Abstract
Benign breast disease (BBD) is diagnosed in 1–2 million women/year in the US, and while these patients are known to be at substantially increased risk for subsequent development of breast cancer, existing models for risk assessment perform poorly at the individual level. Here, we describe a DNA-microarray-based transcriptional model for breast cancer risk prediction for patients with sclerosing adenosis (SA), which represent ¼ of all BBD patients. A training set was developed from 86 patients diagnosed with SA, of which 27 subsequently developed cancer within 10 years (cases) and 59 remained cancer-free at 10 years (controls). An diagonal linear discriminate analysis-prediction model for prediction of cancer within 10 years (SA TTC10) was generated from transcriptional profiles of FFPE biopsy-derived RNA. This model was tested on a separate validation case–control set composed of 65 SA patients. The SA TTC10 gene signature model, composed of 35 gene features, achieved a clear and significant separation between case and control with receiver operating characteristic area under the curve of 0.913 in the training set and 0.836 in the validation set. Our results provide the first demonstration that benign breast tissue contains transcriptional alterations that indicate risk of breast cancer development, demonstrating that essential precursor biomarkers of malignancy are present many years prior to cancer development. Furthermore, the SA TTC10 gene signature model, which can be assessed on FFPE biopsies, constitutes a novel prognostic biomarker for patients with SA.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-015-3513-1) contains supplementary material, which is available to authorized users.
Keywords: Benign breast disease, Breast cancer, Formalin-fixed paraffin-embedded, Transcriptional profiling, Transcriptional model, Risk prediction
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer in women in the US, with estimated incidence of more than 230,000 new cases and 40,000 deaths expected in 2014 [1]. BC is most effectively treated when identified at early stages of development; better identification of which women are at increased risk for developing breast cancer would have considerable benefit for optimal use of surveillance resources. Currently, the most commonly used method for assessment of breast cancer is the Breast Cancer Risk Assessment Tool (BCRAT, also referred to as the Gail model) [2], which works well to identify populations of women at increased BC risk, but which performs less well when used to predict risk for individual women [3, 4].
More than 1 million women per year in the US have breast biopsies with benign findings; these women are classified as having benign breast disease (BBD) and are known to have significantly elevated risk for subsequent development of breast cancer [5, 6]. BC risk for women with BBD can be stratified by histological features present in the benign biopsy, including type of lesion and degree of lobular involution [5, 7–9]. An individualized risk assessment model for women with BBD, designated the BBD-BC model, was recently developed for women with BBD and includes histologic features of the biopsy as well as other demographic and clinical features [4]. The BBD-BC model was found to provide improved performance for women with BBD as compared to the BCRAT model; in a cohort of women with BBD identified at the Mayo Clinic, the BBD-BC had a Receiver Operating Characteristic area under the curve (ROC AUC) of 0.665, while the BCRAT had a ROC AUC of 0.567 [4]. The BBD-BC model thus provided improved risk classification compared to the BCRAT; however, the BBD-BC model was designed to predict risk for all women with BBD. It is likely that better discrimination of risk could be obtained by developing models for specific subtypes of BBD.
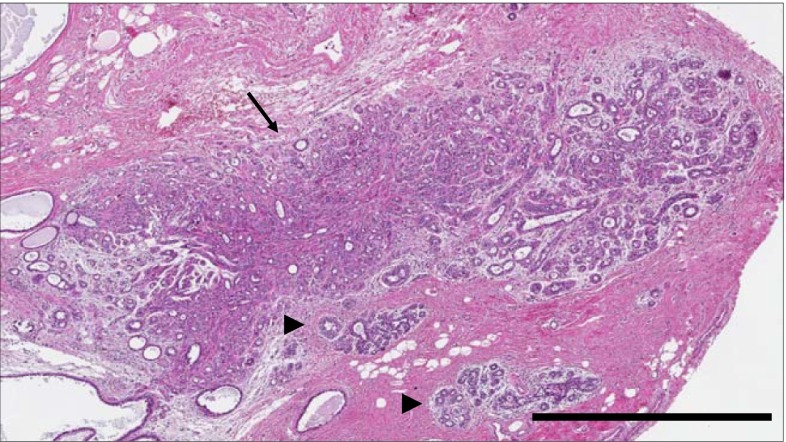
Sclerosing adenosis (SA) is a common BBD lesion that is characterized by epithelial proliferation, disordered acinar architecture, and stromal fibrosis (Fig. 1) [5, 10, 11]. Investigation of the Mayo Clinic BBD cohort revealed that SA was present in 28 % of the cohort and was associated with an approximate doubling of risk of subsequent BC [11]. This increased risk indicates that premalignant changes are likely present in some patients with SA. We obtained RNA from formalin-fixed, paraffin-embedded SA-containing biopsies and sought transcriptional elements that could predict subsequent cancer incidence. Our results demonstrate the development and validation of the first microarray-based gene signature to predict risk of later BC from benign breast tissue and provide a template for a novel clinical assay with considerable translational potential.
Fig. 1.
Histology of sclerosing adenosis (SA). H&E image of SA (arrow) in field containing two normal lobules (arrowheads). Scale bar 1 mm
Methods
Patients and samples
This study sample comprises patients selected from the Mayo BBD Cohort, which has been previously described [5, 9]. The Mayo BBD Cohort includes 9854 women ages 18–85 who had excisional breast biopsy with benign findings between 1967 and 1991 at Mayo Clinic (Rochester, MN). Demographic descriptors and potential breast cancer risk factors were identified via medical record review and from self-response questionnaires [5, 9]. Over a median of 18.9 years of follow-up, 924 of these women have been diagnosed with BC. Informed consent was obtained from each subject. All study procedures have been approved by the Mayo Clinic Institutional Review Board.
Biopsy findings were classified into the following categories: non-proliferative fibrocystic changes (NP), proliferative fibrocystic disease without atypia (PDWA), and proliferative fibrocystic disease with atypia [i.e., atypical hyperplasia (AH)] [12]. Sclerosing adenosis (SA) is a proliferative lesion, without atypia, that consists of enlarged and distorted lobules with prominent myoepithelium and stromal fibrosis. Because of concerns with tissue quality from the older biopsy specimens, eligibility for this study was restricted to the 1486 women diagnosed with PDWA and SA between the years of 1977 and 1991.
Two sets of breast cancer cases and controls from the SA group were formed for study purposes: one for model development (N = 86) and one for model validation (N = 65). For each, an age-stratified random sample of women with breast cancer at any time was selected, with selection probabilities proportional to the size of the age strata. Women from the last 10 years of the cohort recruitment period (1982–1991) were preferentially sampled, again under the assumption that tissue quality would be higher for these women than for those from the earlier years of the cohort. An equal number of controls were then frequency matched to these cases based on 5-year age and year of biopsy categories. The presence of SA lesions in the case/control samples was confirmed and reviewed by a second breast pathologist (AN). Study cohort demographic and clinical characteristics are in Table 1.
Table 1.
Characteristics of study set and comparison to overall SA patient cohort
| Not selected (N = 1316) | Study set (N = 170) | p value | |
|---|---|---|---|
| Age at BBD | 0.3441 | ||
| <45 | 301 (22.9 %) | 40 (23.5 %) | |
| 45–55 | 447 (34.0 %) | 66 (38.8 %) | |
| 55+ | 568 (43.2 %) | 64 (37.6 %) | |
| Year of benign biopsy | 0.2938 | ||
| 1977–1981 | 241 (18.3 %) | 37 (21.8 %) | |
| 1982–1986 | 501 (38.1 %) | 55 (32.4 %) | |
| 1987–1991 | 574 (43.6 %) | 78 (45.9 %) | |
| Breast cancer status | <0.0001 | ||
| Unaffected | 1202 (91.3 %) | 90 (52.9 %) | |
| Breast cancer | 114 (8.7 %) | 80 (47.1 %) | |
| Overall impression | 0.0815 | ||
| PROL. DIS W/O ATYPIA | 1187 (90.2 %) | 146 (85.9 %) | |
| PROL. DIS W/ATYPIA | 129 (9.8 %) | 24 (14.1 %) | |
| Atrophy | 0.0036 | ||
| Missing | 46 | 7 | |
| NO | 148 (11.7 %) | 31 (19.0 %) | |
| 1–74 % TDLU | 996 (78.4 %) | 125 (76.7 %) | |
| >75 % TDLU | 126 (9.9 %) | 7 (4.3 %) | |
| Columnar alteration | 0.0730 | ||
| Missing | 1 | 0 | |
| NO | 614 (46.7 %) | 67 (39.4 %) | |
| Marked | 701 (53.3 %) | 103 (60.6 %) | |
| Family history of breast cancer | 0.0249 | ||
| Missing | 3 | 0 | |
| None | 814 (62.0 %) | 87 (51.2 %) | |
| Weak | 337 (25.7 %) | 56 (32.9 %) | |
| Strong | 162 (12.3 %) | 27 (15.9 %) |
RNA extraction and gene expression profiling
RNA was extracted from FFPE samples using the High Pure RNA Paraffin Kit (Roche Diagnostics, Mannheim, Germany). The amount and quality of RNA were assessed with ND-1000 Spectrophotometer (Nanodrop, Wilmington, DE), and they were considered adequate for further analysis if the optical density 260/280 ratio was ≥1.8, and the total RNA yield was ≥500 ng. Extracted RNA was labeled and hybridized according to manufacturer’s instruction for the Whole Genome DASL assay (Illumina, San Diego, CA). All samples in the model development set had technical replicates. The first replicate was randomized to one 96-well plate for assay preparation, and the second replicate for each sample had a different randomization to a second 96-well plate. For the validation set, only 17 samples were profiled in replicate. Briefly, 200 ng of total RNA was reverse transcribed with biotinylated oligo(dT) and random nonamer primers. The resulting cDNA was annealed to chimeric query oligonucleotides which contain a gene-specific region and a universal primer sequence for PCR amplification, and then bound to streptavidin-conjugated paramagnetic particles. The gene-specific oligonucleotides were extended by second-strand cDNA synthesis and then ligated. Subsequently, the products were sequestered by magnetic separation, washed to remove unbound molecules, and then amplified by PCR with fluorophore-labeled universal primers. The resulting PCR products were purified, applied to Human HT-12 v.4 beadchips (Illumina), and then hybridized for 16 h at 58 °C. The beadchips were washed and then scanned in a BeadArray Reader using BeadScan v3 software (Illumina). Quality control parameters were determined to be within normal ranges before proceeding to the final data analysis. Sample probe gene expression values for the 29,377 probes were exported from Illumina Genomestudio and imported into the software R [13] for normalization, additional quality control, analysis, and prediction model development. The agreement between technical replicates was analyzed for correlation between model metrics in the first and second replicates for the model development set and between the first and second replicates for the validation set respectively (Supplementary Fig. 1).
Gene expression intensities from only the model development set were quantile-normalized in an iterative fashion using the normalization stress metric as described by Mahoney et al. to exclude failed samples [14]. Samples and their respective replicates were kept for further analysis if the normalization stress metric was less than 0.585 = log2(1.5). The model validation set was quantile-normalized separately to the final normalized distribution from the model development set. Normalized gene expression values were transformed to the log2 scale for analysis.
The complete description of the methods involved in SA TTC10 (cancer within 10 years) model development and analysis, including extensive internal model validation, is provided in Supplemental Methods.
Results
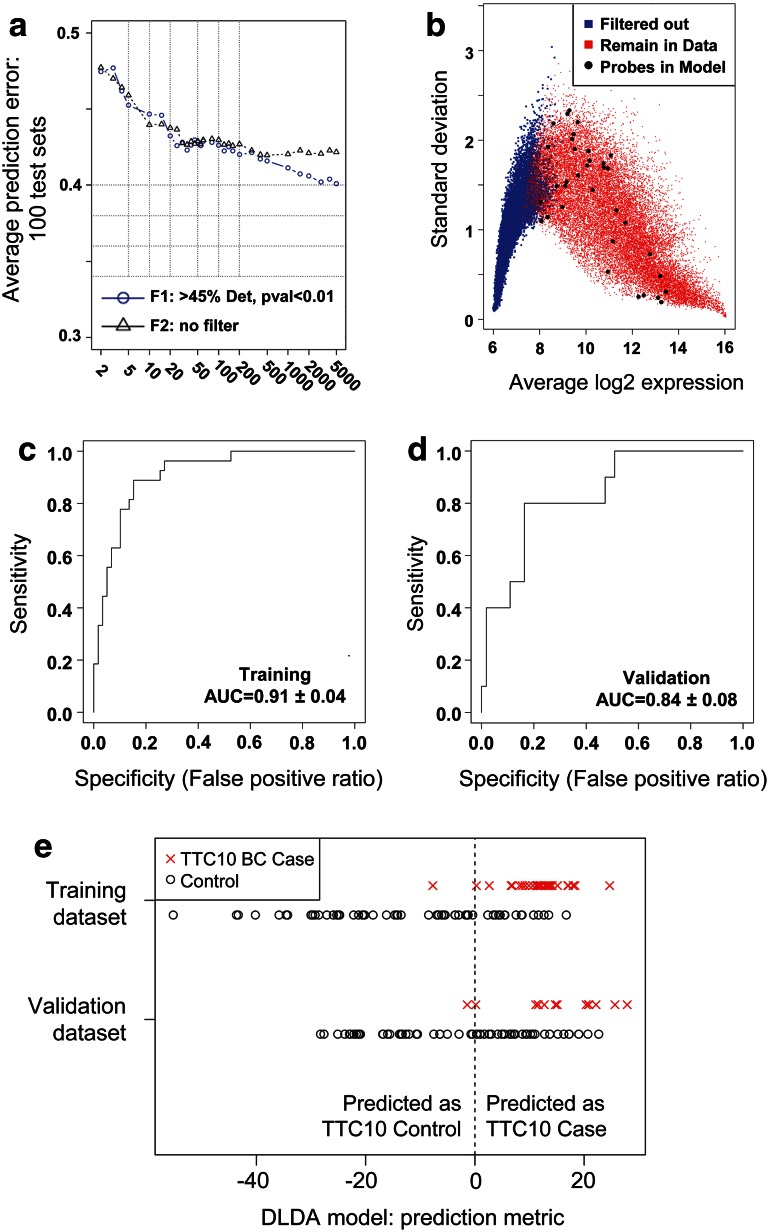
We used the development of BC at 10 years as the primary end point for model development. The 86 patients in the model development set included 27 TTC10-cases and 59 TTC10-controls, and the 65 patients in the validation set included 10 TTC10-cases and 55 TTC10-controls, with 3 and 12 replicates, respectively. The optimum-filtering strategy of 45 % detected-p value <0.01 and 35 probes was chosen based on the local minimum (to prevent over-fitting) of the average prediction error from the 100 Monte Carlo cross-validation test sets (Fig. 2a), which filtered out 8873 probes from further analysis (Fig. 2b). The 35 probes selected as final-model features are shown in Supplementary Table 1.
Fig. 2.
Development and validation of SA TTC10 model. a Mean area above the receiver operating characteristic (ROC) curve plotted against the number of top genes included in the classifiers. b Plot of average gene expression values indicating probes which were used for the model building (>45 % positive expression, p < 0.1 for difference between cases and controls) and locations of which probes were included in the model. Probes passing the filtering threshold are shown in red, those filtered out are shown in blue, and those probes selected as final-model features are shown as large black dots. c, d ROC for SA TTC10 model applied to training set (c; N = 86) and validation set (d; N = 65). e SA TTC10 predictions for training and validation dataset cases and controls. The vertical dashed line separates the samples into those predicted to be a TTC10-control (prediction metric ≤0) or TTC10-case (prediction metric >0)
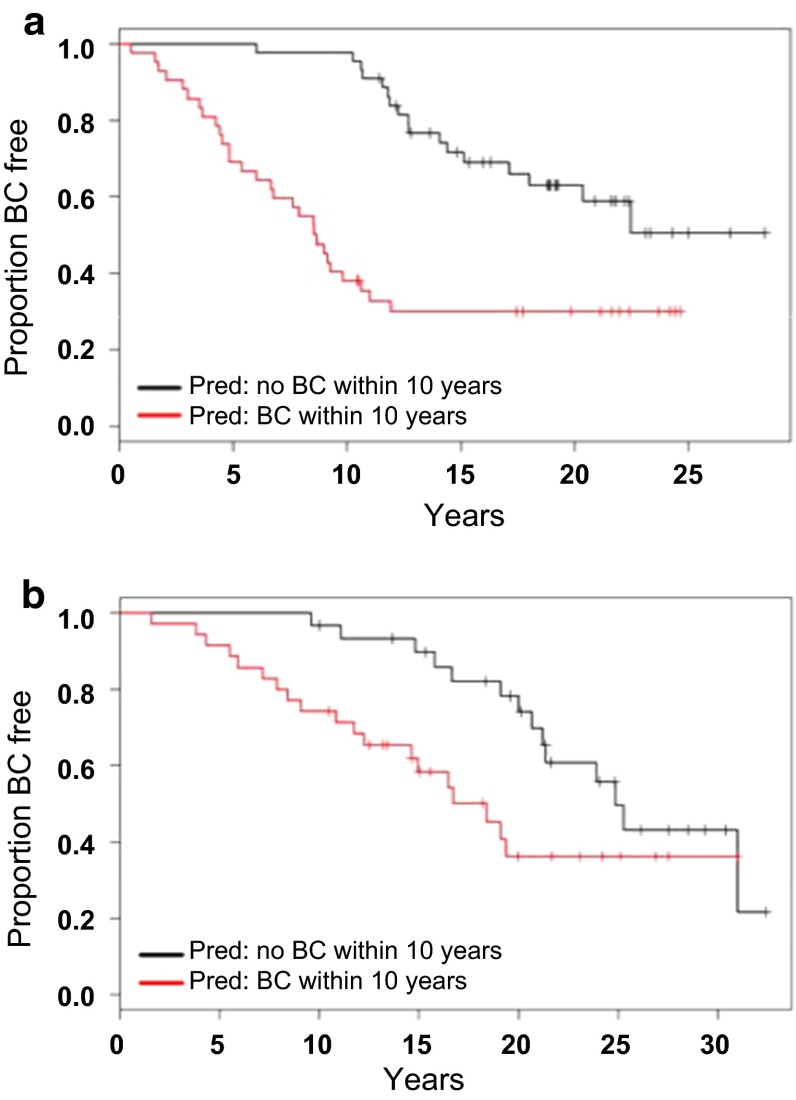
Prediction performance for the model development set was ROC AUC = 0.91 (95 % CI, 0.87, 0.95) with a prediction accuracy of 80 % (95 % CI 70, 88 %; Fig. 2c). The independent validation set had an ROC AUC = 0.84 (95 % CI 0.75, 0.92) with a prediction accuracy of 58 % (95 % CI 46, 71 %; Fig. 2d). Actual TTC10 case/control status and the continuous version of the DLDA prediction model metric for both the model development and validation sets are displayed (Fig. 2e). Full prediction performance and accuracy metrics are shown in Table 2. Actual time-to-cancer based on the predicted group for each sample is displayed in Kaplan–Meier plots (Fig. 3).
Table 2.
Model metrics for the TTC10 model, unless otherwise specified
| Cases/controls | TTC10 | |
|---|---|---|
| Model development | Validation set | |
| 27/59 | 10/55 | |
| Number of final-model features | 35 | 35 |
| TTC10 AUC: only samples with BCRAT and BBD-BC risk scores | 0.91 (0.87, 0.95) | 0.80 (0.71, 0.89) |
| AUC: BCRAT alone | 0.64 (0.58, 0.71) | 0.56 (0.45, 0.66) |
| AUC: BCRAT + TTC10 | 0.91 (0.87, 0.95) | 0.79 (0.70, 0.88) |
| AUC: BBD-BC alone | 0.63 (0.56, 0.70) | 0.75 (0.66, 0.84) |
| AUC: BBD-BC + TTC10 | 0.93 (0.89, 0.96) | 0.82 (0.73, 0.91) |
| True positives | 26 | 9 |
| True negatives | 43 | 29 |
| False positives | 16 | 26 |
| False negatives | 1 | 1 |
| Accuracy | 0.80 (0.70, 0.88) | 0.58 (0.46, 0.71) |
| Sensitivity | 0.96 (0.81, 1.0) | 0.90 (0.56, 1.0) |
| Specificity | 0.73 (0.60, 0.84) | 0.53 (0.39, 0.66) |
| Technical replicates spearman correlation Rho |
0.86 p ≤ 0.0001 |
0.98 p ≤ 0.0001 |
Fig. 3.
Time-to-Cancer Distributions within SA TTC10 prediction groups. a, b Kaplan–Meier plots visualizing the distribution of actual time-to-cancer within predicted case/control groups in training (a) and validation (b) cohorts
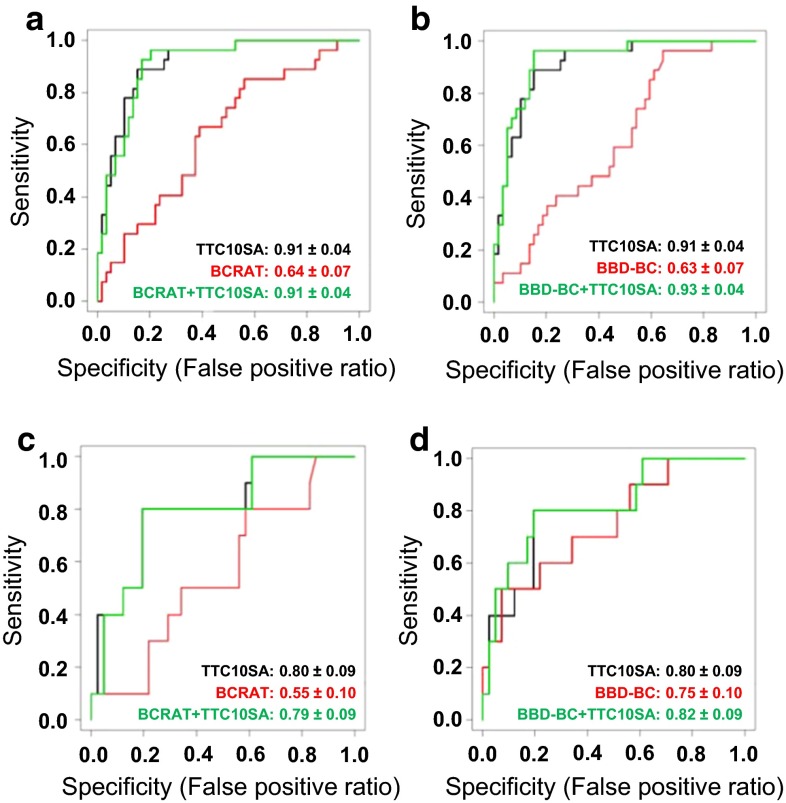
Combined BCRAT + TTC10 and BBD-BC + TTC10 models were built for the samples where both the BCRAT and BBD-BC model metrics were available (Fig. 4). All the patient samples in the model development set could be used, and 10 TTC10 cases and 41 TTC10 controls were available from the validation set. For the training set, the TTC10 model improved discrimination of the BCRAT model, with AUC = 0.64 (95 % CI 0.57–0.71) for BCRAT alone, 0.91 (95 % CI 0.87–0.95) for BCRAT combined with TTC10, and 0.91 (95 % CI 0.87–0.95) for TTC10 alone (Fig. 4a). The TTC10 model also improved discrimination of the BBD-BC model, with AUC of 0.63 (95 % CI 0.56–0.70) for BBD-BC alone, 0.93 (95 % CI 0.89–0.97) for BBD-BC combined with TTC10, and 0.91 (95 % CI 0.87–0.95) for TTC10 alone (Fig. 4b). Similarly, for the validation set, the TTC10 model also improved discrimination: AUC = 0.55 for BCRAT alone, 0.79 (95 % CI 0.70–0.87) for BCRAT combined with TTC10, and 0.80 (95 % CI 0.71–0.89) when TTC10 is used alone (Fig. 4c). Improvements of TTC10 with the BBD-BC model are as follows: AUC = 0.75 (95 % CI 0.65–0.85) for BBD-BC alone, 0.82 (95 % CI 0.73–0.91) for BBD-BC combined with TTC10, and 0.80 (95 % CI 0.71–0.89) for TTC10 alone (Fig. 4d). Thus, in women with SA, the SA TTC10 model provides improved and independent risk assessment compared to those of the BCRAT and the BBD-BC models.
Fig. 4.
Combination of SA TTC10 model with BCRAT or BBD-BC models improves performance of each. a, b ROC for BCRAT (a) and BBD-BC (b) models with SA training set. c, d ROC for SA TTC10 combined with BCRAT (c) and BBD-BC (d) models for SA validation set
Discussion
We have developed a microarray-based gene signature in benign breast tissue that accurately discriminates between patients that will and will not develop breast cancer within 10 years. Specifically, the signature pertains to women with sclerosing adenosis, a common proliferative finding present in about 25 % of all benign biopsies. The signature also performed well in an independent validation set of women with sclerosing adenosis. This molecular-based model, based in a specific subtype of BBD, performs substantially better than traditional models containing clinical and histologic features only; in the validation set, the TTC10 signature provided an improvement in AUC of 0.07 above the BBD-BC model alone and 0.23 above BCRAT alone.
There are multiple advantages to a microarray-based signature based on FFPE tissue. The risk signature is not dependent on pathology interpretation and provides risk information that is independent of risk stratification resulting from standard clinical models. Furthermore, the use of FFPE biopsy tissue ensures widespread applicability of this model, as FFPE is the most common method for tissue preservation and is the standard for tissue archiving. Compared to assays requiring fresh/frozen tissue, an FFPE-based assay will facilitate clinical implementation since no changes in sample collection and processing are needed.
Analysis of the 35 individual probes that compose the TTC10 model revealed a number of genes that have been previously implicated in breast cancer progression. NDRG3, encoding the N-Myc Downstream-Regulated Gene 3 Protein, had a negative coefficient, indicating that increased relative expression was associated with reduced BC risk; NDRG3 has been identified as activated by the estrogen receptor-β (ERβ) and as a component of an antiproliferative response induced by ERβ in the T47D breast cancer cell line [15, 16]. Other probes with a negative coefficient include NPNT1, encoding the protein nephronectin, which has been found to be downregulated in breast cancer cells that acquire the ability to metastasize to the liver [17], and PSMB1, encoding the proteasome macropain subunit-β1, has been identified as a component of a protein interaction network prognostic for BC outcome [18]. GEMIN2, encoding gem (nuclear organelle) associated protein 2, also designated as SIP1, has a positive coefficient and thus increased expression of this gene was associated with increased BC risk; GEMIN2/SIP1 has been extensively studied as a mediator of the epithelial-mesenchymal transition and as a suppressor of the differentiated epithelial phenotype [19–21]. Other probes with a positive coefficient include MTHFD2, encoding methylenetetrahydrofolate dehydrogenase-2, a metabolic regulator associated with poor prognosis for BC patients and as an inducer of BC cell invasion and metastasis [22–25], and UFL1, encoding UFM1-specific ligase 1, recently identified as a component of a protein complex critical for ERα transactivation and breast cancer development [26].
Strengths of our study include our focus on a common proliferative lesion, use of a large cohort as a sampling frame for cases and controls, use of FFPE tissues which should translate well to current clinical practice, and validation in an independent dataset. The threshold used to determine case status from the predicted score can be optimized in future studies to minimize false positives and false negatives while balancing the consequences of each. We chose to use whole tissue sections rather than laser-microdissected lesions, since SA reflects perturbations in both the epithelial and stromal compartments, and thus, a model incorporating both contains information about how elements of the tumor microenvironment contribute to cancer development. As a weakness, our focus on SA limits the application of our signature to this particular patient group, but we estimate that about 250,000 women per year receive a diagnosis of SA on their benign breast biopsies; molecular predictive models for patients with BBDs containing other common proliferative lesions can also be studied.
Furthermore, our sample selection procedure included over-sampling cases in order to improve power to discriminate between patients that would go on to develop breast cancer from those that would not. As a result, the prevalence of breast cancer events within the development and validation samples do not reflect the population prevalence of breast cancer, and therefore, this gene signature cannot be directly used to obtain individualized risk predictions, which will be the subject of future studies. However, this study importantly provides a proof of principle, where we have now identified genes that discriminate with a high level of accuracy, which will form the basis of a future risk prediction model combined with clinical information, as in (Ref [4]). Additionally, the use of the endpoint of time-to-cancer within 10 years provided improved discriminative ability relative to previously developed models. Unsurprisingly, this suggests that a molecular signature derived from a benign biopsy will be a better predictor of earlier cancer occurrences.
In conclusion, this is the first predictive transcriptional model derived from benign breast tissue that can distinguish between patients that developed BC within 10 years from those who did not. We show that this molecular-based profile, focused on patients with SA, provides improvement over previous models that do not include molecular data. We now plan to conduct studies designed to accurately estimate risk with this model and to develop additional signature models for other categories of BBD. By identifying higher risk women among those with SA, this model can help to stratify women into groups who would benefit from more intensive surveillance strategies. Additionally, as our results demonstrate that transcriptional features associated with BC are present in SA-containing biopsies more than a decade before cancer development, our study provides insight into novel potential targets for chemoprevention specific for patients with SA.
Electronic supplementary material
Acknowledgments
This study was supported by the National Cancer Institute (NCI RO1 CA 132879 to LCH; CA187112 to ACD); Bankhead-Coley Foundation (5BC02 to DCR and AN); Mayo Clinic Breast Cancer-Specialized Program of Research Excellence (SPORE CA116201 to DWV, DCR, AN and LCH); Martha and Bruce Atwater (to DWV, DCR, and LCH); Jimmy V Foundation (to DCR and LCH); and the Susan B Komen Foundation (to DCR and LCH).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Amy C. Degnim and Aziza Nassar contributed equally.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Gail MH, Mai PL. Comparing breast cancer risk assessment models. J Natl Cancer Inst. 2010;102(10):665–668. doi: 10.1093/jnci/djq141. [DOI] [PubMed] [Google Scholar]
- 3.Rockhill B, Spiegelman D, Byrne C, Hunter DJ, Colditz GA. Validation of the Gail et al. model of breast cancer risk prediction and implications for chemoprevention. J Natl Cancer Inst. 2001;93(5):358–366. doi: 10.1093/jnci/93.5.358. [DOI] [PubMed] [Google Scholar]
- 4.Pankratz VS, Degnim AC, Frank RD, Frost MH, Visscher DW, Vierkant RA, Hieken TJ, Ghosh K, Tarabishy Y, Vachon CM, Radisky DC, Hartmann LC. Model for individualized prediction of breast cancer risk after a benign breast biopsy. J Clin Oncol. 2015;33(8):923–929. doi: 10.1200/JCO.2014.55.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann LC, Sellers TA, Frost MH, Lingle WL, Degnim AC, Ghosh K, Vierkant RA, Maloney SD, Pankratz VS, Hillman DW, Suman VJ, Johnson J, Blake C, Tlsty T, Vachon CM, Melton LJ, 3rd, Visscher DW. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 6.Allred DC, Mohsin SK, Fuqua SA. Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer. 2001;8(1):47–61. doi: 10.1677/erc.0.0080047. [DOI] [PubMed] [Google Scholar]
- 7.Cichon MA, Degnim AC, Visscher DW, Radisky DC. Microenvironmental influences that drive progression from benign breast disease to invasive breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(4):389–397. doi: 10.1007/s10911-010-9195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupont WD, Parl FF, Hartmann WH, Brinton LA, Winfield AC, Worrell JA, Schuyler PA, Plummer WD. Breast cancer risk associated with proliferative breast disease and atypical hyperplasia. Cancer. 1993;71(4):1258–1265. doi: 10.1002/1097-0142(19930215)71:4<1258::AID-CNCR2820710415>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 9.Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD, Pankratz VS, Degnim AC, Vachon CM, Reynolds CA, Thompson RA, Melton LJ, 3rd, Goode EL, Visscher DW. Age-related lobular involution and risk of breast cancer. J Natl Cancer Inst. 2006;98(22):1600–1607. doi: 10.1093/jnci/djj439. [DOI] [PubMed] [Google Scholar]
- 10.Jensen RA, Page DL, Dupont WD, Rogers LW. Invasive breast cancer risk in women with sclerosing adenosis. Cancer. 1989;64(10):1977–1983. doi: 10.1002/1097-0142(19891115)64:10<1977::AID-CNCR2820641002>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Visscher DW, Nassar A, Degnim AC, Frost MH, Vierkant RA, Frank RD, Tarabishy Y, Radisky DC, Hartmann LC. Sclerosing adenosis and risk of breast cancer. Breast Cancer Res Treat. 2014;144(1):205–212. doi: 10.1007/s10549-014-2862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont WD, Page DL. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med. 1985;312(3):146–151. doi: 10.1056/NEJM198501173120303. [DOI] [PubMed] [Google Scholar]
- 13.Team RC (2013) R: a language and environment for statistical computing. http://www.R-project.org/
- 14.Mahoney DW, Therneau TM, Anderson SK, Jen J, Kocher JP, Reinholz MM, Perez EA, Eckel-Passow JE. Quality assessment metrics for whole genome gene expression profiling of paraffin embedded samples. BMC Res Notes. 2013;6:33. doi: 10.1186/1756-0500-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Li Y, Hong A, Wang J, Lin B, Li R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int J Cancer. 2009;124(3):521–530. doi: 10.1002/ijc.23961. [DOI] [PubMed] [Google Scholar]
- 16.Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008;27(7):1019–1032. doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- 17.Erin N, Wang N, Xin P, Bui V, Weisz J, Barkan GA, Zhao W, Shearer D, Clawson GA. Altered gene expression in breast cancer liver metastases. Int J Cancer. 2009;124(7):1503–1516. doi: 10.1002/ijc.24131. [DOI] [PubMed] [Google Scholar]
- 18.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 19.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126(Pt 2):393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 20.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 22.Lehtinen L, Ketola K, Makela R, Mpindi JP, Viitala M, Kallioniemi O, Iljin K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4(1):48–63. doi: 10.18632/oncotarget.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Liu Y, He C, Tao L, He X, Song H, Zhang G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. 2014;35(9):8685–8690. doi: 10.1007/s13277-014-2111-x. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287(35):29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo HM, Kang SH, Kim JY, Lee JE, Seong MW, Lee SW, Ka SH, Sou YS, Komatsu M, Tanaka K, Lee ST, Noh DY, Baek SH, Jeon YJ, Chung CH. Modification of ASC1 by UFM1 is crucial for ER alpha transactivation and breast cancer development. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.