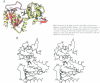Abstract
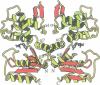
NMP kinases catalyse the phosphorylation of the canonical nucleotides to the corresponding diphosphates using ATP as a phosphate donor. Bacteriophage T4 deoxynucleotide kinase (DNK) is the only member of this family of enzymes that recognizes three structurally dissimilar nucleotides: dGMP, dTMP and 5-hydroxymethyl-dCMP while excluding dCMP and dAMP. The crystal structure of DNK with its substrate dGMP has been determined at 2.0 A resolution by single isomorphous replacement. The structure of the ternary complex with dGMP and ATP has been determined at 2.2 A resolution. The polypeptide chain of DNK is folded into two domains of equal size, one of which resembles the mononucleotide binding motif with the glycine-rich P-loop. The second domain, consisting of five alpha-helices, forms the NMP binding pocket. A hinge connection between the domains allows for large movements upon substrate binding which are not restricted by dimerization of the enzyme. The mechanism of active centre formation via domain closure is described. Comparison with other P-loop-containing proteins indicates an induced-fit mode of NTP binding. Protein-substrate interactions observed at the NMP and NTP sites provide the basis for understanding the principles of nucleotide discrimination.
Full text
PDF
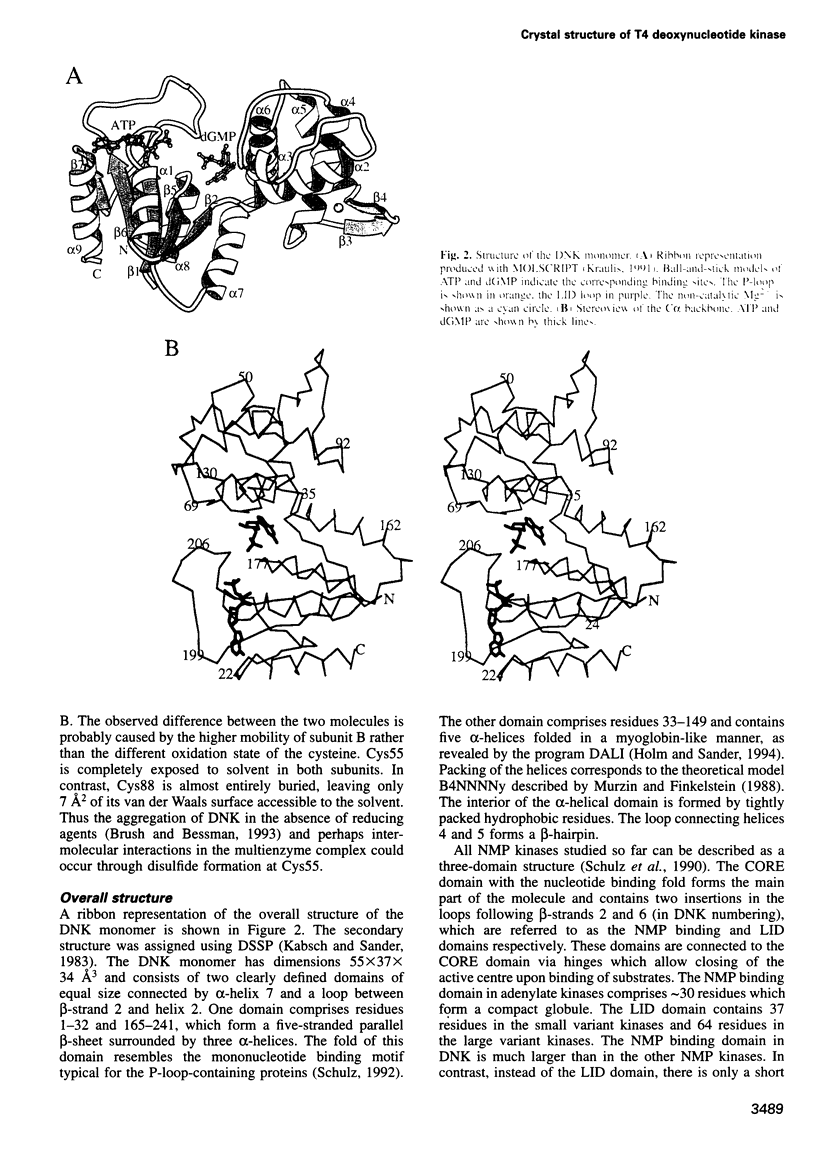

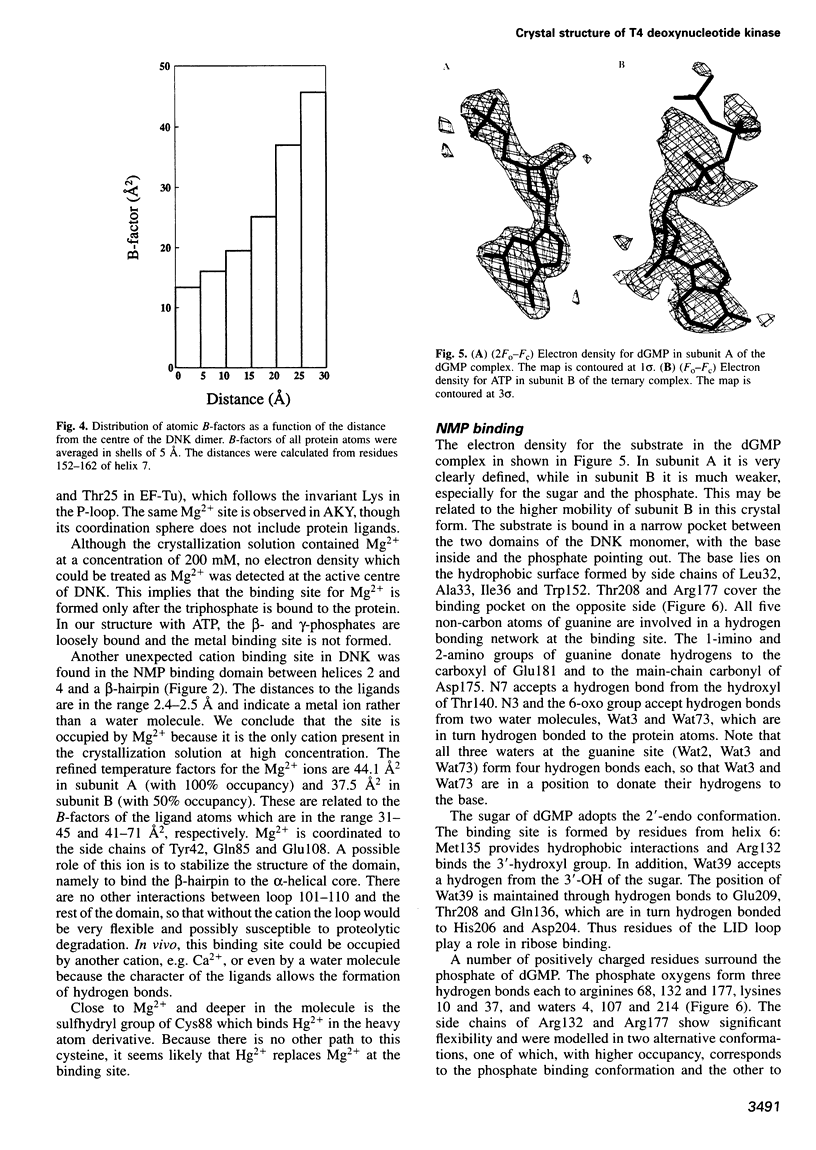
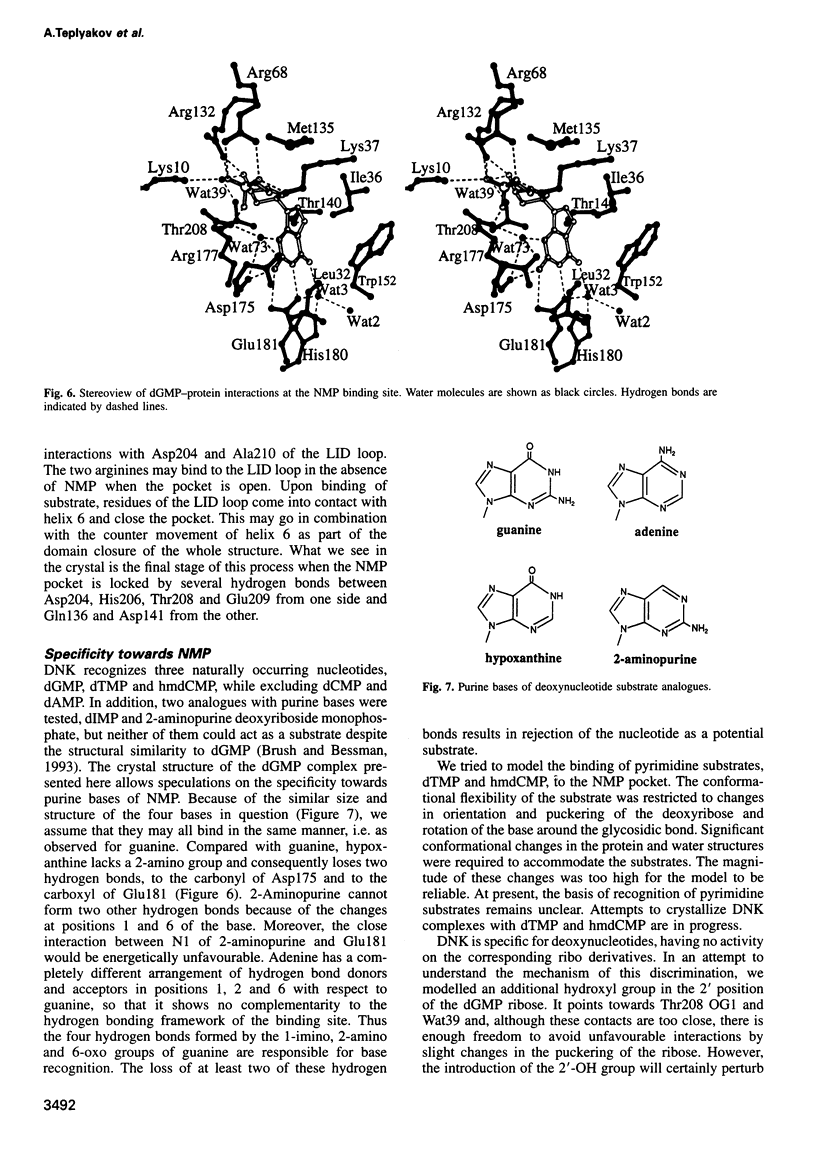
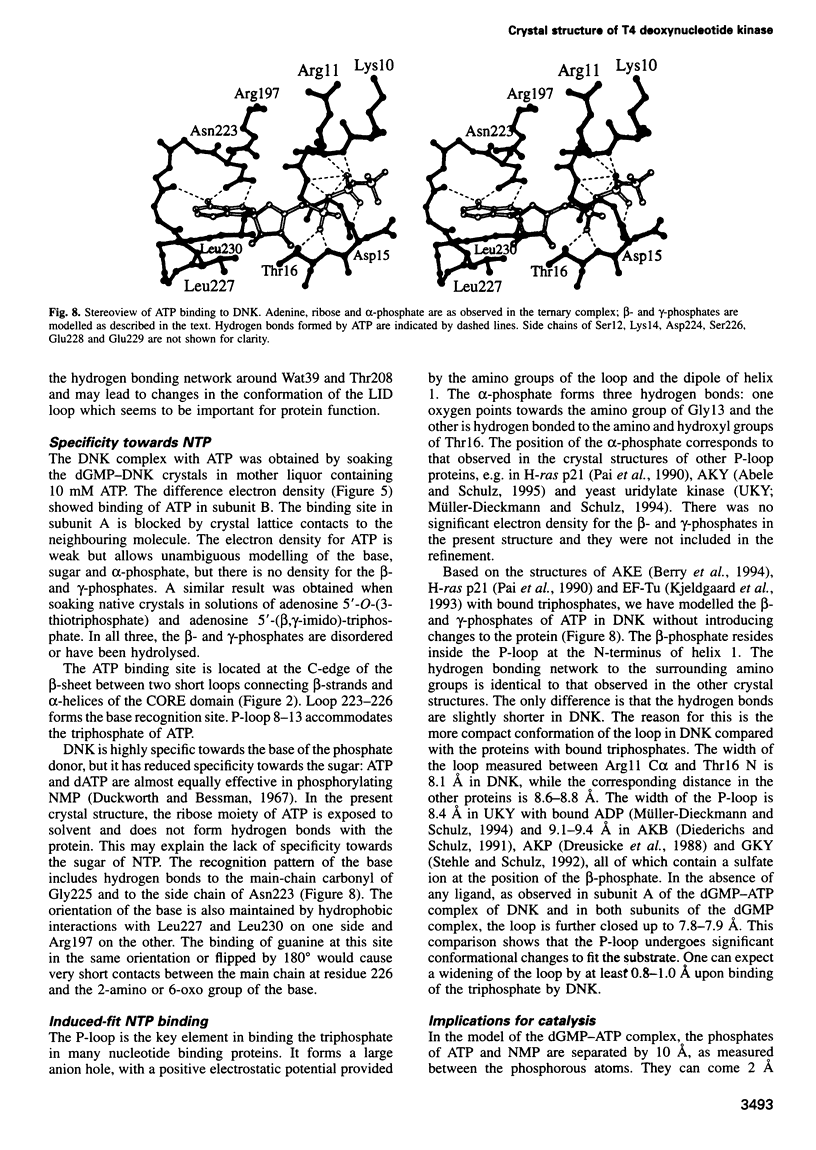




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abele U., Schulz G. E. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995 Jul;4(7):1262–1271. doi: 10.1002/pro.5560040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELLO L. J., BESSMAN M. J. The enzymology of virus-infected bacteria. IV. Purification and properties of the deoxynucleotide kinase induced by bacteriophage T2. J Biol Chem. 1963 May;238:1777–1787. [PubMed] [Google Scholar]
- Berry M. B., Meador B., Bilderback T., Liang P., Glaser M., Phillips G. N., Jr The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP. Proteins. 1994 Jul;19(3):183–198. doi: 10.1002/prot.340190304. [DOI] [PubMed] [Google Scholar]
- Broida J., Abelson J. Sequence organization and control of transcription in the bacteriophage T4 tRNA region. J Mol Biol. 1985 Oct 5;185(3):545–563. doi: 10.1016/0022-2836(85)90071-3. [DOI] [PubMed] [Google Scholar]
- Brown D. G., Visse R., Sandhu G., Davies A., Rizkallah P. J., Melitz C., Summers W. C., Sanderson M. R. Crystal structures of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and ganciclovir. Nat Struct Biol. 1995 Oct;2(10):876–881. doi: 10.1038/nsb1095-876. [DOI] [PubMed] [Google Scholar]
- Brush G. S., Bessman M. J. Chemical modification of bacteriophage T4 deoxynucleotide kinase. Evidence of a single catalytic region. J Biol Chem. 1993 Jan 25;268(3):1603–1609. [PubMed] [Google Scholar]
- Brush G. S., Bhatnagar S. K., Bessman M. J. Bacteriophage T4 deoxynucleotide kinase: gene cloning and enzyme purification. J Bacteriol. 1990 Jun;172(6):2935–2939. doi: 10.1128/jb.172.6.2935-2939.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahnke T., Tsai M. D. Mechanism of adenylate kinase. The conserved aspartates 140 and 141 are important for transition state stabilization instead of substrate-induced conformational changes. J Biol Chem. 1994 Mar 18;269(11):8075–8081. [PubMed] [Google Scholar]
- Diederichs K., Schulz G. E. The refined structure of the complex between adenylate kinase from beef heart mitochondrial matrix and its substrate AMP at 1.85 A resolution. J Mol Biol. 1991 Feb 5;217(3):541–549. doi: 10.1016/0022-2836(91)90756-v. [DOI] [PubMed] [Google Scholar]
- Dreusicke D., Karplus P. A., Schulz G. E. Refined structure of porcine cytosolic adenylate kinase at 2.1 A resolution. J Mol Biol. 1988 Jan 20;199(2):359–371. doi: 10.1016/0022-2836(88)90319-1. [DOI] [PubMed] [Google Scholar]
- Dreusicke D., Schulz G. E. The glycine-rich loop of adenylate kinase forms a giant anion hole. FEBS Lett. 1986 Nov 24;208(2):301–304. doi: 10.1016/0014-5793(86)81037-7. [DOI] [PubMed] [Google Scholar]
- Duckworth D. H., Bessman M. J. The enzymology of virus-infected bacteria. X. A biochemical-genetic study of the deoxynucleotide kinase induced by wild type and amber mutants of phage T4. J Biol Chem. 1967 Jun 25;242(12):2877–2885. [PubMed] [Google Scholar]
- Gerstein M., Schulz G., Chothia C. Domain closure in adenylate kinase. Joints on either side of two helices close like neighboring fingers. J Mol Biol. 1993 Jan 20;229(2):494–501. doi: 10.1006/jmbi.1993.1048. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J Mol Biol. 1990 Jun 20;213(4):583–591. doi: 10.1016/S0022-2836(05)80243-8. [DOI] [PubMed] [Google Scholar]
- Janin J., Miller S., Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988 Nov 5;204(1):155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen P., Thirup S., Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993 Sep 15;1(1):35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- Milner-White E. J., Coggins J. R., Anton I. A. Evidence for an ancestral core structure in nucleotide-binding proteins with the type A motif. J Mol Biol. 1991 Oct 5;221(3):751–754. doi: 10.1016/0022-2836(91)80170-y. [DOI] [PubMed] [Google Scholar]
- Murzin A. G., Finkelstein A. V. General architecture of the alpha-helical globule. J Mol Biol. 1988 Dec 5;204(3):749–769. doi: 10.1016/0022-2836(88)90366-x. [DOI] [PubMed] [Google Scholar]
- Müller-Dieckmann H. J., Schulz G. E. The structure of uridylate kinase with its substrates, showing the transition state geometry. J Mol Biol. 1994 Feb 11;236(1):361–367. doi: 10.1006/jmbi.1994.1140. [DOI] [PubMed] [Google Scholar]
- Müller C. W., Schulz G. E. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. J Mol Biol. 1992 Mar 5;224(1):159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G. P., Mathews C. K. Functional compartmentation of DNA precursors in T4 phage-infected bacteria. J Biol Chem. 1978 May 25;253(10):3461–3467. [PubMed] [Google Scholar]
- Schulz G. E., Müller C. W., Diederichs K. Induced-fit movements in adenylate kinases. J Mol Biol. 1990 Jun 20;213(4):627–630. doi: 10.1016/S0022-2836(05)80250-5. [DOI] [PubMed] [Google Scholar]
- Sebastiao P., Obmolova G., Brush G. S., Bessman M. J., Teplyakov A. Crystallization and preliminary X-ray analysis of bacteriophage T4 deoxynucleotide kinase. Acta Crystallogr D Biol Crystallogr. 1996 Jan 1;52(Pt 1):226–228. doi: 10.1107/S0907444995007281. [DOI] [PubMed] [Google Scholar]
- Stehle T., Schulz G. E. Refined structure of the complex between guanylate kinase and its substrate GMP at 2.0 A resolution. J Mol Biol. 1992 Apr 20;224(4):1127–1141. doi: 10.1016/0022-2836(92)90474-x. [DOI] [PubMed] [Google Scholar]
- Tsai M. D., Yan H. G. Mechanism of adenylate kinase: site-directed mutagenesis versus X-ray and NMR. Biochemistry. 1991 Jul 16;30(28):6806–6818. doi: 10.1021/bi00242a002. [DOI] [PubMed] [Google Scholar]
- Vonrhein C., Schlauderer G. J., Schulz G. E. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure. 1995 May 15;3(5):483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Wild K., Bohner T., Aubry A., Folkers G., Schulz G. E. The three-dimensional structure of thymidine kinase from herpes simplex virus type 1. FEBS Lett. 1995 Jul 17;368(2):289–292. doi: 10.1016/0014-5793(95)00680-8. [DOI] [PubMed] [Google Scholar]