Abstract
Hundreds of variants in autosomal genes associated with the limb girdle muscular dystrophies (LGMDs) have been reported as being causative. However, in most cases the proof of pathogenicity derives from their non-occurrence in hundreds of healthy controls and/or from segregation studies in small families. The limited statistics of the genetic variations in the general population may hamper a correct interpretation of the effect of variants on the protein. To clarify the meaning of low-frequency variants in LGMD genes, we have selected all variants described as causative in the Leiden Open Variation Database and the Human Gene Mutation Database. We have systematically searched for their frequency in the NHLBI GO Exome Sequencing Project (ESP) and in our internal database. Surprisingly, the ESP contains about 4% of the variants previously associated with a dominant inheritance and about 9% of those associated with a recessive inheritance. The putative disease alleles are much more frequent than those estimated considering the disease prevalence. In conclusion, we hypothesize that a number of disease-associated variants are non-pathogenic and that other variations are not fully penetrant, even if they affect the protein function, suggesting a more complex genetic mechanisms for such heterogeneous disorders.
Introduction
Limb girdle muscular dystrophies (LGMDs) are a highly heterogeneous group of non-congenital neuromuscular disorders, characterized predominantly by a progressive weakness and wasting of the pelvic or shoulder girdle musculature.1 Classic pathological features of muscular dystrophy are typically observed on muscle biopsy that shows necrosis, regeneration, a variation in fiber size with various levels of fibrosis and adipose tissue infiltration.2 The age of onset, severity and rate of progression vary greatly among the subtypes of this condition3 and may be different within the same family, because of the variable expressivity. In fact, the clinical course ranges from severe forms with a rapid onset and progression to very mild forms allowing affected people to have normal lifespans and activity levels.1 These disorders have been considered monogenic and are classified further according to their inheritance pattern: limb girdle muscular dystrophy type 1 (LGMD1) indicates an autosomal dominant inheritance pattern, whereas limb girdle muscular dystrophy type 2 (LGMD2) includes recessively inherited disorders. Nevertheless, clinically, it is quite difficult to distinguish among recessive, sporadic and dominant forms.4 As their features vary and overlap with those of other muscle disorders, such as congenital muscular dystrophies, myofibrillar myopathies, metabolic myopathy and inflammatory myopathy, determining the prevalence of LGMDs is very difficult.5 Moreover, the prevalence of each specific subtype differs considerably from study to study. However, the estimated prevalence of all LGMD cases is between 1/20 000 individuals4, 6, 7 and 1/50 000. According to the higher estimate, the LGMD2 carrier frequency can be estimated at 1/72.
To date, hundreds of variants in autosomal genes associated with LGMDs have been reported as being causative. However, in most cases, the proof of pathogenicity derives from their non-occurrence in hundreds of healthy controls and/or from segregation studies in small families. Until recently, the limited knowledge on the genetic variation in the general population made the meaning of low-frequency variants unclear. Since June 2011, the publication of whole-exome data from the NHLBI GO Exome Sequencing Project (ESP)8 has been improving the understanding of the pathogenicity of variants. Recently, several studies have demonstrated a high prevalence of genetic variants previously associated with LQT syndrome9 and cardiomyopathy10 in new population-based exome data.
To clarify the meaning of low-frequency LGMD variants, listed in the Leiden Open Variation Database (LOVD)11 and the Human Gene Mutation Database (HGMD),12 we have systematically searched for their frequency in the new ESP data and compared the results with the expected prevalence of monogenic LGMDs in the same population. As a further step, we have evaluated the frequency of these previously described variations in our internal database, which contains data from next-generation sequencing experiments.
Methods
The LOVD11 and the HGMD12 were searched for missense and nonsense LGMD-associated variants. We evaluated variants in genes associated with the autosomal dominant LGMDs (CAV3, DNAJB6, LMNA, MYOT, TNPO3) and genes for which genetics variants are responsible for autosomal recessive LGMDs (ANO5, CAPN3, DAG1, DYSF, FKRP, FKTN, POMGNT1, POMT1, POMT2, SGCA, SGCB, SGCD, SGCG, TCAP, TRIM32, TTN). The variants classified by at least one of the two databases (LOVD and HGMD) as being pathogenic/disease causing were included in the analyses. To adopt a conservative approach, variants of unknown pathogenicity were excluded from our analysis. In particular, we considered intronic variations only if positioned within 10 nucleotides upstream and downstream of the exons.
Additionally, we used three bioinformatic tools (PolyPhen2,13 SIFT14 and MutationTaster15) to predict the effect of all LGMD-associated variants on their related protein. We classified the variants as ‘benign' or ‘damaging', when at least two of the three tools were in agreement. Otherwise, the variations were categorized as ‘unknown'. As nonsense and synonymous variants cannot be evaluated by bioinformatic tools, we classified the synonymous variants as of ‘unknown pathogenicity', whereas all the nonsense changes were considered damaging.
All identified variants were then systematically searched for in the ESP data. In ESP, whole-exome sequencing (WES) data of 6500 individuals, including both European Americans (4300 individuals) and African Americans (2203 individuals), from different population studies were analyzed. As not all the protein coding regions are properly sequenced by WES, we evaluated the minimum read depth at all coding regions of the investigated genes to be sure that all the selected variations would have been reported in ESP, if present in the analyzed samples.
The literature was searched for functional data of all the LGMD-associated variants identified in the ESP population. In particular, if any in vivo or in vitro experiments had demonstrated differences between the wild-type and mutated models, functional data were considered positive. Family cosegregation was evaluated investigating the LOVD database.
Additionally, we evaluated differences in distributions of the three categories of pathogenicity (benign, damaging and unknown) between the variants identified in ESP versus variants not identified in ESP by using the χ2 test. A P-value of 0.05 was considered as statistically significant.
Finally, we searched for all selected LGMD-associated variants in TIGEM Database (TDB), our internal variation database,16 which stores all the variations found in the resequencing projects performed so far in our institute.
In particular, it contains the NGS data from 530 individuals: 277 patients with a clinical diagnosis of LGMDs or myopathies were analyzed with MotorPlex, a custom NGS-based platform (myopathy MP);17 253 individuals were tested by WES, comprising 44 patients with clinical diagnosis of LGMDs or myopathies (myopathy wes) and 209 with unrelated disorders (e.g., dysplastic nevus, prognathism, ciliopathies, metabolic and psychiatric diseases) (other wes).
Results
Autosomal dominant LGMD1
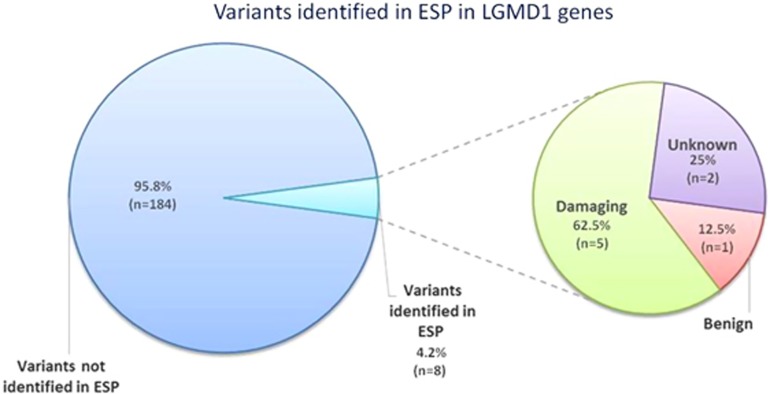
In the LOVD and HGMD databases, we identified 192 single-nucleotide variations previously associated with autosomal dominant LGMD1. In particular, we selected 30 variations in the CAV3 gene, all of them being missense variants; in the LMNA gene, we identified 147 variants, of which 128 are nonsynonymous, 3 are nonsense, 5 are synonymous and 11 are intronic changes. In the MYOT gene, we found 10 missense variations, whereas in TNPO3 and DNAJB6, 1 and 4 nonsynonymous variants were selected, respectively (Table 1). We evaluated the effect of each variant on the related protein: 156 variants were predicted to be damaging, 13 to be benign and 23 unknown (Table 2). In the ESP population, we identified 8 variations previously associated with LGMD1 (4.2%) (Supplementary Table 1 and Figure 1), of which 5 (62.5%) were predicted to be damaging and 1 (12.5%) to be benign. Two variations (25%) were found in ESP and classified as being of unknown pathogenicity. For 5 out of 8 variants, both segregation analyses and functional studies had demonstrated a putative causative effect. In particular, only variations in the CAV3, LMNA and MYOT genes were identified in the ESP population, as expected, because of the low number of variants described until now in the TNPO318 and DNAJB6 genes19 (Table 3).
Table 1. Identified variants in LGMD1 genes in LOVD and HGMD databases.
| LGMD1 | |||||
|---|---|---|---|---|---|
| Gene | Missense | Nonsense | Silent | Intronic | Total |
| CAV3 | 30 | 0 | 0 | 0 | 30 |
| LMNA | 128 | 3 | 5 | 11 | 147 |
| MYOT | 10 | 0 | 0 | 0 | 10 |
| TNPO3 | 1 | 0 | 0 | 0 | 1 |
| DNAJB6 | 1 | 3 | 0 | 0 | 4 |
| Total | 170 | 6 | 5 | 11 | 192 |
Table 2. In silico prediction of identified variants in LGMD1 genes.
| LGMD1 | ||||
|---|---|---|---|---|
| Gene | Benign | Damaging | Unknown | Total |
| CAV3 | 1 | 27 | 2 | 30 |
| LMNA | 7 | 119 | 21 | 147 |
| MYOT | 1 | 9 | 0 | 10 |
| TNPO3 | 0 | 1 | 0 | 1 |
| DNAJB6 | 4 | 0 | 0 | 4 |
| Total | 13 | 156 | 23 | 192 |
Figure 1.
Percentage of variants identified in ESP in LGMD1 genes. In the ESP population, we identified 8 out of 292 variations previously associated with LGMD1 (4.2%), of which 5 (62.5%) were predicted to be damaging, 1 (12.5%) to be benign and 2 (25%) as being of unknown pathogenicity.
Table 3. LGMD1-associated variants identified in ESP.
| LGMD1 | ||||
|---|---|---|---|---|
| Gene | Benign | Damaging | Unknown | Total |
| CAV3 | 0 | 3 | 0 | 3 |
| LMNA | 0 | 1 | 2 | 3 |
| MYOT | 1 | 1 | 0 | 2 |
| Total | 1 | 5 | 2 | 8 |
Of the remaining 184 LGMD1-associated variants not present in ESP, 156 (84.7%) were predicted to be damaging, 9 (4.9%) to be benign and 19 (10.4%) to be of unknown pathogenicity.
Moreover, we studied the genotypes of the 8 variants in ESP population, finding 1 homozygous and 149 heterozygous genotypes, for a total of 150 individuals potentially affected by LGMD1, which is equivalent to an LGMD1 allelic frequency of 0.01 and to an LGMD1 prevalence of 1:43 (Supplementary Table 1). In addition, we studied the genotypes of these variants in our internal database population, the TDB: we have identified in heterozygosity the variant c.51 C>T (p.(=)) in LMNA gene,20 responsible for a splice defect, in a total of 14 subjects, in particular, 9 myopathic patients analyzed by MotorPlex, a targeted NGS approach17 and 5 patients with unrelated phenotypes. In both the cases, the LGMD1 allelic frequency and the LGMD1 prevalence are very similar to those obtained from ESP population. In fact, the allelic frequency is 0.01, whereas the prevalence is between 1:31 and 1:42 (Supplementary Table 1). In the ESP population, the variant c.51 C>T (p.(=)) in LMNA gene has 1 homozygous and 120 heterozygous genotypes. However, if this variant was excluded, the genotype prevalence in ESP would be 1:224. No other variation was identified in the TDB population, as expected, because of the low number of patients sequenced until now.
Autosomal recessive LGMD2
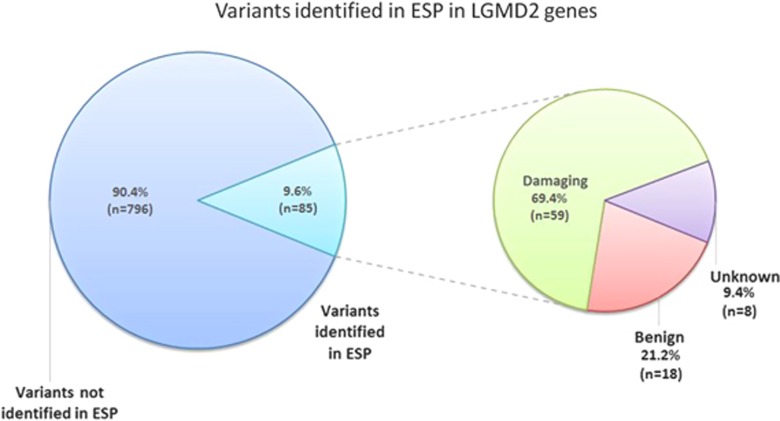
In the LOVD and HGMD databases, we selected 881 single-nucleotide variations previously associated with autosomal recessive LGMD2. In particular, we identified 591 missense, 155 nonsense, 8 synonymous and 127 intronic variations in all 16 genes analyzed (Table 4). Bioinformatic analysis predicted 541 variants to be damaging and 147 to be benign, whereas 293 variations were classified as being of unknown pathogenicity (Table 5). In the ESP population, we identified 85 variations (Table 6, Supplementary Table 2 and Figure 2) previously associated with LGMD2 (9.6%), of which 59 (69.4%) were predicted to be damaging and 18 (21.2%) to be benign. Eight variations (9.4%) were found in ESP and classified as being of unknown pathogenicity. For 19 out of 85 variants, both segregation analyses and functional studies had demonstrated a putative causative effect.
Table 4. Identified variants in LGMD2 genes in LOVD and HGMD databases.
| LGMD2 | |||||
|---|---|---|---|---|---|
| Gene | Missense | Nonsense | Silent | Intronic | Total |
| ANO5 | 19 | 5 | 0 | 5 | 29 |
| CAPN3 | 214 | 29 | 4 | 42 | 289 |
| DAG | 1 | 1 | 0 | 0 | 2 |
| DYSF | 105 | 60 | 3 | 45 | 213 |
| FKRP | 82 | 12 | 1 | 0 | 95 |
| FKTN | 12 | 9 | 0 | 0 | 21 |
| POMGNT1 | 19 | 7 | 0 | 9 | 35 |
| POMT1 | 20 | 8 | 0 | 3 | 31 |
| POMT2 | 22 | 2 | 0 | 6 | 30 |
| SGCA | 50 | 5 | 0 | 8 | 63 |
| SGCB | 21 | 5 | 0 | 5 | 31 |
| SGCD | 5 | 3 | 0 | 0 | 8 |
| SGCG | 10 | 4 | 0 | 3 | 17 |
| TCAP | 0 | 3 | 0 | 0 | 3 |
| TRIM32 | 3 | 0 | 0 | 0 | 3 |
| TTN | 8 | 2 | 0 | 1 | 11 |
| Total | 591 | 155 | 8 | 127 | 881 |
Table 5. In silico prediction of identified variants in LGMD2 genes.
| LGMD2 | ||||
|---|---|---|---|---|
| Gene | Benign | Damaging | Unknown | Total |
| ANO5 | 3 | 16 | 10 | 29 |
| CAPN3 | 12 | 201 | 76 | 289 |
| DAG | 0 | 1 | 1 | 2 |
| DYSF | 10 | 95 | 108 | 213 |
| FKRP | 7 | 75 | 13 | 95 |
| FKTN | 0 | 12 | 9 | 21 |
| POMGNT1 | 1 | 19 | 15 | 35 |
| POMT1 | 2 | 18 | 11 | 31 |
| POMT2 | 0 | 20 | 10 | 30 |
| SGCA | 9 | 41 | 13 | 63 |
| SGCB | 1 | 19 | 11 | 31 |
| SGCD | 0 | 5 | 3 | 8 |
| SGCG | 0 | 10 | 7 | 17 |
| TCAP | 0 | 0 | 3 | 3 |
| TRIM32 | 1 | 2 | 0 | 3 |
| TTN | 1 | 7 | 3 | 11 |
| Total | 47 | 541 | 293 | 881 |
Table 6. LGMD2-associated variants identified in ESP.
| LGMD 2 | ||||
|---|---|---|---|---|
| Gene | Benign | Damaging | Unknown | Total |
| ANO5 | 2 | 5 | 1 | 8 |
| CAPN3 | 5 | 16 | 2 | 23 |
| DAG | 0 | 0 | 0 | 0 |
| DYSF | 4 | 12 | 5 | 21 |
| FKRP | 2 | 4 | 0 | 6 |
| FKTN | 0 | 1 | 1 | 2 |
| POMGNT1 | 0 | 2 | 1 | 3 |
| POMT1 | 1 | 1 | 0 | 2 |
| POMT2 | 0 | 3 | 0 | 3 |
| SGCA | 3 | 7 | 0 | 10 |
| SGCB | 0 | 1 | 0 | 1 |
| SGCD | 0 | 1 | 0 | 1 |
| SGCG | 0 | 0 | 0 | 0 |
| TCAP | 0 | 0 | 0 | 0 |
| TRIM32 | 0 | 0 | 0 | 0 |
| TTN | 1 | 2 | 0 | 3 |
| Total | 18 | 55 | 10 | 83 |
Figure 2.
Percentage of variants identified in ESP in LGMD2 genes. In the ESP population, we identified 85 variations previously associated with LGMD2 (9.6%), of which 59 (69.4%) were predicted to be damaging, 18 (21.2%) to be benign and 8 (9.4%) were classified as being of unknown pathogenicity.
In particular, no variations in the DAG1, SGCG, TCAP and TRIM32 genes were identified in the ESP population, but a very low number of variants had previously been described in these genes.
Of the remaining 796 LGMD2-associated variants not present in ESP, 637 (80%) were predicted be damaging, 29 (3.6%) to be benign and 130 (16.3%) to be of unknown pathogenicity. This difference in the distribution of the three categories of pathogenicity comparing the proportion of variants identified in ESP versus variants not identified in ESP was statistically significant (P<0.0001). This result indicates that the distribution of variants identified in ESP and variants not identified in ESP is correlated to the effect of the variation on the protein. We compared the proportion of benign variants identified in ESP versus the benign ones not present in ESP: also in this case, the χ2 test was statistically significant (21.2% versus 3.6%, respectively, P<0.005).
Moreover, we studied the genotypes of the 85 variants in the ESP population, obtaining 8 homozygous and 1081 heterozygous genotypes, which is equivalent to a LGMD2 allelic frequency of 0.08 and to an LGMD2 prevalence of 1:167, whereas the LGMD2 carrier prevalence was 1:7. If the variants predicted to be benign (21.2% n=18) were additionally excluded, the allelic frequency would be 0.05, the genotype prevalence would be 1:400, whereas the LGMD2 carrier prevalence would be 1:10. The putative disease alleles are much more frequent than the calculated disease prevalence.
In addition, we analyzed the genotypes of the 85 variants in the TDB population, detecting 10 homozygous and 100 heterozygous genotypes. In particular, we identified 8 homozygous and 65 heterozygous myopathic patients, which is equivalent to an LGMD2 allelic frequency of 0.12 and to an LGMD2 prevalence of 1:71, whereas the LGMD2 carrier prevalence was 1:5 (Supplementary Table 2). If the variants predicted to be benign were excluded, the allelic frequency would be 0.052, the genotype prevalence would be 1:370, whereas the LGMD2 carrier prevalence would be 1:11.
In the population with unrelated phenotype, we obtained 2 homozygous and 35 heterozygous genotypes, which results in an LGMD2 allelic frequency of 0.09 and in an LGMD2 prevalence of 1:125, whereas the LGMD2 carrier prevalence was 1:6 (Supplementary Table 2). These results are very similar to those from the ESP database. If the variants predicted to be benign were excluded, the allelic frequency would be 0.028, the genotype prevalence would be 1:1428, whereas the LGMD2 carrier prevalence would be 1:19. In all the cases, the putative disease alleles are much more frequent than the calculated disease prevalence.
Discussion
Sequencing of the human genome and exome in many thousands of individuals is changing our view about the interpretation of the role of causative variants and their true penetrance. In particular, for autosomal dominant disorders, discrepancies may be highlighted by comparing the allele frequency with the disease incidence. This has been demonstrated by pivotal studies on long QT syndrome9 and cardiomyopathy genes.10 We applied a somewhat similar methodology to study frequencies of variants associated with LGMDs. As in cardiac studies,9, 10 hundreds of variants in several autosomal genes also associated with the LGMDs have been described as being causative, based on their absence in a small number of controls or on weak studies of familial cosegregation.3 To re-evaluate the meaning of all the variants previously described as causative, we selected a number of them in the LGMD genes in two databases (LOVD and HGMD).11, 12 We estimated their frequency in the ESP and in our internal databases, respectively.
Our study identified a high prevalence of LGMD-associated genetic variants in the population-based exome data. In the ESP, we identified about 4% of the variants previously associated with an autosomal dominant inheritance and about 9% of those associated with a recessive inheritance. Thus, a much higher frequency of LGMD-associated genetic variants was identified in ESP than expected.
Based on the frequency of LGMD in the general population (~1:20 000),4, 6, 7 no individual in the ESP population would be expected to be affected, whereas the estimated prevalence of individuals in the ESP data who would be expected to be carriers of LGMD2 would be 91. Based on the prevalence of the LGMDs in the general population, we established an allelic frequency of 0.00004 for an autosomal dominant inheritance and 0.0063 for autosomal recessive LGMD.
Our study evidenced a total of 158 putative LGMD-affected individuals and 1081 LGMD2 carriers in ESP. In particular, 150 individuals would be affected by a dominant form and 8 by a recessive one. However, our analysis is not able to identify the presence of compound heterozygosity for recessive variants, hampering the correct calculation of individuals in ESP potentially affected by LGMD2.
The most striking finding is that 84 variants with cosegregation evidence and 24 validated by functional studies were identified in ESP. Twenty four of them (5 in dominant genes and 19 in recessive ones) have both these proofs. For LGMD1, also considering only variants satisfying both these criteria, the number of potentially affected individuals (n=23) remains too high. This strongly suggests the presence of an incomplete penetrance. For LGMD2, most of the variants (66 out of 87) are not functionally validated, since a demonstration of compound heterozygosity has always been considered a sufficient proof of pathogenicity. For this reason, a subset of these variants could be erroneously annotated. On the other hand, the interpretation of our results is complicated by additional genetic mechanisms, such as hypomorphic and low penetrant alleles.21 Dissecting these genetic mechanisms is beyond the scope of this paper.
In conclusion, considering this study and the previous papers on the LQT syndrome9 and the cardiomyopathy,10 statistical data, including the frequency of identified variants in unbiased databases containing NGS data, should be considered. In fact, although functional characterization and family cosegregation analyses are useful tools in determining the pathogenicity of identified sequence variants, small family sizes and a reduced penetrance interfere with segregation analyses. In addition, in vitro functional characterization may not be representative of the human pathology.
A non-biased testing of all variants in patients will be necessary both for therapeutic approaches and genetic counseling. In fact, misclassified genetic variants in LGMD patients could have devastating consequences in the diagnosis of family members. It is therefore important that variations described as being causative of LGMDs are really disease causing.
Acknowledgments
We acknowledge Jonathan Cole for manuscript proofreading. We also thank the TIGEM Bioinformatics Core for support in data analysis. This study was entirely supported by grants from Telethon, Italy (TGM11Z06 to VN). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Nigro V, Aurino S, Piluso G: Limb girdle muscular dystrophies: update on genetic diagnosis and therapeutic approaches. Curr Opin Neurol 2011; 24: 429–436. [DOI] [PubMed] [Google Scholar]
- Nigro V, Savarese M: Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol 2014; 33: 1–12. [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Kang PB: Update on the genetics of limb girdle muscular dystrophy. Semin Pediatr Neurol 2012; 19: 211–218. [DOI] [PubMed] [Google Scholar]
- van der Kooi AJ, Barth PG, Busch HF et al: The clinical spectrum of limb girdle muscular dystrophy. A survey in The Netherlands. Brain 1996; 119 (Part 5): 1471–1480. [DOI] [PubMed] [Google Scholar]
- Rocha CT, Hoffman EP: Limb-girdle and congenital muscular dystrophies: current diagnostics, management, and emerging technologies. Curr Neurol Neurosci Rep 2010; 10: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urtasun M, Saenz A, Roudaut C et al: Limb-girdle muscular dystrophy in Guipuzcoa (Basque Country, Spain). Brain 1998; 121 (Part 9): 1735–1747. [DOI] [PubMed] [Google Scholar]
- Guglieri M, Magri F, D'Angelo MG et al: Clinical, molecular, and protein correlations in a large sample of genetically diagnosed Italian limb girdle muscular dystrophy patients. Hum Mutat 2008; 29: 258–266. [DOI] [PubMed] [Google Scholar]
- Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP), Seattle, WA. Available at: http://evs.gs.washington.edu/EVS (last accessed August 2014).
- Refsgaard L, Holst AG, Sadjadieh G et al: High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet 2012; 20: 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen C, Nielsen JB, Refsgaard L et al: New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet 2013; 21: 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT: LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 2011; 32: 557–563. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M et al: Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 2003; 21: 577–581. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- Mutarelli M, Marwah V, Rispoli R et al: A community-based resource for automatic exome variant-calling and annotation in Mendelian disorders. BMC Genomics 2014; 15: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese M, Di Fruscio G, Mutarelli M et al: MotorPlex provides accurate variant detection across large muscle genes both in single myopathic patients and in pools of DNA samples. Acta Neuropathol Commun 2014; 2: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torella A, Fanin M, Mutarelli M et al: Next-generation sequencing identifies transportin 3 as the causative gene for LGMD1F. PLoS One 2013; 8: e63536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarparanta J, Jonson PH, Golzio C et al: Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet 2012; 44: 450–455, S451–S452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorova A, Halliger-Keller B, Walter MC, Dabauvalle MC, Lochmuller H, Muller CR: A synonymous codon change in the LMNA gene alters mRNA splicing and causes limb girdle muscular dystrophy type 1B. J Med Genet 2003; 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Polychronakos C et al: Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet 2013; 132: 1077–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.