Abstract
We recently showed that spinal cord injury (SCI) leads to a decrease in mRNA editing of serotonin receptor 2C (5-HT2CR) contributing to post-SCI spasticity. Here we study post-SCI mRNA editing and global gene expression using massively parallel sequencing. Evidence is presented that the decrease in 5-HT2CR editing is caused by down-regulation of adenosine deaminase ADAR2 and that editing of at least one other ADAR2 target, potassium channel Kv1.1, is decreased after SCI. Bayesian network analysis of genome-wide transcriptome data indicates that down-regulation of ADAR2 (1) is triggered by persistent inflammatory response to SCI that is associated with activation of microglia and (2) results in changes in neuronal gene expression that are likely to contribute both to post-SCI restoration of neuronal excitability and muscle spasms. These findings have broad implications for other diseases of the Central Nervous System and could open new avenues for developing efficacious antispastic treatments.
Severe Spinal Cord Injury (SCI) causes an immediate, devastating loss of motor function. This effect is triggered not only by damage to brain-derived axons that use fast glutamatergic synaptic transmission and facilitate voluntary initiation of movement, but also by loss of descending brainstem tracts which provide the spinal cord with its major source of neuromodulators such as serotonin (5-HT)1,2. Many of the neurons that coordinate rhythmic locomotor movements in mammals are located in the spinal cord. By activating specific receptors, serotonin facilitates voltage-gated persistent inward currents (PICs) of Ca2+ and Na+ in these spinal motoneurons, thus setting them into an excitable state3. Because of their unusually low threshold, the PICs are easily activated by brief synaptic inputs and play a crucial role in amplifying and prolonging the action of these inputs, ultimately enabling sustained muscle contractions4. When SCI eliminates brainstem-derived serotonin, motoneurons are left in an unexcitable state with small PICs, consistent with the flaccid paralysis and areflexia that are observed early after SCI5,6.
Surprisingly, despite the continued absence of serotonin, over the weeks after SCI, rudimentary locomotor-like movements spontaneously emerge, coinciding with recovery of motoneuron excitability, large PICs and associated sustained firing7. Unlike before injury, however, the powerful depolarizing action of PICs is difficult to terminate, because motoneurons have weaker inhibitory inputs from spinal interneurons that are themselves regulated by descending tracts6,8. Thus, in both humans9 and rats4,6, the PICs can lead to excessive motoneuron activity that produces uncontrolled and debilitating muscle spasms, which are readily triggered by SCI-enhanced synaptic inputs arising from normally innocuous cutaneous stimulation or muscle stretch10. The biological basis for the post-SCI recovery of spinal motoneuron activity remains unknown; however, recent studies have suggested that the serotonin receptors 2C (5-HT2CR) and 2A (5-HT2AR) on motoneurons become constitutively active (i.e., active in the absence of serotonin) to compensate for the loss of the brainstem neurotransmitter, ultimately facilitating the recovery of motoneuron excitability and related motor function, but also contributing to muscle spasms11,12.
5-HT2A/2CRs are two highly similar G-protein-coupled receptors that exhibit considerable constitutive activity and mostly signal by activating phospholipase C (PLC)-associated cascade13. Whereas the constitutive activity of 5-HT2AR is determined by the level of its expression, 5-HT2CR is regulated by editing of its pre-mRNA via adenosine-to-inosine (A-to-I) deamination catalyzed by RNA-specific adenosine deaminases (ADAR1 and ADAR2)14,15,16. In mammals, site-specific mRNA editing resulting in re-coding is known to occur in only a few transcripts that mostly encode CNS-expressed proteins involved in neurotransmission, including 5-HT2CR, several glutamate receptor subunits, α3 subunit of GABAA receptor, voltage-gated potassium (Kv1.1) and calcium (Cav1.3) channels17,18. Notably, the editing occurs at functionally critical positions of these proteins and hence substantially affects neuronal excitability and signal transduction.
The 5-HT2CR mRNA can be edited at up to five closely-spaced (within a 15 nucleotide sequence) adenine positions (denoted A, B, C, D and E sites). Because inosine is decoded as guanosine during translation, editing can alter codons for three amino acids in the second intracellular loop of the receptor19,20, a region involved in the coupling with G-proteins21. Combinatorial editing at the five sites can generate up to 32 mRNA variants encoding 24 receptor isoforms (sites A and B as well as sites E and C are in the same codons). The extent of editing is inversely correlated with the 5-HT2CR activity such that the more highly edited isoforms are less active than less extensively edited ones16. The unedited Ile156-Asn158-Ile160 (INI) isoform possesses considerable constitutive and agonist-stimulated activity. Conversely, when the mRNA is edited, the coupling of 5-HT2CR to G-proteins and its affinity for serotonin are drastically reduced. For example, compared to the INI isoform, the VSV (Val156-Ser158-Val160) isoform that can be encoded by two different mRNA variants, namely ACD (edited at sites A, C, and D) or ABCD (edited at sites A, B, C, and D), shows a 4-to-5-fold decreased coupling to signaling pathways and 4-fold reduced constitutive activity19,22.
Our recent study that used a chronic spinal rat model of spasticity5 has suggested that, months after sacrospinal transection, there is an alteration in 5-HT2CR mRNA editing which leads to increased expression of constitutively active receptor isoforms coinciding with restoration of large Ca2+ PICs in the motoneurons11. Blocking constitutively active 5-HT2CRs with inverse agonists attenuates these Ca2+ currents as well as muscle spasms11. This remarkable adaptation of motoneurons via editing potentially opens new possibilities for antispastic drug therapy.
The present report aimed to characterize the nature and the extent of RNA editing alterations after SCI and to elucidate the molecular pathways that trigger and follow these editing alterations. We present evidence that the post-SCI decrease of 5-HT2CR editing is due to down-regulation of ADAR2 (but not ADAR1) expression, and that SCI triggers editing changes in at least one additional ADAR2 target, namely Kv1.1 channel. To elucidate the pathways that are related to alteration of mRNA editing following SCI, we examine the genome-wide transcriptional response to SCI using RNA-seq in conjunction with sophisticated bioinformatic approaches, including weighted gene co-expression network and Bayesian network analyses.
Results
5-HT2CR Editing Is Altered in the Chronic Spinal Rat Model of Spasticity
We performed a detailed assessment of the SCI-induced alterations in 5-HT2CR mRNA editing in spinal cord samples obtained below the site of injury from sacrospinal (8 weeks post-SCI) and sham-operated (control) rats (N = 8 per group) using massively parallel sequencing (MPS) of PCR-amplified DNA fragments in the region of editing23,24. On average, 1,300,000 ± 80,000 (Mean ± SE) reads were obtained for each sample, enabling a reliable detection of all 32 possible mRNA variants (including the rarest ones25) in each sample (Fig. 1a). The frequencies of the variants within the sacral region were similar to those detected in the thoracic cord23: the ABD variant (edited at sites A, B, and D) accounted for ~36%, ABCD ~18%, and AB (edited at A, and B) ~14%, whereas the unedited version (NONE) accounted for only ~3% of all 5-HT2CR transcripts. Only five variants, ABD, ABCD, AB, ABC, and A, were observed at >5% frequencies. The principal component analysis (PCA) of the 5-HT2CR variants showed that the ABCD and AB aligned along the first PC, which by itself explained 76.3% of the total data variance (Fig. 1b). With the exception of one outlier, the SCI and control animals were perfectly segregated along the first PC, with SCI rats having lower values of ABCD and higher values of AB compared to controls.
Figure 1. Analysis of 5-HT2CR editing in the spinal cord.
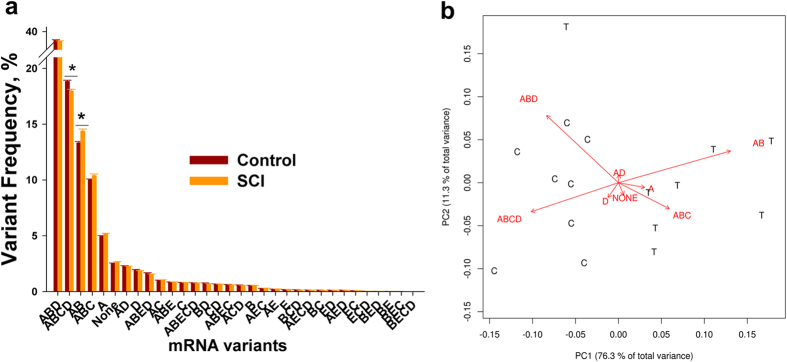
(a) Shown are Means ± SE of the 5-HT2CR editing variant frequencies in the spinal cord of the SCI and control rats (N = 8 rats per group) obtained by MPS. *indicates a significant difference between the groups (see Fig. 2A for details). (b) PCA of the data in (a). Shown are first two PCs of all 5-HT2CR editing variants; ABCD and AB variants are aligned along the first principal component, which by itself explains ~ 76.3% of the total data variability. T - treatment (SCI), C - control.
We found that the frequency of ABCD (which encodes a low-functioning receptor isoform VSV) was significantly lower, and the frequency of AB (which encodes a high-functioning isoform VNI) was significantly higher in the SCI vs. control rats (Figs 1a and 2a). We also found that, although there were no differences in editing efficiencies at the A or B as well as at the E or C sites (not-shown), significantly lower editing efficiency was observed at the D site of the SCI rats (Fig. 2b). Thus, SCI leads to a decrease in 5-HT2CR mRNA editing below the site of injury that is mostly accounted for by the lower editing level at the D site.
Figure 2. Alteration of 5-HT2CR RNA editing after SCI.
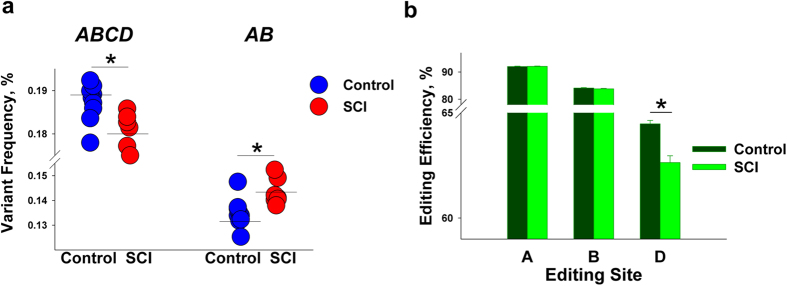
(a) 5-HT2CR mRNA variants: ABCD is decreased and AB is increased in SCI rats (p-values < 3.2e-4). Mean frequency values in each group are shown as horizontal lines. (b) 5-HT2CR editing sites: Editing efficiency is decreased at site D in SCI rats (p = 2e-4). (a,b) N = 8 per group; *indicates a significant difference between the groups.
SCI Induces Alterations of ADAR2 Expression and Kv1.1 mRNA Editing
In 5-HT2CR, sites A and B are predominantly edited by ADAR1 whereas site D is mostly edited by ADAR216. Therefore, we investigated whether the observed SCI-induced editing alterations correlate with changes in the expression of editing enzymes. In line with our finding of decreased editing at the D site, but not at the A or B sites, we detected significantly lower expression of Adarb1 (that encodes ADAR2) but not of Adar (that encodes ADAR1) in the SCI rats (Fig. 3a).
Figure 3. Alterations of ADARs’ mRNA expression and Kv.1.1 mRNA editing after SCI.
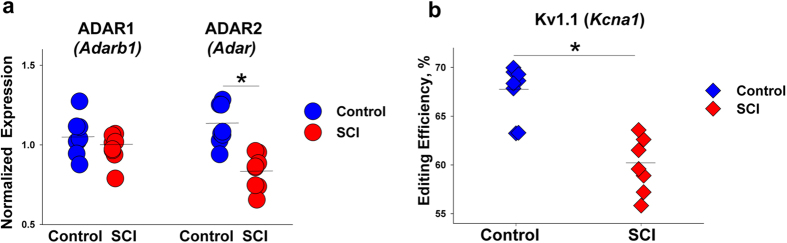
(a) ADAR2 (encoded by Adarb1) mRNA expression was decreased (p = 2.2E-4) and ADAR1 (encoded by Adar) mRNA expression did not differ between SCI vs. control rats. The measurements were performed by qPCR. Expression values were normalized to geometric mean of 3 endogenous control genes—Gapdh, Actb, and Ppia. (b) Kv1.1 (encoded by Kcna1) mRNA editing is decreased in SCI vs. control rats (p = 3.3E-4). Measurements of Kcna1 editing were performed using Mfe1 restriction assay (see Methods). (a,b) N = 8 per group; *indicates a significant difference between the groups. Mean expression (a) or editing efficiency (b) values in each group are shown as horizontal lines.
We next tested if SCI triggers editing changes in another protein, Kv1.1 channel encoded by Kcna1, which could contribute to the recovery of motoneuron excitability and the concurrent spasticity. The Kv1.1 mRNA is edited by ADAR2 at a single site resulting in a Val-to-Ile substitution26. We detected a significant decrease of the Kcna1 mRNA editing in the SCI rats (Fig. 3b). Thus, in addition to 5-HT2CR, SCI induces editing changes in at least one other protein whose mRNA is edited by ADAR2 and that plays an important role in neurotransmission.
SCI Induces Alteration of 5-HT2AR Expression
Because 5-HT2AR mRNA does not undergo editing, its constitutive activity is mostly determined by the expression level. Using qPCR, we investigated if mRNA expression of 5-HT2CR or 5-HT2AR (encoded by Htr2c and Htr2a, respectively) is influenced by SCI. Whereas there were no differences in the expression of Htr2c (which represented the total expression of all 5-HT2CR mRNA variants), we detected significantly higher expression of Htr2a in SCI animals (Fig. 4). This finding was in line with the results of previous studies which reported post-SCI up-regulation of 5-HT2AR mRNA and protein12,23.
Figure 4. Expression of 5-HT2AR and 5-HT2CR following SCI.
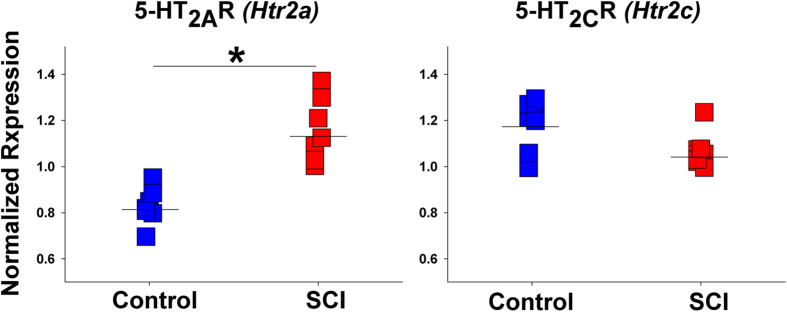
5-HT2AR (encoded by Htr2a) mRNA expression was increased (p = 4.6e-5) and 5-HT2CR (encoded by Htr2c) mRNA expression did not differ between SCI vs. control rats. N = 8 rats per group. The measurements were performed by qPCR as in Fig. 3. *indicates a significant difference between the groups. Mean expression values in each group are shown as horizontal lines.
Global Transcriptional Alterations Following SCI
We then assessed post-SCI transcriptional alterations genome-wide, using RNA-seq of the same RNA preparations that were used in the analysis of editing described above. A total of 14,638 genes were found to be expressed at a detectable level in all animals. Among these genes, 981 were differentially expressed (DE) between SCI vs. control rats: 245 transcripts were down-regulated and 736 were up-regulated (see Supplementary Table1 and examples in Fig. 5a). Although not significant after the adjustment for multiple comparison, expression of Adarb1 was decreased (adjusted p = 0.06) and expression of Htr2a was increased (adjusted p = 0.1) in the SCI animals, compatible with our qPCR data.
Figure 5. Transcriptional alterations following SCI.
(a) The RNA-Seq data in the vicinity of loci for Trem2 and Phox2a. The traces of RNA abundance show a significantly higher expression of Trem2 and significantly lower expression of Phox2a in SCI vs. control rats. For visualization, the data were uploaded to Integrative Genomics Viewer (IGV) visualization tool57. (b) WGCNA analysis of DE genes. Clustering of the DE genes by topological overlap reveals 17 gene modules that are characterized by distinct expression patterns comprised of highly co-regulated genes. The color intensity in the heatmap is proportional to the interaction strength between two genes with red color for the strongest interaction and white color for no interaction. The 17 modules are represented by the color bars along the X- and Y-axes. (C) Functional enrichment for genes within Turquoise and Blue Modules shown in (b). Depicted are five most significant GO Processes identified by MetaCore that are enriched within the Turquoise and Blue modules and most significant networks within several GO processes (encircled).
Because of the large number of the DE transcripts, we next applied weighted gene co-expression network analysis (WGCNA) to characterize functional organization of the SCI-induced transcriptional alterations27. WGCNA identifies putative biologically relevant changes in gene expression by grouping genes into modules with strongly covarying patterns across the samples. Consequently, WGCNA can distinguish gene expression networks associated with specific cell types (e.g., neurons, oligodendrocytes, astrocytes, or microglia) that are present in a heterogeneous sample (e.g., brain or spinal cord) thanks to the distinct transcriptional profiles of these cell types and variation in their relative proportions across the samples28. The gene co-expression modules can be examined for cell-type specificity using cell type-enriched gene sets identified in previous studies29,30,31. WGCNA also enables the identification of modules of functionally related, highly coexpressed genes within different cellular populations28, and has been successfully employed to identify novel pathways and target genes for complex human diseases such as cancer, Alzheimer’s disease, obesity and diabetes32,33,34.
Among the DE genes, we identified 17 co-expression modules, 8 of which included more than 50 genes (Fig. 5b, Supplementary Table2). We examined the two major modules (Turquoise and Blue), which contained 197 and 148 DE genes, respectively. The majority of the Turquoise Module genes (189 of 197) were up-regulated whereas the majority of the Blue Module genes (144 from 148) were down-regulated following SCI (Bonferroni corrected enrichment p-values < 1e-100). Examination of functional annotations using GeneGo MetaCore software revealed that the Turquoise Module was significantly enriched in genes related to immunity, probably reflecting the SCI-induced inflammatory response, whereas the Blue Module was enriched in genes related to neurotransmission and lipid metabolism (Fig. 5c; Supplementary Table3). More detailed analysis revealed several networks associated with neurotransmission-related Gene Ontology categories including potassium ion transport, PLC-activating serotonin receptor signaling pathway, and positive regulation of cytosolic calcium ion concentration (all p values < 4.0 × 10−12) (Fig. 5c).
We then explored cell-specificity of the WGCNA modules by measuring the overlap between the module genes and cell type-specific gene signatures for neurons, oligodendrocytes, astrocytes, and microglia from the Allen Brain Atlas (http://www.brain-map.org/). The signature of neuronal and oligodendrocyte genes was significantly enriched only in the Blue Module, whereas microglia and astrocyte genes were enriched in multiple modules (Turquoise, Brown, Grey, Yellow, Green, and Red) (Supplementary Table4).
In summary, transcriptional changes following SCI clearly revealed at least two distinct components. The first component is related to the long-lasting inflammatory response (that is still present 8 weeks post-SCI) and is enriched in microglia- and astrocyte-specific genes. The second component is related to post-SCI dysregulation of neurotransmission and is represented by genes that are mostly expressed in neurons and oligodendrocytes.
Persistent spinal microgliosis 8 weeks post-SCI
We then assessed the involvement of neuroinflammation in the observed transcriptional alterations using Iba-1 immunohistochemistry in the sacral spinal cord 8 weeks after SCI. Iba-1 expression is up-regulated upon activation of microglia due to inflammation; Iba-1 is also expressed by infiltrating macrophages. We detected increased staining intensity, rounder cell bodies, and shorter and thicker processes in SCI rats both rostral and caudal to the site of injury, which is indicative of substantial microgliosis (Fig. 6). Notably, the entire caudal cord expressed similar gliosis, whereas the rostral cord showed gliosis up to 2.6 mm above the injury. Also, caudal to the injury, the enhanced Iba-1 expression was not changed for at least 6 months (longest point tested) after SCI, whereas rostral to the injury, normal Iba-1 expression was detected at 6 months (not-shown). Approximately, twice as many Iba-1 expressing cells were found rostral and caudal to the injury compared to the intact spinal cord. However, we could not determine from these data whether the cells were resident microglia or infiltrating macrophages from the periphery.
Figure 6. Persistent spinal microgliosis 8 weeks post-SCI.
Fluorescence microscope images of longitudinal spinal cord tissue sections in a control (intact) (a,c,e) and an SCI (injured) rat 8 weeks after complete transection injury (b,d,f). In the SCI rats, the images were taken 0.6 mm above and below injury. Sections were stained with Iba-1 (green). (a) and (b) were taken at 50x magnification and encompass an entire section through the middle of the spinal cord, near the central canal. (c–f) were taken at 200x magnification in the grey matter. Microglia in the intact spinal cord (c,e) have elaborate thin processes which extend in various directions (arrow heads). Note increased staining intensity, rounder cell bodies and shorter and thicker processes (arrows) rostral (d) and caudal (f) to the site of injury. Insets show microglia cell morphology at 400x magnification. Scale bar (a,b), 250 μm. Scale bar (c–f), 75 μm.
Association between 5-HT2CR editing and genome-wide gene expression
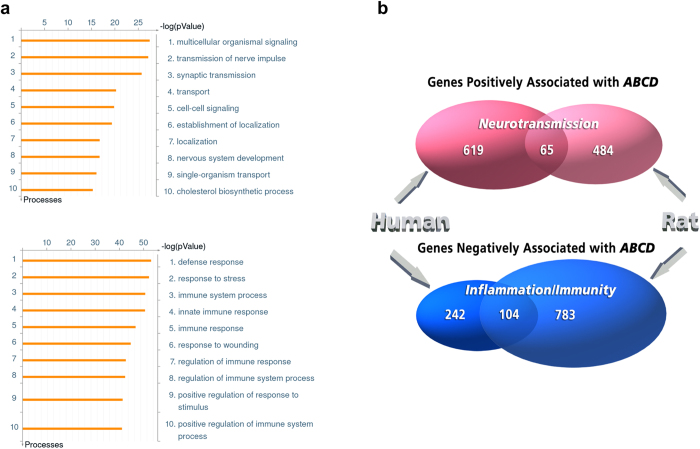
To investigate the pathways that are linked to 5-HT2CR editing, we performed an analysis of the association between editing and genome-wide gene expression using the RNA-seq data obtained from SCI and control rats (N = 16). Because >75% of the total variability of 5-HT2CR editing was explained by PC1 (dominated by the ABCD vs. AB axis) (Fig. 1), we used the frequency of the ABCD variant (hereinafter “ABCD”) as a proxy of the editing level in each animal. Among the 14,638 genes detected in all samples, expression of 1,436 genes showed a significant correlation with ABCD (FDR ≤ 0.1): 549 genes were positively and 887 were negatively correlated (Supplementary Table5). Functional annotation analysis of the positively associated genes revealed significant enrichment in genes implicated in transmission of nerve impulse, synaptic transmission and cholesterol biosynthesis (FDR < 1e-12), whereas negatively associated genes showed significant enrichment for genes involved in stress and immune response (FDR < 1e-50), (Fig. 7a and Supplementary Table6). Expression of Adarb1, but not Adar, was positively associated with ABCD.
Figure 7. Analysis of association between 5-HT2CR editing and genome-wide gene expression.
(a) Functional enrichment for genes that are negatively (upper panel) or positively (lower panel) associated with ABCD. Depicted are ten most significant GO Processes identified by MetaCore for each list of genes. (b) Venn diagrams depicting the overlap among genes that are positively or negatively associated with ABCD in the human PFC35 and in the rat model of SCI (this study). Shown are the numbers of genes as well as GO categories of enrichment.
We have recently reported an analysis of the association between genome-wide gene expression and ABCD in the autopsy specimens of the human prefrontal cortex (PFC)35. We identified 684 genes whose expression was positively and 346 genes whose expression was negatively associated with ABCD (positive and negative ABCD signatures, respectively). Similar to the present report, the positive signature was enriched in genes involved in synaptic transmission, whereas the negative signature was enriched in genes involved in inflammation and immunity. Also similar to the present study in rats, expression of ADARB1 (encodes ADAR2), but not ADAR (encodes ADAR1) was positively associated with ABCD in the human data set35.
Notably, 11,997 genes were expressed in both human PFC and rat spinal cord. We observed a significant overlap between the genes that were positively (N = 65; odds ratio = 3.34, 95% C.I: 2.49–4.42, p = 2.6e-14) or negatively (N = 104; odds ratio = 8.55, 95% C.I.: 6.59–11.05, p < 1e-15) associated with ABCD in both data sets (Fig. 7b Supplementary Table7). Functional annotation of these “positive” and “negative” overlapping genes showed enrichment in genes implicated in neurotransmission (whose products are localized in neurons, axons, dendrites and synapses) (FDR < 1e-4), and genes implicated in immune response (whose products are localized to lysosomes, lytic vacuoles, and phagocytic vesicles) (FDR < 1e-6), respectively (Supplementary Table8). Collectively, these findings suggest that 5-HT2CR editing in human and rat CNS is regulated via common pathways that at least in part converge at ADAR2.
Association between Adarb1 and genome-wide gene expression
We then sought to determine whether the observed association between ABCD and global patterns of gene expression also held for the expression of Adarb1 (the gene for ADAR2), under the premise that post-SCI changes in editing were likely to involve not only 5-HT2CR regulation but a larger network of ADAR2-related pathways. The frequency of ABCD was found to strongly correlate with Adarb1 expression measured by qPCR (r = 0.743, p = 3.1e-4), and the qPCR measurement of Adarb1 strongly correlated with its RNA-seq profile (r = 0.774, p = 6.8e-4). Analysis of the correlation between Adarb1 qPCR measurements and genome-wide gene expression yielded 667 genes that were positively and 1027 genes that were negatively correlated with Adarb1 (Supplementary Table9). Not surprisingly, given the strong correlation between Adarb1 and ABCD, there was a substantial overlap between the sets of genes whose expression was positively or negatively associated with both ABCD and Adarb1 (322 and 676 genes, respectively; hypergeometric p-values < 1e-100). Accordingly, functional annotation of the genes that were positively or negatively associated with Adarb1 revealed significant enrichment for genes in similar categories that were detected for ABCD vs. gene expression analysis described above (Supplementary Table10).
Thus, we detected two large, substantially overlapping sets of genes for which expression was associated with the 5-HT2CR editing (ABCD) and/or Adarb1 level. Within each set, the genes that were positively associated with ABCD or Adarb1 showed enrichment for functional categories related to neurotransmission, whereas the negatively associated genes were enriched in categories related to immune response.
Bayesian Network analysis
We then applied Bayesian network analysis (BNA), which reveals causal relationships among variables36, to identify genes that are “upstream” or “downstream” of Adarb1, using RNA-seq and qPCR Adarb1 expression data obtained from SCI and control animals. The “upstream” genes might affect Adarb1 expression (and consequently ADAR2-dependent mRNA editing); conversely, the “downstream” genes could be affected by Adarb1 and editing. Among 14,638 analyzed genes, 10,071 showed no direct relationship with Adarb1 and were classified as “independent”, 902 genes were identified as “upstream” and 2,001 genes as “downstream” of Adarb1, whereas 1,664 genes could not be assigned to any of these 3 categories and remained “ambiguous” (Supplementary Table11).
In an attempt to identify functionally relevant genes, we restricted further analysis to those “Adarb1 upstream” and “Adarb1 downstream” genes whose expression was significantly (FDR < 0.1) correlated with Adarb1 expression (Supplementary Table10). Among the 902 “upstream” genes, 57 were positively and 270 were negatively correlated with Adarb1 (hereinafter, “upstream positive” and “upstream negative” genes, respectively) (Supplementary Table12–13). Among the 2,001 genes “downstream” of Adarb1, 258 were positively and 305 were negatively correlated with Adarb1 (hereinafter, “downstream positive” and “downstream negative” genes, respectively) (Supplementary Table14–15).
Functional annotation of these 4 sets of genes yielded information on the pathways that appear to be upstream or downstream of Adarb1 expression (Supplementary Tables12-15). The “upstream positive” gene set was enriched in genes involved in the metabolism of cholesterol and other lipids as well as in categories related to transmission of nerve impulse and action potential. Along with Adarb1, many of these genes were down-regulated after SCI, e.g. oligodendrocyte-specific proteins (Mag, Mbp), proteins related to cholesterol biosynthesis (Dhcr24, Fdft1d, Hmgcs1, Dhcr7) and a potassium voltage gated channel, Kcns3. Altogether, 35 of the 57 “upstream positive” genes were down-regulated post-SCI, whereas none of them were up-regulated. Because myelin has the highest cholesterol content in the CNS37, these findings might reflect shutdown of lipid biosynthesis caused by the post-SCI oligodendrocyte death and demyelination resulting in increased concentration of free cholesterol as well as by the subsequent axonal conductance block38.
The “upstream negative” set was enriched in genes involved in immune response and its regulation. This finding reflects the long-lasting cellular and molecular inflammatory response induced by SCI46, which was exemplified by genes that encode neutrophil cytosolic factors (e.g., Ncf1, Ncf4), cytokines, cytokine receptors and related proteins (e.g., Tnf, Tnfrsf9, Tnfrsf1b, Il16, Il10ra, Il4ra, Ifitm1, Irf8, Irf5, Ifi30, Ifi47, Ifngr1), neurotrophic factors and their receptors (e.g., Tgfbr1, Tgfb1, Ptafr), matrix metalloproteinases (e.g., Mmp19, Mmp3), complement components and proteins that are involved in regulation of complement activation (e.g., Cfh, C1r, C1s, C2, C1qc, C1qa, C1qb), and other proteins that are expressed by activated microglia and monocyte-derived macrophages (e.g., Cd86, Trem2, Tlr7, Tlr1, Tlr2) and T lymphocytes (Cd4). The majority of the “upstream negative” genes (249 of 270), including the mentioned above, were up-regulated following SCI, and none of them were down-regulated.
The “downstream positive” set was enriched in genes involved in synaptic transmission and its regulation as well as synapse organization, and regulation of ion transmembrane transport. These categories are exemplified by genes that encode calcium channels (Cacna2d2, Cacng2, Cacng7), potassium channels (Kcnab3, Kcnc1, Kcnc3, Kcnj12, Kcnj9, Kcnk10, Kcnt1, Hcn3), glutamate receptors (Grin2d, Grin3b), adaptor protein in the postsynaptic density of excitatory synapses (Shank1), several guanine nucleotide-binding proteins (Gnal, Gnao1, Gnaz, Gng7, Gng8), and a voltage gated sodium channel (Scn4b). Expression of only 10 of the 258 “downstream positive” genes was altered post-SCI, and all these 10 genes (including Kcnc3, Grin3b, Gnal, Scn4b, Panx2) were down-regulated. Notably, Grin3b encodes a motoneuron-specific NMDA receptor modulatory subunit that reduces the NMDA conductance as well as calcium permeability39. The “downstream negative” set was enriched in genes involved in broad categories of response to wounding and wound healing, response to stress as well as in more specific categories of polysaccharide catabolism. Among the 305 “downstream negative” genes, 49 were up-regulated following SCI, and none were down-regulated. Interestingly, one of these 49 genes, Maob, encodes the mitochondrial monoamine oxidase which is found in neurons and astrocytes and catalyzes the oxidation of monoamines, including serotonin and epinephrine40. This finding suggests that the expression of Maob could be influenced by Adarb1 levels.
Discussion
In more than 80% of individuals with SCI, the injury is followed by development of debilitating spasms and spasticity in muscles innervated by spinal motorneurons below the site of injury5. Treatment of spasms with conventional antispastic drugs (e.g., baclofen) is often not efficacious or is not tolerated because of adverse side effects such as lethargy and weakness41. Post-SCI spasticity has multiple underlying mechanisms, and serotonin and its receptors play an important role in the etiology of this condition42. Recent reports suggest that 5-HT2A and 5-HT2C receptors on spinal motoneurons become constitutively active after SCI to compensate for the loss of brainstem serotonin, thus helping to recover motoneuron excitability and rudimentary locomotor function, yet contributing to muscle spasms11,12.
Whereas the post-SCI increase in 5-HT2AR constitutive activity has been reported to be achieved by up-regulation of its expression12, as also confirmed in this work, our recent study has presented evidence that the constitutive activity of 5-HT2CR was increased via alterations of mRNA editing11. Here we examined post-SCI changes of 5-HT2CR mRNA editing using MPS that yields numerous sequencing reads per editing region and therefore shows substantially increased precision and sensitivity25 compared to the cloning and sequencing method that was used in the initial study11. Although MPS revealed a pattern of the receptor variants that was significantly different from those reported previously11, the major findings of the initial study are corroborated in that the additive effect of the alterations we detected should be a substantially higher constitutive 5-HT2CR activity in the SCI rats. Most importantly, MPS demonstrated that the post-SCI decrease of 5-HT2CR editing involves only one of the 5 editing sites on the receptor mRNA, namely site D. Because site D is mostly edited by ADAR2, we predicted and indeed detected post-SCI down-regulation of ADAR2 but not ADAR1. Although correlation is not equal to causal relationship, it seems most likely that the decrease of ADAR2 activity in SCI rats indeed causes the drop in editing at site D.
We also demonstrated that, in addition to 5-HT2CR, SCI triggers editing changes in at least one more ADAR2 target, potassium voltage-gated channel Kv1.1. This channel gives rise to a rapidly activated sustained outward current and plays an essential role in initiation and shaping of action potentials43. In spinal motoneurons, Kv1.1 is localized mostly to axon initial segments, where it is critical for dampening neuronal excitability44. In addition to membrane potential, Kv1.1 is regulated by its accessory inhibitory β subunit45. Kv1.1 editing dramatically affects the binding of β subunit to Kv1.1 protein, thus accelerating its recovery from inactivation at negative potentials and reducing the frequency of motoneuron firing26. Therefore, a post-SCI decrease in Kv1.1 editing would enhance motoneuron excitability, which might contribute to spasms and spasticity in individuals with SCI. This prediction is in line with the earlier findings showing that the voltage threshold for the action potential is lower in motoneurons of chronic vs. acute spinal rats45 which could be, at least in part, explained by reduced Kv1.1 delayed rectifier current due to the decrease in Kv1.1 editing after SCI. Given the post-SCI down-regulation of ADAR2, it seems likely that in addition to 5-HT2CR and Kv1.1, SCI triggers editing changes in other receptors and/or channels which collectively contribute to the recovery of motoneurons and the concurring development of spasticity. Future work is expected to identify the entire spectrum of effects and the cellular specificity of ADAR2 down-regulation as well as its role in the pathophysiology of the SCI.
We detected numerous genes whose expression is altered 8 weeks after SCI. Analysis of these DE genes using WGCNA (which enables inference of the functional relevance of genes based on their network position27) revealed at least two distinct components. The first component appears to reflect transcriptional alterations associated with the profound and pervasive consequences of lesional inflammation caused by SCI, including activation and proliferation of resident microglia and astrocytes, infiltration of circulating innate immune cells (i.e., neutrophils, monocytes and lymphocytes), and enhanced intraspinal synthesis and release of cytokines, chemokines and other vasoactive substances (e.g., complement proteins) by neurons and non-neuronal cells46. This component shows enrichment for microglia- and astrocyte-specific genes, which are mostly up-regulated following SCI.
The second component apparently reflects post-SCI dysregulation of neurotransmission and lipid metabolism, and is represented by genes that are expressed in neurons or oligodendrocytes and are mostly down-regulated following SCI. Distinctly, the network analysis of the neurotransmission-related genes (see Fig. 5c) are compatible with the prediction that post-SCI adaptations of motoneurons are, at least in part, driven by the increased constitutive activity of 5-HT2A/2CRs, which would enhance signaling through the PLC pathway13, thus mobilizing intracellular Ca2+, activating L-type Ca2+ channels and facilitating Ca2+ PICs47. The changes in the expression of lipid biosynthesis genes most likely are associated with the extensive oligodendrocyte cell death and demyelination after SCI.
We also detected numerous genes whose expression is positively or negatively associated with the level of 5-HT2CR editing in the rat spinal cord. The negatively correlated genes (which included Adarb1, but not Adar) are mostly related to inflammatory/immune response, whereas the positively correlated genes are mostly related to neurotransmission. These findings are fully compatible with the results of our recent 5-HT2CR editing study in the human PFC35. We also found a significant overlap between genes from the human and rat data sets that were positively or negatively associated with ABCD as well as significant overlap between genes that were correlated with both ABCD and Adarb1 in the rat study. Taken together, these findings suggest that down-regulation of ADAR2 and the consequent decrease of 5-HT2CR and Kv1.1 editing are triggered by persistent post-SCI inflammatory response that is mediated by activation and proliferation of resident microglia and astrocytes and/or by post-SCI changes in neurotransmission which are associated with neurons and oligodendrocytes. In an attempt to distinguish between these mechanisms, we applied BNA that is designed to test for cause-and-effect relationships among variables. The BNA results strongly suggest that down-regulation of ADAR2 is mostly caused by inflammatory response triggered by SCI as well as by processes related to oligodendrocyte death and demyelination. The latter, at least in part, could be exacerbated by inflammation (Fig. 8). Notably, down-regulation of ADAR2 and the associated decrease of GluR2 editing is a profound pathological change that is relevant to the death of spinal cord motorneurons in amyotrophic lateral sclerosis (ALS)48. Although ALS had been once considered a motoneuron disease, it has been shown that motoneuron death is driven by a convergence of damaging mechanisms, including glial cell pathology and inflammatory conditions, such as microglial activation49. Oligodendrocyte dysfunction is also prevalent in ALS as indicated by gray matter demyelination that is observed in motor cortex and spinal cord of ALS patients50. Thus, similar mechanisms could be involved in down-regulation of ADAR2 in ALS and spinal cord injury. Furthermore, the association between ADAR2 and inflammation might provide a link to other CNS diseases (e.g., multiple sclerosis) in which inflammation is paralleled by spasticity.
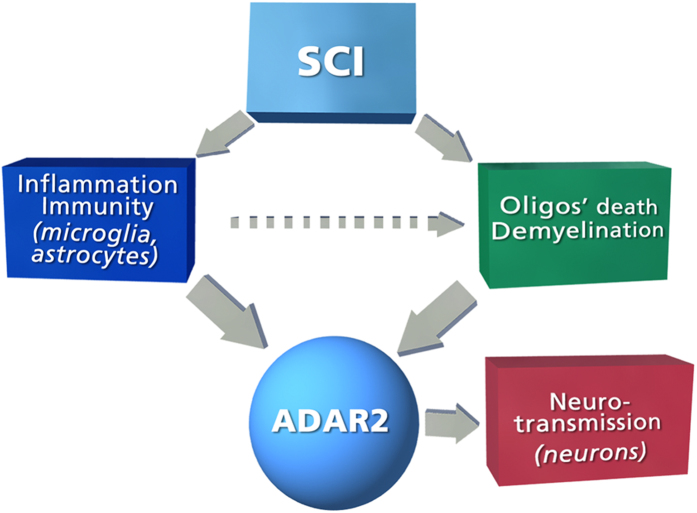
Figure 8. Schematic representation of the BNA results.
SCI causes persistent inflammatory response which is maintained after 8 weeks in the spinal rats and includes activation and proliferation of resident microglia and astrocytes46. It also causes oligodendrocyte death and demyelination that are exacerbated by the inflammation38. Our BNA analysis suggests that SCI-induced down-regulation of the editing enzyme ADAR2 (and consequent decrease in editing of ADAR2 targets) is caused by these processes. BNA also suggests that post-SCI alterations in neuron-specific processes are downstream from ADAR2 and its targets. These downstream processes are likely to contribute to post-SCI recovery of motoneuron excitability but also to muscle spasms and spasticity.
BNA also indicates that post-SCI alterations in neuron-specific processes (e.g., synaptic transmission, synapse organization, and regulation of ion transmembrane transport) are mostly downstream of ADAR2 and consequently its editing targets. RNA editing, including ADAR2-dependent editing, is thought to act as a homeostatic mechanism that modulates the effect of a wide range of environmental and genetic factors that impair neuronal function35,51,52. Such a buffering function of ADAR2 is compatible with our observation that expression of many downstream genes is correlated with ADAR2 expression but independent of SCI. Conversely, SCI-associated changes in the expression of certain downstream genes (i.e., Kcnc3, Grin3b, Gnal, Scn4b, Panx2, Vegf), which are apparently mediated by the down-regulation of ADAR2 expression, likely reflect SCI-induced perturbations of the homeostasis. These downstream genes could belong to the network that contributes to post-SCI recovery of motoneuron excitability and related motor function but also to muscle spasms and spasticity. This network is apparently regulated by altered editing of ADAR2 targets (5-HT2CR, Kv1.1, and possibly, others), and characterization of this network is a goal of our future work.
Conclusions
In this study we present evidence that the decrease of 5-HT2CR editing after SCI is most likely mediated by down-regulation of the editing enzyme ADAR2. We additionally found that SCI causes a substantial decrease in the mRNA editing level of at least one more ADAR2 target, Kv1.1 channel. Bioinformatic analysis of editing and genome-wide transcriptome data indicates that down-regulation of ADAR2 and the ensuing decrease of editing in its targets is triggered by the persistent inflammatory response to SCI that is manifest in a long-term activation of microglia. The results of this analysis further suggest that down-regulation of ADAR2 after SCI results in significant changes of the expression of multiple genes in neurons. The known functions of the affected genes implicate them in the restoration of neuronal excitability that is also associated with post-SCI spasms, a finding that could have broad implications for other diseases and injuries of the nervous system. The results of this work start to elucidate the specific molecular mechanisms that are associated with alterations of RNA editing in SCI and ultimately could lead to effective anti-spasticity treatments.
Methods
Sacral spinal injury model in rat and relation to human spasticity
In our sacral spinal injury model, only tails (not bladder or hindlimbs) are affected in injured animals5. Previously, we have shown that the spasticity that emerges in the tail of chronic spinal rats closely mimics the human spasticity syndrome seen in the legs after SCI, with clonus, hypertonus, hyperreflexia, spasms, and general delayed onset of symptoms5. In particular, flexor spasms emerge first, followed by extensor spasm (months), as also seen in humans. Finally, we have recently shown that the same ionic mechanism that mediates spasms in the tail of rats also mediates spasms in humans (PICs in motoneuron)9, thus justifying the use of the sacral spinal rat as a model of SCI-induced spasticity in humans.
In the current study we used both normal and spastic (with chronic spinal cord injury) female Sprague-Dawley rats. For the spastic rat, a complete spinal cord transection was made at the S2 sacral level when the rats were 7–8 weeks old, as previously described5. Briefly, under general anesthetic (sodium pentobarbital, 58.5 mg·kg–1) and sterile conditions, a laminectomy was performed on the L2 vertebrae to expose the S2 spinal cord. The dura was slit transversely, and 0.1–0.3 ml Xylocaine (1%) was applied topically. Under a surgical microscope, the spinal cord was transected by holding the pia with forceps and sucking under the pia with a fine suction tip. Caution was needed to avoid damaging the anterior artery or posterior/dorsal vein, since the sacrocaudal spinal cord dies without this midline vasculature. The dura was closed with two 8-0 silk sutures, and the muscle layers and skin were tightly sutured over the cord, and the rat allowed to recover. Usually within 30 days dramatic spasticity in the tail muscles (which are innervated by sacrocaudal motoneurons below the level of the injury) was developed in the injured rats. Only animals with clear signs of spasticity were included in the study (see5 for details of spasticity assessment).
Spinal rats were euthanized 8 weeks post injury and control animals were euthanized when they were 15 weeks old using urethane (0.18 mg per 100 g). The lumbosacral spinal cord was then exposed by scissor laminectomy, excised and transferred to a dissection dish containing RNA stabilizer (RNAlater, Life Technologies) for removal of roots and blood. The cord extraction procedure was limited to ~ 5min in order to preserve the integrity of RNA. To further minimize possible RNase contamination, all tools, dishes, and gloves were autoclaved and treated with RNaseZap (Life Technologies). Finally, the cords were cut into lumbar and sacral sections, placed in Eppendorf tubes, frozen in liquid nitrogen, and stored at 75 °C. All animal use and procedures were performed in accordance with Canadian Council for Animal Care guidelines, and all experimental protocols were approved by the University of Alberta animal welfare committee
RNA extraction and cDNA synthesis
Total RNA was extracted using Invitrogen ToTally RNA Kit (Life Technologies) that includes two rounds of phenol-based extractions and DNase treatment Only samples that yielded high quality RNA [RNA integrity number (RIN) ≥ 8.0] by 2100 Bioanalyzer (Agilent)] were used. The average RIN for the specimens was 8.73 ± 0.09 (Mean ± SEM). cDNA was synthesized from equal quantities of RNA from each animal (500 ng of RNA per 10 μl of reaction) using High Capacity cDNA Reverse Transcription kit (Life Technologies).
Analysis of 5-HT2CR editing
5-HT2CR RNA editing was assessed using massively parallel sequencing (MPS) of PCR-amplified DNA fragments in the region of editing as previously described23,35. Because inosine is read as guanine during reverse transcription, MPS provided the number of reads for each of the 32 5-HT2CR mRNA editing variants in each spinal cord sample. The relative frequency of each variant in each sample was calculated as the ratio of the number of reads detected for a particular variant to the total number of reads for all variants in this sample.
Adarb1 and Adar mRNA expression was measured by real time quantitative PCR (qPCR) using an ABI 7900 thermocycler and TaqMan gene-specific assays (Life Technologies) as described53. Expression of the target transcripts were normalized to the geometric mean of three endogenous control genes (ECG): Glyceraldehyde 3-phosphate dehydrogenase (Gapdh), beta-actin (Actb), and cyclophilin A (Ppia). The relative expression of the target transcripts and ECGs were determined using the Relative Standard Curve Method, which provides accurate quantitative results by accounting for differences in the efficiencies between target and ECG amplifications53.
Analysis of Kcna1 editing
To assess the level of Kcna1 editing, we employed a modified Mfe1 restriction assay that is commonly used for measuring mRNA editing when it occurs at a single site26. Kcna1 editing results in re-coding from Ile to Val. The editing event destroys an MfeI restriction enzyme recognition site. A constitutive MfeI site is also found 50 bp upstream of the editing site. Amplification products resulting from RT-PCR digested with MfeI will give two products differing by 50 bp. Specifically, we used forward CACTCCAAGGGCCTTCAGATCCTG and reverse CTGTCAGAGGCTAAGTTAGGAGAACTAACA primer pair which results in 390bp amplicon. Digestion of the “unedited” amplicon with Mfe1 generates 220 bp, 51 bp, and 119 bp DNA fragments, whereas digestion of “edited” amplicon yields only two fragments (220 bp and 170 bp). For each spinal cord sample quantified, two independent PCRs were carried out each time in duplicate. The resulting amplicon was cut with MfeI and analyzed using Bioanalyzer 2100 (Agilent). The ratio between the 171 bp and 120 bp fragments defined the efficiency of Kcna1 editing.
Iba-1 Immunohistochemistry
SCI and control rats were euthanized with a pentobarbital overdose (at 240 mg/kg) 8 weeks post-SCI. Spinal cords were dissected, post-fixed in 4% paraformaldehyde for 24 hrs, and cryoprotected in 30% sucrose in 0.1 M phosphate buffer for 48 hrs. Then, lumbosacral spinal cord segments were embedded in Tissue Tek (Sakura Finetek), mounted onto filter paper and frozen at −40 °C in 2-methylbutane (Fisher Scientific). Longitudinal tissue sections containing the lesion were cut at 25 μm in a Cryostar NX70 (Fisher Scientific) and staggered across four sets. The slides were stored at −20 °C until further processing. For the labelling, slides were warmed at 34 °C for 1 hr and rehydrated 3 × 10 min in TBS, which was followed by 1 hr blocking in 10% NGS in 0.5% TBS-TX at room temperature. Next, the tissue was incubated with the primary antibody (rabbit anti-iba-1; 1:1000; Wako) in 1% NGS and 0.5% TBS-TX overnight at 4 °C. The tissue was then washed 3 × 10 min in 0.5% TBS-TX and incubated with the secondary antibody (AF488 goat anti-rb; 1:400; Molecular Probes) for 2 hrs at room temperature in 1% NGS and 0.5% TBS-TX. Finally, slides were washed 2 × 10 min in 0.5% TBS-TX and 2 × 10 min in TBS, and cover-slipped wet with Fluoromount G (SouthernBiotech). The images were obtained using a Leica DM6000 B fluorescence microscope. Number of Iba-1 positive cells in the central grey matter was counted at 200x magnification. Images were opened on ImageJ and cells counted with the cell counter plugin. Criterion for counting included sharp focus and the presence of clear (round or oval) cell bodies.
RNA-seq
RNA-seq uses MPS to allow transcriptome analyses of genomes at a far higher resolution that is available with Sanger sequencing or microarray-based methods. RNA-seq involves direct sequencing of complementary DNA generated from the RNA of interest using next-generation sequencing technologies. The obtained reads are then aligned to a reference genome in order to construct a whole-genome transcriptome map.
Total RNA (1 μg) extracted from spinal cord specimens (see above) was processed using the TruSeq RNA library preparation kit V2 (Illumina) (which generates mRNA-focused libraries from total RNA) according to manufacturer’s instructions. The libraries were sequenced using an Illumina HiSeq2000. After de-multiplexing of each library pool according to its barcode sequence, 27.5 × 106 ± 6.4 × 106 (MEAN ± SEM) total reads per sample were available for analysis. Alignment, assembly and quantification of the data were performed using TopHat54 against rat genome version rn4 obtained from UCSC browser.
Gene Expression Analysis
The numbers of reads for each gene were obtained using the aligned reads generated by TopHat. Among the 17,233 refseq gene transcripts annotated for the rat genome (version rn4), 1,985 transcripts were removed, maintaining only those with at least one read in at least one sample. This resulted in a total of 15,248 transcripts. For each gene that was represented by several transcripts, only one transcript that resulted in a signal with the highest SD was retained. After this filtering, the data for 14,638 unique gene symbols were retained, and those genes were used in subsequent analyses. The counts data are normalized by the function varianceStabilizingTransformation in the Bioconductor DESeq package55.
Differential Expression Analysis
RNA-Seq data were compared between SCI vs. control rats using the Bioconductor DESeq package55. The differentially expressed (DE) genes were defined as those with fold change (SCI vs. controls) of >1.2 or <0.8 (for positively and negatively DE genes, respectively) and false discovery rate (FDR) adjusted p values < 0.05.
Weighted gene co-expression analysis (WGCNA)
Gene coexpression relationships among the DE genes were analyzed by WGCNA, and the co-expression modules were identified. WGCNA begins with a matrix of the Pearson correlations between all gene pairs, then converts the correlation matrix into an adjacency matrix using a power function f(x) = x^β. The parameter β of the power function is determined in such a way that the resulting adjacency matrix (i.e., the weighted co-expression network), is approximately scale-free. To explore the modular structures of the co-expression network, the adjacency matrix is further transformed into a topological overlap matrix for module detection using a dynamic cut-tree27. To distinguish between modules, each module is assigned a unique color identifier, with the remaining, poorly connected genes colored grey.
Functional enrichment for genes within the identified coexpression modules was assessed using the GeneGo MetaCore software.
Correlational and overlap analyses
Spearman correlation test was used to test the correlations between ABCD frequencies as quantified by MPS (or Adarb1 as quantified by qPCR) and an expression profile of each gene detected by RNA-seq. Multiple testing adjustment was performed using the Benjamini and Hochberg method.
Significance of the overlap between the ABCD signature genes in the rat spinal cord developed in this study and the ABCD signature in the human PFC35 was assessed using Fisher exact test, separately for the positively and negatively correlated genes. First, we identified symbols of gene detected in both studies; this yielded 11,997 gene symbols. From this list of genes, we built the contingency table counting the genes associated with ABCD in both studies, and performed Fisher exact test using this table.
Bayesian Network analysis (BNA)
A Bayesian Network consists of a graphical structure and a probabilistic description of the relationships among variables in a system56. The graphical structure explicitly represents cause-and-effect assumptions that allow a complex causal chain linking actions to outcomes to be factored into an articulated series of conditional relationships. Each of these relationships can then be independently quantified using a sub-model suitable for the type and scale of information available.
BNA of the relationship between genome-wide gene expression (measured by RNA-seq) and Adarb1 (measured by qPCR) was performed using the ‘deal’ R package (http://www.R-project.org/. The genes were analyzed one at a time. For each gene, an analysis was conducted including the following variables: ‘treatment’ (discrete: SCI vs. control), “gene expression” (continuous: normalized read counts), and “Adarb1” (continuous: qPCR quantification). For each gene, there were 2 possible relationships (links) between the treatment (SCI) and the expression of this gene (no link or a link directed from the treatment to gene expression but not vice-versa), and there were 3 possible links between Adarb1 and the expression of this gene (no link, a link directed from a gene to Adarb1, and a link directed from Adarb1 to a gene), resulting in a total of 12 possible networks (3 possible networks between a gene and Adarb1 × 2 possible networks between a gene and the treatment × 2 possible networks between Adarb1 and the treatment). Based on this, for each gene we estimated 12 possible networks and ranked then according to their score. Then, we categorized each gene according to its relationship with Adarb1 based on the following criteria:
if the relative score of the second best network compared to the first was above 0.9, a gene was classified as “ambiguous”;
if the top network showed an outgoing edge from a gene to Adarb1, a gene was classified as “upstream”;
if the top network showed an outgoing edge from Adarb1 to the gene, a gene was classified as “downstream”;
if none of the above 3 conditions were met, a gene was classified as “independent” (with regard to Adarb1).
A schematic of the workflow is presented in Fig. 9.
Figure 9. Bayesian Network Analysis (BNA).
For each expressed gene, prior knowledge indicates 12 possible arrangements of causal links between treatment (SCI), gene expression of each gene, and Adarb1. By performing BNA, we can define the relationship between each gene and Adarb1 by placing it within one of 4 distinct categories: upstream downstream, independent, and ambiguous (genes for which the distinction is unclear). “X “depicts the absence of a link.
Additional Information
How to cite this article: Narzo, A. F. D. et al. Decrease of mRNA Editing after Spinal Cord Injury is Caused by Down-regulation of ADAR2 that is Triggered by Inflammatory Response. Sci. Rep. 5, 12615; doi: 10.1038/srep12615 (2015).
Supplementary Material
Acknowledgments
The work was supported with resources and the use of facilities at the James J Peters VA Medical Center, Bronx, New York and funded by: VISN3 Mental Illness Research and Education Clinical Center (MIRECC) and VA Merit Review Award (BX002343 to S.D.). E.V.K. is supported by intramural funds of the US Department of Health and Human Services to National Library of Medicine.
Footnotes
Author Contributions S.D., D.J.B., A.F.D.N. and A.K. designed the study. S.C.I. injury surgeries and spinal cord dissections were performed by L.S. and Y.L. RNA editing, RNA-seq, and qPCR studies were performed by A.K. Immunohistochemistry was performed by Z.M. Bioinformatic analysis was performed by A.F.D.N., Y.G. and B.Z. S.D., E.V.K., C.C. and K.F. contributed to the analysis of the results and to the writing of the paper.
References
- Rekling J. C., Funk G. D., Bayliss D. A., Dong X. W. & Feldman J. L. Synaptic control of motoneuronal excitability. Physiol Rev 80, 767–852 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultborn H., Denton M. E., Wienecke J. & Nielsen J. B. Variable amplification of synaptic input to cat spinal motoneurones by dendritic persistent inward current. J Physiol 552, 945–52 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElBasiouny S. M., Schuster J. E. & Heckman C. J. Persistent inward currents in spinal motoneurons: important for normal function but potentially harmful after spinal cord injury and in amyotrophic lateral sclerosis. Clin Neurophysiol 121, 1669–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gorassini M. A. & Bennett D. J. Role of persistent sodium and calcium currents in motoneuron firing and spasticity in chronic spinal rats. J Neurophysiol. 91, 767–783 (2004). [DOI] [PubMed] [Google Scholar]
- Bennett D. J. et al. Spasticity in rats with sacral spinal cord injury. J Neurotrauma 16, 69–84 (1999). [DOI] [PubMed] [Google Scholar]
- Bennett D. J., Sanelli L., Cooke C. L., Harvey P. J. & Gorassini M. A. Spastic long-lasting reflexes in the awake rat after sacral spinal cord injury. J Neurophysiol. 91, 2247–2258 (2004). [DOI] [PubMed] [Google Scholar]
- Button D. C. et al. Does elimination of afferent input modify the changes in rat motoneurone properties that occur following chronic spinal cord transection? J Physiol 586, 529–544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J. A., Bennett D. J., Knash M. E., Murray K. C. & Gorassini M. A. Changes in sensory-evoked synaptic activation of motoneurons after spinal cord injury in man. Brain 131, 1478–1491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorassini M. A., Knash M. E., Harvey P. J., Bennett D. J. & Yang J. F. Role of motoneurons in the generation of muscle spasms after spinal cord injury. Brain 127, 2247–2258 (2004). [DOI] [PubMed] [Google Scholar]
- Dietz V. & Sinkjaer T. Spastic movement disorder: impaired reflex function and altered muscle mechanics. Lancet Neurol. 6, 725–733 (2007). [DOI] [PubMed] [Google Scholar]
- Murray K. C. et al. Recovery of motoneuron and locomotor function after spinal cord injury depends on constitutive activity in 5-HT2C receptors. Nat Med 16, 694–700 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. Y., Wienecke J., Hultborn H. & Zhang M. Robust upregulation of serotonin 2A receptors after chronic spinal transection of rats: an immunohistochemical study. Brain Res. 1320, 60–68 (2010). [DOI] [PubMed] [Google Scholar]
- Hannon J. & Hoyer D. Molecular biology of 5-HT receptors. Behav.Brain Res. 195, 198–213 (2008). [DOI] [PubMed] [Google Scholar]
- Gott J. M. & Emeson R. B. Functions and mechanisms of RNA editing. Annu.Rev.Genet. 34, 499–531 (2000). [DOI] [PubMed] [Google Scholar]
- Valente L. & Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog.Nucleic Acid Res.Mol.Biol. 79, 299–338 (2005). [DOI] [PubMed] [Google Scholar]
- Werry T. D., Loiacono R., Sexton P. M. & Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol.Ther. 119, 7–23 (2008). [DOI] [PubMed] [Google Scholar]
- Rosenthal J. J. & Seeburg P. H. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron 74, 432–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn A. C., Balik A. & Greger I. H. Reciprocal regulation of A-to-I RNA editing and the vertebrate nervous system. Front Neurosci 7, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. M. et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 (1997). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 74, 1290–1300 (2000). [DOI] [PubMed] [Google Scholar]
- Moro O., Lameh J., Hogger P. & Sadee W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J.Biol.Chem. 268, 22273–22276 (1993). [PubMed] [Google Scholar]
- Niswender C. M., Copeland S. C., Herrick-Davis K., Emeson R. B. & Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J.Biol.Chem. 274, 9472–9478 (1999). [DOI] [PubMed] [Google Scholar]
- Navarrett S., Collier L., Cardozo C. & Dracheva S. Alterations of serotonin 2C and 2A receptors in response to T10 spinal cord transection in rats. Neurosci Lett. 506, 74–78 (2012). [DOI] [PubMed] [Google Scholar]
- Lyddon R., Dwork A. J., Keddache M., Siever L. J. & Dracheva S. Serotonin 2c receptor RNA editing in major depression and suicide. World J Biol.Psychiatry 14, 590–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabito M. V. et al. High-throughput multiplexed transcript analysis yields enhanced resolution of 5-hydroxytryptamine 2C receptor mRNA editing profiles. Mol.Pharmacol. 77, 895–902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla T., Rosenthal J. J., Holmgren M. & Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat.Struct.Mol.Biol. 11, 950–956 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang B. & Horvath S. A general framework for weighted gene co-expression network analysis. Stat.Appl.Genet.Mol.Biol. 4, Article17 (2005). [DOI] [PubMed] [Google Scholar]
- Oldham M. C. et al. Functional organization of the transcriptome in human brain. Nat.Neurosci 11, 1271–1282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden K. D. et al. The organization of the transcriptional network in specific neuronal classes. Mol.Syst.Biol. 5, 291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet 41, 415–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. M. et al. Inferring causal genomic alterations in breast cancer using gene expression data. BMC Syst Biol 5, 121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Narzo A. F. et al. A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. Hum.Mol.Genet. 23, 4801–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er D. Bayesian network analysis of signaling networks: a primer. Sci STKE 281, pl4 (2005). [DOI] [PubMed] [Google Scholar]
- Saher G. et al. High cholesterol level is essential for myelin membrane growth. Nat Neurosci 8, 468–75 (2005). [DOI] [PubMed] [Google Scholar]
- Plemel J. R. et al. Remyelination after spinal cord injury: is it a target for repair? Prog Neurobiol 117, 54–72 (2014). [DOI] [PubMed] [Google Scholar]
- Matsuda K., Kamiya Y., Matsuda S. & Yuzaki M. Cloning and characterization of a novel NMDA receptor subunit NR3B: a dominant subunit that reduces calcium permeability. Brain Res Mol Brain Res 100, 43–52 (2002). [DOI] [PubMed] [Google Scholar]
- Vitalis T. et al. Developmental expression of monoamine oxidases A and B in the central and peripheral nervous systems of the mouse. J Comp Neurol 442, 331–47 (2002). [DOI] [PubMed] [Google Scholar]
- Taricco M., Pagliacci M. C., Telaro E. & Adone R. Pharmacological interventions for spasticity following spinal cord injury: results of a Cochrane systematic review. Eura.Medicophys. 42, 5–15 (2006). [PubMed] [Google Scholar]
- Roy R. R. & Edgerton V. R. Neurobiological perspective of spasticity as occurs after a spinal cord injury. Exp Neurol 235, 116–22 (2012). [DOI] [PubMed] [Google Scholar]
- Chi X. X. & Nicol G. D. Manipulation of the potassium channel Kv1.1 and its effect on neuronal excitability in rat sensory neurons. J Neurophysiol. 98, 2683–2692 (2007). [DOI] [PubMed] [Google Scholar]
- Robbins C. A. & Tempel B. L. Kv1.1 and Kv1.2: similar channels, different seizure models. Epilepsia 53 Suppl 1, 134–141 (2012). [DOI] [PubMed] [Google Scholar]
- Harvey P. J., Li Y., Li X. & Bennett D. J. Persistent sodium currents and repetitive firing in motoneurons of the sacrocaudal spinal cord of adult rats. J Neurophysiol. 96, 1141–1157 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab J. M., Zhang Y., Kopp M. A., Brommer B. & Popovich P. G. The paradox of chronic neuroinflammation, systemic immune suppression, autoimmunity after traumatic chronic spinal cord injury. Exp Neurol 258, 121–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier J. F. & Hounsgaard J. 5-HT2 receptors promote plateau potentials in turtle spinal motoneurons by facilitating an L-type calcium current. J Neurophysiol. 89, 954–959 (2003). [DOI] [PubMed] [Google Scholar]
- Hideyama T. et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol Dis 45, 1121–8 (2012). [DOI] [PubMed] [Google Scholar]
- Brites D. & Vaz A. R. Microglia centered pathogenesis in ALS: insights in cell interconnectivity. Front Cell Neurosci 8, 117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. H. et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci 16, 571–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. & Mehler M. F. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 31, 227–233 (2008). [DOI] [PubMed] [Google Scholar]
- Eran A. et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol.Psychiatry 18, 1041–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S. et al. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology 34, 2237–2251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N., Linial M., Nachman I. & Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol 7, 601–20 (2000). [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T. & Mesirov J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.