Gelernter Study but with P=0.17; this cohort was smaller than the others and thus had less statistical power than the rest of the cohorts. The three other tested SNPs were not replicated (Table S7).
Figure 2.
Forest plot of association results for the rs3778150-C allele. Results are shown across all heroin addiction case-control cohorts (Urban Health Study [UHS], CIDR – Gelernter Study, and Australian Heroin Dependence Study) and ancestry groups (European Americans, African Americans, Australians of European ancestry).
The replicated SNP rs3778150 was originally implicated as a cis-eQTL SNP at P=0.042 in BrainCloud European Americans (Table 1): the C allele being associated with reduced OPRM1 expression (Figure S4A). Rs3778150 was not significantly implicated in the African Americans only (P=0.38, Figure S4B) or all BrainCloud participants (P=0.089, Figure S4C).
Testing rs3778150-rs1799971 haplotypes for association with heroin addiction
We next evaluated linkage disequilibrium with the widely-studied SNP rs1799971 (r2 values shown in Figure S5 and D’ values shown in Figure S6). Rs3778150 and rs1799971 had r2=0 and D’=0.05 in the AFR panel. In the EUR panel, rs3778150 and rs1799971 had D’=1; the rs3778150 minor allele (C, frequency=15%) occurring on the same haplotype background with the rs1799971 major allele (A, frequency=84%) but a weak r2 value (r2=0.03) reflecting the discrepant allele frequencies.
Rs1799971 was not significantly associated with heroin addiction in any of the cohorts or ancestry groups individually or in a meta-analysis (P=0.12), although its direction of association generally suggested that the major allele (A) conferred an increased risk with heroin addiction (Figure S7). Given the indication of a shared haplotype background between rs3778150 and rs1799971 in the EUR panel, we constructed the two-SNP haplotypes and tested their associations with heroin addiction across all case-control cohorts and ancestry groups. As shown in Table 2, the haplotype carrying both the rs3778150-C and rs1799971-A alleles was consistently and significantly associated with increased risk of heroin addiction for each cohort and for both European Americans and African Americans: meta-analysis P=1.5×10−6. The haplotype carrying rs1799971-A without rs3778150-C was not associated with heroin addiction in any of the cohorts or ancestry groups: meta-analysis P=0.79.
Table 2.
Haplotype analyses of rs3778150 and the widely-studied missense SNP rs1799971 across all study cohorts and ancestry groups. The single SNP analyses suggested that the rs3778150 minor allele (C) and rs1799971 major allele (A) conferred increased risks for heroin addiction.
| rs3778150 allele | rs1799971 allele | Urban Health Study, European Americans (N=7,095) | Urban Health Study, African Americans (N=3,662) | CIDR – Gelernter Study, African Americans (N=852) | Australian Heroin Dependence Study, Australians of European ancestry (N=5,120) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | P | OR (95% CI) | Freq | P | OR (95% CI) | Freq | P | OR (95% CI) | Freq | P | OR (95% CI) | |||
| C | A | 0.16 | 0.0070 | 1.29 (1.07-1.55) | 0.19 | 0.0056 | 1.24 (1.06-1.44) | 0.18 | 0.40 | 1.17 (0.82-1.66) | 0.17 | 0.0045 | 1.26 (1.07-1.47) | 1.5×10−6 |
| T | A | 0.70 | 0.10 | 0.79 (0.60-1.05) | 0.79 | 0.13 | 1.32 (0.92-1.87) | 0.79 | 0.81 | 0.91 (0.41-2.01) | 0.71 | 0.80 | 1.02 (0.89-1.16) | 0.79 |
| T | G | 0.13 | - | ref. | 0.03 | - | ref. | 0.02 | - | ref. | 0.12 | - | ref. | - |
Testing previously implicated OPRM1 intron 1 SNPs for association with heroin addiction
Rs3778150 has varying linkage disequilibrium patterns with six other SNPs located in OPRM1 intron 1, which were previously reported for their associations with heroin/opioid addiction phenotypes (12, 19) (Figures S8 and S9). The intron 1 location of these SNPs is designated according to the longest principal isoform for OPRM1 (58). Levran et al. reported rs510769 and rs3778151 as having nominally significant associations (12), whereas Zhang et al. reported rs524731 as having a nominally significant association and rs511435, rs3823010, and rs495491 as having statistically significant associations that surpassed multiple testing correction (19). Nelson et al. (10), using a subset of the Australian replication cohort reported here, tested three of these SNPs (rs510769, rs3778151, and rs495491) and found a nominal association for rs3778151. To our knowledge, the six prior SNPs have not been tested elsewhere for independent replication. In a meta-analysis across all of our heroin addiction case-control cohorts, all six of the previously reported SNPs or their proxies were significantly replicated at P<0.01 (overall α=0.05 corrected for 5 independent tests). Their meta-analysis P values ranged from 7.0×10−4 for rs510769 to 2.7×10−8 for rs3823010 (Table 3). Similarly to rs3778150, the minor alleles were all associated with increased heroin addiction risk.
Table 3.
Associations of previously implicated OPRM1 intron 1 SNPs with heroin addiction across the cohorts used in the current study (Urban Health Study, CIDR – Gelernter Study, and the Australian Heroin Dependence Study). These SNPs were reported for having nominally to statistically significant associations with heroin addiction (N=412 cases and 184 controls, comprised of European Americans or Israelis with Jewish ancestry) (12), opioid dependence (N=91 cases and 338 controls, comprised of all European Americans) (19), or heroin dependence (N=1,459 cases and 531 controls, comprised of Australians of European ancestry) (10). None of these SNPs were previously tested for independent replication. SNPs are sorted by meta-analysis P value.
| SNP (Min or Allele) | Ref. | Previously reported P | Urban Health Study, African Americans (N=1,293 cases and 2,369 controls) | Urban Health Study, European Americans (N=711 cases and 6,384 controls) | CIDR – Gelernter Study, African Americans (N=307 cases and 545 controls) | Australian Heroin Dependence Study, Australians of European ancestry (N=1,293 cases and2,369 controls) | Meta-analysis P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | P | OR (95% CI) | MAF | P | OR (95% CI) | MAF | P | OR (95% CI) | MAF | P | OR (95% CI) | ||||
| rs3823010 (A) |
(19) | 0.001 | 0.12 | 0.025 | 1.19 (1.02-1.37) |
0.16 | 8.6 ×10−4 |
1.28 (1.11-1.49) |
0.13 | 0.17 | 1.25 (0.91-1.72) |
0.17 | 2.4× 10−4 |
1.23 (1.10-1.37) |
2.7× 10−8 |
| rs3778151 (C)a |
(10, 12) |
0.003 (12) 0.031 (10) |
0.29 | 0.029 | 1.12 (1.01-1.27) |
0.16 | 3.0 ×10−3 |
1.25 (1.08-1.45) |
0.31 | 0.33 | 1.12 (0.89-1.41) |
0.17 | 3.2× 10−4 |
1.23 (1.10-1.37) |
2.4× 10−7 |
| rs524731 (A)a |
(19) | 0.011 | 0.12 | 0.065 | 1.15 (0.99-1.33) |
0.21 | 1.4 ×10−3 |
1.25 (1.09-1.43) |
0.12 | 0.17 | 1.25 (0.91-1.75) |
0.21 | 3.7× 10−3 |
1.16 (1.05-1.29) |
1.2× 10−6 |
| rs511435 (T)a |
(19) | 0.002 | 0.20 | 0.15 | 1.10 (0.97-1.23) |
0.21 | 1.4 ×10−3 |
1.25 (1.09-1.43) |
0.19 | 0.17 | 1.20 (0.93-1.59) |
0.21 | 3.7× 10−3 |
1.16 (1.05-1.29) |
3.0× 10−6 |
| rs495491 (G)a,b |
(19) | 0.002 | 0.52 | 0.015 | 1.14 (1.02-1.25) |
0.26 | 8.2 ×10−3 |
1.19 (1.04-1.35) |
0.52 | 0.25 | 1.14 (0.91-1.43) |
not available |
not available |
not available |
1.8 ×10−4,c |
| rs510769 (T) |
(12) | 0.0008 | 0.24 | 0.97 | 1.00 (0.89-1.12) |
0.26 | 1.0 ×10−2 |
1.19 (1.04-1.35) |
0.24 | 0.29 | 1.15 (0.88-1.52) |
0.26 | 8.4× 10−3 |
1.14 (1.03-1.25) |
7.0× 10−4 |
These SNPs were not available as genotyped SNPs in the Heroin Dependence GWAS, so the following SNPs in high linkage disequilibrium were used: rs514980 as a proxy SNP for both rs511435 and rs524731 (r2=1 and D′=1 in 1000 Genomes EUR) and rs3778153 as a proxy SNP for rs3778151 (r2=0.98 and D′=1 in 1000 Genomes EUR). The best proxy for rs495491 was the other previously reported SNP rs510769 (r2=0.98 and D′=1 in 1000 Genomes EUR).
For rs495491, G is the minor allele in populations of European ancestry, but A is the minor allele in African Americans. The presented allele frequencies correspond to the G allele.
Meta-analysis was computed using the results that were available.
Rs3778150 and the top prior SNP rs3823010 were in perfect linkage disequilibrium (r2=1 and D’=1) with equivalent minor allele frequencies at 15% in EUR, and although D’=1 in AFR, their moderate r2 value of 0.54 reflected varied minor allele frequencies (20% for rs3778150 and 12% for rs3823010). In the African American cohorts from the current study where the SNPs were not collinear, both SNPs were associated with heroin addiction (African American-specific meta-analysis P=0.013 for rs3778150 and 0.0087 for rs3823010), but in follow-up regression analyses with both rs3778150 and rs3823010 included as predictors, neither SNP association remained (African American-specific meta-analysis P=0.48 for rs3778150 and 0.38 for rs3823010), suggesting that their associations with heroin addiction reflect the same underlying signal.
Discussion
The focus of our study was to identify OPRM1 polymorphisms consistently associated with heroin addiction. To increase the likelihood of detecting such associations, we followed the strategy of mapping cis-eQTLs altering OPRM1 expression in human brain from non-addicted participants, nominating this relatively small number of potentially regulatory polymorphisms, and testing them for association with heroin addiction. Among 16 putative cis-eQTL SNPs, we found that rs3778150 had a highly significant association with heroin addiction across three independent cohorts, even surpassing the standard genome-wide significance P value threshold of 5×10−8. Follow-up haplotype analyses showed that the widely-studied missense SNP rs1799971 was associated with heroin addiction only in the presence of the rs3778150 risk allele, suggesting that rs3778150 and its nearby correlated SNPs may explain the inconsistent associations of rs1799971 observed across numerous studies of heroin/opioid addiction.
The rs3778150 minor allele (C) was associated with both reduced OPRM1 expression and increased risk of heroin addiction, with its associations being strongest with European ancestry. It is common for cis-eQTL and disease risk associations to differ by ancestry, possibly due to unmeasured genetic or environmental modifiers (62). The direction of association between OPRM1 expression and risk of heroin addiction has not been conclusively shown. Nonetheless, our observed patterns for the rs3778150 minor allele are consistent with one prior report noting reduced OPRM1 expression levels in thalamus and secondary somatosensory cortex samples of heroin addicts compared to controls and postulating that down-regulation of OPRM1 may play a role in increasing risk of heroin addiction by disrupting the opioid system of a key compensatory response to heroin (25). More specifically, heroin exposure results in reduced μ receptor function, and in response, the opioid system upregulates OPRM1 expression to make more μ receptors available. It is plausible that genetic polymorphisms attenuating OPRM1 expression could hinder this compensatory mechanism, leading to a greater quantity of heroin being needed to trigger a physiological effect and thus increasing risk of becoming addicted to heroin.
One prior study reported a nominal association between rs3778150 and heroin dependence. Using a subset of the Australian Heroin Dependence Study reported here (N=1,459 cases and 531 general population controls), Nelson et al. found that rs3778150 was associated with DSM-IV defined heroin dependence at P=0.024, but this finding did not pass multiple testing correction in that study (10). The rs3778150 association signal was greatly strengthened with the larger sample size in this study: P=1.7×10−4 in the Australian Heroin Dependence Study (N=5,120) and meta-analysis P=4.3×10−8 across all cohorts (total N=16,729). Beyond rs3778150, six other OPRM1 intron 1 SNPs have been reported for their associations with heroin or other opioid addiction phenotypes. Levran et al. reported that rs510769 and rs3778151 were nominally associated in 596 European American severe heroin addicts and healthy controls, but these findings did not surpass multiple testing correction (12). Zhang et al. found that rs524731, rs511435, rs3823010, and rs495491 were nominally to significantly associated in 429 European American DSM-defined opioid dependent cases and controls (19), who were ascertained in parallel to the African Americans from the CIDR – Gelernter Study that we included in our study. We tested the prior SNPs or their proxies across the cohorts used in the current study and found statistically significant evidence to support their associations (smallest meta-analysis P=2.7×10−8 for rs3823010, Table 3). Rs3823010 was not associated with OPRM1 expression (smallest P=0.12, Table S5). Nonetheless, the directions of association for the previously reported SNPs were consistent with our rs3778150 finding: the minor alleles being associated with increased risk. The regulatory potentials of the prior SNPs were hypothesized given their positions in a noncoding region (12, 19, 63).
Our findings show, for the first time, that OPRM1 intron 1 contains SNPs that have replicable associations with heroin addiction. Our findings further suggest that rs3778150 and possibly other intron 1 SNPs may tag a cis-eQTL that influences OPRM1 expression and underlies the disease associations. Our tested SNPs also included rs563649, which is located in intron 1 and was previously reported for being associated with OPRM1 expression in vitro (64); we did not corroborate this association with OPRM1 expression (smallest P=0.26, Table S5) and did not find evidence for association with heroin addiction in our discovery cohort (meta-analysis P=0.57). Fine-mapping in the intron 1 region, particularly in diverse ancestry groups with varied linkage disequilibrium patterns, is needed to delineate the variants that influence both OPRM1 expression and heroin addiction.
Our findings were made possible by mapping putative cis-eQTL SNPs in prefrontal cortex, a highly relevant brain region when studying addiction and other psychiatric diseases (65). The prefrontal cortex is the center for mammalian purposive action including self-control/response inhibition, emotional regulation, flexibility/control of attention, and planning and goal formation. Dysregulation of this region can increase impulsivity and risk taking, stress reactivity, and bias attention and reward- anticipation toward immediate rather than delayed gratification: all characteristics associated with greater risk of addiction (65). Differences in prefrontal cortex activity have been observed in numerous human brain imaging studies comparing healthy controls to those with heroin/opioid or other drug addictions (65).
Limitations of our study include reliance on SNP genotyping and gene expression microarrays for the cis-eQTL mapping means that structural or rare genetic variants were not captured, coverage of common genetic variants was more complete for the participants of European ancestry compared to the African Americans, and only a single OPRM1 probe was represented. Future studies capturing more complete genetic variation and more localized expression patterns and measuring other brain regions relevant to addiction (66) may lead to the identification of other important regulatory polymorphisms associated with heroin addiction.
When testing the putative cis-eQTL SNPs for association with heroin addiction, many of the cases used in our study were derived from the UHS, where no study controls were available, and they were compared to a set of population controls obtained via dbGaP. This study design enabled us to create a large sample size for heroin addiction association testing. Stringent quality control was implemented to reduce the chances of introducing artifactual biases, and our top findings were corroborated in independent cohorts including DSM-IV-assessed cases and study controls. Greater power for identifying variants associated specifically with heroin addiction could be achieved with a large sample of controls who had used heroin but never became addicted. No such large sample of exposed but never addicted controls exists, but their absence does not diminish the observed associations of rs3778150 and other intron 1 SNPs with heroin addiction.
A more complete understanding of the biological factors that underlie the risk of developing heroin addiction is needed to combat its high prevalence and societal impact. To date, no specific genetic variants have been conclusively identified despite much research focused on OPRM1 and its missense SNP rs1799971. Our hypothesis-generating approach of converging cis-eQTL mapping and SNP-disease association testing led to the identification of minor alleles at two OPRM1 intron 1 SNPs (rs3778150 and rs3823010) that are consistently associated with increased risk of heroin addiction across independent cohorts with P values that even exceed the stringent genome-wide significance threshold (P<5×10−8). Our findings highlight the importance of OPRM1 intron 1 and its underlying haplotype structure that provides an explanation for the widely-studied but largely inconsistent association observed for rs1799971, giving credence to the prior suggestion that haplotypes carrying noncoding regulatory SNPs may explain the rs1799971 inconsistencies (19, 63). Consideration of these intronic SNPs and their regulatory effects on OPRM1 expression in disease-relevant human brain tissues is needed to better understand the role of OPRM1 in influencing risk of heroin and other addictions.
Supplementary Material
Acknowledgments
The BrainCloud dataset used for the eQTL mapping described in this manuscript was obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap) via accession number phs000417.v1.p1. Submission of the data to dbGaP was provided by Drs. Barbara Lipska and Joel Kleinman. Collection of the data was through a collaborative study sponsored by the National Institute of Mental Health (NIMH) Intramural Research Program. The BrainCloud applications were downloaded from http://braincloud.jhmi.edu/.
This genetic study of heroin addiction cases from the Urban Health Study was supported by the National Institute of Drug Abuse (NIDA) grant numbers R33 DA027486 and R01 DA026141. Genotyping was conducted at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University.
Controls used for comparison to the Urban Health Study cases were drawn from the following six cohorts in dbGaP: (1) Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the GWAS funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the NCBI. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research (CIDR), was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, NIDA, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained via dbGaP accession number phs000092.v1.p1.
(2) The authors acknowledge the contribution of data from “Genetic Architecture of Smoking and Smoking Cessation” accessed through dbGaP accession number phs000404.v1.p1. Funding support for genotyping, which was performed at CIDR, was provided by X01 HG005274. CIDR is fully funded through a federal contract from the NIH to The Johns Hopkins University, contract number HHSN268200782096C. Assistance with genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Funding support for collection of datasets and samples was provided by COGEND (P01 CA089392) and the University of Wisconsin Transdisciplinary Tobacco Use Research Center (P50 DA019706 and P50 CA084724).
(3) Funding support for the GWAS of Ischemic Stroke study was provided through the NIH GEI (U01 HG004436). The GWAS of Ischemic Stroke study is one of the GWAS funded as part of GENEVA under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the NCBI. Funding support for genotyping, which was performed at The Johns Hopkins University CIDR, was provided by the NIH GEI (U01HG004438) and NIH contract number HHSN268200782096C. Field work for this project was supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention; the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH Office of Research on Women's Health (ORWH) (R01 NS45012); Office of Research and Development, Medical Research Service, Department of Veterans Affairs; and the University of Maryland General Clinical Research Center (M01 RR165001), National Center for Research Resources, NIH. This study used samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds). The datasets used for the analyses described in this manuscript were obtained via dbGaP accession number phs000292.v1.p1.
(4) Funding support for the GENEVA Prostate Cancer study was provided through the National Cancer Institute (R37 CA54281, R01 CA63464, P01 CA33619, U01 CA136792, U01 CA98758, and RC2 CA148085) and the National Human Genome Research Institute (U01 HG004726). Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination, and general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004789). The datasets used for the analyses described in this manuscript were obtained from dbGaP at phs000306.v2.p1.
(5) This work utilized in part data from the NINDS dbGaP database from the CIDR NeuroGenetics Research Consortium (NGRC) Parkinson's disease study (accession number phs000196.v2.p1).
(6) For the “High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation,” research support to collect data and develop an application to support this project was provided by P50 CA093459, P50 CA097007, R01 ES011740, and R01 CA133996 (dbGaP accession number phs000187.v1.p1).
Two additional cohorts with heroin addiction cases and controls were used for replication testing. For the first replication cohort, funding support was provided through the Center for Inherited Disease Research (CIDR) and the Genetics of Alcohol Dependence in American Populations. This CIDR-Gelernter Study is a genome-wide association study (GWAS) funded as part of the “Genetics of Alcohol Dependence in American Populations” cohort, and its investigators provided assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples were provided by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) grant no. R01 AA011330. Funding support for genotyping, which was performed at the Johns Hopkins University CIDR, was provided by the NIH GEI (U01 HG004438), the NIAAA, and the NIH contract number HHSN268200782096C. The datasets used for the analyses described in this manuscript were obtained via dbGaP accession number phs000425.v1.p1.
The second replication cohort included cases, who met lifetime DSM-IV criteria for heroin dependence, and compared to general population controls. Cases and controls, many of which are available in dbGaP under “A Genome-Wide Association Study of Heroin Dependence” (accession number phs000277.v1.p1), were derived from the following datasets. (1) The Comorbidity and Trauma Study (CATS) was funded by NIDA grant number R01 DA17305. (2) The Western Australia Study on Heroin Dependence was funded by the Australia Government's National Health and Medical Research Council (grant number 513862). CATS (3) The Twin Study of Mole Development in Adolescence (PI: Nick Martin) was funded by the Australian Government's National Health and Medical Research Council (grant number 389891). (4) Support for the Hunter Community Study has been previously described (67). Lastly, GWAS genotyping services for “A Genome-Wide Association Study of Heroin Dependence” at CIDR, located at The Johns Hopkins University, were supported by NIH contract number N01 HG65403.
Financial Disclosures
Financial support for this study was provided by the following grants and contracts by the National Institutes of Health, an agency of the United States Department of Health and Human Services: R33 DA027486 (Principal Investigator [PI]: EOJ), R01 DA026141 (PI: EOJ), U01 HG004422 (PI: LJB), U01 HG004446, U10 AA008401, P01 CA089392 (PI: LJB), R01 DA013423 (PI: LJB), U01 HG004438, HHSN268200782096C, X01 HG005274, P50 DA019706, P50 CA084724, U01 HG004436, R01 NS45012, M01 RR165001, R37 CA54281, R01 CA63464, P01 CA33619, U01 CA136792, U01 CA98758, RC2 CA148085, U01 HG004726, U01 HG004789, P50 CA093459, P50 CA097007, R01 ES011740, R01 CA133996, R01 AA011330, R01 DA17305 (PI: ECN), and N01 HG65403. This study was also supported by the Australia Government's National Health and Medical Research Council (grant number 513862; PI: SS) and National Health and Medical Research Council (grant number 389891; PI: NGM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Degenhardt L, Charlson F, Mathers B, Hall WD, Flaxman AD, Johns N, et al. The global epidemiology and burden of opioid dependence: results from the global burden of disease 2010 study. Addiction. 2014 doi: 10.1111/add.12551. [DOI] [PubMed] [Google Scholar]
- 2.Administration SAaMHS . Services USDoHaH. Substance Abuse and Mental Health Services Administration; Rockville, MD: 2013. Results from the 2012 National Survey on Drug Use and Health: Summary of National Findings. [Google Scholar]
- 3.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. The American journal of psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 4.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 5.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. American journal of medical genetics. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.van den Bree MB, Johnson EO, Neale MC, Pickens RW. Genetic and environmental influences on drug use and abuse/dependence in male and female twins. Drug and alcohol dependence. 1998;52:231–241. doi: 10.1016/s0376-8716(98)00101-x. [DOI] [PubMed] [Google Scholar]
- 7.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-Wide Association Study of Opioid Dependence: Multiple Associations Mapped to Calcium and Potassium Pathways. Biological psychiatry. 2013 doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen DA, Ji F, Yuferov V, Ho A, He C, Ott J, et al. Genome-wide association study identifies genes that may contribute to risk for developing heroin addiction. Psychiatric genetics. 2010;20:207–214. doi: 10.1097/YPG.0b013e32833a2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Guo X, Jiang Y, Zhang H. NCK2 is significantly associated with opiates addiction in African-origin men. TheScientificWorldJournal. 2013;2013:748979. doi: 10.1155/2013/748979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addiction biology. 2014;19:111–121. doi: 10.1111/j.1369-1600.2012.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA psychiatry. 2013;70:325–333. doi: 10.1001/jamapsychiatry.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes, brain, and behavior. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith RJ, Doyle GA, Han AM, Crowley JJ, Oslin DW, Patkar AA, et al. Novel exonic mu-opioid receptor gene (OPRM1) polymorphisms not associated with opioid dependence. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:105–109. doi: 10.1002/ajmg.b.30105. [DOI] [PubMed] [Google Scholar]
- 14.Coller JK, Beardsley J, Bignold J, Li Y, Merg F, Sullivan T, et al. Lack of association between the A118G polymorphism of the mu opioid receptor gene (OPRM1) and opioid dependence: A meta-analysis. Pharmacogenomics and personalized medicine. 2009;2:9–19. [PMC free article] [PubMed] [Google Scholar]
- 15.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug and alcohol dependence. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Haerian BS, Haerian MS. OPRM1 rs1799971 polymorphism and opioid dependence: evidence from a meta-analysis. Pharmacogenomics. 2013;14:813–824. doi: 10.2217/pgs.13.57. [DOI] [PubMed] [Google Scholar]
- 17.Glatt SJ, Bousman C, Wang RS, Murthy KK, Rana BK, Lasky-Su JA, et al. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug and alcohol dependence. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr., Foroud T, et al. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:877–884. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charbogne P, Kieffer BL, Befort K. 15 years of genetic approaches in vivo for addiction research: Opioid receptor and peptide gene knockout in mouse models of drug abuse. Neuropharmacology. 2014;76(Pt B):204–217. doi: 10.1016/j.neuropharm.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beyer A, Koch T, Schroder H, Schulz S, Hollt V. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. Journal of neurochemistry. 2004;89:553–560. doi: 10.1111/j.1471-4159.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 23.Kroslak T, Laforge KS, Gianotti RJ, Ho A, Nielsen DA, Kreek MJ. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. Journal of neurochemistry. 2007;103:77–87. doi: 10.1111/j.1471-4159.2007.04738.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. The Journal of biological chemistry. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 25.Oertel BG, Doehring A, Roskam B, Kettner M, Hackmann N, Ferreiros N, et al. Genetic-epigenetic interaction modulates mu-opioid receptor regulation. Hum Mol Genet. 2012;21:4751–4760. doi: 10.1093/hmg/dds314. [DOI] [PubMed] [Google Scholar]
- 26.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy A, Chu JH, Xu M, Carey VJ, Lazarus R, Liu A, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, Festen EA, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet. 2010;19:3482–3488. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- 30.Qiu W, Cho MH, Riley JH, Anderson WH, Singh D, Bakke P, et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS One. 2011;6:e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diekstra FP, Saris CG, van Rheenen W, Franke L, Jansen RC, van Es MA, et al. Mapping of gene expression reveals CYP27A1 as a susceptibility gene for sporadic ALS. PLoS One. 2012;7:e35333. doi: 10.1371/journal.pone.0035333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, et al. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biological psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 37.Kral AH, Lorvick J, Gee L, Bacchetti P, Rawal B, Busch M, et al. Trends in human immunodeficiency virus seroincidence among street-recruited injection drug users in San Francisco, 1987-1998. Am J Epidemiol. 2003;157:915–922. doi: 10.1093/aje/kwg070. [DOI] [PubMed] [Google Scholar]
- 38.Morral AR, McCaffrey D, Iguchi MY. Hardcore drug users claim to be occasional users: drug use frequency underreporting. Drug and alcohol dependence. 2000;57:193–202. doi: 10.1016/s0376-8716(99)00048-4. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes W, Layne M, Johnston P, Hozik L. What America's Users Spend on Illegal Drugs 1988-1998. Office of National Drug Control Policy. 2000 [Google Scholar]
- 40.Gossop M, Griffiths P, Powis B, Strang J. Severity of dependence and route of administration of heroin, cocaine and amphetamines. British journal of addiction. 1992;87:1527–1536. doi: 10.1111/j.1360-0443.1992.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 41.Strang J, Griffiths P, Powis B, Gossop M. Heroin chasers and heroin injectors: differences observed in a community sample in London, UK. The American journal on addictions / American Academy of Psychiatrists in Alcoholism and Addictions. 1999;8:148–160. doi: 10.1080/105504999305956. [DOI] [PubMed] [Google Scholar]
- 42.Association AP. Diagnostic and statistical manual of mental disorders: DSM-IV. 4th ed. American Psychiatric Association; Washington DC: 1994. [Google Scholar]
- 43.Luca D, Ringquist S, Klei L, Lee AB, Gieger C, Wichmann HE, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson J, Griffiths H, De Salvo G, Cole M, Jacob A, Macleod A, et al. Genome-wide association study of primary open angle glaucoma risk and quantitative traits. Molecular vision. 2012;18:1083–1092. [PMC free article] [PubMed] [Google Scholar]
- 47.Teerlink C, Farnham J, Allen-Brady K, Camp NJ, Thomas A, Leachman S, et al. A unique genome-wide association analysis in extended Utah high-risk pedigrees identifies a novel melanoma risk variant on chromosome arm 10q. Hum Genet. 2012;131:77–85. doi: 10.1007/s00439-011-1048-z. [DOI] [PubMed] [Google Scholar]
- 48.Genovese G, Tonna SJ, Knob AU, Appel GB, Katz A, Bernhardy AJ, et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int. 2010;78:698–704. doi: 10.1038/ki.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hancock DB, Levy JL, Gaddis NC, Bierut LJ, Saccone NL, Page GP, et al. Assessment of genotype imputation performance using 1000 Genomes in African American studies. PLoS One. 2012;7:e50610. doi: 10.1371/journal.pone.0050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson EO, Hancock DB, Levy JL, Gaddis NC, Saccone NL, Bierut LJ, et al. Imputation across genotyping arrays for genome-wide association studies: assessment of bias and a correction strategy. Hum Genet. 2013;132:509–522. doi: 10.1007/s00439-013-1266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyles AL, Harris SF, Rooney AA, Thayer KA. Forest Plot Viewer: a new graphing tool. Epidemiology. 2011;22:746–747. doi: 10.1097/EDE.0b013e318225ba48. [DOI] [PubMed] [Google Scholar]
- 54.Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, Keating BJ. Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum Mol Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanktree MB, Guo Y, Murtaza M, Glessner JT, Bailey SD, Onland-Moret NC, et al. Meta-analysis of Dense Genecentric Association Studies Reveals Common and Uncommon Variants Associated with Height. Am J Hum Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez JM, Maietta P, Ezkurdia I, Pietrelli A, Wesselink JJ, Lopez G, et al. APPRIS: annotation of principal and alternative splice isoforms. Nucleic acids research. 2013;41:D110–117. doi: 10.1093/nar/gks1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spivakov M, Akhtar J, Kheradpour P, Beal K, Girardot C, Koscielny G, et al. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome biology. 2012;13:R49. doi: 10.1186/gb-2012-13-9-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levran O, Awolesi O, Linzy S, Adelson M, Kreek MJ. Haplotype block structure of the genomic region of the mu opioid receptor gene. J Hum Genet. 2011;56:147–155. doi: 10.1038/jhg.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet. 2009;18:1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nature reviews Neuroscience. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. The American journal of psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 67.McEvoy M, Smith W, D'Este C, Duke J, Peel R, Schofield P, et al. Cohort profile: The Hunter Community Study. International journal of epidemiology. 2010;39:1452–1463. doi: 10.1093/ije/dyp343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.