Abstract
Genotyping efforts in hemophilia A (HA) populations in many countries have identified large numbers of unique mutations in the Factor VIII gene (F8). To assist HA researchers conducting genotyping analyses, we have developed a listing of F8 mutations including those listed in existing locus-specific databases as well as those identified in patient populations and reported in the literature. Each mutation was reviewed and uniquely identified using Human Genome Variation Society (HGVS) nomenclature standards for coding DNA and predicted protein changes as well as traditional nomenclature based on the mature, processed protein. Listings also include the associated hemophilia severity classified by International Society of Thrombosis and Haemostasis (ISTH) criteria, associations of the mutations with inhibitors, and reference information. The mutation list currently contains 2,537 unique mutations known to cause HA. HA severity caused by the mutation is available for 2,022 mutations (80%) and information on inhibitors is available for 1,816 mutations (72%). The CDC Hemophilia A Mutation Project (CHAMP) Mutation List is available at http://www.cdc.gov/hemophiliamutations for download and search and will be updated quarterly based on periodic literature reviews and submitted reports.
Keywords: hemophilia A, mutation database, locus-specific database, F8 gene
INTRODUCTION
Hemophilia A (HA) is an X-linked inherited disorder that results in impaired blood clotting with subsequent bleeding. The disorder is a consequence of a mutation in the Factor VIII gene (F8; MIM# 300841) that leads to reduced production or inadequate function of Factor VIII (FVIII), a protein necessary for clotting. Because of its inheritance pattern, HA occurs almost exclusively in males, and it is estimated that more than 20,000 people are living with HA in the United States (CDC, 2012; Soucie, et al., 1998).
Severity of HA differs based on FVIII plasma level, with <1% of the normal protein level conferring the most severe disease (White, et al., 2001). HA can result in joint swelling and damage, intracranial hemorrhage, organ damage, and death if bleeding is not prevented or stopped (Berntorp and Shapiro, 2012). Complications of treatment of HA include development of antibodies (inhibitors) to treatment products that decrease the effectiveness of the products to stop hemorrhages and, rarely today, infection resulting from blood product contamination (Berntorp and Shapiro, 2012).
Because HA is an inherited disorder, knowledge of the mutation carried in a family can aid in prenatal diagnosis and identification of familial carriers (Goodeve, 2008). Also, certain mutation types have been shown to increase the risk of developing an inhibitor (Miller, et al., 2012), suggesting that knowledge of the mutation could be used to in the development of an individualized treatment strategy to reduce the chance of an inhibitor.
F8 spans 186,000 base pairs on the X chromosome. The gene includes 26 exons and produces an 8 kilobase mRNA and a mature FVIII protein of 2,332 amino acids (Gitschier, et al., 1984). Sequencing of the F8 cDNA has revealed a repeating domain structure (A1-A2-B-A3-C1-C2). FVIII is sensitive to proteolytic cleavage, with the majority of circulating FVIII consisting of heavy chains (A1, A2, and B domains) and light chains (A3, C1, and C2 domains) (Graw, et al., 2005). During clot formation, FVIII is proteolytically cleaved at the A2/B junction in order to open binding sites and allow for interaction with other proteins important in clot formation. Thus, the B domain is believed unnecessary for protein activity (Ogata, et al., 2011; Pipe, 2009).
Because F8 is located on the X chromosome, HA predominately affects males who either inherit a mutation in F8 from their mother or develop a de novo mutation. Rarely, females are affected with HA as a result of skewed X-chromosome inactivation or inheritance of two aberrant F8 alleles from their mother and father.
Mutations reported to cause HA may be characterized by 6 mechanisms: deletion, duplication, insertion, insertion/deletion, inversion, and substitution. These mutation mechanisms can lead to a shift in the reading frame (frameshift mutation), a large structural change in the gene, the replacement of an amino acid with a different amino acid (missense mutation), the replacement of an amino acid with a premature stop codon (nonsense mutation), a small structural change in the gene, an alteration of a splice site, a change in the promoter region of the gene, or a synonymous change whose effects on the mature protein remain unclear. It is estimated over 40% of severe HA cases are caused by inversions within the F8 gene, with the next most-common mutation mechanism being substitution (Miller, et al., 2012).
The CHAMP Mutation list was developed in response to a need to easily identify recurrent and novel mutations. Several locus-specific databases for F8 are currently available, such as the Haemophilia A Mutation, Structure, and Test Site (HAMSTeRS), Human Gene Mutation Database (HGMD), and Hemobase (Kemball-Cook and Tuddenham, 1997; Stenson, et al., 2009; Vidal F, 2010). However, these databases are not easily searchable, comprehensive in their scope, specific to HA, or regularly updated. We sought to systematically compile mutations known to cause HA as reported in the literature and elsewhere, to document them according to standardized nomenclature, to link them with relevant phenotypic information, and to make them easily searchable. A simple, comprehensive mutation list was therefore developed with information on each specific mutation reported to cause HA. This report describes the CDC Hemophilia A Mutation Project (CHAMP) Mutation List, a mutation list containing more than 2,500 F8 mutations reported to cause HA.
MATERIALS AND METHODS
The CHAMP Mutation List is hosted on the CDC website and can be freely accessed and downloaded at http://www.cdc.gov/hemophiliamutations. The mutation list is updated quarterly with recently-published mutations reported to cause HA.
Data Collection
In order to compile a comprehensive list of mutations reported to cause HA, F8 locus-specific databases (HAMSTeRS, Hemobase, and HGMD) were examined. Mutations reported to cause HA in these databases were identified, and publications cited for these mutations were reviewed in order to better characterize the reported mutations. Subsequently, a systematic literature search was conducted to identify additional reports not included in any of the databases. PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) was searched using the following terms: hemophilia A AND mutation, factor VIII AND mutation, F8 AND mutation. Any studies published in peer-reviewed journals that reported HA-associated mutations were eligible for inclusion. Finally, mutations identified as part of a large inhibitor surveillance effort conducted in our lab were compared to those already published in the literature in order to identify novel mutations. Each month, PubMed will be searched using the search terms described above to identify recently-reported mutations. The results of these searches will be combined into quarterly CHAMP Mutation List updates.
Data Extraction and Preparation
Only single mutations reported to cause HA in males were included in the CHAMP Mutation List, as inclusion of reports of multiple mutations causing disease and affected females could lead to phenotypic ambiguity. In the mutation list, each mutation is uniquely identified by its corresponding HGVS-standardized cDNA change. When available, the mutation list also provides the following additional information for each mutation: HGVS-standardized predicted protein changes as well as a link to the widely used mature protein nomenclature; the exonic, codon, and domain location of the mutation; the type and subtype of the mutation and mechanism underlying it; the phenotypic severity associated with the mutation; whether or not the mutation has been reported in association with inhibitors; a link to an identifier of the reference reporting the mutation; the year the mutation was first reported; and any additional comments, such as predicted functional changes caused by the mutation. Each of these items is described in more detail below.
Each mutation was assigned unique identifiers for cDNA and predicted protein changes using HGVS-standardized nomenclature, based on cDNA reference sequence NM_000132.3 and protein reference sequence NP_000123.1 (www.hgvs.org/mutnomen/). Naming was facilitated by use of the Mutalyzer program (Wildeman, et al., 2008). Each mutation listing also includes historical protein nomenclature, in which numbering begins at the codon that represents the first amino acid of the mature, processed protein.
The type of mutation was categorized using the following terms: frameshift, large structural change, missense, nonsense, promoter, small structural change, splice site change, or synonymous. A frameshift mutation was defined as any mutation that resulted in the shift of the reading frame. Large structural changes were defined as changes in F8 that involved an area of the gene that was >50 base pairs in length. Missense mutations were defined as any mutations that resulted from a single amino acid substitution for another amino acid. Nonsense mutations were defined as any mutation that resulted in the substitution of an amino acid with a stop codon. Promoter mutations were defined as those mutations occurring before the initiation codon. Small structural changes were defined as in-frame changes <50 base pairs in length. Splice site changes were defined as mutations located within the intronic regions of F8 that were reported by the researcher to be associated with splice site alterations. Synonymous changes were defined as mutations that caused no predictable change in the protein sequence.
The subtype of each mutation identifies whether or not a mutation occurred on the heavy chain or light chain of F8, a mutation occurring across multiple domains or a single domain, or mutations within a poly A chain. The mechanism was characterized as deletion or duplication if a portion of the F8 sequence was deleted or duplicated. The mechanism was characterized as insertion if extra base pairs were inserted into the F8 sequence. If a portion of the F8 sequence was deleted and extra base pairs were inserted in their place, the mechanism was characterized as insertion/deletion. If a large region of the gene was inverted, the mechanism was classified as inversion. If one base was substituted for another, the mechanism was characterized as substitution.
Using the recommendation of the Scientific and Standardization Committees of the International Society on Thrombosis and Haemostasis, a phenotypic severity classification was assigned to each report from factor levels provided in each publication, with <1% as severe, 1–5% as moderate, and >5% as mild (White, et al., 2001). The severity described in the original report is also provided in a separate field, even if this description does not match the assigned severity.
The presence or absence of inhibitors reported in association with each mutation is listed if these data were provided in the original publication. Otherwise, the mutation’s link with inhibitor development is listed as ‘Not Reported’.
A Reference ID is provided for each mutation, linking to information on the first publication to report the mutation. If the mutation was identified through searches of the other locus-specific databases and was listed as ‘unpublished’, the Reference ID links to a publication on that database.
Mutation List Design and Implementation
The CHAMP Mutation List is a freely downloadable file. The mutation list was developed in Microsoft Excel 2010 but is compatible with all versions of Microsoft Excel. The file contains tabs providing the Table of Contents, the Mutation List, Field Definitions, References, Figures, Tables, Instructions for Use, and Instructions for Submissions. The Table of Contents tab outlines the contents of all of the tabs in the Excel workbook. The CHAMP Mutation List tab contains a listing of mutations reported to be associated with HA. The Field Definitions tab contains an explanation of all of the fields in the CHAMP Mutation List tab. The References tab provides a link between the Reference ID provided in the CHAMP Mutation List and the citation for the reporting publication. The Figures tab provides several summary figures produced using data from the CHAMP Mutation List. Similarly, the Tables tab provides several standard summary tables produced from data in the mutation list. Instructions for sorting the data, finding a specific mutation, and filtering results are provided in the Instructions tab for all versions of Excel. Instructions for submitting either updates to existing mutations or novel mutations are provided in the Submissions tab. This simple structure allows the user to download and analyze the mutations quickly and easily.
RESULTS AND DISCUSSION
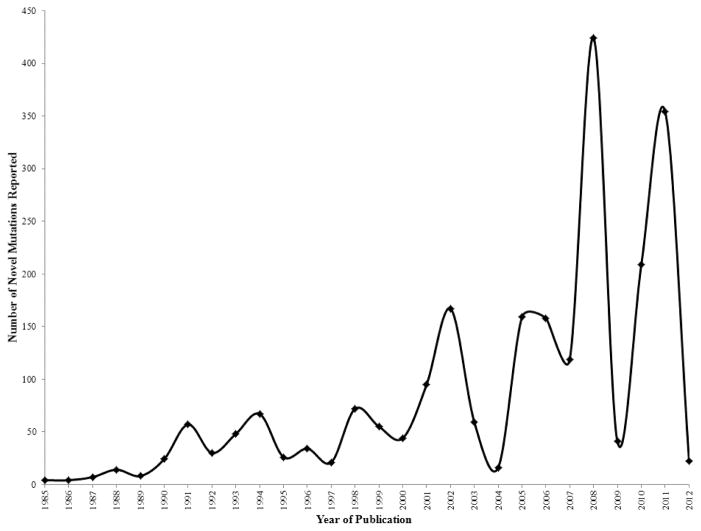
The CHAMP Mutation List was last updated in July of 2012. The mutation list currently contains 2,537 unique mutations reported to cause HA. Figure 1 shows the number of novel mutations reported by year of publication. The number of novel mutations per year peaked at 424 in 2008, a year when results from several large HA studies were published.
Figure 1.
Novel F8 mutations by year of publication.
Mutation Distributions
The distribution of mutation types is outlined in Table 1. The majority of unique mutations were missense mutations (49%), followed by frameshift (24%) and nonsense (11%) mutations. Splice site changes, large structural changes, synonymous mutations, and promoter mutations were the least reported mutation types (8%, 6%, 1%, and <1%, respectively).
Table 1.
Frequency of Mutation Types in CHAMP Mutation List
| Mutation Type | Mechanism | No. of Mutations | % of Mutations |
|---|---|---|---|
| Missense | Substitution | 1237 | 48.8% |
|
| |||
| Nonsense | Substitution | 290 | 11.4% |
|
| |||
| Frameshift | All | 595 | 23.5% |
| Deletion | 419 | 16.5% | |
| Duplication | 127 | 5.0% | |
| Insertion | 25 | 1.0% | |
| Insertion/deletion | 24 | 0.9% | |
|
| |||
| Splice site change | All | 191 | 7.5% |
| Substitution | 167 | 6.6% | |
| Deletion | 19 | 0.7% | |
| Duplication | 1 | 0.04% | |
| Insertion | 1 | 0.04% | |
| Insertion/deletion | 3 | 0.1% | |
|
| |||
| Large Structural Change (>50 bp) | All | 148 | 5.8% |
| Deletion | 121 | 4.8% | |
| Duplication | 19 | 0.7% | |
| Insertion | 5 | 0.2% | |
| Insertion/deletion | 1 | 0.04% | |
| Inversion | 2 | 0.1% | |
|
| |||
| Small Structural Change (in-frame, <50 bp) | All | 57 | 2.2% |
| Deletion | 34 | 1.3% | |
| Duplication | 5 | 0.2% | |
| Insertion | 7 | 0.3% | |
| Insertion/deletion | 11 | 0.4% | |
|
| |||
| Synonymous | Substitution | 13 | 0.5% |
|
| |||
| Promoter | Substitution | 6 | 0.2% |
|
| |||
| Total | 2537 | ||
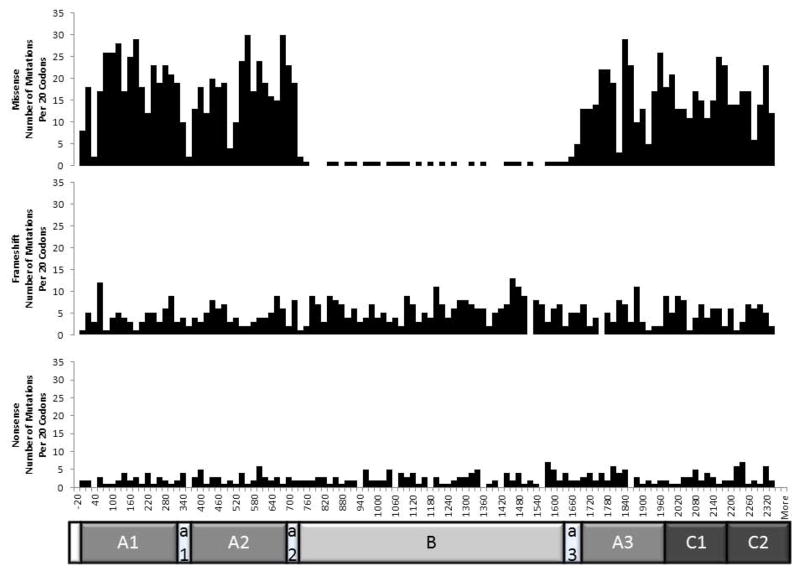
The distribution of missense, frameshift, and nonsense mutations throughout F8 are shown in Figure 2. Each bar represents 20 codons. Nonsense and frameshift mutations are evenly distributed throughout F8. However, missense mutations are relatively uncommon in the B domain of the gene. This is to be expected, since this portion of the gene is not necessary for proper function of the protein. Therefore, changes to the gene that are not expected to affect the downstream portion of the gene should not cause loss of function (Ogata, et al., 2011). Supp. Figure S1 shows the number of mutations per codon across each of the F8 domains. Again, it can be noted that missense mutations are relatively infrequent in the B domain. In the other major domains missense mutations are reported to occur at a frequency approaching 1 per codon.
Figure 2.
Distribution of missense, frameshift, and nonsense mutations in F8.
As stated above, 6% of mutations are reported to be large structural changes. A large percentage of those (82%) are large deletions that span one or more FVIII domains, with the remaining mutations being inversions and large duplications. The distribution of these mutations throughout the gene is shown in Figure 3. These large deletions range from small, single-domain deletions to large, whole-gene deletions.
Figure 3.
Reported large deletions in F8.
Mutations and Severity
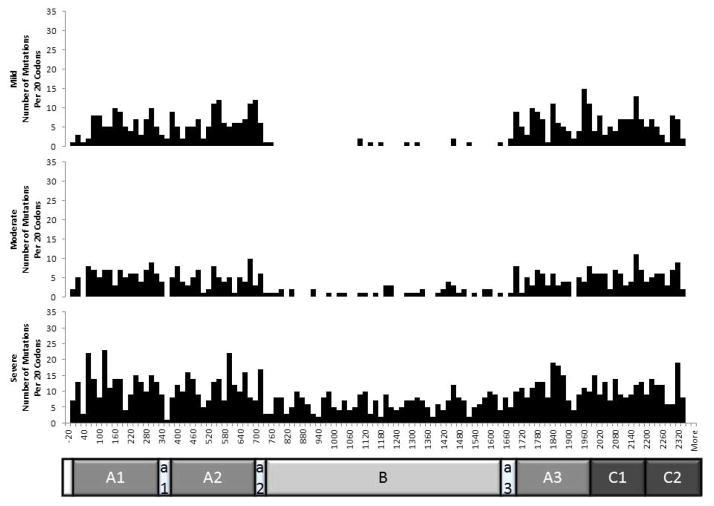
Factor level associated with the HA caused by the mutation was reported for 2,022 mutations (80%). The majority of mutations (n=1,178) were associated only with severe HA. Only moderate HA was reported for 294 mutations, and only mild HA for 398 mutations. Variable severity was reported for 152 mutations (7.5%). The distribution of mutations associated with mild, moderate, and severe HA throughout F8 is shown in Figure 4. The distributions of mild- and moderate-associated mutations are similar, with few mutations associated with mild or moderate disease mapped to the B domain. However, severe-associated mutations are relatively evenly distributed throughout the gene. This is likely due to the fact that many of the mutations causing severe disease are nonsense or frameshift mutations that prematurely truncate the gene product.
Figure 4.
Distribution of mutations associated with mild, moderate, and severe HA throughout F8. Classes of mutations include missense, nonsense, frameshift, promoter, in-frame, and synonymous mutations.
Mutations and Inhibitors
Information on whether or not a mutation was associated with inhibitor development was recorded for 1,816 mutations (72%) (Table 2). Of these, 353 (19%) have been observed in patients with inhibitors. Large structural changes are disproportionately associated with inhibitors. For example, 70% of large deletion mutations have been associated with inhibitors.
Table 2.
F8 mutation types with reported inhibitors
| Classification | No. of Mutations* | No. (%) with Inhibitors | |
|---|---|---|---|
| Missense | All | 895 | 84(9) |
|
| |||
| C1 and C2 | 181 | 25(14) | |
| Non-C1 and C2 | 714 | 59(8) | |
|
| |||
| Nonsense | All | 220 | 64(29) |
|
| |||
| C1 and C2 | 42 | 14(33) | |
| Non-C1 and C2 | 178 | 50(28) | |
|
| |||
| Frameshift | All | 420 | 101(24) |
|
| |||
| Poly A run | 92 | 25(27) | |
| Non-poly A run | 328 | 76(23) | |
|
| |||
| Large Structural Change (>50 bp) | All | 114 | 72(63) |
|
| |||
| Large deletions | 98 | 69(70) | |
| Multiple domain | 56 | 45(80) | |
| Single domain | 57 | 27(47) | |
|
| |||
| Small Structural Change (in-frame, <50 bp) | All | 33 | 5(15) |
|
| |||
| Splice site change | All | 119 | 24(20) |
|
| |||
| Promoter | All | 4 | 0 |
|
| |||
| Synonymous | All | 11 | 3(27) |
|
| |||
| Total | 1816 | 353(19) | |
Mutations with inhibitor status reported, 1,816 (72%) of total records
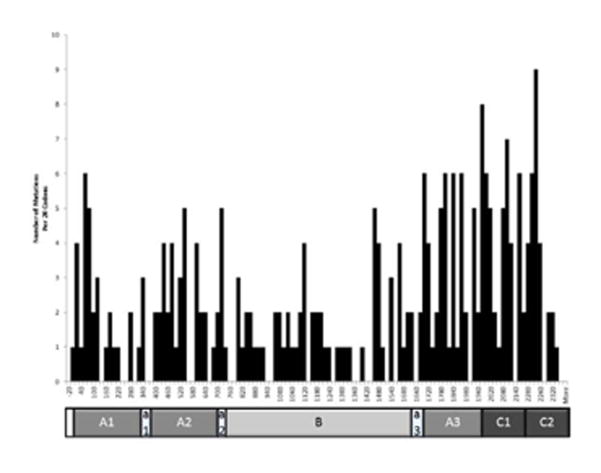
The distribution of the inhibitor-associated mutations throughout F8 is shown in Figure 5. Mutations associated with inhibitors are more heavily clustered in the light chain of the gene. Of 293 mutations located in the C1 and C2 domains, 61 (20.8%) have been reported in association with inhibitors versus 204 of 1,302 mutations located outside the C1 and C2 domains (15.7%) (P=0.03).
Figure 5.
Distribution of mutations associated with inhibitors across F8.
CONCLUSIONS
The CHAMP Mutation List is the most comprehensive, freely-accessible listing of F8 mutations reported to cause HA available to date. It has approximately 1,300 more novel mutations than the current version of HAMSTeRS (Kemball-Cook and Tuddenham, 1997). This new resource allows researchers to easily search and analyze mutations in F8 that are known to be associated with HA by providing them with information regarding their location within F8, their associated severity of disease and their association with disease-complicating inhibitors. Researchers can also use the resource to study mutation distributions, such as the analyses provided in this report.
After its launch in 1996, HAMSTeRS became a useful tool for investigators to assess the novelty of identified mutations or their previous associations with disease. However, the structure of the database has not been updated since 2007, one year before the peak of novel mutation reporting. The database does not use unique identifiers for mutations, such as HGVS cDNA nomenclature. This leads to some ambiguity, particularly regarding large deletions and point mutations occurring within the same codon. Summary analyses of mutation distributions become difficult because of this ambiguity as well as the ambiguity arising from multiple reports of the same mutation. Because the CHAMP Mutation List has been designed to include only a single listing of a mutation with a unique identifier, it is hoped that this ambiguity will be diminished.
Several non-locus-specific databases such as the Database for Single Nucleotide Polymorphisms (dbSNP) (Sherry, et al., 2001) and HGMD (Stenson, et al., 2009) compile reported mutations within F8. However, the location of the mutations within F8 and their relative location to codons and domains is not always intuitive. Limited information regarding the severity of HA associated with mutations or their association with inhibitor development is available, unless one wishes to link to the original publication.
The CHAMP Mutation List provides comprehensive information regarding mutation location, severity of disease reported to be associated with mutations, and whether or not the mutations have been associated with inhibitor development, as well as providing a reference to the original publication. The CHAMP Mutation List does have some limitations. In the majority of large deletions and duplications reported, exact break points have not been determined. It is therefore not possible to distinguish which reported mutations are identical and which are only similar. We have included in the Mutation List a single record for each deletion or duplication described only at the exon level. The true number of unique deletions and duplications is likely to be underestimated. Similarly, we have excluded mutation reports in which more than one mutation was reported to be associated with a patient’s HA, as the severity and inhibitor associations would be ambiguous. It is possible this could lead to an under-reporting of mutations. However, reporting of more than one mutation causing a patient’s hemophilia was rare. For example, fewer than 30 mutations out of close to 1200 mutations reported in HAMSTeRS were seen in combination with another mutation. Also, because updates to the mutation list are based on literature reviews, unpublished mutations would not be included in the Mutation List. However, we have provided a mechanism for researchers to provide information on unpublished mutations. Finally, since the frequencies given in this report are frequencies of mutations and not of patients, they cannot be used as predictors of patient outcomes. As previously stated, it is estimated over 40% of severe HA cases are caused by inversions; however, the CHAMP Mutation List only includes 2 listings for inversions, illustrating the limitations of extrapolating the frequency of a particular mutation type to frequencies of patients affected by that mutation type. We are currently working to coordinate a list of F8 mutations observed in the United States as well as those observed worldwide in population-based studies of HA.
The CHAMP Mutation List was designed to combine information on the location of mutations throughout F8 as well as information on their association with severity of HA and complicating inhibitors. It is hoped this resource will help researchers and clinicians better understand the molecular etiology of HA and foster scientific collaboration.
Supplementary Material
Acknowledgments
This work was supported by the CDC Foundation through a grant from Pfizer Inc. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
The authors have no conflicts of interest to declare.
References
- Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379(9824):1447–56. doi: 10.1016/S0140-6736(11)61139-2. [DOI] [PubMed] [Google Scholar]
- CDC. Hemophilia Data and Statistics. 2012 http://www.cdc.gov/ncbddd/hemophilia/data.html.
- Gitschier J, Wood WI, Goralka TM, Wion KL, Chen EY, Eaton DH, Vehar GA, Capon DJ, Lawn RM. Characterization of the human factor VIII gene. Nature. 1984;312(5992):326–30. doi: 10.1038/312326a0. [DOI] [PubMed] [Google Scholar]
- Goodeve A. Molecular genetic testing of hemophilia A. Semin Thromb Hemost. 2008;34(6):491–501. doi: 10.1055/s-0028-1103360. [DOI] [PubMed] [Google Scholar]
- Kemball-Cook G, Tuddenham EG. The Factor VIII Mutation Database on the World Wide Web: the haemophilia A mutation, search, test and resource site. HAMSTeRS update (version 3.0) Nucleic Acids Res. 1997;25(1):128–32. doi: 10.1093/nar/25.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CH, Benson J, Ellingsen D, Driggers J, Payne A, Kelly FM, Soucie JM, Craig Hooper W The Hemophilia Inhibitor Research Study I. F8 and F9 mutations in US haemophilia patients: correlation with history of inhibitor and race/ethnicity. Haemophilia. 2012;18(3):375–382. doi: 10.1111/j.1365-2516.2011.02700.x. [DOI] [PubMed] [Google Scholar]
- Ogata K, Selvaraj SR, Miao HZ, Pipe SW. Most factor VIII B domain missense mutations are unlikely to be causative mutations for severe hemophilia A: implications for genotyping. J Thromb Haemost. 2011;9(6):1183–90. doi: 10.1111/j.1538-7836.2011.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipe SW. Functional roles of the factor VIII B domain. Haemophilia. 2009;15(6):1187–96. doi: 10.1111/j.1365-2516.2009.02026.x. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucie JM, Evatt B, Jackson D. Occurrence of hemophilia in the United States. The Hemophilia Surveillance System Project Investigators. Am J Hematol. 1998;59(4):288–94. doi: 10.1002/(sici)1096-8652(199812)59:4<288::aid-ajh4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Howells K, Phillips AD, Mort M, Cooper DN. The Human Gene Mutation Database: providing a comprehensive central mutation database for molecular diagnostics and personalized genomics. Hum Genomics. 2009;4(2):69–72. doi: 10.1186/1479-7364-4-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal FGD. Hemobase. Catalonia, Spain: 2010. http://www.hemobase.com/EN/ [Google Scholar]
- White GC, 2nd, Rosendaal F, Aledort LM, Lusher JM, Rothschild C, Ingerslev J, Factor V, Factor IXS. Definitions in hemophilia. Recommendation of the scientific subcommittee on factor VIII and factor IX of the scientific and standardization committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 2001;85(3):560. [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29(1):6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.