Abstract
Microbial secretion is integral for regulating cell homeostasis as well as releasing virulence factors during infection. The genes encoding phosphatidylserine synthase (CHO1) and phosphatidylserine decarboxylase (PSD1 and PSD2) are Candida albicans genes involved in phospholipid biosynthesis, and mutations in these genes affect mitochondrial function, cell wall thickness, and virulence in mice. We tested the roles of these genes in several agar-based secretion assays and observed that the cho1Δ/Δ and psd1Δ/Δ psd2Δ/Δ strains manifested less protease and phospholipase activity. Since extracellular vesicles (EVs) are surrounded by a lipid membrane, we investigated the effects of these mutations on EV structure, composition, and biological activity. The cho1Δ/Δ mutant releases EVs comparable in size to wild-type EVs, but EVs from the psd1Δ/Δ psd2Δ/Δ strain are much larger than those from the wild type, including a population of >100-nm EVs not observed in the EVs from the wild type. Proteomic analysis revealed that EVs from both mutants had a significantly different protein cargo than that of EVs from the wild type. EVs were tested for their ability to activate NF-κB in bone marrow-derived macrophage cells. While wild-type and psd1Δ/Δ psd2Δ/Δ mutant-derived EVs activated NF-κB, the cho1Δ/Δ mutant-derived EV did not. These studies indicate that the presence and absence of these C. albicans genes have qualitative and quantitative effects on EV size, composition, and immunostimulatory phenotypes that highlight a complex interplay between lipid metabolism and vesicle production.
INTRODUCTION
Candida albicans resides on the skin and mucosal surfaces of most individuals in a state of commensalism but can cause disease in individuals with impaired host defenses. Depending on the type of immune impairment, these infections can range from superficial mucosal infections to life-threatening systemic infections. Fungal infections are an increasingly important medical problem, and the majority of these are caused by commensal Candida species (1, 2). C. albicans is the most common species associated with systemic candidiasis.
In its association with the human host, C. albicans uses a number of virulence factors to colonize surfaces and cause disease, including secreted factors such as proteases, lipases, and mannoproteins (3). These secreted proteases are required for pathogenicity, as they take part in fungal attachment, invasion, and subsequent tissue damage (4–7). These factors are secreted by mechanisms that include traditional signal peptide-based secretion using systems homologous to those in the model yeast Saccharomyces cerevisiae as well as nontraditional secretion (8, 9).
Nontraditional secretion systems provide a mechanism for a microbe to circumvent the requirement of a signal peptide for protein secretion and include release of protein cargo in extracellular vesicles (EVs). Like many other fungi, C. albicans produces EVs (10). Recently, C. albicans EVs were found to carry several of these virulence factors and act as immunostimulants for murine immune cells (11). In addition, the membrane lipid bilayer of EVs contains fungal sterols and glucosylceramides (10, 12, 13). EVs from C. albicans stimulate immune responses from macrophage and dendritic cells and can contribute to a protective host effect in a waxworm model of infection (11). EVs therefore are believed to play a role in C. albicans pathogenesis.
A series of C. albicans mutants in phospholipid biosynthesis pathways have been characterized, and the mutations have been found to affect a number of biological characteristics, including pathogenesis. Phosphatidylserine synthase (encoded by CHO1) and phosphatidylserine decarboxylases (encoded by PSD1 and PSD2) are required for phosphatidylserine (PS) and phosphatidylethanolamine (PE) de novo syntheses, respectively (Table 1), which are required for cell wall homeostasis, mitochondrial function, and virulence for mice (14). Based on the requirement of phospholipid building blocks for cell membrane synthesis, we predicted that these mutations would affect the generation and release of virulence-associated EVs. In this work, we isolate and characterize EVs from these mutants and show that these mutations lead to divergent cargo packaging with subsequent immune cell activation differences.
TABLE 1.
Strains used in this studya
| Strain | Relevant genotype | Gene deletion | Function | Source |
|---|---|---|---|---|
| sc5314 | Wild type | NA | Gillum et al. (37) | |
| YLC337 | cho1Δ/cho1Δ | CHO1 | Synthesizes PS from CDP-DAG and serine | Chen et al. (14) |
| YLC344 | cho1Δ/cho1Δ::CHO1-SAT1 | NA | Chen et al. (14) | |
| YLC280 | psd1Δ/psd1Δ | PSD1 | Decarboxylates PE+ from PS (mitochondrial) | Chen et al. (14) |
| YLC294 | psd1Δ/psd1Δ PSD1-SAT1 | NA | Chen et al. (14) | |
| YLC375 | psd2Δ/psd2Δ psd1Δ/psd1Δ | PSD1 and PSD1 | Decarboxylates PE from PS (mitochondrial and endosomal) | Chen et al. (14) |
Abbreviations: PS, phosphatidylserine; CDP-DAG, cytidyldiphosphate-diacylglycerol; PE+, phosphatidylethanolamine; NA, not applicable.
MATERIALS AND METHODS
Strains and media.
All strains were generated from the parental strain SC5314. The mutant strains used in this study have each been characterized previously (14) (Table 1). All strains were propagated in yeast extract-peptone-dextrose (YPD; Difco) and grown with shaking at 37°C. For secretion assays, YPD with 2% agar was supplemented with 8% egg yolk or 1% bovine serum albumin (BSA) (final concentrations), supplemented in some cases with 1 M sorbitol or 0.1 M CaCl2. Agar plates were incubated 2 days at 37°C.
Vesicle preparation.
EVs were prepared from 3-day-old 100-ml YPD cultures as previously described (11, 15). Briefly, cells were removed by centrifugation followed by filtration with a 0.45-μm filter. Cell-free supernatant was concentrated to less than 20 ml using Amicon concentration units. EVs were collected from the concentrate for 1 h at 141,000 × g. EVs were resuspended in 100 to 1,000 μl, depending on their intended use, and quantified using a microprotein measurement kit (Pierce), a phospholipid fluorescent enzyme-linked immunosorbent assay (ELISA; Biosystems Assay), and a sterol fluorescent ELISA (Molecular Probes).
SDS-PAGE separation of proteins.
A volume containing 80 μg of protein was run on a 4 to 20% gradient gel (Bio-Rad). Protein was stained using silver stain reagents (Pierce).
Dynamic light scattering.
EVs were resuspended in phosphate-buffered saline (PBS) and measured with a 90Plus/BI-MAS multiangle particle sizing analyzer (Brookhaven Instruments). A monochromatic laser illuminated the samples and was scattered by the Brownian motion of the EVs. Light fluctuations were detected at a 90° angle and then analyzed by the autocorrelation function, C(t), calculated as follows: C(t) = Ae2Γt + B, where t is time delay, A and B are optical constants, and Γ is related to the relaxation of the fluctuations by the formula Γ = Dq2. D is derived from the formula D = (KBT)/3πη(t)d, which assumes each scattering particle to be a sphere; KB is Boltzmann's constant (1.38054E−23 J K−1); T is the temperature (in K) (303 K); η(t) is the viscosity of the liquid in which the particles are moving; and d is the particle diameter. The parameter q is derived from the scattering angle (ϴ), the laser light wavelength (λ0), and the solvent refractive index (n) from the equation q = (2πn/λ0)2sin(ϴ/2). Data are expressed as the average results from 10 runs of 1 min per run.
Electron microscopy.
For transmission electron microscopy, samples were fixed with 4% glutaraldehyde–2% paraformaldehyde in 0.1 M sodium cacodylate buffer. They were postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries, Burlington VT). Ultrathin sections were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV. For scanning electron microscopy, samples were first fixed in 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 0.2 M sucrose, and 5 mM MgCl2 (pH 7.4). Next, they were dehydrated through a graded series of ethanol and then critical-point dried using liquid carbon dioxide in a Tousimis Samdri 795 Critical Point Drier (Rockville, MD). Finally, they were sputter coated with chromium in a Quorum EMS 150T ES (Quorum Technologies Ltd., United Kingdom). Samples were viewed on a Zeiss Supra field emission scanning electron microscope (Carl Zeiss Microscopy, LLC North America), using an accelerating voltage of 1.5 kV.
Proteomics.
Protein cargo in vesicle suspensions was identified as previously described (16). Briefly, proteins from EVs in 100 μl PBS were precipitated by adding 6 volumes of ice-cold acetone and incubating the mixtures overnight at −20°C. Proteins were then reduced with 10 mM dithiothreitol (DTT), and the cysteine residues were subsequently alkylated with 10 mM iodoacetamide. Protein digestion was carried out with Mass Spec-grade trypsin (1:20, wt/wt) overnight at 37°C. The peptide mixture was desalted, concentrated on ZipTip (Millipore), and then analyzed using nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS). The peptides were loaded onto a 0.3- by 5-mm C18 precolumn and then eluted with a linear gradient of 5 to 90% acetonitrile in a 0.1% aqueous solution of formic acid. The gradient was performed over 120 min using a NanoLC 1D Plus (Eksigent) at a flow rate of 200 nl/min through a 75-μm by 15-cm fused silica capillary C18 high-performance liquid chromatography column (LC Packings) to a stainless steel nanobore emitter (Proxeon). The peptides were scanned and fragmented with an LTQ XL linear ion trap mass spectrometer (ThermoFinnigan) operated in data-depended and MS/MS switching mode using the 3 most intense precursor ions detected in a survey scan from 400 to 1,600 atomic mass units (amu). A database containing the NCBI Candida albicans sequences was searched using Mascot Software (version 2.3; Matrix Science) for protein identification. Search criteria included trypsin specificity with one missed cleavage allowed, methionine oxidation as variable modification, a minimum precursor and fragment-ion mass accuracy of 1.2 and 0.3 Da, respectively, and a requirement of at least one bold red peptide (i.e., the highest-score peptide matching the protein with the highest total score). Cutoff values for Mascot protein scores were set at 29 (P > 0.05) to be considered an accurate identification. Proteins identified with only one peptide were inspected manually.
Macrophage NF-κB staining.
J774.14 macrophage-like murine cells (ATCC) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10% NCTC-109 Gibco medium (Life Technologies), and 1% nonessential amino acids (CellGro). Bone marrow-derived macrophage cells (BMDM) were prepared from female 6- to 8-week-old BALB/c mice (National Cancer Institute). BMDM were extracted from hind leg bone marrow and then developed for 6 to 8 days in DMEM containing 20% L-929 cell-conditioned medium, 10% FBS, 2 mM l-glutamine (CellGro), 1% nonessential amino acids (CellGro), 1% HEPES buffer (CellGro), and 2-ME (Life Technologies). After 7 days, 2 × 105 cells were seeded onto a MatTek dish and incubated overnight. The next day, the cells were treated with fresh medium, 10 μg/ml lipopolysaccharide (LPS; Sigma), or 0.5 μg vesicle preparations (by sterol concentration) and incubated 3 h at 37°C, 10% CO2. Cells were then fixed with 4% formaldehyde and blocked with blocking buffer (PBS with 10% fetal calf serum, 1% bovine serum albumin, and 0.1% Triton X-100). Anti-RelA/p65 antibody (R&D Systems) was diluted to 2 μg/ml in dilution buffer (PBS with 1% fetal calf serum, 1% bovine serum albumin, and 0.1% Triton X-100) and incubated 1 h at room temperature (RT). Cells were washed and incubated 5 m with DAPI (4′,6-diamidino-2-phenylindole), after which they were examined on an Axiovert 200 M inverted microscope (Zeiss). Images were captured using AxioVision software (Zeiss) and analyzed using ImageJ.
Statistics.
The Student t test and/or analysis of variance (ANOVA) tests were applied when indicated using GraphPad Prism software. For comparing population ratios by size, the Z test was used with the calculator found online at Social Science Statistics. For all assays, P values of <0.05 were considered to indicate statistically significant results.
RESULTS
Secretion plays an essential role during C. albicans pathogenesis, with fungal cells releasing a variety of proteases and lipases and other proteins that can mediate tissue damage and assist host cell invasion (3). Recently, several lipid biosynthetic mutants were described with multiple virulence defects, including in an in vivo mouse model (11). Alteration of the sphingolipid concentration negatively affects cell wall integrity and influences virulence factor secretion (17), and thus we hypothesized that phospholipid homeostasis was also required for proper regulation of secretion. We therefore characterized these mutants for several known secreted virulence factors. The strains used include the cho1Δ/Δ strain, which is deficient in de novo phosphatidylserine (PS) synthesis, the psd1Δ/Δ strain, which is deficient in synthesizing phosphatidylethanolamine (PE) from phosphatidylserine in the mitochondria, but not in the endosome, where it synthesizes PE from PS using the Psd2 enzyme, and the psd1Δ/Δ psd2Δ/Δ mutant, which is completely deficient in PE synthesis from PS. In the case of all three mutants, they are viable so long as they have sufficient ethanolamine in the environment, which allows them to synthesize PE via enzymes of the Kennedy pathway (18).
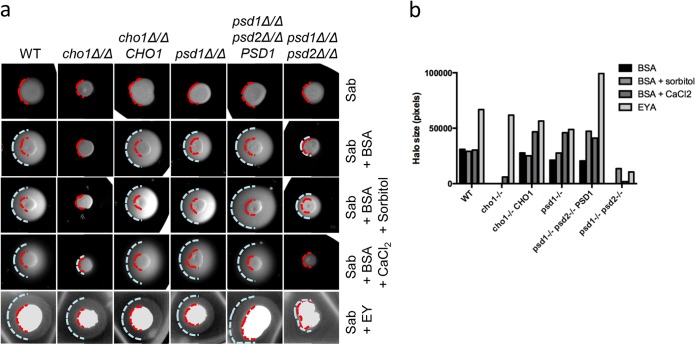
We first investigated the secretion of proteases by plating the strains on Sabouraud agar supplemented with 1% BSA. This resulted in a halo of precipitated protein around the wild type (WT) after 24 h at 37°C (Fig. 1a and b). Neither the colonies from the cho1Δ/Δ mutant nor those from the psd1Δ/Δ psd2Δ/Δ mutant strains produced this halo, indicating a protease secretion defect on this medium. The psd1Δ/Δ mutant had at most a slight defect, with a very slight decrease in halo size relative to the wild type. Next, we investigated the secretion of phospholipase by plating the strains on Sabouraud agar supplemented with 8% egg yolk. This produced a halo of precipitated lipids around colonies from all strains except the psd1Δ/Δ psd2Δ/Δ mutant (Fig. 1a and b). These results suggest that both PS and PE syntheses have effects on virulence factor secretion to various degrees.
FIG 1.
Secretion profiles of phospholipid biosynthetic gene mutants. (a) A 3-μl overnight culture was spotted onto each of the indicated media to test for protease (BSA, BSA-containing plates) or phospholipase (EY, egg yolk-containing plates) release. In some protease detection plates, osmotic stabilizers were added to control for cell wall integrity defects. Red dashed lines indicate the perimeter of colony growth, and blue dashed lines indicate the perimeter of the secretion halo. Sab, Sabouraud medium. (b) Quantification of halo size by ImageJ software.
The lipid biosynthetic mutants have cell wall defects, including susceptibility to SDS, suggesting increased permeability (14). We thus hypothesized that these defects could affect proper secretion. We looked at protease secretion in the presence of osmotic stabilizer sorbitol (1 M) or CaCl2 (0.1 M). Both stabilizers can rescue growth defects of the mutant strains on YPD (14). While a slight rescue of protease secretion was seen in the cho1Δ/Δ strain with CaCl2 and in the psd1Δ/Δ psd2Δ/Δ strain with sorbitol, neither solute was sufficient to entirely rescue protease secretion (Fig. 1a and b). Therefore, we concluded that these mutations affected secretion through a mechanism independent of cell wall-derived defects and considered the possibility that EVs are involved, since these are vehicles for nonclassical secretion.
We suspected that phospholipid synthesis may play a role in EV production, as fungal EVs are surrounded by a membrane constituted mainly of phospholipids and sterols (11, 12, 19). We therefore evaluated the production of EVs by the mutant strains relative to WT. Because of severe growth defects in defined media, we used YPD to grow strains to the stationary phase. We incubated the cultures at 37°C, and all mutants reached comparable OD600 measurements except the cho1Δ/Δ mutant, which consistently grew to 70 to 80% of wild-type levels.
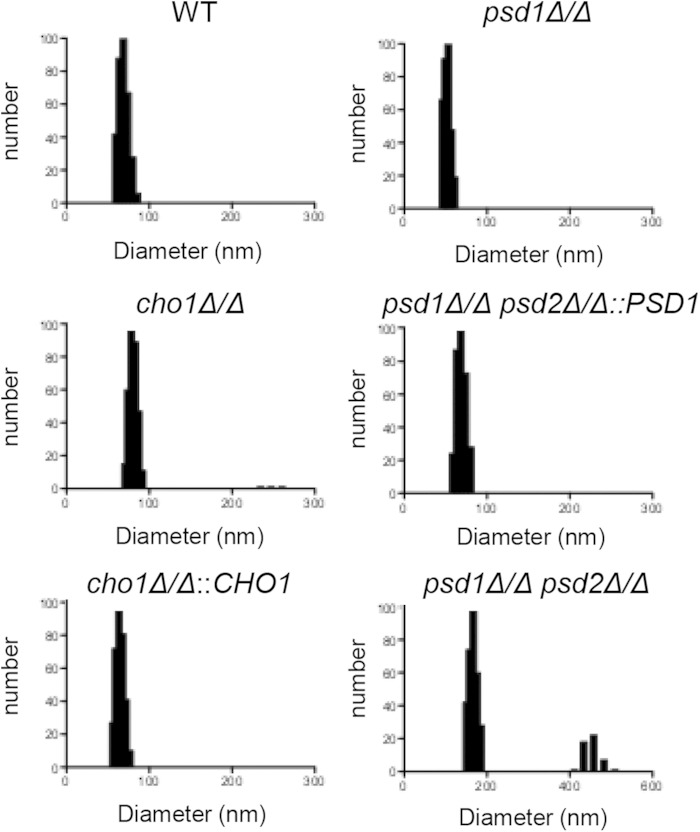
We isolated EVs using a well-defined protocol that has been used in vesicle isolation from various other microbes, including C. albicans. EVs were then studied by dynamic light scattering (DLS) to estimate their sizes. All culture samples except the psd1Δ/Δ psd2Δ/Δ mutant produced similarly sized vesicles, with an average diameter between 50 and 100 nm. The cho1Δ/Δ mutant had a slightly larger average size (78.3 nm) than those of the WT and reconstituted strains (68.9 and 63.3 nm, respectively). The psd1Δ/Δ psd2Δ/Δ mutant produced slightly larger EVs, with an average of 167 nm in this peak. We also observed a population of larger EVs in the cho1Δ/Δ and psd1Δ/Δ psd2Δ/Δ mutant strains (Fig. 2). YPD alone did not produce any measurable signal (data not shown), demonstrating that the particles were not found in the medium alone. We concluded that all strains tested were able to produce particles consistent with the size of EVs, although there was some deviation in size compared to the WT.
FIG 2.
Light scattering measurements of EV preparations. Dynamic light scattering was used to determine size estimates of EV preparations resuspended in PBS.
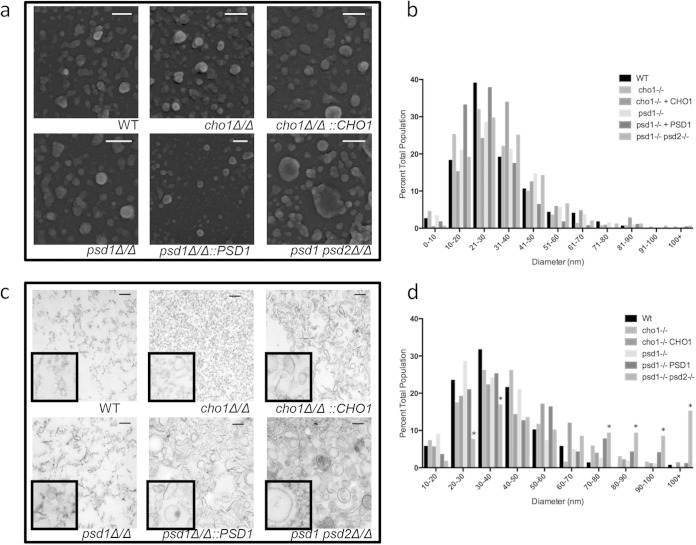
To further characterize EV morphology, we used electron microscopy. We first imaged the surface of EVs using scanning electron microscopy (SEM), which revealed a variety of vesicle sizes, consistent with EV produced in other fungal species (10, 15) (Fig. 3a). Like the WT strain, the mutant strains also produced a variety of sizes, but a marked size difference was most apparent in the psd1Δ/Δ psd2Δ/Δ mutant strain. Imaging software was used to measure the diameter of several hundred EVs from each strain, which revealed no statistical difference in size between any of the strains (Fig. 3c). This is likely due to the many smaller-sized EVs that bring the average for all the samples to a similar range or the poor ability of SEM to fully differentiate true EVs.
FIG 3.
Electron micrographs of EV preparations. EV preparations were visualized either by scanning electron microscopy (a) or by transmission electron microscopy (c). EVs were measured using ImageJ software and plotted by size on corresponding histograms (b and d). Scale bars, 200 nm in all panels. *, P < 0.05 by z test for population ratios.
We also imaged the EVs using transmission electron microscopy (TEM), which facilitates identification of EVs by their double-membrane border. Again, the wild-type sample contained a variety of vesicle sizes and densities, as previously reported (10, 15, 20) (Fig. 3c). This technique allows us to identify true EVs and revealed that the cho1Δ/Δ mutant preparation contained many fewer double-membraned EVs, despite releasing extracellular material that stained. These cho1Δ/Δ EVs were in the same size range as the wild-type EVs, but identifying them was more difficult among the debris on the field. The stained debris was not observed when the mutant was complemented with an exogenous CHO1 allele, suggesting that it is a function of lipid metabolic dysregulation (Fig. 3c). While the psd1Δ/Δ mutant strain did not produce EVs that were very different from those of the wild type, the psd1Δ/Δ psd2Δ/Δ double mutant strain produced EVs significantly larger than those observed in the wild type (Fig. 3c). Analysis using imaging software revealed a significant population of EVs with diameters above 100 nm, which was not observed in the wild type or in other mutant strains (Fig. 3d). TEM therefore provided two additional insights into the vesicle samples: the cho1Δ/Δ mutant generated fewer EVs than the stained debris, and the psd1Δ/Δ psd2Δ/Δ-derived EVs are indeed significantly larger than those derived from the wild type. Although the samples were collected identically, differences in processing for TEM versus SEM may account for the identification of this larger psd1Δ/Δ psd2Δ/Δ population (also identified by DLS) by TEM but not SEM.
We hypothesized that the lipid makeup of the cho1Δ/Δ-derived EVs and the psd1Δ/Δ psd2Δ/Δ-derived EVs would differ from the lipid makeup of the WT-derived EVs. We tested the phosphatidylcholine concentration using a fluorometric ELISA to measure vesicle preparation lipid (Fig. 4b). This revealed an increase in the vesicle-associated PC content of both the cho1Δ/Δ and psd1Δ/Δ psd2Δ/Δ mutant strains compared to the content in the wild-type strain (Fig. 4a). We observed no significant differences in sterol composition (Fig. 4b), however, suggesting that vesicle sterol composition is not affected by phospholipid concentration. As noted above, all strains but one grew in density to comparable OD600 in YPD broth, suggesting that strain differences in vesicle dimensions and composition were not due to different growth states or total culture biomass. The slight growth defect observed in the cho1Δ/Δ mutant suggests that the per-cell lipid output of this strain was greater in this strain than in the other strains studied here.
FIG 4.
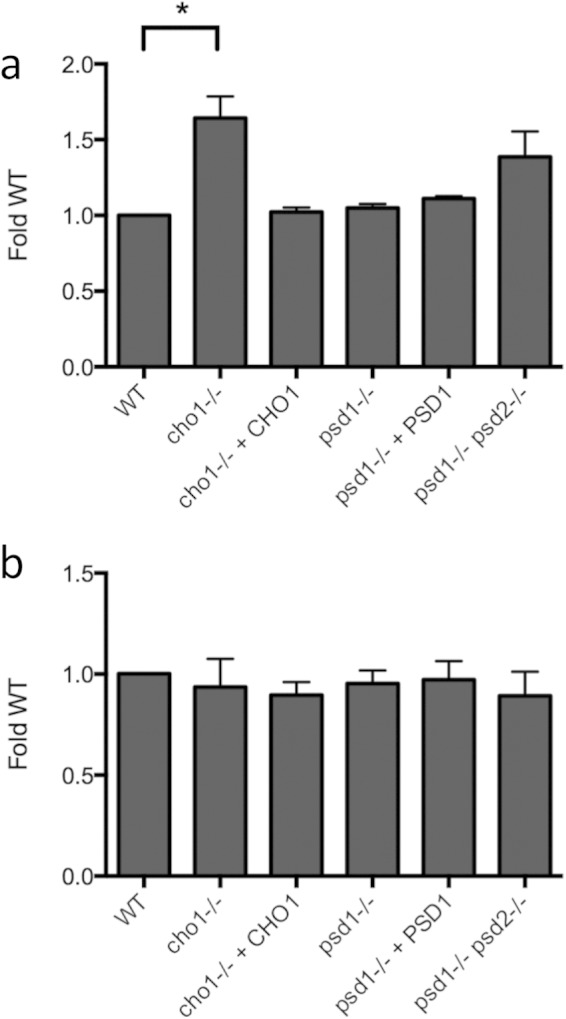
EV lipid profiles. Phosphatidylcholine (a) and sterol (b) content of EV preparations as normalized to the WT content. *, P < 0.05.
We suspected that differences in lipid composition might be reflected in the protein cargo associated with the vesicle preparations. A silver-stained SDS gel confirmed our suspicions, with several bands in the wild type not observed in the mutant strain samples and, conversely, some bands not found in the wild type observed in the mutant strain samples (Fig. 5). To gain a more complete picture of the protein components, we used proteomic analysis.
FIG 5.
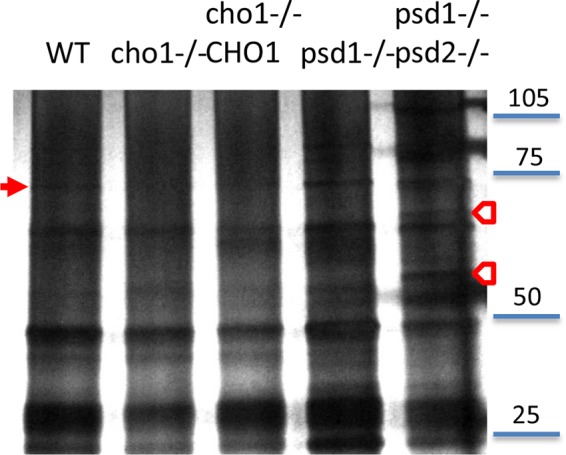
EV protein profiles. Equivalent protein concentrations were run on a 4 to 20% gradient gel and silver stained. Arrows indicate bands found in WT but not mutant EV preparations (filled arrow) or bands found not in WT but in mutant EV preparations (empty arrows).
Proteomic analysis revealed a wide variety of proteins in each sample (Table 2; see also Table S1 in the supplemental material). We identified between 179 and 278 unique proteins per strain. As with previous extracellular vesicle proteomics analyses, the proteins identified were associated with a wide variety of cellular functions and localizations (10, 13, 15, 21, 22). The 179 proteins identified in the wild-type EVs were in line with previously described C. albicans proteins (11). We noted differences in the detected protein content between the mutant and wild-type strains. Notably, several virulence factors were identified in wild-type EVs that were not in cho1Δ/Δ EVs (among them, phospholipase Plb3 and adhesin Sim1) or in psd1Δ/Δ psd2Δ/Δ EVs (including proteinase Prd1) (Table 2). Thus, phospholipid biosynthetic genes are involved in EV size, lipid composition, and protein cargo.
TABLE 2.
Partial list of proteomic results representing the strongest differences between WT and mutant EV preparationsa
| Protein name | Protein abbreviation | No. of peptide hits |
|||||
|---|---|---|---|---|---|---|---|
| WT strain | cho1Δ/Δ mutant | cho1Δ/Δ::CHO1 mutant | psd1Δ/Δ psd2Δ/Δ mutant | psd1Δ/Δ mutant | psd1Δ/Δ psd2Δ/Δ:: PSD1 mutant | ||
| Fatty acid synthase subunit beta | FAS1 | 52 | 15 | 53 | 51 | 63 | 66 |
| Fatty acid synthase subunit alpha | FAS2 | 30 | 9 | 37 | 34 | 51 | 52 |
| ATP alpha synthase chain, mitochondrial precursor, putative | ATP1 | 18 | 16 | 16 | 17 | ||
| Heat shock protein 60 | HSP60 | 14 | 13 | 13 | 21 | 19 | 16 |
| Clathrin heavy chain | CHC1 | 10 | 5 | 10 | |||
| Beta-1,3-glucan synthase catalytic subunit | GSC1 | 9 | |||||
| Fructose-1,6-bisphosphatase | FBP1 | 8 | 8 | 5 | 6 | 8 | |
| ATP-dependent 6-phosphofructokinase subunit beta | PFK2 | 5 | 12 | 9 | 5 | ||
| GPI-anchored cell surface phospholipase B | PLB3 | 5 | 3 | 1 | 5 | ||
| Putative peptide:N-glycanase | PNG2 | 5 | 5 | 9 | |||
| Vacuolar H(+)-ATPase | VMA2 | 5 | 9 | 9 | 5 | 4 | |
| Adenyl succinate synthetase | ADE12 | 4 | 2 | 6 | |||
| Putative F0-ATP synthase subunit 4 | ATP4 | 4 | 5 | 2 | |||
| Drug resistance protein 2 | CDR2 | 4 | |||||
| Cytochrome c oxidase polypeptide VI, mitochondrial precursor | COX6 | 4 | 1 | 5 | |||
| ATP-dependent RNA helicase SUB2 | SUB2 | 4 | 4 | ||||
| Exportin-1 | CRM1 | 3 | 6 | 4 | 3 | ||
| YEF3-subfamily ABC family protein | KRE30 | 3 | 7 | 6 | |||
| Putative dihydrolipoamide acetyltransferase component (E2) of pyruvate dehydrogenase complex | LAT1 | 3 | 2 | 4 | 5 | ||
| Adhesin-like protein | SIM1 | 1 | 2 | 1 | |||
| Phosphoacetylglucosamine mutase (N-acetylglucosamine-phosphate mutase) | AGM1 | 3 | |||||
| Catalase | CAT1 | 3 | 5 | ||||
| Putative cofilin | COF1 | 6 | |||||
| NADPH oxidoreductase | EBP1 | 4 | |||||
| Glutathione S-transferase | GST3 | 3 | |||||
| Essential 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (cobalamin-independent methionine synthase) | MET6 | 20 | 23 | 18 | 23 | ||
| Cell surface mannoprotein | MP65 | 3 | 2 | 5 | |||
| Predicted acyl-CoA oxidase | POX1-3 | 17 | 10 | 21 | 10 | 4 | |
| Putative proteinase | PRD1 | 8 | |||||
| Alpha7 (C8) subunit of the 20S proteasome | PRE10 | 4 | |||||
| Putative proteasome beta-5 subunit | PRE2 | 3 | |||||
| Putative beta-1 proteasome subunit | PRE3 | 3 | |||||
| Alpha6 subunit of the 20S proteasome | PRE5 | 8 | 5 | ||||
| Putative alpha-2_sc subunit of proteasome | PRE8 | 8 | |||||
| Alpha3 (C9) subunit of the 20S proteasome | PRE9 | 7 | |||||
| Thioredoxin peroxidase | PRX1 | 3 | 2 | ||||
| Essential protein | SEC14 | 6 | |||||
| Plasma membrane t-SNARE | SSO2 | 2 | |||||
| Putative transketolase | TKL1 | 17 | 4 | 8 | |||
| H+ transporting ATPase E chain | VMA4 | 3 | 3 | 3 | |||
| Secreted yeast wall protein | YWP1 | 1 | 3 | ||||
For a full list of results, see the supplemental material. Abbreviations: GPI, glycosylphosphatidylinositol; acyl-CoA, acyl coenzyme A.
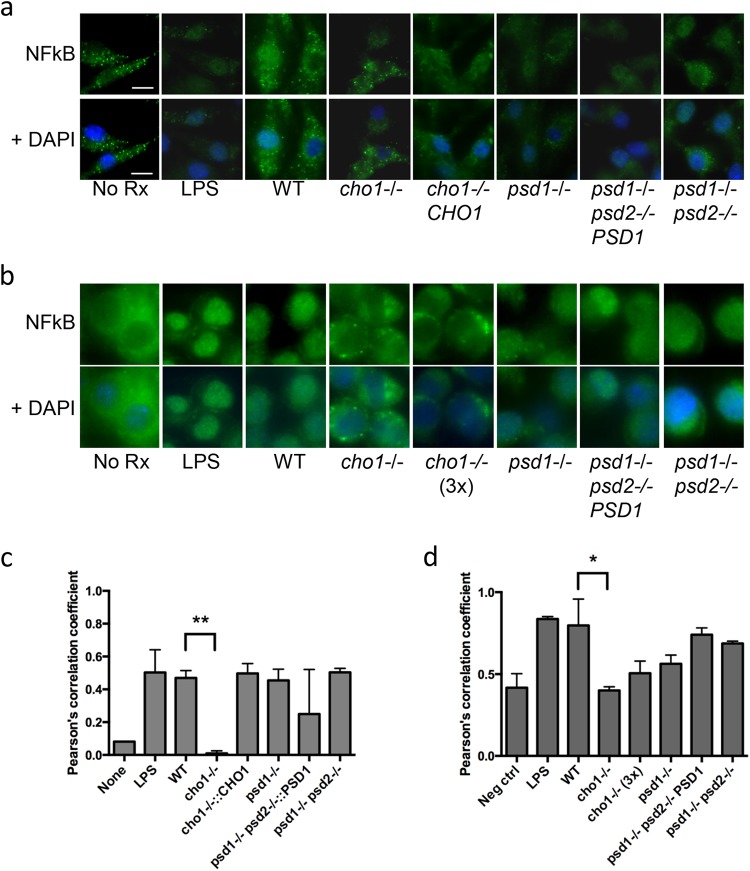
To test whether the cargo and/or lipid differences would reflect a change in host cell activation, we incubated the EVs with bone marrow-derived macrophage (BMDM) cells or with the murine macrophage-like J774.14 cell line. C. albicans activates NF-κB activation through Toll-like receptor 2 (TLR2) and TLR4 interactions, leading to transactivation of the activated transcription factor, NF-κB p65, also called RelA (23, 24). We observed nucleus-specific staining of NF-κB p65 in both cell types treated with wild-type EVs, based on colocalization of NF-κB p65 and DAPI, which marks the nucleus (Fig. 6a and b). Colocalization quantification via ImageJ analysis confirmed these findings (Fig. 6c and d). Of the strains tested, only the cho1Δ/Δ EVs failed to show any NF-κB activation in either cell type via nuclear staining, while all other strains showed no statistical difference from the wild type (Fig. 6a and b). When we added three times the amount of sterol to the cho1Δ/Δ EV-treated J774.14 cells, we saw no significant difference in the results despite a continuing trend for decreased signaling in this sample (Fig. 6b and d). Further, when we exposed BMDM to EVs prepared with a larger pore size (0.8 μm instead of 0.45 μm), we observed a staining pattern similar to that seen with the smaller filter, with cho1Δ/Δ-derived EVs consistently not stimulating transcription factor translocation (see Fig. S1 in the supplemental material). Thus, the differences observed in lipid and proteomic makeup impact host cell stimulation.
FIG 6.
Activation of NF-κB by EV preparations. BMDM were stimulated by equal sterol concentrations of EV preparations. (a) NF-κB p65 staining of BMDM; (b) NF-κB p65 staining of J774.14 cells. (c and d) Quantification of NF-κB colocalization with DAPI by ImageJ imaging software in BMDM (c) and J774.14 cells (d). **, P < 0.001; *, P < 0.05.
DISCUSSION
Our studies demonstrate that lipid biosynthesis plays a role in the secretion of virulence factors and in vesicle size and makeup. These differences in EV release likely contribute to a different secretion profile of these strains for reasons independent from their effects on cell wall stability. Our results indicate that the cho1Δ/Δ mutant strain, deficient in PS de novo synthesis, generates EVs with higher PC content than, and different protein cargo content from, that of the wild-type strain. Further, while the single gene deletion psd1Δ/Δ does not play a large role in vesicle biology, the psd1Δ/Δ psd2Δ/Δ mutant generates EVs with higher PC content, increased size, and different protein cargo content from both the wild-type and cho1Δ/Δ mutant strains.
How could phospholipid biosynthesis affect EV size and lipid composition? Both PS and PE are constituents of Paracoccidioides brasiliensis EV and are present in compositional ratios different from those in whole-cell lipid composition, suggesting a role of specific organelles in EV biogenesis (19). Membrane composition will be dictated in part by the phospholipids available for the cell incorporation into plasma membrane and membrane-bound organelles. With depleted PS and decreased PE, the cell may compensate with other phospholipids, such as phosphatidylcholine (PC), which accumulates at abnormal levels in these cells (14). The geometrical characteristics of phospholipid molecules may affect the overall EV structure: PC and PS have a cylindrical shape, while PE is conical (25). This conical nature means that PE cannot form lipid bilayers itself but often plays a role in disrupting the naturally planar bilayers formed by PC and PS, required for biological functions such as budding (26). Thus, a decrease in cellular PE concentrations may lead to delayed budding, manifesting in the large EVs observed in the psd1Δ/Δ psd2Δ/Δ mutant. The decrease of inner-leaflet members PE and PS in the cho1Δ/Δ mutant may alter the lipid stoichiometry such that the cell sheds outer-leaflet PC, resulting in some of the hydrophobic, non-EV material observed in the EV electromicrographs (Fig. 3). Different lipid head groups may also change the geometry of the EVs as they are forming, either internally as intraluminal EVs in multivesicular bodies or while budding from the plasma membrane itself. Alternatively, the change in plasma membrane lipid composition may affect exocytosis. Studies of the Schizosaccharomyces pombe phosphatidylserine synthase gene PPS1 demonstrated a role for PS in F actin homeostasis (27). In S. cerevisiae, Cdc42 accumulates at PS-rich microdomains (28), where Cdc42 can interact with Bni1 and/or Exo70 to mediate localized actin assembly and vesicle exocytosis (29). Cdc42 is mislocalized in the C. albicans cho1Δ/Δ mutant (T. Reynolds, unpublished data), supporting a role for phospholipid homeostasis in Cdc42 regulation. We observed no difference in the phalloidin staining of mutant versus wild-type actin in rich medium (data not shown), but more detailed studies of cortical actin staining patterns may illuminate cytoskeletal differences between strains. Interestingly, the absence of many true EVs in the cho1Δ/Δ mutant and the presence of larger EVs in the psd1Δ/Δ psd2Δ/Δ mutant correlate with the cellular levels of PS. The cho1Δ/Δ lacks PS altogether, while the psd1Δ/Δ psd2Δ/Δ mutant has a higher level of PS than the wild type. The mechanism driving this is not clear, but it could be regulated by PS levels.
Many of the proteins identified here as EV associated were previously found in association with the cell wall proteome (30), where the EVs may have been captured en route to release into the extracellular environment. It is of note that both the cho1Δ/Δ mutant and the psd1Δ/Δ psd2Δ/Δ mutant have cell wall defects (14), which may be related to different cell wall-modifying content, such as Gsc1, of EVs from these strains (Table 2). How could phospholipid synthesis genes affect vesicle protein cargo? One model is that the changes affecting membrane bending also affect protein loading kinetics. A second model would suggest that a specific protein-lipid interaction may be altered due to a change in lipid composition. However, both of these models predict a phenotype based on different membrane structures. PS is concentrated on the outer endosomal membrane and the cytoplasmic side of the plasma membrane, which could potentially impact the loading of such vesicles. Of course, we cannot dismiss the possibility that altering various lipid membrane homeostasis may in turn increase cell stress levels, which in turn may alter EV release. Stressful environments have been shown to increase EV output in other organisms, such as Escherichia coli (31).
Given the lipid and protein differences, it is perhaps not surprising that EVs from mutant strains have altered host cell interactions relative to wild-type strains. While the proteomics data did not reveal vast differences between the strains, we did note a few potential virulence-related proteins that may modify the biological activity of these EVs. Among these differences, some were specific to the cho1Δ/Δ mutant, including the absence of phospholipase 3b (Plb3) and a putative adhesin-like protein (Sim1). Several virulence-related proteins that were identified as associated with the cho1Δ/Δ mutant were not found in the wild type, including cofilin (Cof1), catalase (Cat1), and a secreted yeast wall protein (Zwf1). The presence or absence of this cargo may play a role in the differential activation of macrophage by EVs derived from the cho1Δ/Δ mutant. We cannot conclude that this is the case for the psd1Δ/Δ psd2Δ/Δ mutant, which did contain Plb3. Enzymatic cargo differences can be too low in concentration to measure by proteomics and are more readily detected through biochemical assays (21).
Lipid composition differences may compound the stimulation defect. Phospholipomannan is a released phosphatidylinositol-based C. albicans antigen that stimulates TLR2 activation (24). It is found in vesicle-like structures near the cell wall (32), and its proper secretion may be affected by phospholipid synthesis. Recently, the cho1Δ/Δ mutant was shown to display more β(1-3)-glucan on its surface, promoting proinflammatory cytokine secretion from macrophage but without a change in antimicrobial activity (33). The differential stimulatory effects of cells and their EVs suggest that C. albicans EVs do not contain cell wall particles, unlike EVs from the fungus Paracoccidioides brasiliensis (34).
We note that although no single gene has been associated with a null phenotype for vesicle production in either bacteria or fungi, several genes have now been shown to modify aspects of EV composition and structure (35, 36). In this study, we report that genes involved in phospholipid synthesis can affect a variety of secretion phenotypes based not only on cell wall integrity but also on release of EVs carrying microbial cargo. Phospholipid synthesis impacts the fungal components that interact with host cells, and thus the genes in this study play a role in mediating host-pathogen interactions. Factors that affect lipid synthesis have the potential to affect EV structure and constitution and are an important consideration in future research investigating secreted virulence factors.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00054-15.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 2.Pappas PG. 2006. Invasive candidiasis. Infect Dis Clin North Am 20:485–506. doi: 10.1016/j.idc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson BE, Wilhelmus KR, Hube B. 2007. The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci 48:3559–3565. doi: 10.1167/iovs.07-0114. [DOI] [PubMed] [Google Scholar]
- 5.Lian CH, Liu WD. 2007. Differential expression of Candida albicans secreted aspartyl proteinase in human vulvovaginal candidiasis. Mycoses 50:383–390. doi: 10.1111/j.1439-0507.2007.01384.x. [DOI] [PubMed] [Google Scholar]
- 6.Stehr F, Felk A, Gacser A, Kretschmar M, Mahnss B, Neuber K, Hube B, Schafer W. 2004. Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res 4:401–408. doi: 10.1016/S1567-1356(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 7.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonzi WA. 2009. The protein secretory pathway of Candida albicans. Mycoses 52:291–303. doi: 10.1111/j.1439-0507.2008.01673.x. [DOI] [PubMed] [Google Scholar]
- 9.Nombela C, Gil C, Chaffin WL. 2006. Non-conventional protein secretion in yeast. Trends Microbiol 14:15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, Zancope-Oliveira RM, Almeida IC, Nosanchuk JD. 2008. Vesicular transport in Histoplama capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol 10:1695–1720. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vargas G, Rocha JD, Oliveira DL, Albuquerque PC, Frases S, Santos SS, Nosanchuk JD, Gomes AM, Medeiros LC, Miranda K, Sobreira TJ, Nakayasu ES, Arigi EA, Casadevall A, Guimaraes AJ, Rodrigues ML, Freire-de-Lima CG, Almeida IC, Nimrichter L. 2015. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol 17:389–407. doi: 10.1111/cmi.12374. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues ML, Nakayasu ES, Almeida IC, Nimrichter L. 2014. The impact of proteomics on the understanding of functions and biogenesis of fungal extracellular vesicles. J Proteomics 97:177–186. doi: 10.1016/j.jprot.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YL, Montedonico AE, Kauffman S, Dunlap JR, Menn FM, Reynolds TB. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol Microbiol 75:1112–1132. doi: 10.1111/j.1365-2958.2009.07018.x. [DOI] [PubMed] [Google Scholar]
- 15.Wolf J, Espadas-Moreno J, Luque-Garcia JL, Casadevall A. 2014. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot Cell 13:1484–1493. doi: 10.1128/EC.00111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR Jr, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest 121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardo SM, Lee SA. 2010. Candida albicans SUR7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol 10:133. doi: 10.1186/1471-2180-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibellini F, Smith TK. 2010. The Kennedy pathway—de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 19.Vallejo MC, Nakayasu ES, Longo LV, Ganiko L, Lopes FG, Matsuo AL, Almeida IC, Puccia R. 2012. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One 7:e39463. doi: 10.1371/journal.pone.0039463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. 2009. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. 2008. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell 7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJ, Longo LV, Ganiko L, Almeida IC, Puccia R. 2012. Vesicle and vesicle-free extracellular proteome of Paracoccidiodes brasiliensis: comparative analysis with other pathogenic fungi. J Proteome Res 11:1676–1685. doi: 10.1021/pr200872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, Kimberly RP, Underhill D, Cruz PD Jr, Ariizumi K. 2006. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J Biol Chem 281:38854–38866. doi: 10.1074/jbc.M606542200. [DOI] [PubMed] [Google Scholar]
- 24.Jouault T, Ibata-Ombetta S, Takeuchi O, Trinel PA, Sacchetti P, Lefebvre P, Akira S, Poulain D. 2003. Candida albicans phospholipomannan is sensed through toll-like receptors. J Infect Dis 188:165–172. doi: 10.1086/375784. [DOI] [PubMed] [Google Scholar]
- 25.Suetsugu S, Kurisu S, Takenawa T. 2014. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev 94:1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- 26.Janmey PA, Kinnunen PK. 2006. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol 16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo Y, Fisher E, Patton-Vogt J, Marcus S. 2007. Functional characterization of the fission yeast phosphatidylserine synthase gene, pps1, reveals novel cellular functions for phosphatidylserine. Eukaryot Cell 6:2092–2101. doi: 10.1128/EC.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairn GD, Hermansson M, Somerharju P, Grinstein S. 2011. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat Cell Biol 13:1424–1430. doi: 10.1038/ncb2351. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. 2008. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol 180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitarch A, Sanchez M, Nombela C, Gil C. 2002. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol Cell Proteomics 1:967–982. doi: 10.1074/mcp.M200062-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poulain D, Slomianny C, Jouault T, Gomez JM, Trinel PA. 2002. Contribution of phospholipomannan to the surface expression of beta-1,2-oligomannosides in Candida albicans and its presence in cell wall extracts. Infect Immun 70:4323–4328. doi: 10.1128/IAI.70.8.4323-4328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis SE, Hopke A, Minkin SC Jr, Montedonico AE, Wheeler RT, Reynolds TB. 2014. Masking of beta(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect Immun 82:4405–4413. doi: 10.1128/IAI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallejo MC, Matsuo AL, Ganiko L, Medeiros LC, Miranda K, Silva LS, Freymuller-Haapalainen E, Sinigaglia-Coimbra R, Almeida IC, Puccia R. 2011. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-galactosyl epitopes. Eukaryot Cell 10:343–351. doi: 10.1128/EC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveira DL, Rizzo J, Joffe LS, Godinho RM, Rodrigues ML. 2013. Where do they come from and where do they go: candidates for regulating extracellular vesicle formation in fungi. Int J Mol Sci 14:9581–9603. doi: 10.3390/ijms14059581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues ML, Franzen AJ, Nimrichter L, Miranda K. 2013. Vesicular mechanisms of traffic of fungal molecules to the extracellular space. Curr Opin Microbiol 16:414–420. doi: 10.1016/j.mib.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.