Abstract
The microneutralization assay is commonly used to detect antibodies to influenza virus, and multiple protocols are used worldwide. These protocols differ in the incubation time of the assay as well as in the order of specific steps, and even within protocols there are often further adjustments in individual laboratories. The impact these protocol variations have on influenza serology data is unclear. Thus, a laboratory comparison of the 2-day enzyme-linked immunosorbent assay (ELISA) and 3-day hemagglutination (HA) microneutralization (MN) protocols, using A(H1N1)pdm09, A(H3N2), and A(H5N1) viruses, was performed by the CONSISE Laboratory Working Group. Individual laboratories performed both assay protocols, on multiple occasions, using different serum panels. Thirteen laboratories from around the world participated. Within each laboratory, serum sample titers for the different assay protocols were compared between assays to determine the sensitivity of each assay and were compared between replicates to assess the reproducibility of each protocol for each laboratory. There was good correlation of the results obtained using the two assay protocols in most laboratories, indicating that these assays may be interchangeable for detecting antibodies to the influenza A viruses included in this study. Importantly, participating laboratories have aligned their methodologies to the CONSISE consensus 2-day ELISA and 3-day HA MN assay protocols to enable better correlation of these assays in the future.
INTRODUCTION
Following infection with influenza viruses, most people develop antibodies specific to the infecting virus that can be measured by serological assays. These antibodies can be detected in the majority of people 2 to 3 weeks after symptom onset and can persist for months (1–4). Thus, serology can confirm past infection in the absence of clinical symptoms or virological data, detecting most symptomatic and asymptomatic infections (5).
In 2011, an international partnership termed CONSISE (the Consortium for the Standardization of Influenza Seroepidemiology) was created in recognition of a need identified during the 2009 pandemic for timely seroepidemiological data to better estimate pandemic virus infection severity and attack rates and to inform policy decisions. CONSISE is comprised of individuals from various organizations, with free membership. The activities of CONSISE are performed by two interlinked working groups, the Laboratory Working Group and the Epidemiology Working Group, and a Steering Committee. The focus of the Laboratory Working Group is to improve serological assay comparability and standardization through consensus assay development, comparative laboratory testing, and quality assurance (6) (https://consise.tghn.org).
The main serological assays to detect antibodies to influenza virus are the hemagglutination (HA) inhibition (HI) assay and the microneutralization (MN) assay. The HI assay detects antibodies that block the influenza virus hemagglutinin binding to sialic acid-linked residues on red blood cells (RBC), while the MN assay detects functional antibodies primarily directed toward the hemagglutinin that prevent infection of cells in tissue culture (reviewed in references 7 and 8). There are various forms of the MN assay used in laboratories around the world, such as the 2-day enzyme-linked immunosorbent assay (ELISA) protocol (8, 9), 3-day HA protocol (10), and 7-day HA protocol (11, 12). For the purposes of seroepidemiology, the shorter protocols of 2 and 3 days are preferred. The 2- and 3-day MN assays measure antibodies to hemagglutinin and yet differ in their methods of preparation of cell monolayers for infection as well as detection of virus infection. Cells are plated with the virus-serum mixture for the 2-day MN assay, while a preformed cell monolayer is used for the 3-day MN assay. The 2-day MN assay detects nucleoprotein in infected cells (9), while the 3-day assay measures hemagglutinating virus in the culture medium or cytopathic effect (CPE) in the cell monolayer. Although there have been some direct comparisons between serological assays performed by multiple laboratories (12–15), the impact of various MN assay protocols on the determination of serological titers is unknown. Therefore, the aim of this study was to assess the intralaboratory variability and sensitivity of the 2-day ELISA MN assay and the 3-day HA MN assay for detecting antibodies to A(H1N1)pdm09 virus and, as an extension, A(H3N2) and A(H5N1) influenza viruses. The study was performed by the CONSISE Laboratory Working Group members (see Acknowledgments).
MATERIALS AND METHODS
Reagents used in the study.
Laboratories were required to supply their own reagents, virus stocks, MDCK cell lines, and appropriate cell culture media for the study. Wild-type or reassortant viruses were used: the A(H1N1)pdm09 strains were antigenically similar to the A/California/7/2009 vaccine strain, and the A(H3N2) strains were antigenically similar to the A/Perth/16/2009 or the A/Victoria/361/2011 vaccine strain. A representative A(H5N1) virus from a clade that was recognized by the laboratory's serum panel was used. Serum panels contained approximately 10 test samples (sera or plasma), comprising low-, medium-, and high-titer antibody levels. Sera were from seroepidemiology studies and vaccine studies and from ferrets (to obtain high-titer serum in some laboratories) and were supplied by each participating laboratory.
Development of consensus 2-day ELISA and 3-day MN protocols.
Parameters and variables for the 2-day ELISA (8) and the 3-day HA (10, 16) MN assays were listed. Laboratories within CONSISE shared their protocols for either or both of the MN assays and listed their preferred variables for each parameter identified. Data were collected anonymously, collated, and used to develop the consensus protocols.
Consensus 2-day ELISA and 3-day HA MN assays.
The 2-day ELISA MN assay was to be performed as described in references 8 and 9, while the 3-day HA MN assay was to be performed as described in references 10 and 16. Laboratories were required to use the specified parameters listed in the CONSISE consensus protocols (Tables 1 and 2). Cell culture conditions and the virus and serum panels used differed between laboratories. The reciprocal of the highest dilution whereby 50% infection was prevented was recorded as the titer for each serum sample.
TABLE 1.
CONSISE consensus 2-day ELISA MN assay for detecting antibodies to A(H1N1)pdm09 virusa
| Parameter | Required parameter | Recommended parameter |
|---|---|---|
| Stock virus prepn | ||
| Cell substrate for virus growth | Day 10 embryonated eggs | |
| Stock virus infectivity concn and method of determination | At least 106 TCID50/ml, read by ELISA | |
| Stock storage | Aliquots of bulk virus prepn | |
| Serum prepn | ||
| Storage of sera following receipt | −70°C, −20°C, and 4°C, 1 or 2 freeze-thaw cycles in testing laboratory | |
| Preassay treatment of sera | Heat treatment at 56°C for 30 min, without dilution in media | |
| Initial serum dilution | 1:10 | |
| Sample type | Sera only or plasma only | |
| Virus prepn | ||
| Final virus amt per well | 100 TCID50 | |
| Vol of virus solution added per sample | 50 μl | |
| Virus/serum mix incubation | 1 h at 37°C | |
| Calculated starting serum dilution | 1:10, excluding cell culture vol | |
| Cell prepn | ||
| Prepn of cells | Cell suspension | |
| Cell type used | MDCK (“Salisbury”), MDCK-SIAT1 | |
| Assay diluent/culture medium | Coon's/Dulbecco's modified Eagle's medium with 1% BSA–FCS, laboratory-preferred media | |
| Assay setup | ||
| Incubation time of assay to endpoint reading | 18–22 h | |
| Incubation conditions | 35–37°C, 5% CO2 | |
| No. of sample replicates | Replicates preferred if available | |
| Endpoint estimation | ||
| Endpoint determination | Viral antigen detection by ELISA using antinucleoprotein antibody (clone) | |
| Endpoint calculation method | 50% neutralization |
TABLE 2.
CONSISE consensus 3-day HA MN assay for detecting antibodies to A(H1N1)pdm09 virus
| Parameter | Required parameter | Recommended parameter |
|---|---|---|
| Stock virus prepn | ||
| Cell substrates for virus growth | Day 10 embryonated eggs, MDCK cells, MDCK-SIAT1 cells | |
| Stock virus infectivity concn and method of determination | At least 106 TCID50/ml, read by RBC agglutination | |
| Stock storage | Aliquots of bulk virus prepn | |
| Serum prepn | ||
| Storage of sera following receipt | −70°C, −20°C, and 4°C, 1 or 2 freeze-thaw cycles in testing laboratory | |
| Preassay treatment of sera | Heat treatment at 56°C for 30 min, without dilution in media | |
| Initial serum dilution | 1:10 | |
| Sample type | Sera only or plasma only | |
| Virus prepn | ||
| Final virus amt per well | 100 TCID50 | |
| Vol of virus solution added per sample/well | 50 μl, 100 μl, or 200 μl | |
| Virus/serum mix incubation | 1 h at 37°C | |
| Virus/serum mix incubation on cell monolayer | 1 h at 37°C | |
| Calculated starting serum dilution | 1:10, excluding virus vol | |
| Cell prepn | ||
| Prepn of cells | Preformed monolayer | |
| Cell types used | MDCK (ATCC), MDCK (Salisbury), MDCK-SIAT1 | |
| Assay diluent | Coon's/Dulbecco's modified Eagle's medium, with trypsin (1/2 μg/ml), laboratory-preferred media | |
| Cell infection media | Coon's/Dulbecco's modified Eagle's medium, with trypsin (1/2 μg/ml), laboratory-preferred media | |
| Assay setup | ||
| Incubation time of assay to endpoint reading | 3 days | |
| Incubation conditions | 35–37°C, 5% CO2 | |
| No. of sample replicates | Replicates preferred if available | |
| Endpoint estimation | ||
| Endpoint determination | Turkey/guinea pig RBC agglutination, CPE | |
| Endpoint calculation method | 50% neutralization |
Design of study—laboratory assay comparison.
Individuals who were members of CONSISE were invited to participate in the experimental laboratory comparative study. The researchers at 13 laboratories agreed to participate, and each laboratory was assigned a code letter from A to M (the order of assigned letters did not represent the order of the listing of participants in Acknowledgments). Eleven laboratories (laboratories A to K) took part in the initial A(H1N1)pdm09 study, seven (laboratories A, C, D, F, I, K, and L) in the A(H3N2) study, and three (laboratories H, L, and M) in the A(H5N1) study. Overall, 12 laboratories (laboratories A to L) provided data that could be included in the analyses. Each laboratory was required to assay A(H1N1)pdm09, A(H3N2), or A(H5N1) antibody levels in their panel of sera on at least three separate occasions using the CONSISE protocols for both consensus MN assays: the 2-day ELISA and the 3-day HA.
Statistical analysis.
All analyses were based on the titers reported by the participants. To enable comparison of assays for each laboratory, the geometric mean titer (GMT) was calculated across runs and replicates to give a single value for each sample for each MN assay method. To calculate the overall ratios between the assays for each laboratory, the ratio of the 3-day titer (detected by HA or CPE) to the 2-day ELISA MN titer was calculated for each sample. The GMT was then calculated for all samples in the serum panel for each laboratory. For the purpose of calculations, negative titers reported as <10 were assigned a value of 5, while high titers reported as greater than or equal to a given value were assigned a value corresponding to the next 2-fold titer; e.g., the value of ≥1,280 was assigned a value of 2,560. Correlations in results between assay methods for the panels of serum samples were calculated using Spearman rank correlations.
RESULTS
Development of consensus protocols for the MN assays.
We assessed the similarities between the methodologies used in 10 laboratories for the 2-day ELISA and the 3-day HA MN assays. Parameters were highly consistent between laboratories for the 2-day ELISA MN assay method and closely followed published methods (8, 9). There was less consistency between the 3-day HA MN assay methods, particularly in numbers of sample replicates performed and determinations of the endpoint titer (50% or 100% neutralization). There was variability in both assays with respect to cell culture conditions (data not shown). To facilitate greater comparability between laboratories, we developed consensus protocols for the 2-day ELISA and 3-day HA MN assays by discussion and agreement at CONSISE meetings (17) (Tables 1 and 2). Parameters were classified as either required or recommended, based on their importance in the interpretation of the assay titers. Required parameters included serum dilutions and reporting of sample titers, assay incubation times, and endpoint calculation methods. Recommended parameters listed appropriate variables for use.
Data received. (i) A(H1N1)pdm09 MN assays.
Ten laboratories returned data for both assays. ELISA was used for detection for all 2-day MN assays. For the 3-day MN assay, 7 laboratories used HA as the only detection method (turkey or guinea pig RBC were used), 2 laboratories used CPE only, and 2 laboratories used both HA and CPE detection methods. Both HA and CPE detection methods were assessed. Three laboratories used multiple additional detection methods for one or both of the MN assays, but these data have not been included in the analysis. An eleventh laboratory (laboratory K) did not perform the 2-day ELISA MN assay and returned data for only the 3-day HA MN assay. This laboratory shared a serum panel with laboratory F, which performed both assays. The 3-day HA MN assay titers from laboratory K were compared with the 2-day ELISA MN assay titers from laboratory F. Laboratories G and J performed each assay twice, rather than three times; laboratory I performed the 3-day MN assays twice.
(ii) A(H3N2) MN assays.
Data were received from seven laboratories. All laboratories used ELISA for detection in the 2-day MN assay and HA (turkey or guinea pig RBC) for detection in the 3-day HA MN assay, and two laboratories also sent corresponding titers detected by CPE. Laboratory K performed only the 3-day HA MN assay and shared a serum panel with laboratory F.
(iii) A(H5N1) MN assays.
Data were received from three laboratories. The results from laboratory M were negative for all of the serum samples for all tests. No further analysis was possible for this laboratory. Laboratories H and L used horse or goose RBC to read out the 3-day HA MN assay.
Reproducibility within laboratories: comparison of replicate tests.
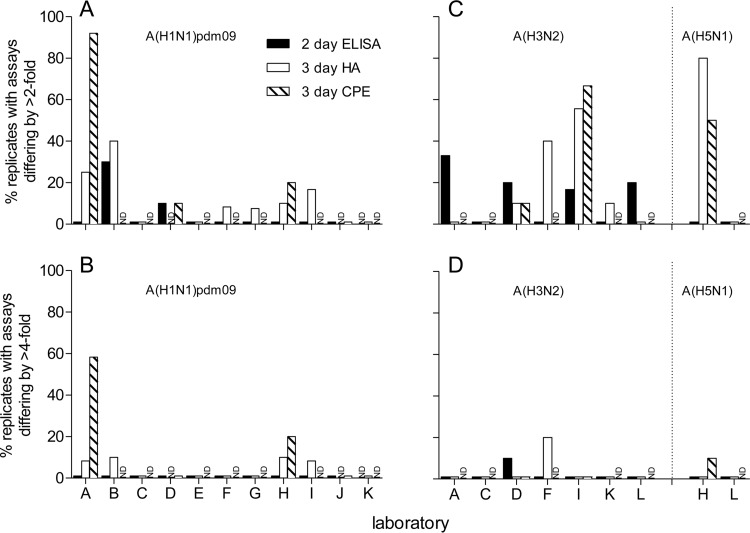
Laboratories performed an internal comparison of assay protocols using their own serum panels. The titers within each laboratory for each sample across replicate tests were compared. Detecting antibodies to A(H1N1)pdm09 virus, there was good reproducibility of the 2-day ELISA MN assays for the majority of the laboratories, with titers from over 80% of the laboratories with replicate tests differing by ≤2-fold, while laboratories B and D had 10 to 30% of replicate titers differing by >2-fold (Fig. 1A). No laboratories had titers from replicate tests differing by >4-fold (Fig. 1B). For the 3-day MN assays, the variability differed depending on the assay detection method. Using detection by HA, four laboratories had replicates differing by >4-fold, and yet this was the case for only a very few samples (1 sample of 10 to 12 samples, 8.3 to 10%), while, using detection by CPE, two laboratories had titers from replicate tests differing by >4-fold (2 samples of 10 [20%] and 7/12 samples [58%]) (Fig. 1B). Six laboratories had titers from replicates differing by >2-fold by HA detection (7.5 to 40%), while three laboratories had titers from tests differing by >2-fold using detection by CPE (10 to 91.7%) (Fig. 1A). Laboratory A showed high variability between replicates using detection of the 3-day MN titers by CPE (91.7%), as the data from two replicate assays were comparable whereas the data from the third assay were inconsistent (data not shown). In seven laboratories (laboratories C, D, E, F, G, J, and K), there were no replicates with results that differed by more than 2-fold.
FIG 1.
Reproducibility within laboratories of serology assay results for assays detecting antibodies to A(H1N1)pdm09 (A and B) and A(H3N2) and A(H5N1) (C and D) viruses. Graphs show the (percent) proportions of replicate assays differing by >2-fold (A and C) and >4-fold (B and D) for the 2-day ELISA MN assay and the 3-day MN assay with detection by HA and CPE for each participating laboratory for all sera. ND indicates instances in which the assay or detection method was not performed.
For studies detecting antibodies to A(H3N2) and A(H5N1) viruses, reproducibility was also good, with three instances of data from replicates that differed by greater than 4-fold, though this was the case for a small number of samples for each laboratory (10 to 20%). All other laboratories had data from replicates that differed by ≤4-fold (Fig. 1C and D).
Relationship between 2-day ELISA MN assay and 3-day HA and CPE MN assays for test serum panels.
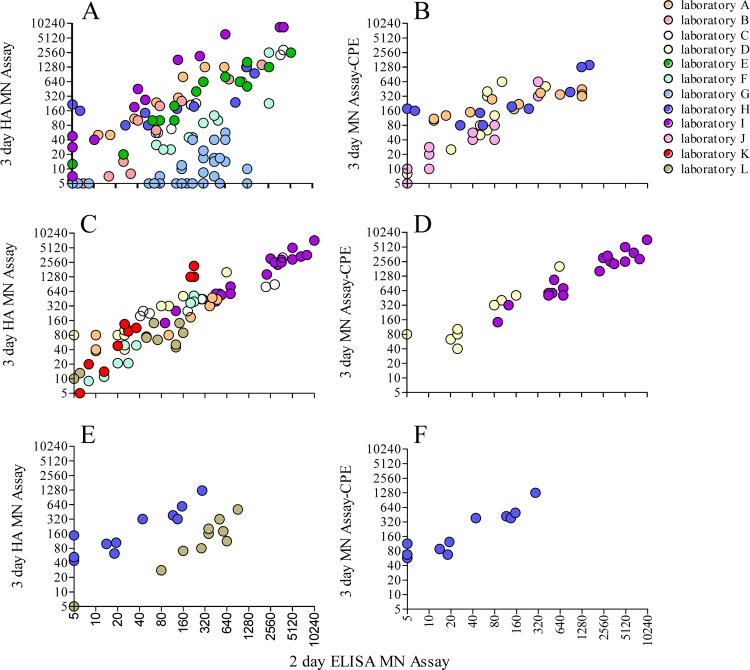
Titers for individual test serum panels were compared for each laboratory between the two assays. Comparing the titers obtained using the 2-day ELISA and the 3-day HA MN assays for studies detecting antibodies to A(H1N1)pdm09 virus (Fig. 2A), seven of the nine laboratories had individual correlation coefficients that were above 0.9 (good) and two of the nine laboratories did not (Table 3). Two laboratories (G and H) had low correlation between the assays (0.580 and 0.638, respectively), as the 3-day MN HA assay gave narrow response ranges compared to the 2-day ELISA MN assay. Comparison of titers between the 2-day ELISA MN assay and the 3-day MN assay detected by CPE showed higher overall consistency between the assays (Fig. 2B and Table 3).
FIG 2.
Relationship between test sample titers for antibodies to A(H1N1)pdm09 (A and B), A(H3N2) (C and D), or A(H5N1) (E and F) viruses determined by the 2-day ELISA MN assay and by the 3-day MN assay with detection by HA (A, C, and E) or CPE (B, D, and F). Each laboratory is represented by a color as indicated in the key.
TABLE 3.
Correlation of titers for test samples between assays by laboratorya
| Laboratory | Correlation of 2-day MN to 3-day MN (Spearman rank correlation coefficient) |
Preferred assay(s) | |||||
|---|---|---|---|---|---|---|---|
| A(H1N1)pdm09 |
A(H3N2) |
A(H5N1) |
|||||
| 3-day HA | 3-day CPE | 3-day HA | 3-day CPE | 3-day HA | 3-day CPE | ||
| A | 0.966 | 0.901 | 0.865 | 2 day | |||
| B | 0.976 | 2 day | |||||
| C | 0.992 | 0.966 | 3 day | ||||
| D | 0.892 | 0.890 | 0.898 | 3 day | |||
| E | 0.944 | 2 day | |||||
| F | 0.965 | 0.966 | 3 day | ||||
| G | 0.580 | 2 day | |||||
| H | 0.638 | 0.738 | 0.883 | 0.908 | 2 day | ||
| I | 0.970 | 0.954 | 0.901 | 2 day | |||
| J | 0.944 | 3 day | |||||
| Kb | 0.931 | 0.942 | 3 day | ||||
| L | 0.439 | 0.833 | Both | ||||
For each laboratory, Spearman rank correlation coefficients were calculated to compare the 2-day ELISA MN data to the 3-day HA or CPE MN assay data.
Data represent the 2-day ELISA MN assay titer data from laboratory F, as laboratories F and K shared serum panels.
The equivalent correlations were determined for comparisons of assays detecting antibodies to A(H3N2) (Fig. 2C and D) and A(H5N1) (Fig. 2E and F). For assays detecting antibodies to A(H3N2) virus, the majority of laboratories obtained good correlation between the 3-day HA and the 2-day ELISA MN assays (range, 0.865 to 0.966) (Table 3). Laboratory L had a poor correlation, with a coefficient of 0.439, as both of the assays gave a narrow titer range (<10 to 160) across the serum panel. There were only two laboratories that had data detecting antibodies to A(H5N1) virus, and they both showed reasonable correlation between the assays (Table 3). Laboratory L had much better correlation with the assays detecting antibodies to A(H5N1) virus than with those detecting antibodies to A(H3N2) virus.
Overall relationship between assays for each laboratory.
The ratio between the titers for the 2-day ELISA and 3-day MN assays for the serum panels was calculated to assess whether a consistent relationship between the performances of assays could be observed for each laboratory (Table 4). Many of the ratios indicated average titers within a 2-fold range for comparisons between methods (i.e., ratios between 0.5 and 2.0), representing reasonable agreement in assay sensitivity. For assays detecting antibodies to A(H1N1)pdm09, laboratories F and G had lower 3-day HA MN titers than 2-day ELISA MN titers. For laboratory G, the 3-day HA MN assay gave negative or low titers for all serum panel samples. Laboratories I and K had 3-day MN titers that were much higher than those measured for the 2-day ELISA MN assay. The ratios between assays were different for the laboratories that also participated in the comparison studies detecting antibodies to A(H3N2) or A(H5N1). For the comparison of the assays detecting A(H3N2) antibodies, laboratories D and K also had much higher 3-day MN assay titers than 2-day ELISA MN assay titers. For comparison of assays detecting A(H5N1) antibodies, laboratory H had much higher 3-day HA MN assay titers than 2-day ELISA MN assay titers (Table 4).
TABLE 4.
Titer ratios between the 2-day ELISA MN assay and the 3-day MN assay detected by HA and CPE
| Laboratory | Avg ratio of 3-day MN titer to 2-day MN titer |
|||||
|---|---|---|---|---|---|---|
| A(H1N1)pdm09 |
A(H3N2) |
A(H5N1) |
||||
| 3-day HA | 3-day CPE | 3-day HA | 3-day CPE | 3-day HA | 3-day CPE | |
| A | 2.4 | 1.5 | 1.9 | |||
| B | 0.9 | |||||
| C | 0.8 | 1.5 | ||||
| D | 2.0 | 3.7 | 3.8 | |||
| E | 1.0 | |||||
| F | 0.3 | 1.3 | ||||
| G | 0.1 | |||||
| H | 1.5 | 1.6 | 5.8 | 6.3 | ||
| I | 5.22 | 0.88 | 0.87 | |||
| J | 1.2 | |||||
| K | 2.4a | 3.2 | ||||
| L | 1.1 | 0.4 | ||||
Titer data for 2-day ELISA MN assay are from laboratory F, as laboratories F and K shared serum panels.
Analysis for bias within the study.
Potential factors for bias were assessed. Although each laboratory had a preferred MN assay (2-day ELISA, 3 day HA, or 3 day CPE, indicated in Table 3), overall, this did not seem to affect the correlation between assays. However, for laboratories where the titers from the two assays did not correlate well [A(H1N1)pdm09 virus for laboratories G and H; A(H3N2) virus for laboratory L] (Table 3), the 3-day MN assay showed less variation in titers (i.e., had less discriminating power) (Fig. 2A and C, respectively). Yet there was also variability in the correlations for different viruses within the same laboratory (Table 3), indicating that this effect may be virus specific or due to experience, as the studies were performed consecutively [the A(H1N1)pdm09 assay was followed by the A(H3N2)/A(H5N1) assay]. However, for laboratories with markedly different overall titers in the comparisons of the assays (laboratories F, H, I, and K; Table 4), there was no relationship between the preferred assay and the titer achieved. Most laboratories used sera from adults (10 of 11) and wild-type influenza viruses (8 of 11); neither showed any effect on assay comparability. The ratio of HA level to 50% tissue culture infective dose (TCID50) for the virus stock from each laboratory was assessed, and there was no clear relationship, suggesting that there was no bias due to the presence of interfering virus particles (HA titer range, 16 to 1,280 [median, 128]; log10 TCID50/ml range, 4.5 to 7 [median, 6]). Overall, there was no clear indication of major bias in the study.
DISCUSSION
Upon the emergence of a novel influenza virus, seroepidemiological data are critical in understanding the spread and attack rate of the virus to form the basis of pandemic risk and severity assessments. Serology can also identify groups susceptible to infection by a novel influenza virus in a population. Understanding the impact of different MN assay protocols would strengthen these estimates for policy decisions. Our comparison of MN assay methodologies indicates that there is good correlation between the 2-day ELISA and 3-day HA MN assays for detection of antibodies to A(H1N1)pdm09 virus in most laboratories. These findings were confirmed in an extension to this study performed with A(H3N2) and A(H5N1) viruses. Overall, there is potential for either assay to be used. Importantly, through participating in this study, laboratories have aligned their methodology to the CONSISE consensus assays described, harmonizing protocols internationally for the 2-day ELISA and 3-day HA MN assays.
Our intralaboratory assessment demonstrated that the results from the 2-day ELISA and 3-day HA MN assays were largely reproducible and comparable. The 2-day MN assay is read out by ELISA using spectrophotometry, which is objective, while the 3-day MN assay, which is read out by HA and CPE, requires more experience and training. In addition, as our study required participating laboratories to perform the assays on three separate occasions, different preparations of RBC for the replicate 3-day HA MN assays were likely. As a 2-fold range between titers for the same sample is considered acceptable for serological studies, the assays were overall highly reproducible on different days, with all laboratories having >97% of samples with titers within a 4-fold difference for the 2-day ELISA, 92% for the 3-day HA, and 92% for the 3-day CPE MN assays.
Importantly, in over half of the laboratories, there was very good correlation between the 2-day ELISA and 3-day HA MN assays when a panel of sera was tested. This suggests that there is no inherent difference in the results from the different assays, despite their different readouts. Thus, there is no underlying scientific reason that the different MN assay formats cannot be compared for detection of antibodies to A(H1N1)pdm09, A(H3N2), or A(H5N1) viruses.
However, three laboratories did have poor correlation between the assays, which may have been related to experience in, and performance of, one particular method. Seven laboratories were experienced in the 2-day ELISA MN assay before commencing this study, and 6 laboratories were experienced in the 3-day HA MN assay. Poor correlation between assays was more likely in those laboratories inexperienced in performing the 3-day HA MN assay, indicating that training in HA or CPE readout might be required. From the MN assay comparison performed here, we anticipate that, as laboratories gain experience in both assays, the correlation between the titers obtained for the 2-day ELISA and 3-day HA MN assays will improve. A mentoring system will be established in subsequent international comparison studies performed by CONSISE whereby personnel in laboratories who are learning an assay will be assisted by local “experienced” laboratory personnel. We anticipate that this collaborative assay development will encourage rapid assay results and will result in data that are more comparable between assays and laboratories in the future.
It is notable that, although we were able to standardize many assay-specific variables, such as virus concentration, incubation times, serum dilution, and endpoint determination, in this study, some factors were impossible to standardize. Cell culture conditions are laboratory specific and may differ with respect to cell lines, medium supplements, and incubation temperatures. Cell culture conditions are often optimized for the variety of viruses and experiments performed in a laboratory as well as for the availability of reagents and thus cannot be prescriptive.
A limitation of the present study is that the test serum panels were not shared among all of the laboratories. As our study compared assay protocols, rather than the performances of the different laboratories, this is acceptable. Thus, the impact of using consensus assay protocols on interlaboratory variability could not be examined thoroughly. In a future study being planned by the CONSISE Laboratory Working Group, shared serum panels will be tested for antibodies to A(H1N1)pdm09 virus using consensus 2-day ELISA and 3-day MN assays and a consensus HI assay in comparison with use of local assay protocols.
ACKNOWLEDGMENTS
Study participants. The study participants and their institutional affiliations were as follows: Jaccqueline Katz, Xiuhua Lu, Min Levine, Vic Veguilla, Feng Liu, and Yaohui Bai, U.S. Centers for Disease Control and Prevention; Pilaipan Puthavathana, Hatairat Lerdsamran, Phisanu Pooruk, and Knnika Nateerom, Department of Microbiology, Faculty of Medicine, Siriraj Hospital, Mahidol University, Thailand; Maria Rita Castrucci, Isabella Donatelli, and Marzia Facchini, Istituto Superiore di Sanita, Italy; Noriko Kishida, Masato Tashiro, and Takato Odagiri, National Institute of Infectious Diseases, Japan; Shailesh D. Pawar and Sadhana S. Kode, National Institute of Virology, Pune, India; Anthony Hawksworth, Ryan Ortiguerra, and Gary Brice, U.S. Naval Health Research Center; Nicholas Martin, Tad Kochel, Jose Sanchez, Michael Cooper, and James Cummings, U.S. Naval Medical Research Center; Katja Hoschler, Public Health England, United Kingdom; Ralf Wagner, Constanze Goepfert, Nina Alex, Joanna Hammann, and Britta Neumann, Paul-Ehrlich-Institut, Germany; Malik Peiris and Mahendra Perera, School of Public Health, The University of Hong Kong, Hong Kong; Emanuele Montomoli, Guilia Lapini, and Sara Sbragi, University of Siena, Italy; Tian Bai, Zaijiang Yu, and Jianfang Zhou, WHO Collaborating Centre for Reference and Research on Influenza, Chinese National Influenza Center, China; and Louise Carolan and Karen Laurie, WHO Collaborating Centre for Reference and Research on Influenza, Victorian Infectious Diseases Reference Laboratory, Australia.
The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by a grant from the Australian Government Department of Health to K.L.L. This work was also supported by a grant from the Area of Excellence Scheme of the University Grants Committee of Hong Kong (AoE/M-12/96) to M.P. at the University of Hong Kong, by grants from the UK Medical Research Council and the Bill and Melinda Gates Foundation to M.D.V.K., and by the United States Armed Forces Health Surveillance Center's Global Emerging Infections Surveillance and Response System for the Naval Medical Research Center and the Naval Health Research Center and the Italian Ministry of Health (virological surveillance of epidemic and pandemic influenza) for the Istituto Superiore di Sanita.
K.L.L., J.W., A.H., M.P., K.H., O.H., W.Z., and M.D.V.K. declare no conflicts of interest. O.G.E. reports grants from IFPMA (International Federation of Pharmaceutical Manufacturers & Associations) outside the submitted work. J.M.K. reports grants from Juvaris Bio-Therapeutics, Inc., and Glaxo SmithKline outside the submitted work. J.M.K. has several patents (18–20).
REFERENCES
- 1.Chen MI, Barr IG, Koh GCH, Lee VJ, Lee CPS, Shaw R, Lin C, Yap J, Cook AR, Tan BH, Loh JP, Barkham T, Chow VTK, Lin RTP, Leo YS. 2010. Serological response in RT-PCR confirmed H1N1-2009 influenza A by hemagglutination inhibition and virus neutralization assays: an observational study. PLoS One 5:e12474. doi: 10.1371/journal.pone.0012474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong L, Bo H, Bai T, Gao R, Dong J, Zhang Y, Guo J, Zou S, Zhou J, Zhu Y, Xin L, Li X, Xu C, Wang D, Shu Y. 2014. A combination of serological assays to detect human antibodies to the avian influenza A H7N9 virus. PLoS One 9:e95612. doi: 10.1371/journal.pone.0095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. 2010. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet 375:1100–1108. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 4.Veguilla V, Hancock K, Schiffer J, Gargiullo P, Lu X, Aranio D, Branch A, Dong L, Holiday C, Liu F, Steward-Clark E, Sun H, Tsang B, Wang D, Whaley M, Bai Y, Cronin L, Browning P, Dababneh H, Noland H, Thomas L, Foster L, Quinn CP, Soroka SD, Katz JM. 2011. Sensitivity and specificity of serologic assays for the detection of human infection with 2009 pandemic H1N1 virus in U.S. populations. J Clin Microbiol 49:2210–2215. doi: 10.1128/JCM.00229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurie KL, Huston P, Riley S, Katz JM, Willison DJ, Tam JS, Mounts AW, Hoschler K, Miller E, Vandemaele K, Broberg E, Van Kerkhove MD, Nicoll A. 30 April 2012. Influenza serological studies to inform public health action: best practices to optimise timing, quality and reporting. Influenza Other Respir Viruses doi: 10.1111/j.1750-2659.2012.0370a.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Kerkhove MD, Broberg E, Engelhardt OG, Wood J, Nicoll A, The CONSISE Steering Committee. 26 December 2012. The Consortium for the Standardization of Influenza Seroepidemiology (CONSISE): a global partnership to standardize influenza seroepidemiology and develop influenza investigation protocols to inform public health policy. Influenza Other Respir Viruses doi: 10.1111/irv.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laurie KL, Engelhardt OG, Wood J, Van Kerkhove M. 2014. Global seroepidemiology: value and limitations, p 51–66. In Oxford J. (ed), Clinical insights: influenza surveillance. Future Medicine Ltd doi: 10.2217/ebo.13.504. [DOI] [Google Scholar]
- 8.World Health Organization Global Influenza Surveillance Network (GISN). 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/gisrs_laboratory/manual_diagnosis_surveillance_influenza/en/. [Google Scholar]
- 9.Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, Fukuda K, Cox NJ, Katz JM. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol 37:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization Global Influenza Programme (GIP). 2002. WHO manual on animal influenza diagnosis and surveillance. WHO/CDS/CSR/NCS/2002.5 Rev.1. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 11.Hehme N, Engelmann H, Kuenzel W, Neumeier E, Saenger R. 2004. Immunogenicity of a monovalent, aluminum-adjuvanted influenza whole virus vaccine for pandemic use. Virus Res 103:163–171. doi: 10.1016/j.virusres.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson I, Das RG, Wood JM, Katz JM. 2007. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine 25:4056–4063. doi: 10.1016/j.vaccine.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 13.Stephenson I, Heath A, Major D, Newman RW, Hoschler K, Junzi W, Katz JM, Weir JP, Zambon MC, Wood JM. 2009. Reproducibility of serologic assays for influenza virus A (H5N1). Emerg Infect Dis 15:1250–1259. doi: 10.3201/eid1508.081754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood JM, Gaines-Das RE, Taylor J, Chakraverty P. 1994. Comparison of influenza serological techniques by international collaborative study. Vaccine 12:167–174. doi: 10.1016/0264-410X(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 15.Wood JM, Major D, Heath A, Newman RW, Höschler K, Stephenson I, Clark T, Katz JM, Zambon MC. 2012. Reproducibility of serology assays for pandemic influenza H1N1: collaborative study to evaluate a candidate WHO International Standard. Vaccine 30:210–217. doi: 10.1016/j.vaccine.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Tannock GA, Paul JA, Herd R, Barry RD, Reid AL, Hensley MJ, Gillett RS, Gillett SM, Lawrance P, Henry RL, Saunders NA. 1989. Improved colorimetric assay for detecting influenza B virus neutralizing antibody responses to vaccination and infection. J Clin Microbiol 27:524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CONSISE Laboratory Working Group. 15 January 2014. Consensus protocols for laboratory assay of antibody to influenza A(H1N1)pdm09. https://consise.tghn.org/articles/consensus-protocols-laboratory-assay-antibody-influenza-h1n1pdm09/. [Google Scholar]
- 18.Frace AM, Klimov AI, Katz JM. January 2001. Preparation and use of recombinant influenza A virus M2 construct vaccine. US patent 6,169,175.
- 19.Crawford PC, Gibbs PJ, Dubovi EJ, Donis RO, Katz JM, Klimov AI, Lakshmanan NP, Lum MA, Goovaerts DGE, Mellencamp MW, Cox NJ, Castleman WL. June 2011. Materials and methods for respiratory disease control in canines. US patent 7,959,929.
- 20.Sambhara S, Katz JM, Hoelscher M, Mittal SK, Bangari DS. April 2012. An effective vaccine against pandemic strains of influenza viruses. US patent 8,163,545.