Abstract
MicroRNAs (miRNAs) are a group of endogenous small non-coding RNAs that play important roles in plant growth, development, and stress response processes. Verticillium wilt is a vascular disease in plants mainly caused by Verticillium dahliae Kleb., the soil-borne fungal pathogen. However, the role of miRNAs in the regulation of Verticillium defense responses is mostly unknown. This study aimed to identify new miRNAs and their potential targets that are involved in the regulation of Verticillium defense responses. Four small RNA libraries and two degradome libraries from mock-infected and infected roots of cotton (both Gossypium hirsutum L. and Gossypium barbadense L.) were constructed for deep sequencing. A total of 140 known miRNAs and 58 novel miRNAs were identified. Among the identified miRNAs, many were differentially expressed between libraries. Degradome analysis showed that a total of 83 and 24 genes were the targets of 31 known and 14 novel miRNA families, respectively. Gene Ontology analysis indicated that many of the identified miRNA targets may function in controlling root development and the regulation of Verticillium defense responses in cotton. Our findings provide an overview of potential miRNAs involved in the regulation of Verticillium defense responses in cotton and the interactions between miRNAs and their corresponding targets. The profiling of these miRNAs lays the foundation for further understanding of the function of small RNAs in regulating plant response to fungal infection and Verticillium wilt in particular.
Keywords: cotton, Verticillium dahliae Kleb., microRNA, high-throughput sequencing, degradome
1. Introduction
MicroRNAs (miRNAs) are a class of endogenous non-coding small RNAs (sRNAs) that regulate gene expression at the transcriptional and post-transcriptional levels via mRNA cleavage or translational repression in plants and animals [1,2,3]. In higher plants, miRNAs play important roles in growth, development, stress responses, and many other biological processes [4,5,6]. In particular, there is increasing evidence that miRNAs are involved in regulating abiotic and biotic stress responses, including disease resistance [7,8,9,10]. Recently, sRNA-mediated gene silencing was found to play a significant role in plant defense against pathogens [8,11,12,13]. In Arabidopsis thaliana, miR393 was found to contribute to basal defense against Pseudomonas syringae by regulating auxin signaling [8]. miR393 can be induced upon perception of flg22 (a 22-amino acid peptide), a PAMP (pathogen-associated molecular pattern) derived from bacterial flagellin, and negatively regulates transcripts of a number of F-box auxin receptors [8]. Moreover, miR160, miR167, and miR393 were identified as highly induced after infection using sRNA expression profiling on Arabidopsis leaves collected at 1 and 3 h post-inoculation of Pseudomonas syringae pv. tomato (DC3000hrcC). [12]. Interestingly, all three miRNAs negatively regulate auxin signaling by either targeting auxin receptor genes or auxin response factors [12]. Another recent study reported that miR162 and miR168 targeted Dicer-like1 and Argonaute proteins, which are likely up-regulated by infection and presumably positively regulated by plant defense responses, although their functions need to be confirmed by experimental data [13].
Cotton is one of the most important economic crops in the world. It is highly susceptible to cotton Verticillium wilt, a disease that significantly affects cotton yield and quality. Verticillium wilt is the primary disease attacking cotton crops and is mainly caused by a soil-borne fungal pathogen, Verticillium dahliae Kleb. (V. dahliae) [14]. The representative symptoms of susceptible cotton include leaf curl, necrosis and defoliation, and stem wilt [15]. It leads to discoloration of cotton leaves and stem vascular bundles, and it inhibits photosynthesis and increases respiration [16]. However, there is no proven control (chemical or cultural) for this disease as the mechanisms of Verticillium wilt remain poorly understood. Despite great efforts in producing wilt-resistant cotton cultivars by traditional breeding, very few Gossypium hirsutum varieties—the main species of cotton currently cultivated in the world—are resistant to the wilt [17]. Therefore, identification of new miRNAs and elucidation of their functions in response to V. dahliae infection will help us understand the regulation of pathogen defense responses. Recently, the Gossypium raimondii genome sequence was completed [18,19], and this will greatly advance biological research on cotton.
Although many cotton miRNAs were identified in previous research, the role of miRNAs in the regulation of Verticillium defense responses is mostly unknown. To date, the majority of miRNA targets in cotton were predicted by bioinformatics approaches, and only a small portion were experimentally validated. Although no cotton cultivar is immune to Verticillium wilt, most cultivars of G. barbadense, such as Hai-7124, show significant advantages in Verticillium wilt resistance [17,20]. Therefore, to detect new miRNAs and their potential targets participating in the regulation of Verticillium defense responses, four sRNA libraries and two degradome libraries using RNAs from mock- and Verticillium-inoculated Gossypium hirsutum L. (G. Hirsutum) and Gossypium barbadense L. (G. Barbadense) roots were constructed and sequenced using a Solexa analyzer. A total of 140 known miRNAs and 58 novel miRNAs were identified and 107 genes sliced by 45 miRNA families were detected via degradome sequencing. The profiling of the miRNAs and their target genes provides novel information about the regulatory network of defense responses in cotton to V. dahliae.
2. Results
2.1. Overview of Small RNA (sRNA) Library Sequencing
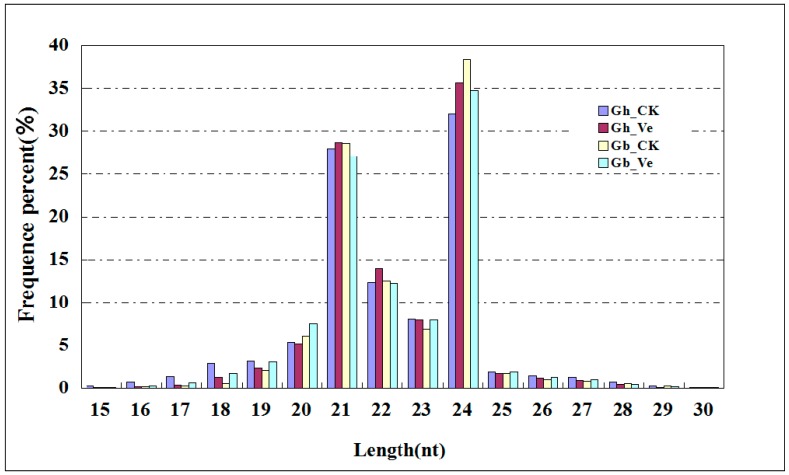
Four sRNA libraries were constructed and deep sequenced with total RNAs from G. hirsutum (Gh_CK: mock-inoculated; Gh_Ve: Verticillium-inoculated) and G. barbadense (Gb_CK: mock-inoculated; Gb_Ve: Verticillium-inoculated) roots. A total of about 26, 20, 21, and 20 million raw reads were obtained from Gh_CK, Gh_Ve, Gb_CK, and Gb_Ve, respectively (Table 1). After filtering out the reads of low quality and the adaptor sequences, there were approximately 26, 19, 20, and 20 million clean reads obtained in the Gh_CK, Gh_Ve, Gb_CK, and Gb_Ve libraries. To simplify the sequencing data, all identical sequence reads in each sRNA library were grouped and converted into unique sequence tags with associated counts of the individual sequence reads. There were 7,181,742; 6,033,991; 6,729,005; and 6,415,595 unique tags in the four sRNA libraries, respectively (Table 1). The length distributions of sRNAs were very similar between the four libraries (Figure 1). In these four libraries, the majority of sRNA sequences were 20–24 nt in size with 21 or 24 nt as the major size classes, which is typical for Dicer-derived products (Figure 1).
Table 1.
Analysis of small RNA (sRNA) reads from four sRNA libraries.
| Category | Library Name | |||
|---|---|---|---|---|
| Gh_CK | Gh_Ve | Gb_CK | Gb_Ve | |
| Total reads | 26,495,780 | 19,511,637 | 20,609,252 | 19,965,211 |
| High quality | 26,409,519 | 19,450,186 | 20,548,220 | 19,903,524 |
| Clean reads | 25,679,650 | 19,296,249 | 20,389,102 | 19,638,276 |
| Unique sRNAs | 7,181,742 | 6,033,991 | 6,729,005 | 6,415,595 |
| Unique sRNA mapping to genome | 2,235,867 | 1,855,605 | 2,078,150 | 1,988,015 |
| miRNA | 37,936 | 29,080 | 32,658 | 34,630 |
| Unannotated | 6,264,557 | 5,281,380 | 5,889,075 | 5,583,553 |
Figure 1.
Length distribution of sRNAs in G. hirsutum roots and G. barbadense roots. Gh_CK: mock-inoculated G. hirsutum roots; Gh_Ve: Verticillium-inoculated G. hirsutum roots; Gb_CK: mock-inoculated G. barbadense roots; Gb_Ve: Verticillium-inoculated G. barbadense roots.
2.2. Identification of Known MicroRNAs (miRNAs) by sRNA Sequencing
To identify known miRNAs in the four libraries, all mappable sRNA sequences were compared with the currently known plant miRNAs in the miRBase database (release 20.0), which contains 78 known cotton miRNAs. In total, approximately 77 known cotton miRNAs belonging to 52 families were identified in the four libraries. The numbers of reads of the 77 known cotton miRNAs in the four sRNA libraries are listed in Table S1. The miRNA families miR156 and miR166 were the most abundant in the four libraries. In addition, a total of the 63 known unique miRNAs with high sequence similarity to the other known plant miRNAs, representing 50 known miRNA families, were identified in the four libraries (Table S2). These known miRNAs, with a minimal folding free energy (MFE) of the predicted hairpins ranging from −21.34 to −130.6 kcal/mol (Table S2), covered almost all the plant-conserved miRNA families. Several known but non-conserved miRNA families that have previously been identified only from one or a few plant species were also found: e.g., miR477, miR530, miR827, miR1448, miR2111, miR2947, miR2950, miR3476, and miR5083. In the present study, most of the newly identified miRNA families, such as miR168, miR403, miR477, miR828, and miR1448, were from G. hirsutum, while most miRNAs from G. barbedense had already been identified previously (Table S2).
2.3. Identification of Novel miRNAs in G. hirsutum and G. barbadense
To predict novel miRNAs in G. hirsutum and G. barbadense, all mappable sRNAs were BLASTed to the G. raimondii genome sequence and known plant miRNAs in the miRBase database (release 20.0). The sRNAs that exactly mapped to the genome sequence and unknown plant miRNAs and their flanking sequences that could be folded into a secondary structure were considered as miRNA candidates. To increase predictive accuracy, five criteria described in the experimental section were mainly used to search for novel miRNAs. In total, 58 novel miRNAs were identified in the present study (Table 2). These new miRNAs were named temporarily in the form of novel_miR_number: e.g., novel_miR_1 and their lengths were 20, 21, 22, or 23 nt (Table 2). Among these miRNAs, 34 were detected in at least two of the four sRNA libraries, and 10 were detected in all four sRNA libraries. The predicted hairpins of their precursors had a MFE ranging from −21.1 to −143.2 kcal/mol with an average of −57.5 kcal/mol. All secondary hairpin structures are listed in Table S3.
Table 2.
Novel microRNAs (miRNAs) identified in G. hirsutum and G. barbadense.
| miRNA | Mature Sequence | LM | Arm | LP | G + C (%) | MFE | Reads Per Million | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Gh_CK | Gh_Ve | Gb_CK | Gb_Ve | |||||||
| novel_miR_1 | TTGACAAGTAAAAGAACATA | 20 | 3p | 147 | 29.25 | −23.44 | 0 | 0.98 | 0.74 | 1.07 |
| novel_miR_2 | CGAAGTCTTGGAAGAGAGTAA | 21 | 3p | 91 | 37.36 | −39.7 | 49.65 | 0 | 24.82 | 30.3 |
| novel_miR_3 | TGGTATTGGAGTGAAGGGAGC | 21 | 5p | 70 | 37.14 | −21.1 | 50.9 | 73.64 | 68.57 | 44.5 |
| novel_miR_4 | TGATTGAGCCGTGCCAATATC | 21 | 3p | 101 | 42.57 | −53.9 | 115.81 | 141.22 | 231.64 | 163.25 |
| novel_miR_5 | GTGGGCGTGCCGGAGTGGTTA | 21 | 5p | 74 | 56.76 | −26.8 | 110.52 | 96.7 | 34.04 | 36.76 |
| novel_miR_6 | CGGACTCTCAAACAGTGGAGGTA | 23 | 5p | 256 | 45.31 | −108.08 | 7.63 | 5.44 | 0 | 12.78 |
| novel_miR_7 | CTGGACTGTCAATGGCCGGCAC | 22 | 3p | 95 | 45.26 | −35.4 | 0 | 0 | 2.45 | 1.63 |
| novel_miR_8 | ATTAGATACTATAGCATGAGACA | 23 | 5p | 289 | 24.57 | −46.7 | 3.66 | 0 | 0 | 9.01 |
| novel_miR_9 | CTGTCGCAGGGGAGATGGCTCGT | 23 | 5p | 141 | 58.16 | −66.1 | 9.27 | 8.24 | 0 | 0.51 |
| novel_miR_10 | TCTAATAGAAGAATGACAAATCA | 23 | 5p | 84 | 20.24 | −22 | 0 | 0.93 | 1.52 | 0.81 |
| novel_miR_11 | TGGGAAATGATGACAGCTTA | 20 | 3p | 193 | 29.53 | −44.9 | 4.24 | 0 | 0 | 1.22 |
| novel_miR_12 | GCAGATGATGATAAGAATAGACA | 23 | 5p | 140 | 40.71 | −33.3 | 2.26 | 2.23 | 1.77 | 0.66 |
| novel_miR_13 | GGGCGCCTCTTACTTGGCAGG | 21 | 5p | 132 | 44.7 | −56.5 | 0.9 | 4.3 | 0 | 7.13 |
| novel_miR_14 | ATTAGATACTATAGCGTGAGACA | 23 | 3p | 226 | 37.17 | −61.1 | 0 | 0 | 0 | 7.69 |
| novel_miR_15 | GCTCACTTCTCTTTCTGTCAGTT | 23 | 3p | 107 | 44.86 | −57.7 | 5.33 | 9.74 | 11.87 | 7.84 |
| novel_miR_16 | TGCCTGGCTCCCTGAATGCCA | 21 | 5p | 104 | 50 | −53.3 | 1.75 | 0.73 | 0.49 | 1.32 |
| novel_miR_17 | AGGTGCAGGTGCAGGCGCAGC | 21 | 3p | 146 | 40.41 | −49.62 | 0 | 0 | 7.9 | 1.53 |
| novel_miR_18 | CGGACCCTCTAACAGTGGAGG | 21 | 5p | 214 | 50.47 | −90.3 | 56.93 | 37.47 | 0 | 38.9 |
| novel_miR_19 | TGAGAAAGTGGAGATGGGTGG | 21 | 3p | 128 | 45.31 | −63.9 | 2.18 | 1.71 | 3.87 | 2.6 |
| novel_miR_20 | CTCTGGTTTGACTCATTTGTA | 21 | 5p | 181 | 33.7 | −66.8 | 3.12 | 0 | 0 | 1.63 |
| novel_miR_21 | CAAATGAGTTAGGCGAGAGGT | 21 | 3p | 170 | 33.53 | −63.56 | 0 | 13.42 | 0 | 7.08 |
| novel_miR_22 | TCAAGCAAATCAAGGAAAGGCC | 22 | 3p | 344 | 38.66 | −113.5 | 0.55 | 0.47 | 1.13 | 0.36 |
| novel_miR_23 | TCTGAAAAGCAATAAAGAACACA | 23 | 3p | 94 | 27.66 | −25.7 | 0 | 0 | 2.55 | 1.78 |
| novel_miR_24 | GTGGAAGTTAGGCTGACTTAGGC | 23 | 5p | 79 | 45.57 | −21.7 | 0 | 0 | 0 | 4.12 |
| novel_miR_25 | AGGCAGTCACCTTGGCTAAC | 20 | 5p | 180 | 45.56 | −64.6 | 0 | 4.98 | 0 | 1.68 |
| novel_miR_26 | TTGGATGGGCGGTGTGTTTACTT | 23 | 3p | 246 | 28.46 | −49.4 | 0 | 0 | 0.64 | 1.93 |
| novel_miR_27 | GGCAAGTTGTCCTCGGCTACA | 21 | 3p | 178 | 39.89 | −59.96 | 0 | 1.87 | 0 | 1.12 |
| novel_miR_28 | TCCATATTTCACTATCTCTTA | 21 | 3p | 105 | 32.38 | −48.6 | 11.25 | 16.01 | 72.54 | 22.66 |
| novel_miR_29 | TTGAACACCGAAGTAAAGCCAT | 22 | 5p | 209 | 41.15 | −82.8 | 0 | 0 | 0 | 1.78 |
| novel_miR_30 | TGCCAAATCAGGGAAGCGAA | 20 | 5p | 148 | 41.22 | −52.32 | 0 | 0 | 43.5 | 0 |
| novel_miR_31 | AGGCTGTGATGACGAGAAATTA | 22 | 3p | 237 | 30.38 | −47.71 | 32.94 | 24.1 | 16.92 | 0 |
| novel_miR_32 | TTTCCAATAGAAGAATGACAAAT | 23 | 5p | 140 | 27.14 | −54.8 | 1.36 | 1.09 | 1.03 | 0 |
| novel_miR_33 | AATCTCCCTCAAACGCTTCCAG | 22 | 5p | 118 | 45.76 | −47.84 | 0 | 0 | 9.66 | 0 |
| novel_miR_34 | TTTGGATTGAAGGGAGCTCTA | 21 | 3p | 203 | 42.36 | −92.45 | 0 | 0 | 141.2 | 0 |
| novel_miR_35 | TAGTGAGGATGGGAAATTTGT | 21 | 5p | 124 | 30.65 | −49.3 | 34.93 | 0 | 18.78 | 14 |
| novel_miR_36 | GAGCTTGGAAGTGCATCCGGC | 21 | 5p | 107 | 45.79 | −54.9 | 0 | 0 | 4.07 | 0 |
| novel_miR_37 | TCGCTTCCCTAATTTGGACGA | 21 | 3p | 148 | 41.22 | −52.32 | 36.33 | 31.09 | 0 | 85.09 |
| novel_miR_38 | TGACTCCTAGTACAACGGCCTC | 22 | 3p | 328 | 32.32 | −123.5 | 0 | 0 | 5.84 | 0 |
| novel_miR_39 | TTGAGCCGTGCCAATATCAATC | 22 | 3p | 108 | 47.22 | −49.81 | 0 | 0 | 226.44 | 0 |
| novel_miR_40 | CAAAGAGTAGAGGTATTGTGC | 21 | 5p | 272 | 41.54 | −112 | 0 | 13.89 | 1.23 | 0 |
| novel_miR_41 | ACAGGTTAGTAGAAATTAAGGTT | 23 | 5p | 115 | 41.74 | −22.6 | 0 | 1.09 | 0 | 0 |
| novel_miR_42 | CAGAATGACCAATTTACTCTTTA | 23 | 3p | 199 | 24.12 | −40.4 | 0 | 4.92 | 0 | 0 |
| novel_miR_43 | ACTCTCTTCCAAAGGCTTCAAG | 22 | 5p | 108 | 35.19 | −46 | 0 | 49.91 | 0 | 0 |
| novel_miR_44 | TGGTGCAGGTCGGGAACTGAT | 21 | 5p | 139 | 47.48 | −57.4 | 0 | 5.86 | 0 | 0 |
| novel_miR_45 | GTTCAATAAAGCTGTGGGAAG | 21 | 3p | 138 | 40.58 | −54.6 | 0 | 100.69 | 0 | 0 |
| novel_miR_46 | CAAAAGCAATGAAGAACTGGCCA | 23 | 5p | 331 | 34.14 | −78.9 | 0 | 1.97 | 0 | 0 |
| novel_miR_47 | ACAGTAGAAATGGATGGAATT | 21 | 3p | 153 | 30.07 | −48 | 0 | 3.99 | 0 | 0 |
| novel_miR_48 | TTGGATGGACGGTGCATTTATCT | 23 | 3p | 295 | 37.29 | −62.4 | 0 | 1.04 | 0 | 0 |
| novel_miR_49 | ATTATTGTTAATGTAGGAGGA | 21 | 5p | 371 | 31 | −61.26 | 0 | 1.92 | 0 | 0 |
| novel_miR_50 | TTGTACTAGGAGTCGGATTGC | 21 | 5p | 344 | 33.72 | −143.2 | 0 | 4.15 | 0 | 1.27 |
| novel_miR_51 | TTAGATCAAAGAGCAAACCGG | 21 | 5p | 210 | 36.67 | −95.5 | 1.32 | 0 | 0 | 0 |
| novel_miR_52 | CGAGACTTGCGGTAGAAACAAA | 22 | 3p | 149 | 40.27 | −35.2 | 6.74 | 0 | 0 | 0 |
| novel_miR_53 | TGGAGGCAGCGGTTCATCGATC | 22 | 5p | 110 | 42.73 | −40.1 | 0 | 0 | 0 | 12.37 |
| novel_miR_54 | GGGACTCTCCGGACTGTTTGGTT | 23 | 3p | 148 | 36.49 | −43.3 | 1.05 | 0 | 0 | 0 |
| novel_miR_55 | TGCCTGGCTCCCTGTATGCCA | 21 | 5p | 103 | 47.57 | −57.4 | 107.56 | 71.93 | 11.92 | 57.64 |
| novel_miR_56 | TTTTGCCAGCTCCGCCCATTCC | 22 | 3p | 122 | 40.98 | −45.01 | 66.24 | 87.89 | 0 | 1.73 |
| novel_miR_57 | CAGCCAAGGATGACTTGCCGG | 21 | 5p | 175 | 35.43 | −57.4 | 0.97 | 1.66 | 0 | 0.92 |
| novel_miR_58 | TTTTTTAGTAGAAGGAGCAAAAT | 23 | 5p | 254 | 32.68 | −56.8 | 0.35 | 0 | 0 | 0 |
LM: length of mature miRNA; LP: length of precursor; MFE: minimal folding free energy; Gh_CK: mock-inoculated G. hirsutum roots; Gh_Ve: Verticillium-inoculated G. hirsutum roots; Gb_CK: mock-inoculated G. barbadense roots; Gb_Ve: Verticillium-inoculated G. barbadense roots.
2.4. Expression Profiling of Differentially Expressed miRNAs in Response to V. dahliae Infection
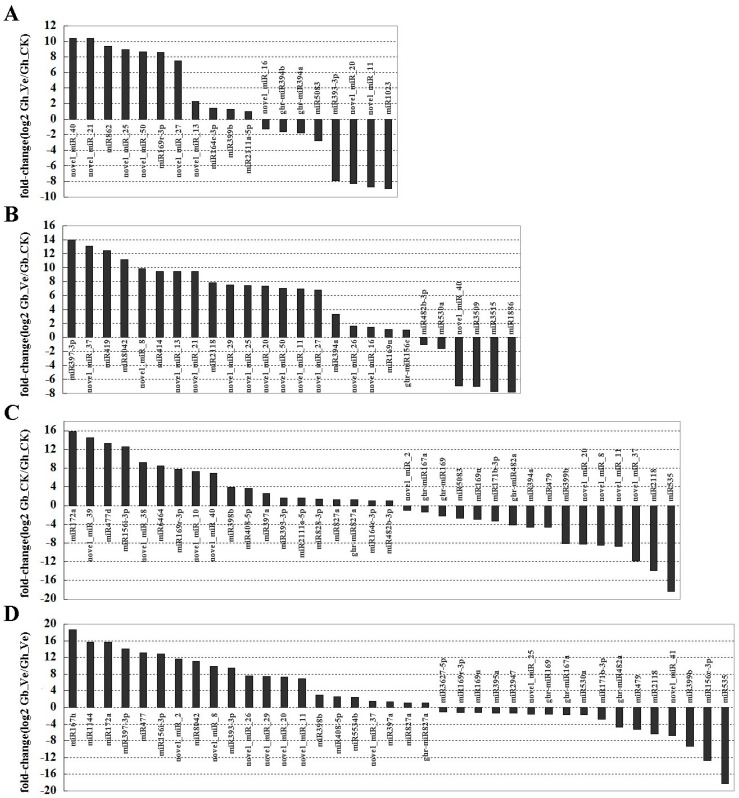
The expression profiles of miRNAs were analyzed and compared between the four libraries based on the number of clean reads generated from the high-throughput sequencing. After normalization, the reads of the tags of each miRNA family as “reads per million”, p-value < 0.01, and the absolute value of |log2Ratio| ≥ 1 were considered to indicate the statistical significance of miRNA expression. Interestingly, many miRNAs were differentially expressed between the libraries. miRNA expression between the mock- and Verticillium-inoculated treatments for the two cotton species was first analyzed. In G. hirsutum roots, a total of 19 miRNAs representing nine novel miRNAs were identified as V. dahliae-responsive miRNAs (Figure 2A). Among them, 11 miRNAs were preferentially expressed in the Gh_Ve treatment and eight were preferentially expressed in the Gh_CK treatment. In G. barbadense roots, a total of 26 miRNAs representing 13 novel miRNAs were identified as V. dahliae-responsive miRNAs (Figure 2B). Among them, 20 were of higher abundance and only six were of lower abundance in the Gb_Ve treatment. Notably, novel_miR_13, novel_miR_21, novel_miR_25, novel_miR_27, and novel_miR_50 were of higher abundance in the roots of both cotton species after 24 h of treatment.
Figure 2.
Expression profiles of differentially expressed miRNAs by high-throughput sequencing. (A) Differentially expressed miRNAs between Gh_Ve and Gh_CK libraries; (B) Differentially expressed miRNAs between Gb_Ve and Gb_CK libraries; (C) Differentially expressed miRNAs between Gb_CK and Gh_CK libraries; (D) Differentially expressed miRNAs between Gb_Ve and Gh_Ve libraries. Gh_CK: mock-inoculated G. hirsutum roots; Gh_Ve: Verticillium-inoculated G. hirsutum roots; Gb_CK: mock-inoculated G. barbadense roots; Gb_Ve: Verticillium-inoculated G. barbadense roots.
The miRNA expressions between the two cotton species with mock- and Verticillium-inoculated treatments were also analyzed. We made a comparative analysis of miRNA expression between the two cotton species with mock-inoculated treatment (Figure 2C). It was shown that a total of 35 miRNAs had a species-specific expression. Among them, 16 miRNAs were preferentially expressed in G. hirsutum roots and 19 were preferentially expressed in G. barbadense roots. Because of the genotype-specific expression of miRNAs under mock-inoculated treatment, it was plausible to assume that some miRNAs had preferential expression in one of the two species when both of them were inoculated by Verticillium. Thus, we compared the expression levels of miRNAs between Gb_Ve and Gh_Ve libraries. We found that 38 miRNAs had species-specific expression in Verticillium-inoculated treatments (Figure 2D). Among them, 17 miRNAs were preferentially expressed in G. hirsutum roots and 21 were preferentially expressed in G. barbadense roots. Interestingly, most of the species-specific-expressed novel miRNAs (e.g., novel_miR_2, novel_miR_8, novel_miR_11, novel_miR_20, novel_miR_26, novel_miR_29, and novel_miR_37) were significantly preferentially expressed in Verticillium-inoculated G. hirsutum roots.
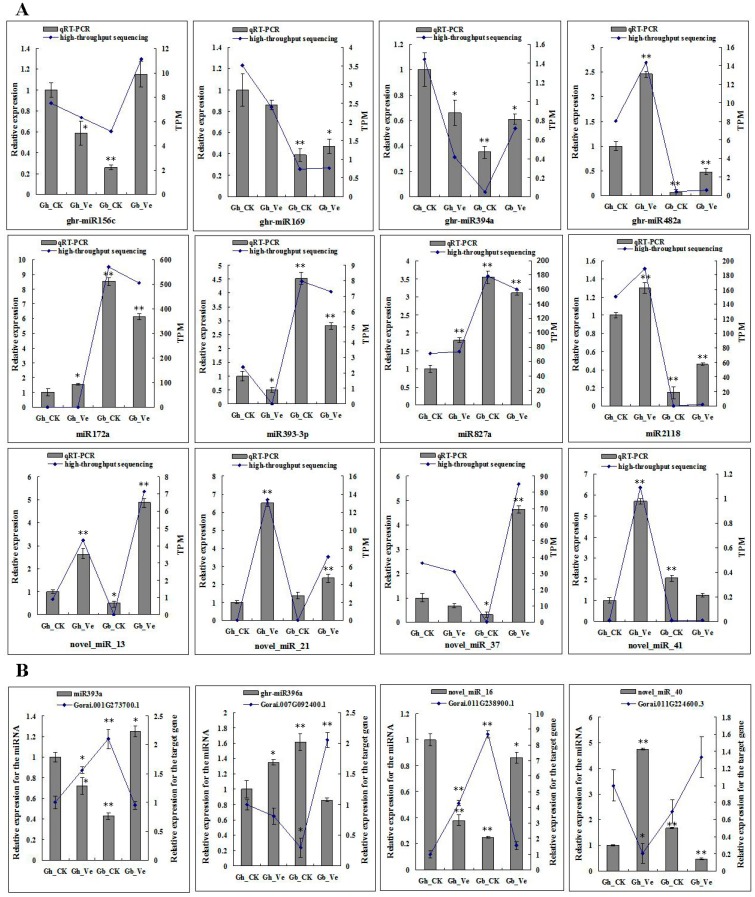
To validate the existence and expression patterns of the predicted miRNAs in mock- and Verticillium-inoculated cotton roots, four novel miRNAs, as well as eight representative known miRNAs, were selected for qRT-PCR analysis. Although some non-conserved and novel miRNAs were identified in low read number or were undetectable in one or two sRNA libraries by Solexa sequencing, the 12 selected miRNAs were detected by qRT-PCR. The qRT-PCR results of those miRNAs were quite consistent with the results from the sequencing data, and confirmed the changes in miRNA expression in response to V. dahliae infection (Figure 3A). Furthermore, to confirm the causality of the miRNA expression patterns and their target gene, we studied the expression of four miRNAs and their target genes by qRT-PCR in G. hirsutum and G. barbadense roots. Among the target mRNAs tested, four targets showed a significant inverse correlation in expression with their corresponding miRNAs (Figure 3B).
Figure 3.
Relative expression analysis of miRNAs and target genes in the G. hirsutum and G. barbadense roots. (A) Relative expression analysis of miRNAs by qRT-PCR analysis and high-throughput sequencing; (B) Relative expression analysis of miRNAs and their target genes by qRT-PCR analysis. Gh_CK: mock-inoculated G. hirsutum roots; Gh_Ve: Verticillium-inoculated G. hirsutum roots; Gb_CK: mock-inoculated G. barbadense roots; Gb_Ve: Verticillium-inoculated G. barbadense roots; TPM: Tags per million clean reads. U6 snRNA and Ubiquitin1 were chosen as endogenous control genes. Error bars indicate standard deviation of three biological replicates.* and ** indicate significant differences relative to the Gh_CK at p < 0.05 and p < 0.001 by Student’s t-test, respectively.
2.5. Target Genes of miRNAs Identified by Degradome Analysis
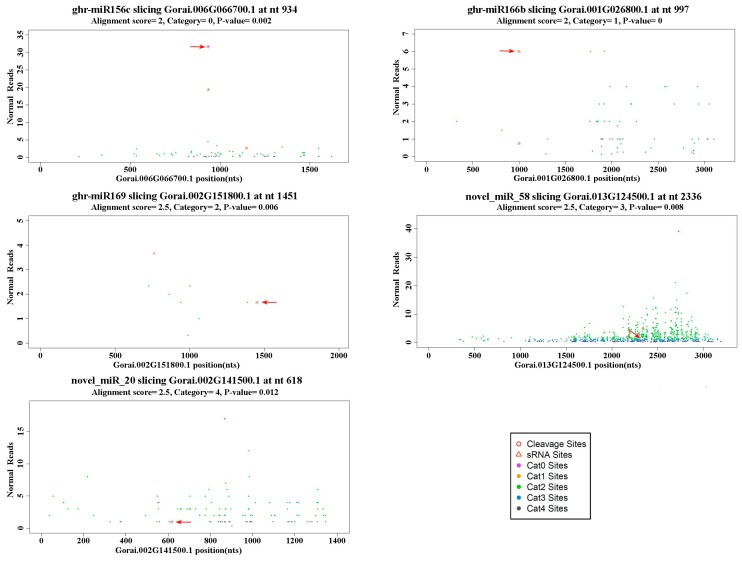
In the present study, degradome sequencing was used to search for the target genes of identified miRNAs in cotton. According to the relative abundance of tags at the predicted miRNA target sites, the identified targets were grouped into five categories (0–4) as described by Yang [21]. Category “0” was defined as >1 raw read at the position, with abundance at the position equal to the maximum on the transcript, and with only one maximum on the transcript. Category “1” was defined as >1 raw read at the position, with abundance at the position equal to the maximum on the transcript, and with more than one maximum position on the transcript. Category “2” included >1 raw read at the position, and abundance at the position less than the maximum but higher than the median for the transcript. Category “3” comprised transcripts with >1 raw read at the position, and abundance at the position equal to or less than the median for the transcript. Category “4” showed only one raw read at the position [21]. The representative miRNAs and corresponding targets are shown in Figure 4, in which the red arrows indicate the cleavage sites. A total of 83 and 24 targets were predicted to be cleaved by 31 known and 14 novel miRNA families, respectively (Table 3 and Table S4). Among the identified targets of known miRNAs, squamosa-promoter binding protein (SBP) and squamosa-promoter binding protein-like (SPL) transcription factor genes, NAM, ATAF, and CUC (NAC) domain-containing protein genes, Class III homeodomain leucine-zipper (HD-Zip) protein genes, APETALA2 (AP2)-like factor genes, F-box/RNI-like superfamily protein (TIR1) genes, and growth-regulating factor (GRF) genes were targeted by several conserved miRNA families (including miR156, miR164, miR166, miR172, miR393, and miR396) that play significant roles in gene regulation (Table S4). ghr-miR7502, ghr-miR7505, ghr-miR7509, ghr-miR7510a, and ghr-miR7514, which were only reported in G. hirsutum, were predicted to cleave Nudix hydrolase, ATP binding and tetratricopeptide repeat-like superfamily protein, glutamate receptor 2 isoform 1, peroxidase superfamily protein, and RHO guanyl-nucleotide exchange factor 7 (Table S4). Among the identified targets of novel miRNAs, ubiquitin carboxyl-terminal hydrolase family protein gene was targeted by novel_miR_12; auxin response factor genes were targeted by novel_miR_16; purine permease 3 and ferrochelatase 2 genes were targeted by novel_miR_20; and TT12-2 (a T-DNA insertion line) MATE (multidrug and toxic efflux) transporter gene was targeted by novel_miR_24 (Table 3). Interestingly, we also found that leucine-rich repeat (LRR) containing protein genes and LRR and NB-ARC (nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4) domains-containing disease resistance-like protein genes that play important roles in plant defense against pathogens were targeted by novel_miR_28 and novel_miR_56, respectively (Table 3). Unfortunately, target genes of some known miRNAs and novel miRNAs identified through deep sequencing were not detected in the present degradome analysis.
Figure 4.
Target plots (t-plots) of the representative miRNA targets in different categories confirmed by using degradome sequencing. Normal Reads: repeat normalized number of 5′ends at that position for target gene. Red arrows indicate the target genes cleavage sites.
Table 3.
Target genes of novel miRNAs identified by degradome sequencing in cotton.
| miRNA | Target Gene | Cleavage Site | Categoty | Norm Reads | Target Description | |
|---|---|---|---|---|---|---|
| Gb | Gh | |||||
| novel_miR_12 | Gorai.005G020900.1 | 1117 | 1 | 0 | 3 | Ubiquitin carboxyl-terminal hydrolase family protein |
| novel_miR_16 | Gorai.003G142500.1 | 1315 | 0 | 58 | 54 | Auxin response factor 10 isoform 1 |
| Gorai.013G267100.1 | 1416 | 0 | 60 | 31 | Auxin response factor 10 isoform 1 | |
| Gorai.010G046000.1 | 1638 | 0 | 16 | 19 | Auxin response factor 17 | |
| Gorai.006G166400.1 | 1773 | 0 | 36 | 12 | Auxin response factor 19 | |
| Gorai.011G238900.1 | 1502 | 0 | 316 | 557 | Auxin response factor 19 | |
| Gorai.012G004800.1 | 1679 | 0 | 110 | 160 | Auxin response factor 19 | |
| novel_miR_20 | Gorai.002G141500.1 | 618 | 4 | 1 | 0 | Purine permease 3 |
| Gorai.003G066400.1 | 1545 | 4 | 1 | 0 | Ferrochelatase 2 isoform 1 | |
| Gorai.008G199200.1 | 1792 | 4 | 1 | 0 | Uncharacterized protein TCM_014264 | |
| novel_miR_21 | Gorai.009G146400.1 | 1251 | 0 | 7 | 0 | Uncharacterized protein TCM_029129 |
| novel_miR_24 | Gorai.006G008600.5 | 496 | 0 | 1 | 0 | TT12-2 MATE transporter |
| novel_miR_26 | Gorai.007G279100.4 | 319 | 2 | 0 | 2 | Intracellular protein transport protein USO1 |
| Gorai.012G074700.1 | 79 | 2 | 0 | 2 | Serine hydroxymethyltransferase 6 isoform 3 | |
| novel_miR_28 | Gorai.003G163700.1 | 1250 | 0 | 13 | 0 | Leucine-rich repeat containing protein |
| novel_miR_29 | Gorai.008G010200.1 | 855 | 2 | 7 | 5 | Glutathione S-transferase 7 isoform 1 |
| Gorai.008G120500.1 | 1587 | 3 | 0 | 2 | Fringe-related protein, putative | |
| novel_miR_30 | Gorai.005G163400.1 | 1744 | 0 | 28 | 0 | Uncharacterized protein TCM_010813 |
| novel_miR_40 | Gorai.011G224600.3 | 1555 | 1 | 0 | 1 | Inositol 1,3,4-trisphosphate 5/6-kinase 4 |
| novel_miR_48 | Gorai.007G279100.4 | 319 | 2 | 0 | 2 | Intracellular protein transport protein USO1 |
| Gorai.012G074700.1 | 79 | 2 | 0 | 2 | Serine hydroxymethyltransferase 6 isoform 3 | |
| novel_miR_56 | Gorai.007G319800.1 | 613 | 0 | 10 | 1 | LRR and NB-ARC domains-containing disease resistance-like protein |
| novel_miR_57 | Gorai.004G172100.1 | 1219 | 0 | 19 | 26 | Nuclear transcription factor Y subunit A-1, putative isoform 1 |
| novel_miR_58 | Gorai.013G124500.1 | 2336 | 3 | 1 | 1 | Bacterial-induced lipoxygenase |
Normal Reads: repeat normalized number of 5′ends at that position for target gene.
3. Discussion
Verticillium dahliae Kleb. is a soil-borne fungal pathogen that causes vascular wilt of more than 200 dicotyledonous plant species, including cotton crops [14]. Sea-island cotton (G. barbadense L.) exhibits relative resistance to Verticillium wilt, but upland cotton (G. hirsutum L.), the main species cultivated on a large scale, is sensitive to this disease [17,22]. Previous study has shown that a number of miRNAs and other small non-coding RNAs were involved in response to V. dahliae infection in G. hirsutum and G. barbadense roots [23]. However, most of these miRNA targets were previously predicted in cotton [23], and only a few miRNA targets have been identified experimentally [23,24]. The functions of most of these miRNAs in relation to the regulation of Verticillium defense responses remain unknown and further studies must be conducted. In the present study, the two cotton species G. hirsutum and G. barbadense were used as models to study the miRNA functions associated with the regulation of Verticillium defense responses. We constructed and sequenced four sRNA libraries and two degradome libraries from mock- and Verticillium-inoculated cotton roots. In total, 140 known miRNAs and 58 novel miRNAs were detected in G. hirsutum and G. barbadense by deep sequencing. In addition, 107 target genes of 45 miRNA families were identified by degradome library sequencing. The present study is the first to report comprehensive identification of miRNAs and their targets involved in cotton response to V. dahliae by using high-throughput sequencing and degradome analysis. This will provide useful information for improving the Verticillium wilt resistance of economically important crops.
miRNAs have been identified experimentally in many plant species, especially in model plants. In plants, some miRNAs seem to be universally expressed, while others are present in only a few species. According to previous reports, conserved miRNAs are present throughout at least one major ancient clade of land plants, and non-conserved miRNAs, with a limited phylogenetic distribution, are characterized by primarily being single-copy genes [25]. In this study, most known miRNAs were conserved in other species and had been previously predicted. Many other studies have shown that a number of the most conserved miRNA targets common among the data sets include many transcription factors: SBP, SPL, auxin response factors (ARF), MYB (Myeloblastosis), NAC, TCP (Teosinte-like 1, Cycloidea, and Proliferating cell factor 1), NF-Y (Nuclear Factor Y), GRF, HD-ZIP, PPR (Pentatricopeptide Repeat), and AP2-like factors. It is possible that conserved miRNAs play a crucial role in universal mechanisms of regulation in different plant species and may help us understand the evolutionary relationships between cotton and other plants. Some known but non-conserved miRNAs (e.g., miR477, miR530, miR827, miR1448, miR2111, miR2947, miR2950, miR3476, and miR5083), which were also detected in the present study, have been only identified in one or few plant species so far. In addition, 58 novel miRNAs with a lower abundance than that of conserved miRNAs were identified by using universal rules for miRNA annotation [26]. They are likely to be cotton-specific miRNAs, which are classified into non-conserved miRNAs. It seems likely that these miRNAs evolved relatively recently [5], and may function only to regulate gene expression during Malvaceae- or cotton-specific biological processes.
miRNAs regulate gene expression at the transcriptional and post-transcriptional levels via mRNA cleavage or translational repression. In higher plants, miRNAs mediate gene silencing mainly by slicing mRNAs [27]. miRNA-directed cleavage leaves a 5ʹ-uncapped 3ʹ-fraction of the sliced genes. Therefore, the powerful tool of degradome sequencing was applied to identify miRNA target genes in many species with greater throughput [28,29,30]. In our study, this experimental approach was performed to identify target genes for known and novel miRNAs in cotton. As expected, a number of target genes were predicted to be cleaved by known and novel miRNA families. Many of the identified target genes of known conserved miRNAs belong to diverse gene families of transcription factors (e.g., MYB, NAC, HD-ZIP and AP2-like factor) which are known to regulate diverse aspects of plant growth and development as well as the response to V. dahliae infection [31,32]. In addition, we found some novel miRNA targets (e.g., ubiquitin carboxyl-terminal hydrolase family protein, purine permease 3, TT12-2 MATE transporter, intracellular protein transport protein USO1, glutathione S-transferase 7, and bacterial-induced lipoxygenase), suggesting a new feature of miRNA regulation in cotton. The novel miRNAs and their targets might offer useful information in potential future studies on how miRNAs and their targets are involved in the response to V. dahliae infection, which should be further investigated. However, the target genes for some known miRNAs and more novel miRNAs were not detected in the present degradome analysis. It is possible that the levels of these sliced targets were too low to detect, or some miRNAs might inhibit target gene expression through translational repression [33,34]. In summary, degradome analysis has greatly accelerated the identification of miRNA targets and has sped up research on miRNA–target interactions.
In our study, a number of the miRNAs exhibited altered expression in G. hirsutum and G. barbadense roots after infection with V. dahliae, indicating that V. dahliae infection could disrupt global gene regulatory networks during cotton development. These V. dahliae–responsive miRNAs might contribute to species-specific regulation, act as “early” regulators of signal transduction or be advantageous for adaptation to stressed environments [23,35]. In Populus, Ptc-miR482 was validated to cleave disease resistance protein genes, which were involved in the resistance of plants to biotic and abiotic stresses [36,37]. Previous studies have shown that the NBS (nucleotide-binding site)-LRR resistance gene might contribute to V. dahliae resistance in cotton [31,38]. In this study, ghr-miR482a, which targets LRR and NB-ARC domain-containing disease resistance–like proteins, was expressed at a very low level in G. barbadense mock- and Verticillium-inoculated roots compared with G. hirsutum roots, suggesting its possible role in relative resistance to Verticillium wilt of G. barbadense. The expression profiles of miR1886, miR3509, and miR3515 were down-regulated, while miR419 and miR2118 were up-regulated in G. barbadense roots after infection with V. dahliae. This was consistent with a previous study that showed that many miRNAs had a species-specific expression after infection of the fungal pathogen Verticillium in G. hirsutum and G. barbadense [23]. In plants, ARFs were involved in regulating the auxin signaling pathway, which plays an important role in growth, development, and environmental responses. In Arabidopsis, repression of auxin signaling could restrict P. syringae growth, implicating auxin in disease susceptibility and miRNA-mediated suppression of auxin signaling in disease resistance [8,39]. novel_miR_16, which targets ARF10, ARF17, and ARF19, was also induced by V. dahliae, indicating that it also played an important role in plant disease resistance, but this requires further experimental confirmation. There are still many other miRNAs involved in Verticillium-infection response; however, their target genes were not detected in the present degradome analysis and their functions in plants are unknown. Future analysis of target genes and molecular components downstream could help us to understand the significance of their roles in the process.
4. Experimental Section
4.1. Plant Material and Total RNA Isolation
The G. barbadense L. variety Hai-7124 (resistant) and G. hirsutum L. variety Yi-11 (susceptible) seeds were grown in pasteurized sand which was placed in a greenhouse (day temperature 28 °C, night temperature 25 °C, and relative humidity 60%) with a photoperiod of 14/10 h of light/dark and watered with Hoagland culture liquid every 3 day.
Verticillium dahliae isolates were provided by the College of Plant Protection, Shan Dong Agricultural University. After in-plate activation, V. dahliae was transferred to Czapek Broth liquid medium and cultured for 15 day (200 rpm, 25 °C). Then, after filtration through four layers of sterile gauze, V. dahliae was diluted to approximately 107 spores per ml of suspension with sterile water before inoculation.
The cotton seedlings with two fully expanded leaves were inoculated with V. dahliae by root dip inoculation into a suspension of fungal spores for 5 min and were then returned to their original pots. Control plants were not inoculated but were otherwise treated and sampled with distilled water in the same way. After 24 h of inoculation, the roots of both pathogen-infected and control seedlings were harvested immediately, frozen in liquid nitrogen, and stored at −80 °C for RNA isolation. In each case, samples were harvested and pooled from 20 individual plants. Total RNA was isolated from each sample using a modified CTAB (cetyltrimethylammonium bromide) method [40].
4.2. sRNA Library and Degradome Library Construction and Sequencing
sRNA library construction and deep sequencing were performed as described by Hafner [41]. A 20 µg aliquot of total RNA was sent to the Beijing Genomics Institute (Shenzhen, China) where the libraries were constructed and sequenced using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA). Briefly, the sRNAs (~18–30 nt) were purified from 10 µg of total RNA by polyacrylamide gel electrophoresis, and ligated first to a 5ʹ-RNA adaptor and then to a 3ʹ-RNA adaptor. A reverse transcription reaction was followed by several cycles of PCR to obtain sufficient product for Sequencing by Synthesis (SBS) sequencing via Solexa technology.
Two cotton degradome libraries (Gh and Gb) were constructed based on a method previously described [28]. Briefly, total RNAs were respectively extracted from Gh_CK and Gh_Ve, and mixed at an equal molar ratio as one sample for Gh degradome library construction. Total RNAs from Gb_CK and Gb_Ve were mixed for Gb degradome library construction in the same way. Approximately 200 µg of the mixed total RNA was used for polyadenylation using the Oligotex mRNA kit (Qiagen, Valencia, CA, USA), and then a 5ʹ-RNA oligonucleotide adaptor containing an MmeI recognition site was ligated to the 5ʹ-phosphate of the poly(A+) RNA by T4 RNA ligase. This was followed by purification of the ligated products using the Oligotex kit. Subsequently, five PCR cycles were performed on the products of a reverse transcription reaction which were then digested with MmeI and ligated to a 3ʹ-double DNA adaptor. Finally, the ligation products were amplified with 20 PCR cycles, gel-purified and subjected to SBS sequencing by the Illumina Genome Analyzer.
4.3. Analysis of Sequencing Data
Bioinformatic analysis of sRNA and degradome sequencing data was based on a method described previously [42]. For the sRNA sequencing data, the unique RNA sequences that perfectly matched the cotton genome were subjected to subsequent analysis. RNA reads showing sequences identical to known miRNAs from the miRBase 20.0 database were picked up as the miRNA dataset of cotton. Sequences matching non-coding rRNA, tRNA, snRNA, and snoRNA in the Rfam database were removed. Reads overlapping with exons of protein-coding genes were excluded to avoid mRNA contamination. The remaining sequences were used to predict their secondary structures by using the mfold web server [43,44]. A potential miRNA precursor must meet certain criteria: (1) Both a candidate miRNA and its corresponding reverse sequence (miRNA*) must be detected in the present high-throughput sequencing; (2) The candidate miRNA and miRNA* sequences must be found on the stem, and the number of mismatched bases between them must be less than four; (3) Within the miRNA/miRNA* duplex, the number of asymmetric bulges must be one or fewer, and the number of bases in the asymmetric bulges must fewer than two; (4) The miRNA and miRNA* should be located in opposite stem-arms and form a duplex with two nucleotide 3′overhangs; (5) The potential miRNA precursor must have higher negative minimal folding energy (MFE) with the MFE <−18 kcal/mol.
To investigate the differentially expressed miRNAs between libraries, each identified miRNA read count was normalized to the total number of miRNA reads in each given sample and multiplied by a million. Then, the Bayesian method was applied to infer the statistical significance value [45]. After the Bayesian test, if the p-value <0.01 and the absolute value of |log2Ratio| ≥ 1, a specific miRNA was considered to be differentially expressed.
For the degradome sequencing data, 20–21 nt sequences of high quality were collected for subsequent analysis. The unique reads that perfectly matched cotton cDNA sequences were retained. The 15-nt of sequence upstream and downstream of the 5ʹ-end of matched reads was extracted to constitute 30-nt sequence tags for searching corresponding miRNA. The CleaveLand pipeline [27] was used to align the 30-nt sequence to cotton-known miRNAs from miRBase20.0 database and our newly identified miRNAs. All alignments with scores up to seven and no mismatches at the cleavage site (between nucleotides 10 and 11) were considered candidate targets.
4.4. qRT-PCR
To validate the presence and expression of the identified miRNAs and target genes, 16 miRNAs and four target genes were selected for qRT-PCR analysis. The expression profile of miRNAs and target genes were assayed in pathogen- and mock-infected roots of Hai-7124 and Yi-11 by SYBR® Premix Ex TaqTM II (TAKARA, Dalian, China) on Bio-RAD iCycler iQ5 Machine. The primers used for qRT-PCR are listed in Table S5. qRT-PCR were performed using the One Step PrimeScript® miRNA cDNA Synthesis Kit (TAKARA) and using 12.5 µL of SYBR®Premix Ex TaqTM II (2×), 1 µL of PCR forward primer (10 µM), 1 µL of PCR reverse primer (10 µM), and 2 μL of fivefold diluted cDNA template in a 25-µL system with the following cycling profile: 95 °C at 30 s, followed by 40 cycles of 95 °C at 15 s and 60 °C at 30 s. All reactions of qRT-PCR were repeated three times for each sample. U6 snRNA and Ubiquitin1 gene were used as the internal control genes. All the gene expression data were obtained from three individual biological replicates and processed according to strict statistical methods [46]. Statistical significance was evaluated using a Student’s t-test analysis.
5. Conclusions
In this study, four sRNA libraries and two degradome libraries were constructed from G. hirsutum and G. barbadense roots with and without V. dahliae infection for deep sequencing. A large number of miRNAs were identified in both species, including 58 novel and 140 known miRNAs. Meanwhile, 107 genes sliced by 45 miRNA families were detected via degradome sequencing. The differential patterns of miRNAs expression are a valuable resource for further studies on post-transcriptional gene regulation in the defense response of cotton to Verticillium wilt. Thus, the present study might provide valuable clues for exploring miRNA-mediated regulatory networks in Verticillium defense response.
Acknowledgments
This work was supported by the China Major Projects for Transgenic Breeding (Grant Nos. 2010ZX08005-002 and 2010ZX005-004) and the China Key Development Project for Basic Research (973) (Grant No. 2007CB116208).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/07/14749/s1.
Author Contributions
Yujuan Zhang and Fafu Shen designed the study. Yujuan Zhang performed the experiments, analyzed the data, and drafted the manuscript. Wei Wang and Jie Chen assisted with bioinformatic analysis and aided in writing the manuscript. Jubo Liu and Minxuan Xia aided in performing the experiments. All authors carefully checked and approved this version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 2.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 3.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 4.Mallory A.C., Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 5.Jones-Rhoades M.W., Bartel D.P., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A., Zhu J.-K., Staskawicz B.J., Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 9.Padmanabhan C., Zhang X., Jin H. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Zhao H., Gao S., Wang W.-C., Katiyar-Agarwal S., Huang H.-D., Raikhel N., Jin H. Arabidopsis argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol. Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellendorff U., Fradin E.F., De Jonge R., Thomma B.P. H.J. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Ferrer V., Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 14.Bhat R.G., Subbarao K.V. Host range specificity in Verticillium dahliae. Phytopathology. 1999;89:1218–1225. doi: 10.1094/PHYTO.1999.89.12.1218. [DOI] [PubMed] [Google Scholar]
- 15.Sink K.C., Grey W.E. A root-injection method to assess verticillium wilt resistance of peppermint (Mentha × piperita L.) and its use in identifying resistant somaclones of cv. Black Mitcham. Euphytica. 1999;106:223–230. doi: 10.1023/A:1003591908308. [DOI] [Google Scholar]
- 16.Hampton R.E., Wullschleger S.D., Oosterhuis D.M. Impact of verticillium wilt on net photosynthesis, respiration and photorespiration in field-grown cotton ( Gossypium hirsutum L.) Physiol. Mol. Plant Pathol. 1990;37:271–280. doi: 10.1016/0885-5765(90)90076-A. [DOI] [Google Scholar]
- 17.Cai Y., Xiaohong H., Mo J., Sun Q., Yang J., Liu J. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: A review. Afr. J. Biotechnol. 2009;8:7363–7372. [Google Scholar]
- 18.Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 19.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 20.Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., Long L., Zhang X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011;62:5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Liu X., Xu B., Zhao N., Yang X., Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genomics. 2013;14:9. doi: 10.1186/1471-2164-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C., Guo W., Li G., Gao F., Lin S., Zhang T. QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 2008;174:290–298. doi: 10.1016/j.plantsci.2007.11.016. [DOI] [Google Scholar]
- 23.Yin Z., Li Y., Han X., Shen F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS ONE. 2012;7:e35765. doi: 10.1371/journal.pone.0035765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang M., Woodward A.W., Agarwal V., Guan X., Ha M., Ramachandran V., Chen X., Triplett B.A., Stelly D.M., Chen Z.J. Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.) Genome Biol. 2009;10:R122. doi: 10.1186/gb-2009-10-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axtell M.J., Bowman J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Meyers B.C., Axtell M.J., Bartel B., Bartel D.P., Baulcombe D., Bowman J.L., Cao X., Carrington J.C., Chen X., Green P.J. Criteria for annotation of plant microRNAs. Plant Cell Online. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Addo-Quaye C., Miller W., Axtell M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y.F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M.J., Zhang W., Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 30.Pantaleo V., Szittya G., Moxon S., Miozzi L., Moulton V., Dalmay T., Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Wang X.F., Ding Z.G., Ma Q., Zhang G.R., Zhang S.L., Li Z.K., Wu L.Q., Zhang G.Y., Ma Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics. 2013;14:637. doi: 10.1186/1471-2164-14-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Q., Jiang H., Zhu X., Wang W., He X., Shi Y., Yuan Y., Du X., Cai Y. Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genomics. 2013;14:852. doi: 10.1186/1471-2164-14-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., Liang Z., Yang B., Tian H., Ma J., Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 34.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 35.Yang L., Jue D., Li W., Zhang R., Chen M., Yang Q. Identification of miRNA from eggplant (Solanum melongena L.) by small RNA deep sequencing and their response to Verticillium dahliae infection. PLoS ONE. 2013;8:e72840. doi: 10.1371/journal.pone.0072840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S., Sun Y.-H., Shi R., Clark C., Li L., Chiang V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell Online. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S., Sun Y.H., Chiang V.L. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 38.He X., Sun Q., Jiang H., Zhu X., Mo J., Long L., Xiang L., Xie Y., Shi Y., Yuan Y., Cai Y. Identification of novel microRNAs in the Verticillium wilt-resistant upland cotton variety KV-1 by high-throughput sequencing. SpringerPlus. 2014;3:564. doi: 10.1186/2193-1801-3-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llorente F., Muskett P., Sánchez-Vallet A., López G., Ramos B., Sánchez-Rodríguez C., Jordá L., Parker J., Molina A. Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant. 2008;1:496–509. doi: 10.1093/mp/ssn025. [DOI] [PubMed] [Google Scholar]
- 40.Carra A., Gambino G., Schubert A. A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Anal. Biochem. 2007;360:318–320. doi: 10.1016/j.ab.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Hafner M., Landgraf P., Ludwig J., Rice A., Ojo T., Lin C., Holoch D., Lim C., Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X., Wang L., Yuan D., Lindsey K., Zhang X. Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 2013;64:1521–1536. doi: 10.1093/jxb/ert013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The mfold Web Server. [(accessed on 1 April 2014)]. Available online: http://mfold.rna.albany.edu/?q=mfold/download-mfold.
- 45.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 46.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.