Abstract
The developing mammalian brain is destined for a female phenotype unless exposed to gonadal hormones during a perinatal sensitive period. It has been assumed that the undifferentiated brain is masculinized by direct induction of transcription by ligand-activated nuclear steroid receptors. We found that a primary effect of gonadal steroids in the highly sexually-dimorphic preoptic area (POA) is to reduce activity of DNA methyltransferase (Dnmt) enzymes, thereby decreasing DNA methylation and releasing masculinizing genes from epigenetic repression. Pharmacological inhibition of Dnmts mimicked gonadal steroids, resulting in masculinized neuronal markers and male sexual behavior in females. Conditional knockout of the de novo Dnmt isoform, Dnmt3a, also masculinized sexual behavior in female mice. RNA sequencing revealed gene and isoform variants modulated by methylation that may underlie the divergent reproductive behaviors of males versus females. Our data show that brain feminization is maintained by the active suppression of masculinization via DNA methylation.
In sexually reproducing species the developing brain is either masculinized or feminized in a manner that assures adult neural physiology and reproductive behavior are in register with the differentiated gonads. In mammals, feminization of the brain is independent of the ovary and therefore considered a default developmental pathway not requiring active secretion of ovarian steroids. Masculinization is the differentiation away from the female phenotype and is mediated by gonadal steroids generated by the fetal testis during a perinatal sensitive period. Chromosome complement and environmental variables either magnify or diminish sex differences in the brain but the actions of testosterone and its potent metabolite, estradiol, are the most robust mediators of rodent brain masculinization1, 2. Steroids bind to nuclear receptors which are members of an extended family of transcription factors3, and the direct induction of gene expression via activation of estrogen (ER) and androgen receptors (AR) has been the presumptive route for brain masculinization4, 5, yet surprisingly few gene candidates have been identified6–8.
The most robust and reliable brain sexual dimorphisms are found in the preoptic area (POA), consistent with its central role in male copulatory behavior and control of gonadotropin secretion from the anterior pituitary9, 10. In the neonatal male rodent, estradiol aromatized within the POA from testicular androgens induces large changes in regional volume and cell number, increases astrocyte stellation and triggers a two-fold increase in putative excitatory synapses as indicated by increased dendritic spine density on POA neurons1. All of these changes are established within the first few days following birth and endure until adulthood, but it has been unknown how these developmental, hormonally-induced changes in the brain are maintained across the lifespan.
Epigenetic processes are a means by which endogenous and exogenous cues exert long-term control over gene expression. DNA methylation, which occurs predominantly at the 5’-position of cytosine residues adjacent to guanines (referred to as CpG sites), is frequently associated with long-term transcriptional repression by altering protein-DNA interactions and ultimately chromatin conformation. The neuronal DNA methylome is highly modifiable, with rapid demethylation and de novo methylation occurring in response to changes in excitability, particularly in genes associated with neural plasticity11. To determine if DNA methylation contributes to estradiol-mediated brain masculinization, we measured the activity and expression of DNA methyltransferase (Dnmt) enzymes in the POA of male, female, and estradiol- treated masculinized female rat pups and disrupted Dnmt activity in the brains of both rats and mice. We also measured the level of DNA methylation globally and via whole genome bisulfite sequencing. Females had higher levels of methylation with significantly more fully methylated CpG sites than males. Our results demonstrate that hormonally-mediated decreases in Dnmt activity result in masculinization of the POA and copulatory behavior, and that feminization must be actively maintained by DNA methylation.
RESULTS
Males have lower Dnmt activity in the neonatal POA than females
Total Dnmt activity in POA tissue from males was significantly lower than activity in females on postnatal day 0 (PN0) and PN2 and was variable at PN4 (Fig. 1a; Supplementary Fig. 1). There were no sex differences outside of the sensitive period, at embryonic day 20 or PN14 (Supplementary Fig. 1). Treatment of newborn females with a masculinizing dose of estradiol decreased Dnmt activity to that of males within 6 hrs and the lower activity was maintained until PN2. By PN4 activity levels dropped in normal females to that of males and masculinized females and dropped still further in all animals by PN7 (Fig. 1a). Treatment with estradiol on PN14, following the close of the sensitive period, no longer impacted Dnmt activity (Supplementary Fig. 1). Within the POA, levels of global DNA methylation measured on PN1 paralleled levels of Dnmt activity (Fig. 1b). Whole genome bisulfite sequencing revealed that this sex difference was restricted to highly methylated CpG sites (>90%), which are the majority, and that females had nearly twice the level of fully (100%) methylated CpG sites as males or masculinized females (Fig. 1c; Supplementary Fig. 2a). Sex differences were generally dispersed across chromosomes, although methylation on chromosome 5 and 13 was biased toward females and males, respectively (Supplementary Fig. 3). Sites methylated 80–90% were more frequent in males and masculinized females, likely revealing a population shift downward from 100% methylation, consistent with reduced or inhibited Dnmt activity (Supplementary Fig. 2a). The overwhelming majority of CpG sites exhibiting a sex difference in methylation were in intergenic regions (~84%), followed by introns (~14%), promoter regions (~2%) and exons (<1%) and were not in CpG islands (Fig. 1d). The proportion of differences in CpG methylation might reflect the distribution of CpG sites throughout different genomic regions and may not necessarily indicate an enrichment of sex differences in a specific genomic compartment. High percentages of CHG methylation were substantially lower in frequency than CpG methylation, as expected. Estradiol significantly reduced the number of CHG sites with less than 10% methylation, compared to males and control females, and males had more CHG sites with < 10% methylation compared to control females (Supplementary Fig. 2b).
Figure 1. Females have higher Dnmt activity and DNA methylation in the POA, which is reduced by estradiol treatment.
(a) On PN0 and PN2, females had significantly higher levels of Dnmt activity in the POA compared to males and females treated with 100 µg estradiol (e) at birth and sacrificed six hours (on PN0) or 48 hours later (on PN2; 2-way ANOVA, Fsex(2, 64) = 10.1, p = 0.0002). Dnmt activity decreased in all groups by PN7 (Fage(3, 64) = 76.85, p < 0.0001, Fint(6, 64) = 3.72, p = 0.0031). (b) On PN1, females had higher percent levels of global DNA methylation in the POA than males. Twenty-four hours of e treatment significantly reduces global DNA methylation in the female POA to the level of the male (ANOVA, F(2,12) = 5.55, p = 0.0197). (c) Whole genome bisulfite sequencing revealed that females have more fully methylated CpG sites across the genome compared to males and e-treated females (ANOVA, F(2,6) = 6.594, p = 0.0306). (d) Pie charts displaying the distribution of sex differences in CpG methylation across genomic compartments and within/outside CpG islands and shores. Sex differences in CpG methylation were most prevalent in intergenic regions, outside of CpG islands and shores (based on 1242 sex differences in CpG methylation). (e) 2-way ANOVA on qPCR analyses of Dnmt1, Dnmt3a and Dnmt3b detected no sex differences or hormonal modulation in the amount of mRNA in the POA at any given age across groups (Dnmt1 Fsex(2,54) = 0.661, p = 0.5205, Dnmt3a Fsex(2,54) = 0.024, p = 0.975, Dnmt3b Fsex(2,54) = 0.013, p = 0.9867), although there was a significant decline between PN0 and PN7 in Dnmt1 (FAge(3,54) = 36.19, p < 0.0001, Fint(6,54) = 0.8851, p = 0.5123), and Dnmt3b (FAge(3,54) = 20.46, p < 0.0001, Fint(6,54) = 0.555, p = 0.7634), whereas Dnmt3a expression increased between PN0 and PN2 (FAge(3,54) = 51.46, p < 0.0001, Fint(6,54) = 1.323, p = 0.2629). PN0 n = 6 male, female, and female + e; PN2 n = 7 male, 4 female, 5 female + e; PN4 n = 6 male, female, and female + e; PN7 n = 7 male, female, and female + e (a), n = 5 male, female, and female + e (b), n = 3 male, female, and female + e (c, d), Dnmt1: PN0 n = 5 male, 6 female, 5 female + e; PN2 n = 6 male, female, and female + e; PN4 n = 5 male, 5 female, 6 female + e; PN7 n = 6 male, 5 female, 5 female + e; Dnmt3a: PN0 n = 5 male, 6 female, 5 female + e; PN2 n = 6 male, female, female + e; PN4 n = 5 male, 5 female, 6 female + e; PN7 n = 6 male, 5 female, 5 female + e. Dnmt3b: PN0 n = 5 male, 6 female, 5 female + e; PN2 n = 6 male, female, female + e; PN4 n = 5 male, 5 female, 6 female + e; PN7 n = 6 male, 5 female, 5 female + e (e). *p < 0.05 in control females vs. other groups, **p<0.01 in control females vs. other groups. Data are represented as mean ± s.e.m. (a, b, e), data are represented as mean ± s.d. (c).
There were no sex differences in mRNA or protein levels of either the maintenance Dnmt1 or de novo Dnmts, Dnmt3a and Dnmt3b (Fig. 1e; Supplementary Fig. 4). Based on relative units, Dnmt1 and Dnmt3b mRNA were most abundant at birth but levels of both of these enzymes sharply decreased within 1 week, while Dnmt3a levels increased by PN2 and remained elevated and substantially more abundant than the other Dnmts until at least PN7 (Fig. 1e). There was no hormonal modulation of mRNA or protein levels of any of the Dnmts (Fig. 1e; Supplementary Fig. 4), suggesting enzyme activity is modulated indirectly via substrate availability, phosphorylation or other means unrelated to transcription.
Low levels of Dnmt activity masculinize the bipotential brain
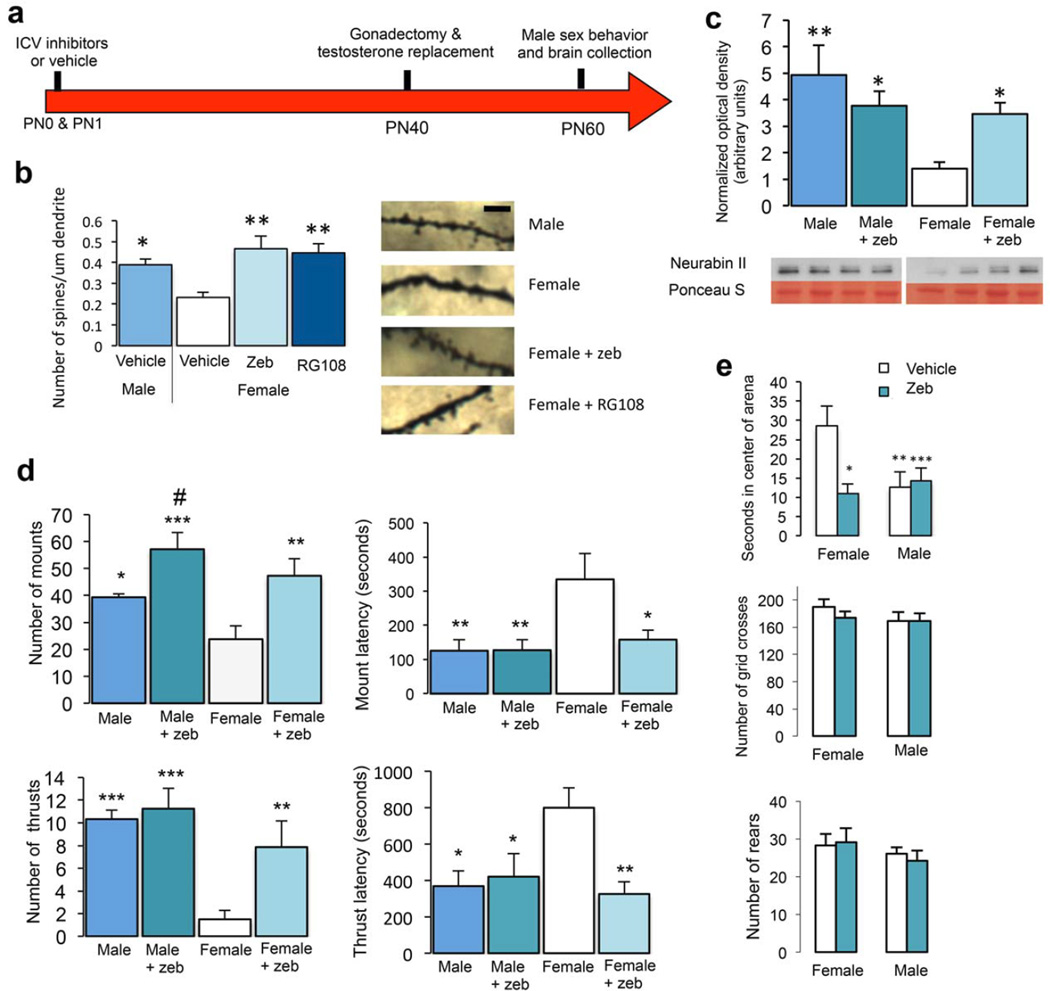
Exogenous estradiol treatment of newborn female rat pups reliably masculinizes the POA and sexual behavior1. The marked decrease in fully methylated CpG’s following estradiol treatment of females led us to hypothesize that high levels of fully methylated CpG’s in the female POA serve to repress gene expression to prevent neural and behavioral masculinization and promote feminization. We predicted that pharmacological inhibition of Dnmt activity in the neonatal female brain would mimic the effects of estradiol, producing masculinized adult neuronal markers and behavior in adulthood (Fig 2a). We used two different Dnmt inhibitors: the low toxicity cytidine analog zebularine (Zeb), which traps the enzyme in a DNA-adduct12 but may also stimulate base-excision repair13, and the small-molecule direct inhibitor, RG108, targeted to Dnmt1 but considered to have broad applicability due to high conservation of the catalytic binding site across the Dnmts14. In males, estradiol induces the formation of dendritic spines on POA neurons and these putative excitatory synapses are then maintained at twice the density of females into adulthood6. Neurabin II is an F-actin binding protein also known as spinophilin due to its enhanced localization to the postsynaptic density of dendritic spines15. We have found both western blot and immunohistochemical quantification of Neurabin II to be an accurate and reliable proxy marker for dendritic spines in the POA, correlating precisely with the relative spine density detected by neuroanatomical measures6. Moreover, Neurabin II levels in the POA of individual adult animals positively correlate with the robustness of male sexual behavior16, which is quantified as the number of times the masculinized animal mounts or attempts to intromit a receptive female and the latency with which these behaviors are initiated following introduction of a sexually receptive female.
Figure 2. Neonatal Dnmt inhibition increases dendritic spine density on POA neurons and masculinizes behavior.
(a) Animals were treated with the Dnmt inhibitors Zeb or RG108 on PN0 and PN1. As adults, animals were gonadectomized and implanted with testosterone-releasing capsules to achieve stable adult male circulating testosterone levels. Male sexual behavior was assessed 20 days later and brains were collected at the completion of behavioral testing. (b) Golgi-Cox impregnated adult POA neurons were quantified for dendritic spine density. Neonatal Dnmt inhibition by either Zeb or RG108 in females masculinized dendritic spine density of adults (ANOVA, F(3,22) = 4.92, p = 0.0091). Scale bar = 10 µm (c) Zeb treatment on PN0 & PN1 significantly increased Neurabin II protein levels in the adult female POA (F(3,32) = 4.69, p = 0.0079). Cropped representative western bands are shown and full length blots are provided in Supplementary Figure 10. (d) Neonatal Zeb treatment increased mounts (F(3,25) = 22.18, p < 0.0001) and thrusts (F(3,25) = 7.87, p < 0.0001) and decreased the latency to mount (F(3,24) = 4.25, p = 0.0153) and thrust (F(3,21) = 4.05, p = 0.0203) in adult females. Neonatal Zeb treatment also increased the number of mounts by adult males. (e) Animals treated with Zeb showed no impairments in locomotor activity as measured by grid crosses (F(3,27) = 0.72, p = 0.5486) and rears (F(3, 27) = 0.6, p = 0.6205), however time spent in the center of the open field arena was masculinized by Zeb treatment of females (F(3,27) = 7.4, p = 0.0009); n = 5 female, 7 male, 7 female + zeb, 7 female + RG108 (b), n = 9 female, 9 male, 9 female + zeb, 9 male + zeb (c), Mounts: n = 8 female, 6 male, 7 female + zeb, 8 male + zeb. Thrusts: n = 8 female, 6 male, 7 female + zeb, 8 male + zeb. Mount latency: n = 8 female, 6 male, 7 female + zeb, 7 male + zeb. Thrust latency: n = 6 female, 6 male, 6 female + zeb, 7 male + zeb (d), Grid crosses: n = 8 female, 8 male, 7 female + zeb, 8 male + zeb. Rears: n = 8 female, 8 male, 7 female + zeb, 8 male + zeb. Time in center of arena: n = 8 female, 8 male, 7 female + zeb, 8 male + zeb (e). *p < 0.05, **p < 0.01, ***p < 0.001 compared to control female. #p < 0.05 compared to control male. Data are represented as mean ± s.e.m.
Intracerebroventricular (ICV) administration of Zeb or RG108 on PN0 and PN1 resulted in increased POA dendritic spine density (Fig. 2b) and higher levels of the post-synaptic protein Neurabin II in the adult female POA (Fig. 2c; Supplementary Fig. 5b). Importantly, treatment of females with either Dnmt inhibitor masculinized adult sexual behavior. Females who received neonatal Dnmt inhibition engaged in significantly more mounts and thrusts (intromission-like behaviors) towards a sexually receptive female, whereas control females treated with vehicle as neonates displayed very few male-like behaviors, as expected of normally feminized animals (Fig. 2d; Supplementary Fig. 5a). Dnmt inhibitor-treated females also had significantly shorter latencies to mount, a measure of male sexual motivation17, and Zeb treatment decreased the latency to display thrust behaviors (Fig. 2d). Analysis of behavior in an open field confirmed that general motor activity was not impaired, however anxiety-related behaviors were masculinized in females following neonatal Dnmt inhibition (Fig. 2e; Supplementary Fig. 5c). There was no impact of Zeb treatment on estrous cyclicity (Supplementary Fig. 6), suggesting the role of DNA methylation in sexual differentiation is not generalizable to other dimorphic endpoints. The efficacy of Zeb treatment at reducing CpG methylation was confirmed using the global methylation assay. Twenty-four hours after ICV Zeb injection methylation was significantly reduced compared to control (t-test; (t(6) = 2.32, p = 0.0417).
Dnmt1 largely maintains DNA methylation patterns following DNA replication18 whereas Dnmt3a induces de novo DNA methylation in response to internal and external stimuli and is implicated in neuronal plasticity19, although a strong interdependence between the maintenance and de novo Dnmts is now emerging20. Current pharmacological tools do not distinguish between Dnmts, therefore to gain some insight into the relative role of a specific Dnmt we used AAV2-driven conditional knockout of Dnmt3a in the neonatal mouse POA. Newborn male and female mice homozygous for a floxed Dnmt3a exon (Dnmt3a2lox/2lox;Fig. 3a) were injected with either AAV2-Cre or AAV2-GFP into the POA on PN0. GFP visualization in a subset of animals confirmed viral transfection was localized to the POA (Fig. 3b). In adulthood, 100% of AAV2-Cre Dnmt3a2lox/2lox females displayed male sexual behavior, compared to 20% of control AAV2-GFP Dnmt3a2lox/2lox females (Fig. 3c). AAV2-Cre Dnmt3a2lox/2lox female mice exhibited extremely fast mount latencies, long mount times, and an increased number of thrusts compared to control AAV2-GFP Dnmt3a2lox/2lox females. Dnmt3a knockout in the POA had no effect on activity in an open field, nor was there a sex difference in this behavior in the mice (Fig. 3d). Significant depletion of Dnmt3a mRNA by AAV2-Cre was confirmed by quantitative real-time PCR (qPCR) in adult POA from animals treated as neonates (sexes mixed; t-test, t(9)=4.691, p= 0.0011).
Figure 3. Conditional Dnmt3a knockout in the POA masculinizes reproductive behavior of female mice.
(a) Schematic diagram of genomic locus, targeting construct and modified alleles of Dnmt3a. The numbered boxes represent exons (redrawn from43) (b) Representative images of GFP staining in the POA at 10X (scale bar indicates 75um). Immunohistochemistry was performed in a subset of animals from each experimental group to verify that the POA was targeted by AAV injection. There were no observable differences in staining intensity or cell number between groups. (c) Quantification of male sexual behavior in adult animals treated neonatally with either AAV2-Cre or AAV-GFP revealed that 100% of the AAV2-Cre Dnmt3a2lox/2lox females displayed male sexual behavior. Appetitive components of male sexual behavior were unaffected by Dnmt3a knockout (ANOVA; Solicitations, F(3,10) = 11.17, p = 0.0016; Anogenital investigation, F(3,9) = 6.372, p = 0.0132), whereas AAV2-Cre females showed a significantly decreased mount latency (F(3,18) = 3.505, p = 0.036), increased mount time (F(3,16) = 7.17, p = 0.0029), and a greater number of thrusts (F(3,13) = 3.385, p = 0.051) compared to AAV2-GFP females in a 30 minute trial across 3 consecutive weeks. (d) There were no significant differences in time spent in the center of (F(3,18) = 1.07, p = 0.387) or total distance travelled in an open field arena (F(3,18) = 1.64, p = 0.274). Solicitations: n = 3 female cre, 4 female gfp, 4 male cre, 3 male gfp. Anogenital investigation: n = 2 female cre, 5 female gfp, 4 male cre, 2 male gfp. Mount latency: n = 5 female cre, 5 female gfp, 8 male cre, 4 male gfp. Mount time: n = 4 female cre, 5 female gfp, 8 male cre, 3 male gfp. Thrusts: n = 4 female cre, 3 female gfp, 7 male cre, 3 male gfp (c), Time in center of arena: n = 5 female cre, 7 female gfp, 6 male cre, 4 male gfp. Total distance: n = 5 female cre, 7 female gfp, 6 male cre, 4 male gfp (d). * p < 0.05 compared to AAV2-GFP female, ** p < 0.01 compared to AAV2-GFP female, # p < 0.05 compared to AAV2-Cre female (c–d). Data are represented as mean ± s.e.m.
DNA methylation maintains the female brain phenotype
AAV2-mediated knockout takes days to weeks to become effective. We injected virus into the POA of mice on the day of birth and therefore knockout of Dnmt3a may not have been fully implemented by the close of the sensitive period for sexual differentiation, suggesting that DNA methylation maintains the female brain phenotype and that masculinization can occur if critical genes are de-repressed outside of the critical period. To directly test this hypothesis, we administered Zeb to female rats outside of the sensitive period (PN10 and PN11; Fig. 4a), and once again observed significant increases in Neurabin II protein levels, as well as mounting behavior in adult female rats (Fig. 4b, c). The same treatment also reduced mount and thrust latencies. Importantly, estradiol treatment of females at the same developmental time point did not masculinize Neurabin II levels or sexual behavior, confirming the previously defined sensitive period for hormonally-mediated sexual differentiation. Moreover, there was no reduction in Dnmt activity by estradiol treatment outside of the sensitive period (Supplementary Fig. 1), suggesting that at least one factor leading to the close of the sensitive period is a loss of hormonally-mediated inhibition of Dnmt activity.
Figure 4. Dnmt inhibition outside of the critical period increases dendritic spine markers in the POA and masculinizes behavior.
(a) Animals were treated with Zeb or estradiol at PN10 and PN11. In adulthood, animals were gonadectomized and implanted with testosterone-releasing capsules and male sexual behavior was assessed 20 days later. Brains were collected and Neurabin II levels were quantified in the POA following the completion of three weeks of behavioral testing. (b) Zeb administration after the close of the critical period for sexual differentiation of the brain masculinized Neurabin II protein levels (a priori t-test between females and females + zeb: t(8) = –2.833, p = 0.0221), although there was no main effect across all treatment groups (ANOVA, F(4,22) = 2.22, p = 0.1002). Cropped representative western bands are shown and full length blots are provided in Supplementary Figure 10. (c) Number of mounts (F(4,27) = 5.23, p = 0.003), the latency to mount (F(4,26) = 2.44, p = 0.0718, a priori t-test between females and females + zeb: t(10) = 2.862, p = 0.0169), and thrust latency (F(4,21) = 6.67, p = 0.0013) were also masculinized by Dnmt inhibition whereas estradiol treatment administered at this same time point did not induce masculinization of brain or behavior. Although there were large differences in the number of thrusts between males and females (F(4,27) = 14.861, p < 0.0001), Zeb did not masculinize number of thrusts in females. n = 7 male, 7 male zeb, 6 female, 4 female + e, 4 female + zeb (b), Mounts: n = 7 male, 8 male zeb, 7 female, 4 female + e, 6 female + zeb. Mount latency: n = 7 male, 8 male zeb, 6 female, 4 female + e, 6 female + zeb. Thrusts: n = 7 male, 8 male zeb, 6 female, 4 female + e, 7 female + zeb. Thrust latency: n = 7 male, 8 male zeb, 4 female, 4 female + e, 3 female + zeb (c). *p < 0.05, **p < 0.01, ***p < 0.001 one-way ANOVA main effect compared to vehicle-treated female. #p<0.05 t-test compared to vehicle-treated female. Data are represented as mean ± s.e.m.
Methylation-dependent and -independent sex differences in gene expression
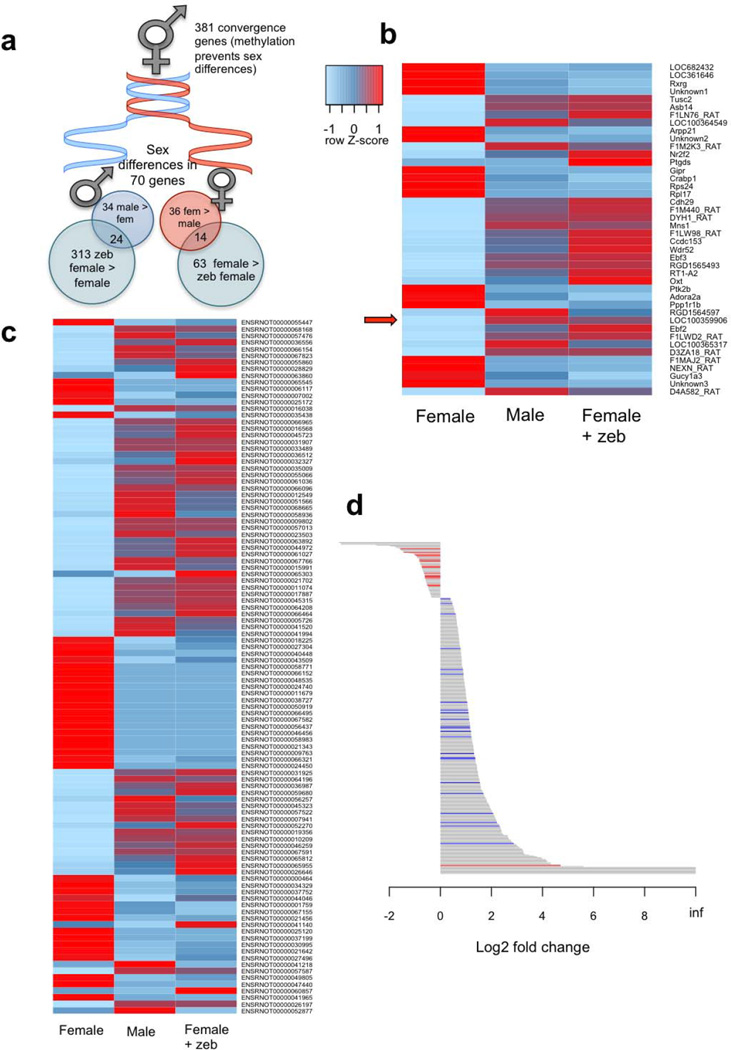
If sex differences in Dnmt activity mediate differential expression of genes necessary for masculinization, then inhibiting Dnmt activity should result in masculinized patterns of gene expression. We used RNA-Seq to compare total gene and genetic isoform expression in the PN2 POA of males and females treated with either vehicle or Zeb on PN0 and PN1. Sex differences were observed in the expression of 70 genes with approximately half higher in males than females and vice versa (Fig. 5a; Supplementary Table 1). Two genes, Mcpt2 and D4AD88, which code for mast cell protease 2 and an uncharacterized protein respectively, were exclusively expressed in the male POA. Mast cells mediate acquired immunity and have been associated with reproductive functions controlled by the brain, including the release of gonadotropin releasing hormone (GnRH) and prostaglandins21,22. Prostaglandins are critical mediators of masculinization of the POA23 and mast cells both respond to and produce prostaglandins, thus this male-specific expression of Mcpt2 may be a critical node initiating the signal transduction cascade leading to masculinization.
Figure 5. Masculinization of POA gene expression by Dnmt inhibition.
Animals were treated on PN0 and PN1 with Zeb. On PN2, mRNA was extracted and subject to RNA-Seq to identify methylation-dependent changes in gene expression in the female POA. (a) Schematic representation of sex differences in methylation-dependent gene expression in the neonatal POA. Only 70 genes were found to be expressed at different levels in males and females, with 34 higher in males and 36 higher in females. Of these, 70% of genes with higher expression in males were increased in females following Dnmt inhibition indicating methylation-dependent expression. Close to half of the genes expressed at higher levels in females were decreased to male levels following Dnmt inhibition. Overall, more genes significantly changed expression in females compared to males following Zeb treatment, consistent with the higher DNA methylation observed in females. Almost 400 genes became differentially expressed in males and females following Dnmt inhibition in box sexes, and are therefore referred to as convergence genes, meaning expression is maintained at equal levels in males and females by DNA methylation. (b) Heat map of genes with basal sex differences in the POA that were also significantly altered by Zeb treatment in females. Light blue indicates lower relative expression and red indicates higher relative expression across groups. Zeb disrupted basal sex-differences in gene expression in the female POA, creating patterns resembling male gene expression. Arrow indicates Cyp19a1 gene, which codes for the enzyme p450 aromatase, a known contributor to POA masculinization. (c) Heat map of genetic isoforms with basal sex differences that were significantly altered by Zeb treatment in females to create a male-like pattern of isoform expression. (d) Bar plot of all changes in total gene expression induced by Zeb treatment in the neonatal female POA. Grey lines indicate genes without sex differences in expression. Blue lines indicate genes with higher expression in the control male POA vs. the control female POA (putative masculinization genes). Red lines indicate genes with higher expression in the control female POA vs. the control male POA (putative feminization genes). Of the genes with basal sex differences that were altered by Zeb in females, all male-biased genes increased in females following Zeb (blue lines), and all but one of the female-biased genes decreased expression in response to Zeb (red lines), suggesting Dnmt inhibition masculinizes gene expression in the female POA by both increasing male-biased genes and decreasing female-biased genes. Based on data from n = 2 female, 3 Zeb female, 3 male, 3 Zeb male (not represented in plots).
Only a subset of genes de-repressed by Dnmt inhibition are relevant to sexual differentiation (Fig. 5a, d). To identify genes involved in masculinization we focused on those that were up-regulated by Zeb in females and higher in males compared to control females. We found that 24 of the 34 genes with higher expression in the male POA were significantly increased by Dnmt inhibition in the female (Fig. 5a,b; Supplementary Table 2). We considered this subset of genes methylation-dependent gene candidates for sexual differentiation of the brain, and regarded the 10 genes highly expressed in males but not Zeb-treated or control females as methylation-independent candidate genes. Among the methylation-dependent set was the gene Cytochrome P450 19A1 (Cyp19a1; LOC100359906), which is the estradiol synthesizing enzyme, aromatase, and one of the few genes previously shown to be expressed at a higher level in the male POA during the first few days after birth24. We confirmed the sex difference and estradiol-induced increase in Cyp19a1 mRNA with qPCR (Supplementary Fig. 7) but our findings indicate that contrary to prior assumptions, the estradiol-mediated increase in Cyp19a1 is not due to induction of transcription by ER but instead secondary to a loss of DNA methylation. Moreover, Cyp19a1 mRNA expression is elevated following estradiol treatment for at least an additional 5 days. Analysis of a large CpG island in the Cyp19a1 promoter revealed unusually high levels of CpG methylation (~50%) compared to most other candidate masculinization genes (5–8%; Supplementary Fig. 8). Expression patterns of additional candidate masculinization genes (Ebf2, Efb3, Nr2f2) were marginally higher in males, confirming our RNA-Seq results (Supplementary Fig. 9).
mRNA levels of a relatively small number of genes decreased after Zeb treatment, as would be expected following demethylation, but a surprisingly large percentage (22%) of them were candidate feminization genes, defined as those that were higher in control females compared to males but decreased following Zeb treatment (Fig. 5a,d). This suggests that feminization genes are maintained at higher levels in females vs. males in a manner that involves methylation but is opposite to that of masculinization genes, which are repressed by methylation.
Also unexpected was that 381 genes became differentially expressed when both sexes were subject to Dnmt inhibition, suggesting that some underlying sex differences in gene expression are actively suppressed by DNA methylation, resulting in a convergence as opposed to divergence of males and females2, 25(Fig. 5a). This methylation-dependent convergence in gene expression may provide an undifferentiated backdrop against which divergence in gene expression is more greatly amplified. One notable example was Foxp2 in which males showed increased expression after Dnmt inhibition thereby making them different from Dnmt inhibitor treated females. We have recently observed a sex difference in Foxp2 mRNA and protein in multiple brain regions, but the POA and hypothalamus were notable for the absence of a sex difference26, suggesting pressure toward convergent expression of this gene in males and females in reproductively relevant brain regions, which is achieved via DNA methylation.
DNA methylation regulates sex-specific gene isoform expression
DNA methylation has been implicated in selective expression of specific transcript isoforms via differential methylation of alternative gene promoters27. We detected 187 sex differences in gene isoform expression in the developing POA, far more than observed for overall gene expression. A total of 100 gene isoforms were higher in the male POA compared to control females, and 87 isoforms were higher in females vs. males (Figure 5c). One gene isoform higher in males compared to females was Ppp1r9b which codes for Neurabin II, suggesting the well-established sex difference in this proxy dendritic spine marker may arise from differential transcript expression. Using qPCR, we again confirmed higher levels of Ppp1r9b in male POA and a significant increase in females following estradiol treatment (Supplementary Fig. 7b). Moreover, 32 gene isoforms were exclusively found in the male POA (Supplementary Table 3), whereas 37 isoforms were exclusive to the female POA (Supplementary Table 3). Some of the differences in isoform expression were regulated by DNA methylation as Dnmt inhibition in females significantly increased the expression level of 58 of the 100 gene isoforms normally higher in males, and induced the expression of 16 of the 32 isoforms that were exclusively detected in males, creating patterns in isoform expression similar to males in Zeb-treated females (Fig. 5c; Supplementary Table 4). One notable isoform codes for drebrin 1 (Dbn1), a synaptic protein which determines dendritic spine morphogenesis in other brain regions28 and therefore may be important for sex differences in dendritic spine density in the POA. Remarkably, we found that Dnmt inhibition in females significantly reduced the expression of 40 of the 87 isoforms higher in females compared to males, and was able to prevent the expression of 16 of the 37 isoforms exclusive to females, which are likely transcripts important for feminization of the POA (Fig. 5c, Supplementary Table 4). Many of theses gene isoforms code for proteins involved in basic cellular structure and physiology, such as Ank1 (Ankyrin 1) and Cacna1a the voltage-gated Ca2+ channel, Cav2.1. The potential sources of sex differences in isoform expression include differential promoter usage, splicing variants and parental imprinting29. Analysis of promoter usage revealed only 25% and 29% of male and female-biased isoforms, respectively, were due to alternative promoter use, suggesting a mix of variables regulating sex-specific isoform expression. Many genes observed to have sex-specific isoform expression were not found to have overall differential expression, greatly amplifying both the amount and nature of sex differences in the transcriptome.
DISCUSSION
The traditional view of hormonally mediated sexual differentiation of the brain holds that steroids induce a male phenotype from the default female phenotype by binding to cognate nuclear transcription factor receptors and directly inducing expression of genes critical to brain development. This occurs during a restricted sensitive period and once complete, the process cannot be undone. Our data challenge both of these views. First, we demonstrate that in the developing POA a principle action of the masculinizing steroid, estradiol, is suppression of Dnmt activity and either prevention or reversal of DNA methylation, particularly at fully methylated CpG sites. This is evident in the ability of estradiol treatment of newborn female pups to both decrease the activity of Dnmts and decrease the number of fully methylated CpGs. The high degree of consistency between measures of global methylation using an antibody based assay and whole genome sequencing detection of fully methylated CpGs suggests the antibody based assay is biased toward fully methylated sites. Pharmacological inhibition of Dnmt activity mimics the actions of estradiol and masculinizes brain and behavior to the same degree as estradiol treatment during the sensitive period. Second, pharmacological inhibition of total Dnmt activity or genetic deletion of the de novo methyltransferase Dnmt3a outside of the sensitive period is effective at masculinizing brain and behavior, confirming that feminization is an active and ongoing repression of masculinization and thus reversible.
Non-CpG methylation is emerging as an important regulator of gene expression in the nervous system, acting as a transcriptional repressor similar to CpG methylation, shown to increase in frequency during periods of synaptogenesis30. The majority of CHG sites had very low levels of methylation yet there were significant sex differences, in particular females had fewer sites with 10–20% methylation than both males and masculinized females. The number of sites that were 90–100% CHG methylated was highly variable in females compared to males and masculinized females and may be reflective of dynamic synaptogenesis at this time. A limitation of our study is the heterogeneity of cell types within our POA tissue samples, given that patterns of both CpG and CHG methylation are markedly different in neurons and glia31.
In this study inhibition of Dnmt activity resulted in decreased DNA methylation in a global methylation assay, an effect previously observed by others32. Zeb is a well-established demethylating agent in cancer therapy and is used to reawaken hypermethylated tumor suppressor genes33. Originally thought to be a cell suicide inhibitor that irreversibly binds the Dnmts and requires cell division to be active, Zeb is now known to reversibly bind Dnmts and enhance their binding to unmethylated DNA. This, combined with a long-half life, acts to sequester the Dnmts and broadly prevent their action, thereby leading to demethylation34. This last step is likely achieved by concomitant activity of additional enzymes such as the TET-family of proteins which convert 5-methyl-cytosines into 5-hydroxymethyl-cytosines. This substrate is then vulnerable to oxidation into a variety of intermediates that are subject to base excision repair and return of the cytosine to an unmethylated state35. TET proteins are highly localized to gene promoters and speculated to maintain CpG islands in a relatively demethylated state and therefore free of epigenetic repression36. Also involved are the Gadd45 enzymes which are integral to base excision repair. Both the TET enzymes and Gadd45 are regulated by environmental factors, such as stress, and recruiters of transcriptional regulatory proteins including ERα37. We did not explore the role of TET or Gadd45 but they are established regulators of DNA demethylation in the brain38,39. Thus, regardless of the final source of demethylation, our data indicate feminization is the active process of suppressing masculinization via DNA methylation, which regulates both overall gene and specific gene isoform expression.
The concept of permanent perinatal organizational effects of steroids on the developing brain is long standing but not without challenges. There are examples of female rodents that exhibit male-like mounting behavior either in response to adult hormone treatment40 or consequent to genetic manipulation41. These examples are used as evidence for the duality of the brain, with some arguing for the simultaneous presence of both male and female circuits or phenotypes. The current data are wholly consistent with that view and shed new light by revealing that rather than the male genetic program simply not being induced, that it is instead actively suppressed in the female by a female-specific pattern of gene methylation. It is possible that adult hormones of appropriate duration and dose unmask the male genetic profile by removing epigenetic marks. Moreover, changes in gene expression profile are relatively modest in the brains of adult females following gonadectomy but robust in males8, consistent with the view that females maintain tight control of sexually relevant gene expression. A similar principle appears to apply to the ovary, where the deletion of a single gene, Foxl2, results in emergence of testicular tissue in the adult42.
With the use of RNA-Seq we identified differences in gene expression in the highly-sexually dimorphic POA. We identified relatively few sex differences in overall gene expression and, contrary to the expectation that the majority of genes would be expressed at higher levels in males (due to predicted direct genomic transcription by ER or AR), we found an almost equal distribution of genes expressed at higher levels in males or in females. Identifying genes regulating feminization of the brain has previously been difficult given that it is the default developmental pathway occurring in the absence of steroid hormone stimulation. Our observation that Dnmt inhibition reduced a substantial number of genes in females reveals an additional layer of complexity for which there is no current explanation. There were also few sex differences in X-linked genes. Only 1 of the 34 genes more highly expressed in males, and 3 of the 36 genes higher in females are X-linked, suggesting that autosomal genes play a large role in sexual differentiation of the POA.
In summary, the proximate mechanism by which females maintain control over neural gene expression clearly involves differential DNA methylation, both for purposes of preventing masculinization and allowing feminization, ultimately enabling the development of sex differences in the brain.
METHODS
Rat treatment and behavioral testing
All experiments were conducted with approval from the University of Maryland School of Medicine Institutional Care and Use Committee. Rat pups used in the following experiments were birthed by timed-pregnant Sprague Dawley rats ordered from Harlan Laboratories (Fredrick, MD). In all experiments, pups from multiple litters were randomly assigned to experimental treatment and then randomly distributed back to dams to control for differences in maternal care. Pregnant females were isolated and allowed to deliver normally. Cages were checked daily for the presence of pups to determine the timing of birth. All animals were allowed food and water ad libitum and were maintained on a reverse 12 hour light/dark cycle. After weaning, same sex animals were caged in groups of 3. One hundred micrograms of estradiol benzoate (Sigma) was delivered subcutaneously and dissolved in sesame oil. The benzoate moiety increases solubility of the steroid and enhances deposition into muscle for slow release, it does not alter the actions of the steroid. The Dnmt inhibitors zebularine (Zeb; 300ng in 1% DMSO; Calbiochem) or RG108 (300ng in 10% DMSO; Tocris) were administered intracerebroventricularly (ICV) to cryoanesthetized pups under bright light illumination, allowing for visualization of the cranial landmark Bregma to approximate the location of the lateral ventricle. A 23-gauge, 1 µl Hamilton syringe was lowered 2mm below the surface of the skull to reach the ventricle. ICV injections were delivered bilaterally. Each hemisphere was infused with a 1µl volume of drug delivered over 60 seconds. Following SC and ICV hormone/drug treatments, animals received small SC injections of ink on the paw for treatment group identification. Tissue used in each experiment was collected at specified time points.
At PN40 male and female rats were gonadectomized and implanted with a 30mm silastic capsule (1.57 mm inner diameter, 3.18 outer diameter) filled with crystalline testosterone (Sigma) at the nape of the neck. Implantation of these capsules mimics the circulating hormone levels typical of an adult male and allows for performance of male sexual behavior in developmentally masculinized females6. Three weeks following surgery and testosterone replacement, male sexual behavior was assessed. Animals were video recorded for 30 minutes between the hours of 12 pm and 4 pm, in a Plexiglas behavioral arena in the presence of a hormonally primed, sexually receptive stimulus female. Parameters of male sexual behavior quantified were the number of mounts, latency to first mount, number of intromission-like behaviors (thrusts), and latency to first intromission-like behavior, previously described44. Behavior in an open field was used to assess anxiety-related behavior (time spent in center of the arena) and locomotor activity (grid crosses and rears). Rats were placed in the arena for 5 minutes each. Assessment of locomotor activity and male sexual behavior was conducted under dim red light during the dark phase of the animal’s light cycle. The observer was blind to the experimental treatment of each animal. Data generated during behavioral testing were analyzed by one-way analysis of variance (ANOVA). All ANOVAs were followed by Tukey’s post-hoc comparisons to determine group differences. D’Agostino & Pearson omnibus normality tests indicated that these and all other parametric data were normally distributed. Statistical significance was set at α = 0.05. Statistical analyses for all behavioral tests were performed in GraphPad Prism for Mac OS X (version 6.00; GraphPad Sofware, La Jolla, CA).
Estrous cyclicity was assessed in a separate group of female rats, injected with Zeb or vehicle on PN0 and PN1 as described above, and raised to adulthood. Around PN40 vaginal smears were performed between 10 and 11 am daily for 15 consecutive days. Cells were viewed at 10× magnification and each animal was described as in diestrus, proestrus, or estrus each day, based on the shape and density of cells as previously described45. T-tests were used to assess differences in the number of days spent in each stage of estrus between groups. Statistical significance was set at α = 0.05. Data were graphed in R46.
Golgi-Cox labeling and quantification
Labeling was based on Glaser and Van der Loos47. Briefly, deeply anesthetized rats were transcardially perfused with ice cold saline for approximately 5 minutes. Brains were removed and placed in COX solution (K2Cr2O7, HgCl2, K2CrO4) for 3 weeks. COX solution was replaced every 3 days. Brains were transferred to Solution C (RapidGolgi Kit, FD) for 3 days at 4°C. Brains were sectioned on a cryostat at 100 um and mounted onto slides. Slides were placed in the dark to dry for 1 week. Slides were rinsed in dH2O, incubated in NH4OH, rinsed again in dH2O, incubated in 1% Kodak Dektol, rinsed again, then fixed in 18% Kodak Fix. Neurons were traced using Neurolucida (MBF Biosciences) at microscope at 40×.
Conditional deletion of Dnmt3a and behavioral testing in mice
All experiments were conducted with approval from the Mt.Sinai School of Medicine Institutional Care and Use Committee. Dnmt3a 2lox/2lox mice were originally generated in the laboratory of En Li at Novartis Institutes of Biomedical Research43. They were backcrossed with 129Jae mice for several generations to generate a common genetic background. At the Ichan School of Medicine at Mt. Sinai, male and female pups from 6 litters were bilaterally infused into the POA with 0.23 ul of AAV (1.5 × 108 infectious units/ ml) expressing control GFP or Cre/GFP on the day-of-birth or postnatal day one. Injections were performed with a fixed needle 1ul Hamilton syringe on cryoanesthetized pups. Bilateral injections were made 0.5mm lateral to Bregma to a depth of 2.5mm and infused over a 60 sec period. Group identification was achieved by injection of India ink into the paws. Animals were warmed by a heating pad and returned to the dam when fully active. At ~70 days of age all animals were gonadectomized under anesthesia (Ketamine/acepromazine 0.25ml) and implanted with a testosterone releasing silastic capsule that produces adult male circulating levels within 2–3 weeks. Animals were group housed, 5/cage, by sex until behavioral testing.
Stimulus females were gonadectomized as above and implanted with estradiol-releasing silastic implants to induce sexual receptivity. On the day of testing subject animals were placed in test cages for 30 min prior to the introduction of a sexual receptive stimulus female and behavioral interactions videotaped for 30 min. Testing was conducted at 3–5 day intervals for a total of 3 tests. All testing started at 6 pm and was performed under red light conditions. Behavioral scoring was done by an investigator blind to experiment group and followed previously established protocols for mice48,49. Briefly, animals were assessed for duration of anogenital investigation, number of solicitations, mounts and thrusts, mount time and latency, thrust rate and % of animals performing male sex behavior. At the completion of behavioral testing a subset of animals was euthanized and perfused for detection of GFP in the POA.
Immunohistochemistry
Mouse brain sections were frozen sectioned at 45 µm, blocked with 10% normal goat serum (NGS) in PBS + 0.4% Triton-X (PBST), incubated overnight at 4°C in antiserum to GFP (Aves #1020; 1:3000; 1DegreeBio ID: 1DB-001-0000745552) in PBST + 5% NGS, subsequently rinsed and incubated for 2 h in goat anti-chicken IgG conjugated to AlexaFluor-488 (Invitrogen, 1:333) in PBST + 5% NGS, mounted, and cover slipped with VectaShield Hard Set (Vector Laboratories). Sections were imaged with an Olympus BX51 microscope coupled to a cooled CCD camera. Injection site accuracy into the POA was confirmed via GFP staining in all mice used for behavioral experiments.
Nuclear extraction and western blot
Nuclear extracts were generated from POA dissections using the EpiQuik ® Nuclear Extraction Kit (Epigentek) using the manufacturer’s protocol. Protein concentrations were determined by Bradford assay. Nuclear extracts were used immediately or frozen at −80°C until use in the Dnmt activity assay or for quantification of Dnmt protein levels by western blot.
Protein concentrations of 10 µg /20µl were electrophoresed in separate lanes on an 8–16% SDS-polyacrylamide gel (Invitrogen) and transferred to a polyvinyl diflouride membrane (Bio-Rad, Hercules, CA). Membranes were blocked in 5% nonfat milk in 0.1% Tween TBS (T-TTBS) for 1 hr at room temperature and then incubated overnight at 4°C in either anti-Dnmt1 (Santa Cruz, Cat.# SC–20701; 1:2000; 1DegreeBio ID: 1DB-001-0001405395), anti-Dnmt3a (Cell Signaling, Cat.# 2160; 1:1000; 1DegreeBio ID: 1DB-001-0000807788), anti-Dnmt3b (Cell Signaling, Cat.# 2161; 1:1500; 1DegreeBio ID: 1DB-001-0000807789), or anti-Neurabin (Millipore, Cat.# 06–852;1:000; 1DegreeBio ID: 1DB-001-0000850617) in 2.5 % milk in TTBS, followed by a 30-minute incubation in goat anti-rabbit HRP conjugated IgG (Cell Signaling; 1:3000) in TTBS. The Phototope chemiluminescence system (New England BioLabs) was used for detection of proteins recognized by the antisera. The blots were exposed to Hyperfilm-ECL (Amersham) for varying amounts of time (1–10 min). After visualization, membranes were washed briefly with distilled water and stained with Ponceau S solution. The dense protein band stained by Ponceau S appearing at approximately 45 kDa was used for standardization to correct errors in sample loading. The band appearing at approximately 45 kDa contains a large number of proteins ranging in size from 43–47 kDa, which reduces the possibility of a treatment effect on any one control protein.
The integrative grayscale pixel area density (IAD) for each protein was captured with a CCD camera and quantified using NIH Image software. Samples were compared across several immunoblots for all experiments with 5–8 samples for each group within each blot. For all western blots, each lane represents one animal. Data were analyzed using a one-way ANOVA followed by a Tukey’s post hoc test when appropriate. All statistical tests used α<0.05 as the criterion for significance. Data in graphical form represent the mean ± SEM for each group.
Dnmt activity assay
One half micrograms of nuclear extracts from the POA were analyzed in triplicate for relative levels of Dnmt activity using the EpiQuik ® Dnmt Activity/Inhibition Assay Ultra Kit (Epigentek #p-3010) according to the manufacturer’s instructions. The EpiQuik™ Dnmt Activity/Inhibition Assay Ultra Kit is designed to measure total Dnmt activity, from all three Dnmt isoforms. In this assay, strip wells are coated with a cytosine-rich DNA substrate. During a 45 min reaction, Dnmt enzymes from the nuclear extract samples transfer a methyl group to cytosines from the methyl-donor molecule, Adomet, to methylate the DNA substrate. The methylated DNA is then recognized with an anti-5-methylcytosine antibody. The amount of methylated DNA, which is proportional to enzyme activity, can then be fluorometrically quantified using 530nm excitation and 590 nm emission readings from a microplate reader. To compare across plates, each plate contained triplicate standards of 1, 0.6, 0.4, 0.2 and 0.1 ug of protein from pooled nuclear extracts, which together were regressed with a quadratic equation and set equal to the regressed equation from the other plates to transform the fluorescent values from each sample. Data were analyzed using a two-way ANOVA followed by a Tukey’s post hoc test. Dnmt activity data in supplementary material were obtained using an older, colormetric version of the same assay kit. All statistical tests used a<0.05 as the criterion for significance. Data in graphical form represent the mean ± SEM for each group.
DNA isolation and quantification of global methylation
DNA from POA samples was extracted using the Wizard Genomic DNA Purification Kit (Promega) following the manufacturer’s instructions. The Methylamp Global DNA Methylation Quantification Ultra Kit (Epigentek) was used to quantify global methylation of DNA collected from POA tissue samples following the manufacturer’s instructions. DNA was bound to plate wells and then probed with an anti-5-methylcytosine antibody and HRP-linked secondary to produce a colorometric reaction read at an absorbance of 450nm using a microplate reader. Percent methylation was calculated using the following equation supplied by the manufacturer:
Where X = 42%, CG content in the rat. Data were analyzed using a one-way ANOVA followed by a Tukey’s post hoc test when appropriate. All statistical tests used α<0.05 as the criterion for significance. Data in graphical form represent the mean ± SEM for each group.
Whole Genome Bisulfite Sequencing
Pups were treated subcutaneously with 100 µg of estradiol benzoate or oil on PN0 and PN1. Genomic DNA was isolated from PN4 preoptic areas using the Qiaamp DNA Micro Kit (Qiagen, Hilden Germany) according to the manufacturer instructions. The concentration and quality of gDNA was verified with absorbance spectroscopy such that the samples had a 260 nm/280 nm ratio of between 1.8 to 2.1. One microgram of gDNA sample was used for library construction. Bisulfite converted DNA libraries were produced and adaptor ligated, and single-end reads were sequenced on an Illumina HiSeq-2500 by Epigentek (Farmingdale, NY), following library QC. Bisulfite conversion efficiency was greater that 98% for each of 9 samples and we obtained an average of 276 million reads per library. Details of libraries are provided in Supplementary Table 5. Quality control on raw reads was performed with FastQC50 (version 0.10.1), and adapter trimming and removal of trimmed reads shorter than 20 bp was performed with Trim Galore51 (version 0.3.7). Trimmed reads were mapped to the UCSC Rattus norvegicus rn5 genome with the methylation-aware mapper, bismark52 (version 0.13.0). Samtools53 (version 0.1.19–96b5f2294a) used to sort SAM files produced by bismark and de-duplicate reads. SAM files were analyzed using MethylKit54 (version 0.9.2) in R. Reads were filtered based on coverage, with a cutoff of at least 3 reads per site, and normalized for coverage prior to analysis. Differential methylation was defined as a difference of at least 10% between groups, with a q value of < 0.01. Additional data analysis was performed in R and graphed in GraphPad Prism for Mac OS X (version 6.00; GraphPad Sofware, La Jolla, CA).
Quantitative Real Time PCR
RNA was extracted using the RNAse easy kit with Qiazol and columns and DNase digestion per Qiagen’s protocol. Single strand complementary DNA was synthesized with the high capacity cDNA kit (Applied Biosystems) by mixing 1 µg total RNA with 4 µL random hexamers, 4 µL of 10X RT Buffer, 4 µL of 25X dNTP Mix, 2 µL of MultiScribe reverse transcriptase and 2 µL of RNase inhibitor and bringing the total volume to 40 µL with nuclease free water. The mixture was then incubated at 25° C for 10 min, 37° C for 2 hrs, and 85° C for 5 min and then was stored at −20 ° C until used. All primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Primers for total Nr2f2 and Gapdh were designed at www.ncbi.nih.gov. All other primers were designed using Primer Express (version 3.0, Applied Biosystems) and. Primer sequences are as follows: Dnmt1 (NM_053354.3) For 5’ GCTTTGACGGTGGCGAGAA, Rev 5’ TCTGCAAGAACTCGACCACAATC; Dnmt3a (NM_001003958.1 & NM_001003957.1 primers designed against all variants) For 5’ TTTCTTGAGTCTAACCCCGTGATG, Rev 5’ TGCAACTCCAGCTTATCATTCACA; Dnmt3b (NM_001003959.1 but primers designed against all variants) For 5’ GATGTGACACCTAAGAGCAGCAGTAC, Rev 5’ CAAACTCCTTGTCATCCTGATACTCA; Adora2a (NM_053294.3) For 5’ CCTCTTCTTCGCCTGTTTTGTC, Rev 5’ ATGCCCTTCGCCCTCACA; Darpp32 (NM_138521.1 and XM_006247344.1 primers designed against all variants) For 5’ AGCCCCAGAGAGATGGAAACTC, Rev 5’ TGAGCTTATGTGCCGGACTCA; Ebf2 (NM_001108383.1 and XM_006252133.1 primers designed against all variants) For 5’ ATGAGACGGTTTCAGGTCGTGTT, Rev 5’ TTTGATGCAGGGTGTAGCTTCTG; Ebf3 (NM_001108506.1, and XM_006230462.1, XM_006230463.1, XM_006230464.1, XM_006230465.1, primers designed against all variants) ACGCTTTGTCTACACTGCCCTTAA, Rev 5’ TGCCGCCCTCTTCAGTAACA; Nr2f2 (NM_080778.2 and XM_006229366.1, primers designed against all variants) For 5’ CACGTCGACTCCGCCGAGTAC, Rev 5’ ACGAAGCAAAGCTTTCCGAACCGT; Ppp1r9b (NM_053474.1) For 5’ AGGCCAAGCGTCTCATCAAG, Rev 5’ GGCGGTCTCTTTTTTCAGGAA; brain specific promoter Cyp19a1 (EF474097.1 for forward primer design, NM_017085.2 in the translated region for reverse primer design) For 5’ GCAAACTCACCACCTTCAAGAGT, Rev 5’ ATCTTGTGCTATTTTGCCTCAGAA; Gapdh (NM_017008.4) For 5’ TGGTGAAGGTCGGTGTGAACGG, Rev 5’ TCACAAGAGAAGGCAGCCCTGGT. Quantitative real-time PCR (qPCR) was quantified using the standard curve method on a ViiA 7 real-time PCR machine (Applied Biosystems) using ViiA 7 software (version 1.1). The standards were generated by pooling an equal amount of cDNA from all the samples and diluting the pool 1:10, 1:30, 1:90, 1:270, and 1:810. Values attained from the 1:30 standard were defined as one genomic equivalent (G.E..). The cDNA from each sample was diluted to 1:30. Performing all reactions in triplicate, 5 µL of each diluted sample or standard was then added to 15 µL of Power SYBR Green PCR Master Mix (Applied Biosystems) containing 100 nM of the primer pairs above and cycled in the real-time machine as follows: 95° C for 10 min, followed by 40 cycles of 95° C for 15 sec and an extension step of 60° C for 60 sec. The protocol for Ebf2 had an additional primer annealing step of 59 °C for 10 sec before the extension step above. Melting curves were generated at 0.1°C increments between 65°C and 95°C after the 40 cycles. Threshold fluorescence was set to a value that generated cycle thresholds from the standard curve with a regressed exponential growth of 2 (R2>0.98). The G.E. for each sample was determined against the standard curve. The G.E. for the gene of interest for each sample was normalized to the G.E.. for Gapdh for each sample.
RNA-Seq
RNA-Seq was performed by the Institute for Genome Sciences (IGS) at the University of Maryland on 12 rat cDNA samples, n=3/experimental group. One vehicle-treated female sample was later removed from the analysis because it showed aberrant read counts compared to all other samples. Animals were treated on PN0 and PN1 with either vehicle (0.1% DMSO) or 300ng/hemisphere Zeb and RNA was extracted on PN2. Illumina mRNA-Seq libraries were prepared with a TruSeq RNA Sample Prep kit (Illumina, San Diego, CA) using the manufacturer’s protocol with IGS-specific optimizations. Adapters containing 6 nucleotide indexes were ligated to the double-stranded cDNA. DNA was purified between enzymatic reactions and the size selection of the library was performed with AMPure XT beads (Beckman Coulter Genomics, Danvers, MA). Libraries were sequenced using the 100bp paired-end protocol on an Illumina HiSeq2000 sequencer. An average of 87.9 million reads were generated per library. Raw data from the sequencer was processed using Illumina’s RTA and CASAVA pipeline software, which includes image analysis, base calling, sequence quality scoring, and index demultiplexing. Data were then processing through both FastQC and IGS in-house pipelines for sequence assessment and quality control. By default, IGS quality control pipeline assesses base call quality and truncates reads where the median Phred-like quality score falls below Q20.
The reads obtained from each rat HiSeq sample were fed into the TopHat alignment tool55 to be aligned to the Rattus norvegicus (rn4) reference genomic sequence, downloaded from the UCSC Genome Browser’s server. In the alignment phase, we allowed up to two mismatches per 30 bp segment and removed reads that aligned to more than 20 locations. BAM files were then forwarded into DESeq56 to analyze count data and test for differential expression. The number of reads mapped to each gene described in the Ensembl gene annotation for rn4 was calculated by the HTSeq read count tool. DESeq utilizes read counts to determine plausibly differentially expressed genes when compared between samples. The read counts of samples were normalized for sequencing depth and distortion caused by highly differentially expressed genes. Genes were considered differentially expressed if they met a fold change (FC) cutoff of >1.0, a false discovery rate (FDR) < 0.1, and a p value < 0.05.
The alignment BAM files next inputted into the Cufflinks transcriptome identification tool. The Cufflinks tool has been developed at University of Maryland Center for Bioinformatics and Computational Biology as a method to assemble aligned RNA-Seq reads into transcripts, estimate their abundances, and test for differential expression and regulation transcriptome-wide. Cufflinks assembles individual transcripts utilizing the spliced read information provided within the TopHat alignment files and produces a measure of abundance of transcripts in Fragments Per Kilobase of exon per Million fragments mapped (FPKM). Cuffdiff was used to estimate the differential expression between samples (grouped by condition) at the transcript level. The statistical model assumes that the number of reads produced by each transcript is proportional to its abundances. Transcript expression from replicates is used to estimate variance and calculate the significance of observed changes in expression. The significance of differential expression of genes or transcripts belonging to the same gene across the conditions was tested using the negative binomial distribution. A cutoff of false discovery rate (FDR) less than 0.1, a p value less than 0.01, and a fold change (FC) greater than 1 were used to select significant differentially expressed transcripts and genes. Heat maps were generated in R using gplots57.
Additional Information on Sample Sizes
Sample sizes for Dnmt activity and global methylation assays were kept small due to technical limitations of these assays, which prevent across assay comparisons in the 96 well plates. Sample size for all other molecular and behavioral analyses was based on a predicted standard deviation that did not exceed 10% of the mean and an anticipated magnitude difference of 0.5–2.0, both of which are based on 30 years of experience in the study of sex differences in the brain. For all experiments, data points were excluded if they fell 2X's outside the standard deviation above or below the mean. There was never more than one outlier per group and overall the occurrence of outliers was rare.
In most behavioral experiments, the n's in each group started out the same. Because these experiments included developmental treatment of animals raised to adulthood, naturally-occurring, random animal loss sometimes altered final group n's. N’s for behavioral measures often varied due to uncertainty during scoring videotapes. If a behavior was not observed it was not scored, i.e. females that did not display thrusts where excluded from thrust latency analysis. A supplementary methods checklist is available.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. B.K. Krueger and S.M. Thompson for their helpful input on this manuscript. We thank Dr. Gouping Fan for kindly providing the Dnmt3a floxed mice. This work was supported by grant R01 MH052716 to MMM and F31NS073545–01 to BMN. This work was conducted as part of the doctoral thesis requirements of BMN.
Footnotes
The authors of this manuscript have no competing financial interests or other interests that might be perceived to influence this article.
DATABASE ACCESSION NUMBERS
RNA-Seq reads (accession: GSE66203) have been deposited in NCBI’s Gene Expression Omnibus (GEO). Whole genome bisulfite sequencing reads can be found in NCBI’s Short Read Archive under the BioProject ID:275796.
AUTHOR CONTRIBUTIONS
BMN and MMM designed the experiments and wrote the manuscript. BMN performed most molecular biology experiments, rat pharmacology and behavioral experiments, analyzed molecular and behavioral data, and performed bioinformatics analysis of WGBS data. CLW conducted the qPCR, some Dnmt activity assays, and extracted DNA for WGBS. ACS analyzed RNA-Seq data, and AM and SD provided additional bioinformatics support for RNA-Seq. SJR provided transgenic mice and GEH and MMM performed mouse experiments. KML performed immunohistochemistry.
References
- 1.McCarthy MM. Estradiol and the developing brain. Physiol. Rev. 2008;88:91–134. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat. Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kudwa A, Michopoulos V, Gatewood J, Rissman E. Roles of estrogen receptors α and β in differentiation of mouse sexual behavior. Neuroscience. 2006;138:921–928. doi: 10.1016/j.neuroscience.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, et al. Brain masculinization requires androgen receptor function. Proc. Natl. Acad. Sci. 2004;101:1673–1678. doi: 10.1073/pnas.0305303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amateau SK, McCarthy MM. A novel mechanism of dendritic spine plasticity involving estradiol induction of prostaglandin-E2. J Neurosci. 2002;22:8586–8596. doi: 10.1523/JNEUROSCI.22-19-08586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakker J, Honda SI, Harada N, Balthazart J. The aromatase knock-out mouse provides new evidence that estradiol is required during development in the female for the expression of sociosexual behaviors in adulthood. J Neurosci. 2002;22:9104–9112. doi: 10.1523/JNEUROSCI.22-20-09104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, et al. Modular Genetic Control of Sexually Dimorphic Behaviors. Cell. 2012;148:596–607. doi: 10.1016/j.cell.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simerly RB. Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu. Rev. Neurosci. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- 10.Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nat. Neurosci. 2004;7:1034–1039. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo CB, Cheng JC, Jones PA. Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans. 2004;32:910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 13.Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siedlecki P, et al. Discovery of two novel, small-molecule inhibitors of DNA methylation. J Med Chem. 2006;49:678–683. doi: 10.1021/jm050844z. [DOI] [PubMed] [Google Scholar]
- 15.Satoh A, et al. Neurabin-II/spinophilin. J. Biol. Chem. 1998;273:3470–3475. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- 16.Wright CL, Burks SR, McCarthy MM. Identification of prostaglandin E2 receptors mediating perinatal masculinization of adult sex behavior and neuroanatomical correlates. Dev. Neurobiol. 2008;68:1406–1419. doi: 10.1002/dneu.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everitt BJ. Sexual motivation: a neural and behavioural analysis of the mechanisms underlying appetitive and copulatory responses of male rats. Neurosci Biobehavl Rev. 1990;14:217–232. doi: 10.1016/s0149-7634(05)80222-2. [DOI] [PubMed] [Google Scholar]
- 18.Cardoso MC, Leonhardt H. DNA methyltrasferase is actively retained in the cytoplasm during early development. J. Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 39:310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Wedemeyer J, Galli SJ. Mast cells and basophils in acquired immunity. Br. Med. Bull. 2000;56:936–955. doi: 10.1258/0007142001903616. [DOI] [PubMed] [Google Scholar]
- 22.Silverman AJ, Asarian L, Khalil M, Silver R. GnRH, brain mast cells and behavior. Prog. Brain Res. 2002;141:315–325. doi: 10.1016/S0079-6123(02)41102-8. [DOI] [PubMed] [Google Scholar]
- 23.Amateau SK, McCarthy MM. Induction of PGE2 by estradiol mediates developmental masculinization of sex behavior. Nat. Neurosci. 2004;7:643–650. doi: 10.1038/nn1254. [DOI] [PubMed] [Google Scholar]
- 24.Lauber M, Sarasin A, Lichtensteiger W. Transient sex differences of aromatase (CYP19) mRNA expression in the developing rat brain. Neuroendocrinology. 1997;66:173–180. doi: 10.1159/000127235. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy MM, et al. Sex differences in the brain: the not so inconvenient truth. J Neurosci. 2012;32:2241–2247. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers JM, Perez-Pouchoulen MR, McCarthy MM. Foxp2 mediates sex differences in ultrasonic vocalization by rat pups and directs order of maternal retrieval. J Neurosci. 2013;20:3276–3283. doi: 10.1523/JNEUROSCI.0425-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H, et al. Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J Neurosci. 2003;23:6586–6595. doi: 10.1523/JNEUROSCI.23-16-06586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luco RF, et al. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JU. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237–1905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatino MA, et al. Zebularine partially reverses GST methylation in prostate cancer cells and restores sensitivity to the DNA minor groove binder brostallicin. Epigenetics. 8:656–665. doi: 10.4161/epi.24916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champion C, et al. Mechanistic insights on the inhibition of C5 DNA methyltransferases by zebularine. PLos One. 2010:e12388. doi: 10.1371/journal.pone.0012388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS one. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO reports. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Bio. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gavin DP, et al. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharm. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodersten P. Mounting behavior in the female rat during the estrous cycle, after ovariectomy, and after estrogen or testosterone administration. Horm. Behav. 1972;3:307–320. [Google Scholar]
- 41.Kimchi T, Xu J, Dulac C. A functional circuit underlying male sexual behaviour in the female mouse brain. Nature. 2007;448:1009–1014. doi: 10.1038/nature06089. [DOI] [PubMed] [Google Scholar]
- 42.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Dodge, et al. Inactivation of dnmt3b in mouse embryonic fibroblasts results in dna hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 44.Todd BJ, Schwarz JM, McCarthy MM. Prostaglandin-E2: a point of divergence in estradiol-mediated sexual differentiation. Horm Behav. 2005;48:512–521. doi: 10.1016/j.yhbeh.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy MM, Schlenker EH, Pfaff DW. Enduring consequences of neonatal treatment with antisense oligodeoxynucleotides to estrogen receptor messenger ribonucleic acid on sexual differentiation of rat brain. Endocrinology. 1993;133:433–439. doi: 10.1210/endo.133.2.8344188. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team. R Foundation for Statistical Computing. Vienna Austria: 2014. R: A language and environment for statistical computing. URL http://www.R-project.org. [Google Scholar]
- 47.Glaser EM, Van der Loos H. Analysis of thick brain sections by obverse-reverse computer microscopy: application of a new, high clarity Golgi-Nissl stain. J Neurosci Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- 48.Kimchi T, Xu J, Dulac C. A functional circuit underlying male sexual behavior in the female mouse brain. Nature. 2007;448:1009–1014. doi: 10.1038/nature06089. [DOI] [PubMed] [Google Scholar]
- 49.Park JH, Bonthuis P, Ding A, Rais S, Rissman EF. Androgen- and estrogen-independent regulation of copulatory behavior following castration in male B6D2F1 mice. Horm Behav. 2009;56:254–263. doi: 10.1016/j.yhbeh.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.FastQC. http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc. [Google Scholar]
- 51.Trim Galore. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore. [Google Scholar]
- 52.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;11:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;16:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akalin A, et al. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. http://dx.doi.org/10.1186/gb-2010-11-10-r106, http://genomebiology.com/2010/11/10/R106/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory R, Warnes GR, et al. gplots. Various R programming tools for plotting data. R package version 2.14.1. 2014 http://CRAN.R-project.org/package=gplots. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.