Abstract
Vibrio splendidus is identified as one of the major pathogenic factors for the skin ulceration syndrome in sea cucumber (Apostichopus japonicus), which has vastly limited the development of the sea cucumber culture industry. In order to screen the immune genes involving Vibrio splendidus challenge in sea cucumber and explore the molecular mechanism of this process, the related transcriptome and gene expression profiling of resistant and susceptible biotypes of sea cucumber with Vibrio splendidus challenge were collected for analysis. A total of 319,455,942 trimmed reads were obtained, which were assembled into 186,658 contigs. After that, 89,891 representative contigs (without isoform) were clustered. The analysis of the gene expression profiling identified 358 differentially expression genes (DEGs) in the bacterial-resistant group, and 102 DEGs in the bacterial-susceptible group, compared with that in control group. According to the reported references and annotation information from BLAST, GO and KEGG, 30 putative bacterial-resistant genes and 19 putative bacterial-susceptible genes were identified from DEGs. The qRT-PCR results were consistent with the RNA-Seq results. Furthermore, many DGEs were involved in immune signaling related pathways, such as Endocytosis, Lysosome, MAPK, Chemokine and the ERBB signaling pathway.
Keywords: sea cucumber (Apostichopus japonicus), Vibrio splendidus, transcriptome sequencing, differentially expressed genes, bacteria-resistant gene, bacteria-susceptible gene
1. Introduction
Currently, the sea cucumber (Apostichopus japonicus) has become one of the most important aquaculture species in China, achieving valuable profits [1]. However, many diseases occurred along with the rapid expansion and intensification of farming, causing serious economic losses and disrupting the sustainable development of this industry [2]. Skin ulceration syndrome with clinical signs of anorexia, shaking head, mouth timidity, viscera ejection and skin ulceration, is the most serious disease and is highly infectious and lethal to the species. Etiological studies [3,4,5] indicated that a bacterial species (Vibrio/Pseudoalteromonas/Aeromonas spp.), a parasite (Parasitic nematode) and virus (a spherical virus) are the major pathogens for this serious disease, including Vibrio splendidus [6].
Like other invertebrates, sea cucumber lacks adaptive immunity and relies solely on innate immunity, which is composed of cellular responses such as phagocytosis and encapsulation, as well as humoral immunity that produces immune-related factors [7,8]. Coelomocytes function as the major sites for the elimination of pathogens [9,10]. Moreover, there are plentiful hydrolases in coelomic fluid such as lysozyme (LSZ), phenoloxidase activity (PO), total nitric oxide synthase (T-NOS), superoxide dismutase (SOD), and alkaline phosphatase (AKP), which could hydrolyze foreign pathogens [11,12]. In recent years, various studies have been conducted to investigate the immunity of sea cucumber, trying to identify the immune factors, clone immune-related genes, and obtain immune-related expressed sequence tags (EST) [13,14,15,16,17,18]. In addition, the studies of different microRNAs and proteins between healthy and natural skin ulceration syndrome Apostichopus japonicus have also been performed [19,20,21]. Nevertheless, the responsive mechanism of sea cucumber to pathogenic bacteria remains unclear.
In the present study, a mid-sensitive full-sib family was chosen for artificial-challenge experiment using Vibrio splendidus. The sea cucumbers were divided into a disease-resistant group and susceptibility group according to a series of symptoms of skin ulcer syndrome [4]. The transcriptome and expression profile of bacterial-resistant and bacterial-susceptible sea cucumber juveniles’ coelomocytes were analyzed using the Illumina sequencing method and bioinformatics analysis. Putative disease-resistant genes and susceptibility genes of sea cucumber were also screened. Additionally, gene-associated markers were screened for potential genetic research.
2. Results and Discussion
2.1. Illumina Sequencing and Assembly
To obtain an overview of the sea cucumber coelomocytes transcriptome, a cDNA library was generated from an equal mixture of RNA from nine individuals of the three groups (control group, bacterial-resistant group, and bacterial-susceptible group) using Illumina Hiseq2500 platform. After cleaning and quality checks, approximately 319 million (319,455,942) trimmed reads from 320 million (320,106,172) raw reads were generated using deep sequencing. Assembling analysis obtained 186,658 contigs with a median of contigs (N50) length of 1245, and 89,891 representative contigs (without isoform) with a N50 length of 791 using the 25-mer parameter in Trinity [22,23] (Table 1). Length statistics of assembled contigs and representative contigs are displayed in Figure 1 and Figure 2. The transcriptome database was next used as a source for the large set of functional genes (disease-resistant genes and susceptibility genes). The abundant data could also be a reference for further study including molecular markers and the genome of sea cucumber.
Table 1.
Summary statistics of assembled transcriptome length for Apostichopus japonicus coelomocytes.
| Assembled Transcriptome | All | Min | Median | Mean | N50 | Max | Total |
|---|---|---|---|---|---|---|---|
| contigs | 186,658 | 201 | 543 | 832 | 1245 | 15,051 | 155,375,852 |
| representative contigs (without isoform) | 89,891 | 201 | 376 | 593 | 791 | 15,051 | 53,310,798 |
N50, contig length—weighted median.
Figure 1.
The length distribution of assembled contigs in the sequenced cDNA library.
Figure 2.
The length distribution of representative contigs in the sequenced cDNA library.
2.2. Gene Annotation
Representative contigs were first annotated by BLAST to protein databases nr, Swiss-prot, Pfam, KEGG and COG separately and then annotated to nucleotide databases Nt with an E-value cut-off of 10−5. The percent values of the representative contigs to these databases are listed in Table 2. Altogether, 20,060 (22.32%) had at least one significant match to these databases (Table 2).
Table 2.
Result of functional annotation of the assembled representative contigs to the databases.
| No. of Representative Contigs | Swiss-Prot | Nr | Pfam | KEGG | COG |
|---|---|---|---|---|---|
| 89,891 | 13,955 | 19,777 | 14,398 | 18,887 | 13,126 |
| Percentage | 15.52% | 22.00% | 16.02% | 21.01% | 14.60% |
2.2.1. Nr Annotation
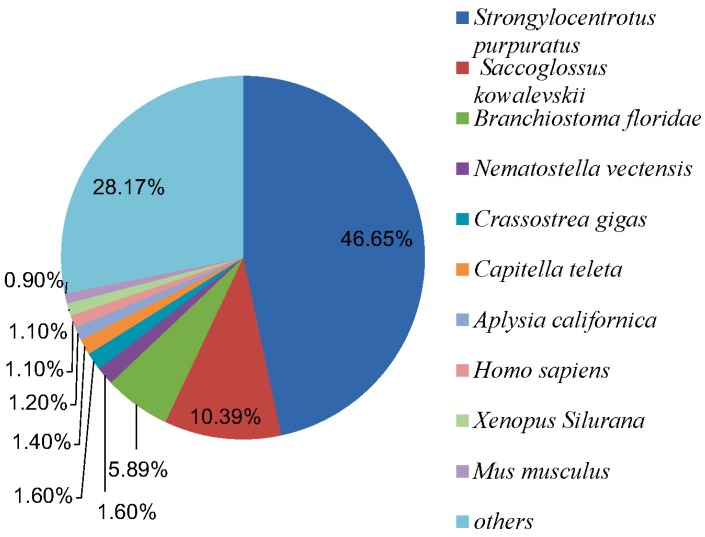
Among the annotated representative contigs to the Nr database, 6724 (46.7%) representative contigs were matched to Strongylocentrotus purpuratus, 1497 (10.4%) to Saccoglossus kowalevskii, 849 (5.9%) to Branchiostoma floridae, 230 (1.6%) to Nematostella vectensis, 230 (1.6%) to Crassostrea gigas, 202 (1.4%) to Capitella teleta, 173 (1.2%) to Aplysiacali fornica, 158 (1.1%) to Homo sapiens, 158 (1.1%) to Xenopus silurana, 130 (0.9%) to Mus musculus and 4047 (28.2%) to other species (Figure 3).
Figure 3.
Species distribution of the BLAST matches of the transcriptome representative contigs.
2.2.2. GO Annotation
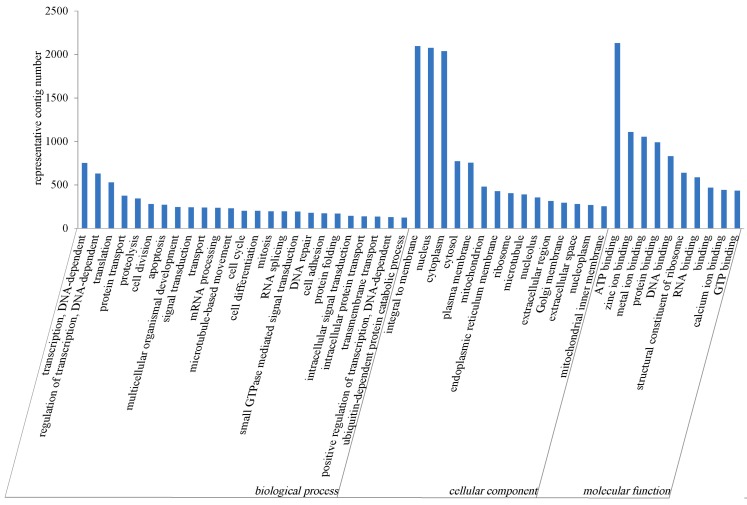
Gene Ontology (GO) [24] analysis was carried out, and for the three major functional categories: biological process, cellular component and molecular function, there were 9574, 11,078 and 10,994 representative contigs, respectively (Figure 4). For biological process, genes involved in transcription and DNA-dependent (752) was highest represented, followed by regulation of transcription DNA-dependent (630), and translation (528). Regarding cellular component, the top three categories were integral to membrane (2095), nucleus (2075) and cytoplasm (2037). For molecular function, ATP binding was the most represented GO term, followed by zinc ion binding.
Figure 4.
Classification of the gene ontology (GO) for the sea cucumber coelomocytes transcriptome representative contigs.
2.2.3. COG Annotation
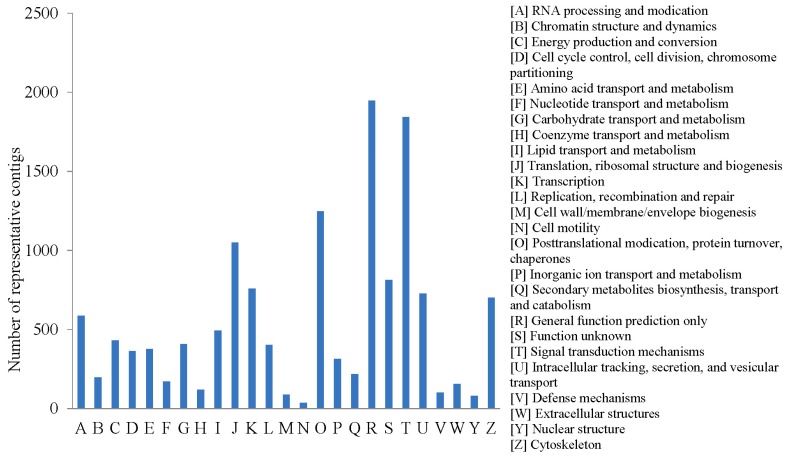
The COG database was used to classify orthologous gene products. 13,126 representative contigs were allocated to 25 COG classifications (Figure 5). Among them, “general function prediction only” (1946, 14.83%) and “signal transduction mechanisms” (1844, 14.05%) was the largest group, which indicated multiple genes were involved in signaling pathways after Vibrio splendidus infection.
Figure 5.
Clusters of Orthologous Groups (COG) classification of the sea cucumber coelomocytes transcriptome representative contigs.
2.2.4. KEGG Annotation
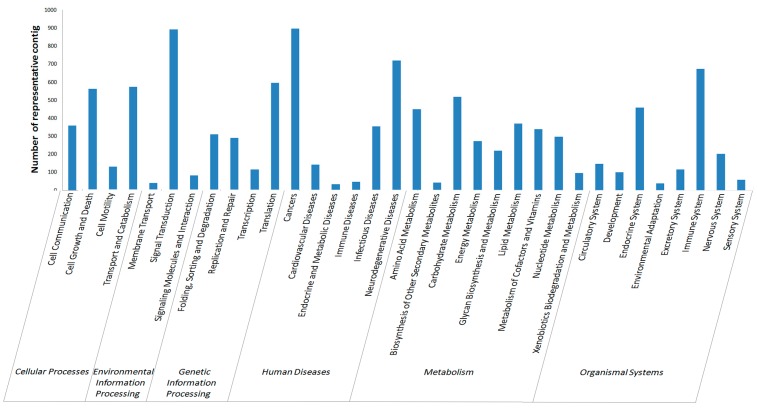
KEGG was used as a powerful tool to analyze biological metabolism and study metabolism networks. In all, 18,887 representative contigs were consequently classified into specific pathways (Figure 6), among which maximum members fell into “metabolism” (2346) and “human diseases” (2151), followed by “organism system” (1779), “cellular processes” (1625) and “genetic information processing” (1308), while the least amount of members were assigned to “environmental information processing” (1009). The highest number of genes were involved in signal transduction and immune system in KEGG, indicating many genes could respond to Vibrio splendidus challenge.
Figure 6.
KEGG classification of the sea cucumber coelomocytes transcriptome representative contigs.
2.3. Single Nucleotide Polymorphism (SNP) and Simple Sequence Repeat (SSR) Detecting
The transcriptome is also an important EST resource for rapid and effective mining of genetic markers, such as SNP and SSR [25]. The molecular markers have also been widely used in identifying functional genes, genetic breeding, genome mapping, and cloning genes.
In total, 149,745 high-quality SNPs were detected using Bowtie and the SAMTOOLS software. The dominant type of variation was transition (86,345, 57.67%), followed by transversion (63,391, 42.33%). The most common transition type was A→G and C→T (Table 3).
Table 3.
Summary of SNP identified from the sea cucumber coelomocytes transcriptome.
| SNP Type | NO. of SNP |
|---|---|
| Transition | 86,354 |
| A-G | 41,651 |
| C-T | 44,703 |
| Transversion | 63,391 |
| A-C | 16,243 |
| A-T | 21,687 |
| C-G | 10,327 |
| G-T | 15,134 |
| Total | 149,745 |
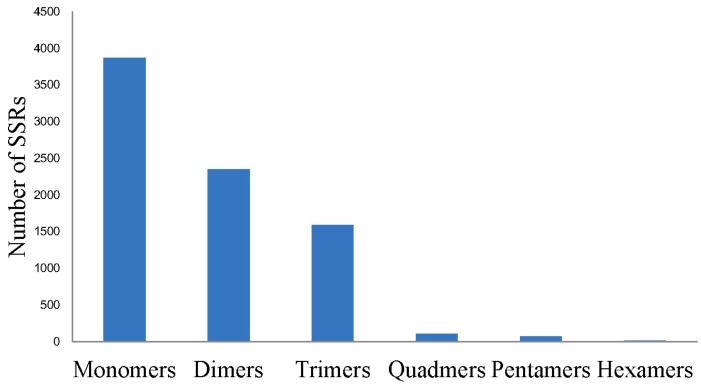
In addition, 8009 SSRs (simple sequence repeats) were identified from the assembled sequences. The most abundant repeat motifs were mono-nucleotides (3869), which accounted for 48.31% of all SSRs, followed by dinucleotides (2350, 29.34%), trinucleotides (1591, 19.87%), tetranucleotides (111, 1.39%), pentanucleotides (70, 0.87%), and hexanucleotides (18, 0.22%) (Figure 7).
Figure 7.
Summary of simple sequence repeat (SSR) identified from the sea cucumber coelomocytes transcriptome.
2.4. Construction of Digital Expression Profiling for Differentially Expressed Genes
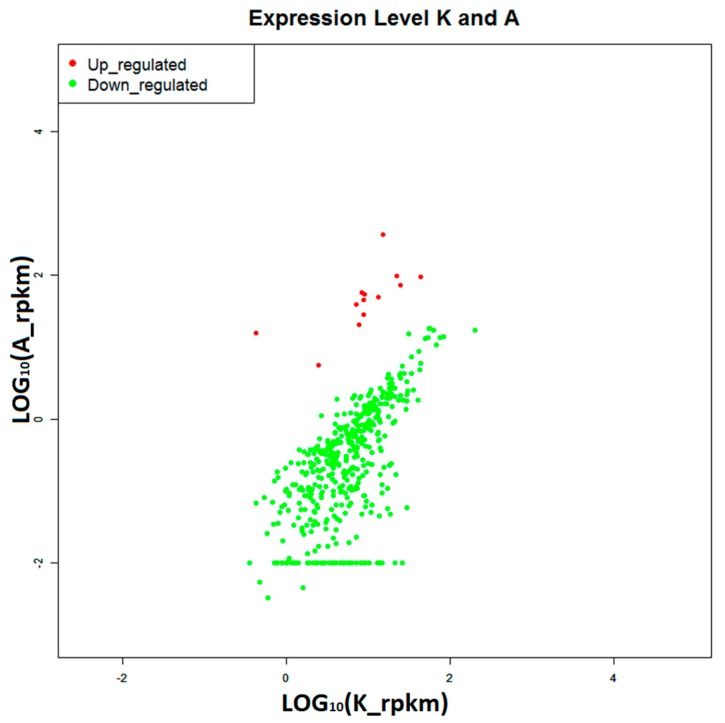
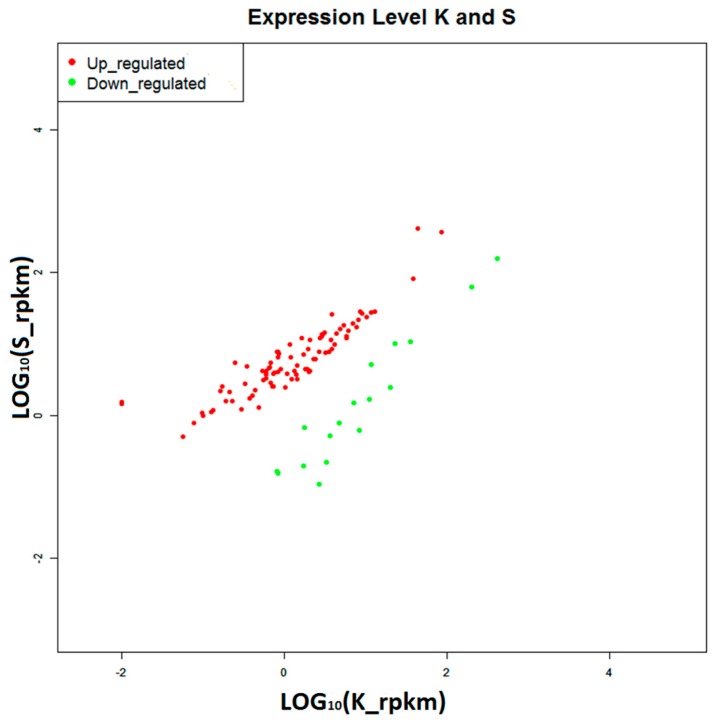
The digital gene expression profiling (DGE) is a rapid and efficient approach for gene expression analysis [26,27]. Many significantly differentially expressed genes (DEGs) were acquired by comparing the gene expressions in disease-resistant or susceptibility group (A or S, respectively) with the control group (K), under the criteria of p-value ≤ 0.01 and |log2 fold-change (FC)| ≥ 1 (FDR ≤ 0.05). As a result, we obtained 358 DEGs in the disease-resistant group (13 up-regulated and 345 down-regulated) (Figure 8) and 102 DEGs in the susceptibility group (86 up-regulated and 16 down-regulated) (Figure 9).
Figure 8.
Differentially expressed genes from disease-resistant group (A), comparing with control group (K). Red points represent 13 up-regulated genes, and green points represent 345 down-regulated genes.
Figure 9.
Differentially expressed genes from susceptibility group (S) comparing with control group (K). Red points represent 86 up-regulated genes, green point represents 16 down-regulated genes.
2.5. Selecting Disease-Resistant and Susceptibility Genes
The information of genes involved in immune response of other species were collected, which were used in combination of the GO, KEGG, NCBI annotation for the identification of potential bacterial-resistant genes, and 30 genes were identified (listed in Table 4 and Table 5). Some of these 30 genes had been reported in sea cucumber, such as heat shock protein70 (HSP70-like), which is a member of the heat shock protein family, stimulates the innate immune response [28,29] and plays crucial roles in environmental stress tolerance and adaptation in sea cucumber [30,31]. HSP70-like expression was significantly up-regulated after Vibrio splendidus infection, which is consistent with the result of LPS challenge [32].
Table 4.
A subset of candidate Vibrio splendidus-resistant and susceptibility genes that are involved in the immune signaling pathway.
| Coding Number | Contig ID | Gene Name | Predict Function | Regulation | Log2 FC | Accession Number in Nr Database | Identities (%) |
|---|---|---|---|---|---|---|---|
| Chemokine signaling pathway | |||||||
| A1 | comp76725_c0_seq6 | FOXO1-like | Fork head box protein (Strongylocentrotus purpuratus) | Down | −2.23 | XP_790591.3 | 78 |
| A2 | comp78415_c0_seq14 | ADCY2-like | Adenylatecyclase type 2 (Strongylocentrotus purpuratus) | Down | −5.91 | XP_780688.3 | 72 |
| A3 | comp79708_c0_seq1 | STAT5B-like | Signal transducer and activator of transcription 5B (Strongylocentrotus purpuratus) | Down | −3.81 | XP_003723422.1 | 70 |
| S1 | comp79328_c1_seq13 | NFKB-like | Nuclear factor NF-κB p105 subunit (Apostichopus japonicus) | Up | 1.8 | AEP33644.1 | 68 |
| S2 | comp74502_c1_seq4 | ADCY2-like | Adenylatecyclase type 2-like (Strongylocentrotus purpuratus) | Up | 4.18 | XP_780688.3 | 75 |
| Lysosome | |||||||
| A4 | comp74062_c0_seq5 | NEU1-like | Sialidase-1 (Strongylocentrotus purpuratus) | Down | −1.88 | DAA35227.1 | 85 |
| A5 | com78701_c0_seq2 | AP-1-like | AP-1 complex subunit mu-1-like (Strongylocentrotus purpuratus) | Down | −2.67 | XP_789616.3 | 77 |
| S3 | comp78293_c0_seq2 | ABCA2-like | ATP-binding cassette sub-family A member 2-like (Cricetulus griseus) | Up | 2.49 | XP_003514719.1 | 70 |
| S4 | comp78293_c0_seq4 | ABCA2-like | ATP-binding cassette sub-family A member 2-like (Cricetulus griseus) | Up | 2.49 | XP_003514719.1 | 70 |
| S5 | comp79570_c0_seq6 | SGSH-like | N-sulphoglucosamine sulphohydrolase-like (Strongylocentrotus purpuratus) | Up | 1.01 | XP_794467.1 | 70 |
| S6 | comp77223_c0_seq3 | ABCA2-like | ATP-binding cassette sub-family A member 2, partial (Strongylocentrotus purpuratus) | Up | 1.77 | XP_798273.3 | 68 |
| S7 | comp80153_c0_seq15 | AP-3-like | Adaptor-related protein complex 3, δ 1 subunit-like (Strongylocentrotus purpuratus) | Up | 1.79 | XP_002733668.1 | 69 |
| S8 | comp78750_c3_seq11 | DNase-II like | Plancitoxin-1 (Capitella teleta) | Up | 1.89 | ELU06802.1 | 75 |
| Endocytosis | |||||||
| A6 | comp76401_c0_seq2 | VPS37-like | ESCRT-I complex subunit VPS37 (Nematostella vectensis) | Down | −3.6 | XP_001624048.1 | 82 |
| S9 | comp77471_c1_seq34 | rabaptin5-like | RabGTPase -binding effector protein 1-like (Strongylocentrotus purpuratus) | Up | 1.8 | XP_789966.3 | 77 |
| S10 | comp80156_c1_seq5 | AP-2-like | AP-2 complex subunit alpha-2 (Rattus norvegicus) | Up | 1.16 | NP_112270.2 | 74 |
| S11 | comp77877_c0_seq1 | CHMP5-like | Charged multivesicular body protein 5-like (Strongylocentrotus purpuratus) | Up | 1.76 | XP_786663.1 | 72 |
| S12 | comp75233_c0_seq13 | PAR6-like | partitioning defective 6 (Hemicentrotus pulcherrimus) | Down | −2.11 | BAF99001.1 | 77 |
| S13 | comp79698_c0_seq6 | EGFR/ RTK-like | Epidermal growth factor receptor (Apostichopus japonicas) | Up | 1.32 | AEY55412.1 | 97 |
| ERBB signaling pathway | |||||||
| A7 | comp76122_c1_seq21 | NCK2-like | Cytoplasmic protein NCK2 (Strongylocentrotus purpuratus) | Up | 3.62 | XP_784072.1 | 72 |
| A8 | comp76122_c1_seq7 | NCK2-like | Cytoplasmic protein NCK2 (Strongylocentrotus purpuratus) | Up | 2.72 | XP_784072.2 | 72 |
| A3 | comp79708_c0_seq1 | STAT5B-like | Signal transducer and activator of transcription 5B (Strongylocentrotus purpuratus) | Down | −3.81 | XP_003723422.1 | 70 |
| S13 | comp79698_c0_seq6 | EGFR/RTK-like | Epidermal growth factor receptor (Apostichopus japonicas) | Up | 1.32 | AEY55412.1 | 97 |
| MAPK signaling pathway | |||||||
| A9 | comp77146_c0_seq3 | MAP3K4-like | Mitogen-activated protein kinase kinase kinase 4 (Strongylocentrotus purpuratus) | Down | −2.23 | XP_784029.3 | 72 |
| A10 | comp78357_c1_seq8 | MAPK10-like | Mitogen-activated protein kinase 10 (Strongylocentrotus purpuratus) | Down | −4.43 | XP_786040.3 | 75 |
| S1 | comp79328_c1_seq13 | NFKB-like | Nuclear factor NF-κB p105 subunit (Apostichopus japonicas) | Up | 1.8 | AEP33644.1 | 68 |
| S13 | comp79698_c0_seq6 | EGFR/RTK-like | Epidermal growth factor receptor (Apostichopus japonicas) | Up | 1.32 | AEY55412.1 | 97 |
| S14 | comp80408_c0_seq17 | FLNA-like | Filamin-A (Strongylocentrotus purpuratus) | Up | 1.75 | XP_792145.3 | 74 |
A, represents disease-resistant gene; S, represents susceptibility gene. Log2 FC (fold change) indicates differential expression level of disease-resistant group (A) relative to the control group (K). “−”, indicates fold change of down-regulation.
Table 5.
Other putative disease-resistant genes.
| Coding Number | Contig ID | Gene Name | Predict Function | Regulation | Log2 FC | Accession Number in Nr Database | Identities (%) |
|---|---|---|---|---|---|---|---|
| A11 | comp71589_c0_seq4 | COX19-like | cytochrome c oxidase assembly protein COX19 (Danio rerio) | Down | −2.64 | NP_001104010.1 | 72 |
| A12 | comp72396_c0_seq2 | DDX47-like | probable ATP-dependent RNA helicase DDX47-like (Strongylocentrotus purpuratus) | Down | −4.9 | XP_786173.3 | 76 |
| A13 | comp72841_c2_seq2 | Trmt1-like | tRNA methyltransferase 1-like (Saccoglossus kowalevskii) | Down | −3.12 | XP_002736321.1 | 67 |
| A14 | comp73256_c0_seq3 | Hrsp12-like | heat-responsive protein 12 (Mus musculus) | Down | −3.8 | EDL08846.1 | 77 |
| A15 | comp74533_c0_seq6 | CNOT10-like | CCR4-NOT transcription complex subunit 10-like (Ornithorhynchus anatinus) | Down | −2.52 | XP_001509062.1 | 76 |
| A16 | comp74754_c1_seq1 | phyhd1-like | phytanoyl-CoA dioxygenase domain-containing protein 1-like (Strongylocentrotus purpuratus) | Down | −3.55 | XP_789562.2 | 69 |
| A17 | comp74908_c0_seq5 | ehhadh-like | Peroxisomal bifunctional enzyme (Branchiostoma floridae) | Down | −3.86 | XP_002593843.1 | 73 |
| A18 | comp75055_c2_seq2 | DHX35-like | probable ATP-dependent RNA helicase DHX35-like (Strongylocentrotus purpuratus) | Down | −3.6 | XP_783015.1 | 66 |
| A19 | comp75531_c0_seq3 | RIOK3-like | Serine/threonine-protein kinase RIO3 (Saccoglossus kowalevskii) | Down | −2.19 | XP_002736242.1 | 69 |
| A20 | comp76071_c1_seq12 | Map2k6-like | Dual specificity mitogen-activated protein kinase kinase 6 (Capitella teleta) | Down | −2.71 | ELT91393.1 | 72 |
| A21 | comp76305_c0_seq6 | Gvin1-like | interferon-induced very large GTPase 1-like isoform X2 (Danio rerio) | Down | −2.67 | XP_684086.4 | 83 |
| A22 | comp76655_c1_seq14 | Ndufb3-like | NADH dehydrogenase (ubiquinone) 1 β subcomplex subunit 3-like (Strongylocentrotus purpuratus) | Down | −1.02 | XP_783578.1 | 81 |
| A23 | comp76725_c0_seq4 | PRPFF19-like | pre-mRNA-processing factor 19 (Strongylocentrotus purpuratus) | Down | −2.97 | XP_787949.3 | 74 |
| A24 | comp77143_c0_seq19 | Mapkap1-like | target of rapamycin complex 2 subunit MAPKAP1-like (Strongylocentrotus purpuratus) | Down | −7.52 | XP_787234.2 | 65 |
| A25 | comp77913_c0_seq1 | V1g163483-like | Inosine triphosphate pyrophosphatase (Rana catesbeiana) | Down | −3.36 | ACO51724.1 | 75 |
| A26 | comp78256_c0_seq1 | SMU1-like | WD40 repeat-containing protein SMU1 (Gallus gallus) | Down | −2.46 | NP_001007980.1 | 76 |
| A27 | comp78900_c0_seq70 | ND5-like | NADH dehydrogenase subunit 5 (Apostichopus japonicas) | Up | 1.15 | YP_002836162.1 | 100 |
| A28 | comp79236_c0_seq23 | YPEL5-like | protein yippee-like 5-like isoform 2 (Strongylocentrotus purpuratus) | Down | −3.51 | XP_786314.1 | 75 |
| A29 | comp80082_c0_seq9 | Usp39-like | tri-snRNP-associated protein 2 (Strongylocentrotus purpuratus) | Down | −4.33 | XP_001185686.2 | 71 |
| A30 | comp80196_c0_seq6 | Hsp70Ab-like | heat shock protein 70 (Apostichopus japonicas) | Up | 4.6 | ACJ54702.1 | 75 |
Log2 FC (fold change) indicates differential expression level of susceptibility group (S) relative to the control group (K). “−”, indicates fold change of down-regulation.
In addition, many disease-resistant genes that had not been previously linked to the immune response, including heat-responsive protein 12 (Hrp12-like), serine/threonine-protein kinase RIO3 (RIOK3-like) and Interferon-induced very large GTPase 1 (Gvin1-like) were involved in the immune response. However, these genes had been investigated in other species. For instance, Hrp12 has significant similarity to Hsp70 [33] and Hrp12-like may also play an important role in protein transport, protein folding and cell signaling. RIOK3 is a novel regulator of the antiviral type I interferon pathway and plays a crucial role in the antiviral type I interferon pathway. However, type I interferon is involved in the innate immune response which functions as the first line of defense and limits infectious pathogens directly [34], therefore we predicted that RIOK3-like is related closely to innate immunity. Mitogen-activated protein kinase kinase 6 (MP2K6), a member of the MAPK family which are signal transduction mediators that have been implicated in cell survival and death [35], is also activated during engagement of the Type I IFN receptor and plays important roles in Type I IFN signaling and generation of IFN responses [36]. Gvin1 that contributes to the cellular response to both type I and type II IFNs could lead to cell-autonomous resistance against various pathogens [37].
Nineteen potential susceptibility genes (Table 4 and Table 6) were identified from the DEGs, including Rho GTPase-activating protein 39 (ARHGAP39-like), scavenger receptor cysteine-rich protein type 12 precursor (DMBT1-like), and nuclear factor NF-κB p105 subunit (NFkB-like). The Rho GTPase-activating proteins (RhoGAPs) are one of the major classes of regulators of Rho GTPase which are important in cell cytoskeletal organization, membrane trafficking, transcriptional regulation, cell growth and differentiation, neuronal morphogenesis, and endocytosis [38,39]. Thus, ARHGAP39-like may be related with the immune response. Members of the scavenger receptor cysteine-rich (SRCR) superfamily have diverse functions, including pathogen recognition and immune-regulation [40], and we inferred that DMBT1-like might be also involved in the immune response.
Table 6.
Other putative susceptibility genes.
| Coding Number | Contig ID | Gene Name | Predict Function | Regulation | Log2 FC | Accession Number in Nr Database | Identities (%) |
|---|---|---|---|---|---|---|---|
| S15 | comp73644_c0_seq2 | ARHGAP39-like | Rho GTPase-activating protein 39 (Capitella teleta) | Up | 3.14 | ELT94447.1 | 66 |
| S16 | comp74218_c0_seq25 | ftsjd2-like | cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1-like (Danio rerio) | Up | 2.04 | XP_003729301.1 | 70 |
| S17 | comp75066_c0_seq3 | DMBT1-like | scavenger receptor cysteine-rich protein type 12 precursor (Strongylocentrotus purpuratus) | Up | 1.38 | NP_999762.1 | 70 |
| S18 | comp73655_c0_seq9 | Calr-like | Calreticulin (Strongylocentrotus purpuratus) | Up | 1.33 | XM_006792233.1 | 77 |
| S19 | comp72192_c0_seq1 | ATG5-like | autophagy-related protein 5 (Strongylocentrotus purpuratus) | Up | 1.32 | XM_011665174.1 | 70 |
Log2 FC (fold change) indicates differential expression level of susceptibility group (S) relative to the control group (K). “−”, indicates fold change of down-regulation.
2.6. Immune Signaling Pathway
2.6.1. MAPK Signaling Pathway
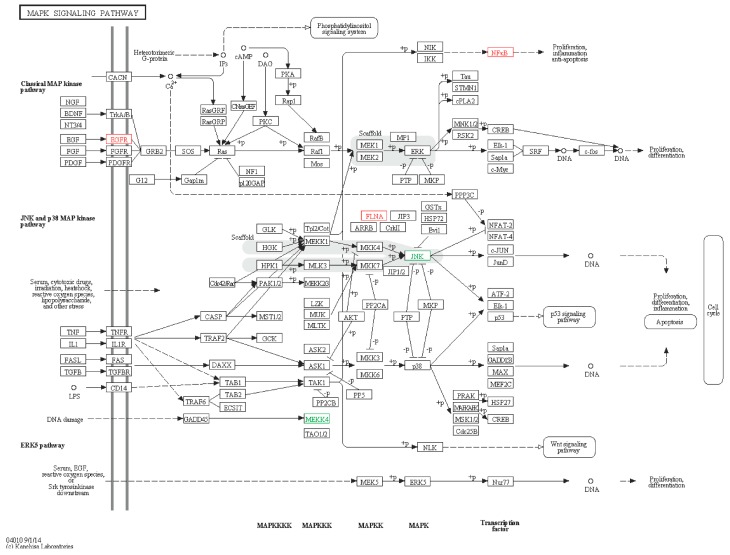
Five immune genes were enriched in the mitogen-activated protein kinase (MAPK) pathway, including JNK-like, MEKK4-like, NFkB-like, FLNA-like and EGFR-like (Figure 10). The MAPK cascade is a highly conserved module that is involved in various cellular functions, such as cell proliferation, differentiation and migration.
Figure 10.
MAPK signaling pathway. Red boxes represent up-regulated genes, and green boxes represent down-regulated genes.
The MAPK signaling pathway exists widely in all eukaryotes from yeast to human. The genes involved in the MAPK pathway in other species have been identified including Eriocheir sinensis [7], Pseudosciaena crocea [41], but not in sea cucumber. The genes found in sea cucumber: EGFR, JNK, MEKK4, NF-κB and FLNA, had been identified in Eriocheir sinensis, but not in Pseudosciaena crocea. EGFR exists on the cell surface and is activated by binding with its specific ligands, including epidermal growth factor and transforming growth factor α (TGFα), and it activates several signaling cascades to convert extracellular cues into appropriate cellular responses, principally the MAPK, protein kinase B (Akt) and Jun N-terminal kinase (JNK) pathways, leading to DNA synthesis and cell proliferation [42]. FLNA gene serves as a scaffold for a wide range of cytoplasmic signaling proteins. Moreover, the lack of filamin A damages cyclinB relevant proteins, and consequently delays the initiation and progression of mitosis [43]. MEKK4 is a member of MAPK family and could activate downstream MAPK kinase [44]. The c-Jun N-terminal kinases (JNKs) belonging to a large group of serine/threonine (Ser/Thr) protein kinase from the MAPK family, could phosphorylate and activate the transcription factor c-Jun [45]. NF-kappa-B gene is a transcription factor, regulating expression of a number of genes that participate in the inflammatory response, immune response, cell growth and apoptosis [46]. NF-κB p105 plays a role in the activation of NF-κB as a MAPK kinase signaling regulatory protein [47].
2.6.2. ERBB Signaling Pathway
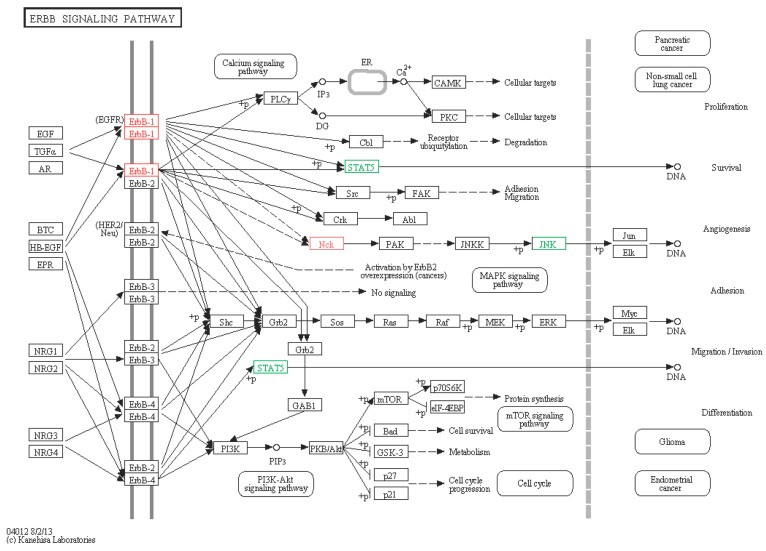
Four genes of sea cucumber were enriched in the ERBB pathway on the basis of our analysis (Figure 11), including ERBB-1-like (EFGR), STAT5-like, JNK-like, and Nck-like. The ERBB family of receptor tyrosine kinases (RTKs) couples binding of extracellular growth factor ligands to intracellular signaling pathways and regulates diverse biologic responses, including proliferation, differentiation, cell motility, and survival.
Figure 11.
ERBB signaling pathway. Red boxes represent up-regulated genes, and green boxes represent down-regulated genes.
The ERBB pathway has been investigated widely in human, however, few studies have been done in other animal species, including sea cucumber. STAT5 is phosphorylated by EGFR stimulation and binds with specific DNA, further activating or inactivating transcription [48,49]. Nck regulates cell cycle arrest after DNA damage in some pathways including a translocation of Nck to the nucleus [50].
2.6.3. Lysosomes
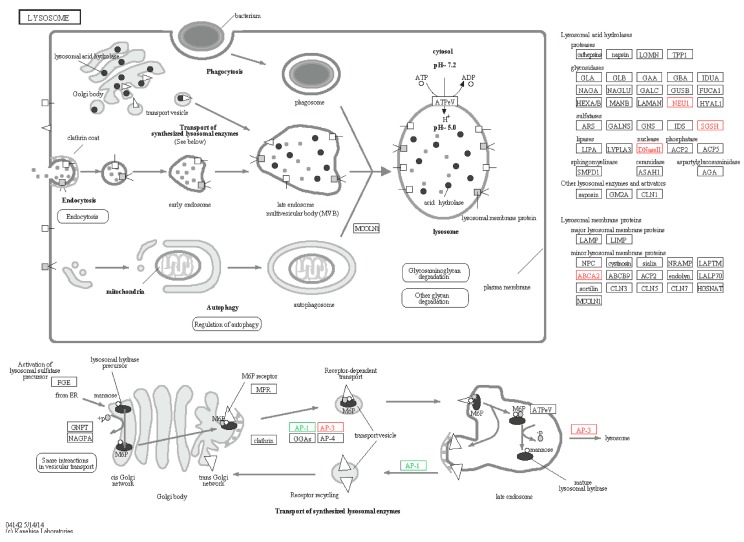
The hydrolysis of lysosomes is an important process of immune response for sea cucumber, which degrades pathogens through hydrolytic enzymes. The acidic hydrolytic enzymes of lysosomes include protease, glycosidase, sulfatase and lipase. Lysosomes include some membrane proteins such as LAMP, LIMP, and ABCA2. SGSH-like, DNaseII-like, ABCA2-like, AP-3-like, NEU1-like and AP-1-like (Figure 12) involved in lysosome were identified, among which NEU1-like belongs to glycosidase, SGSH-like belongs to sulfatase, DNaseII-like belongs to nuclease, ABCA2-like belongs to membrane proteins, and AP-1-like and AP-3-like are members of the clathrin family. The lysosome produces various acidic hydrolysis after Vibrio splendidus infection, therefore SGSH-like, DNaseII-like and NEU1-like could represent up-regulated expression. ABCA2-like may be relevant to transmembrane transport, hence, foreign pathogens are transported into the cell via membrane proteins and further hydrolysis [51]. AP-1 is mainly localized to the trans-Golgi network (TGN) and mediates protein trafficking between the TGN and endosomes [52,53]. AP-3 localized to endosomes and/or the TGN and in mammals, is thought to mediate protein sorting to lysosomes and specialized endosomal-lysosomal organelles [54,55]. Therefore, we inferred that AP-1-like and AP-3-like also change in response to the infection of Vibrio splendidus.
Figure 12.
Lysosome signaling pathway. Red boxes represent up-regulated genes, and green boxes represent down-regulated genes.
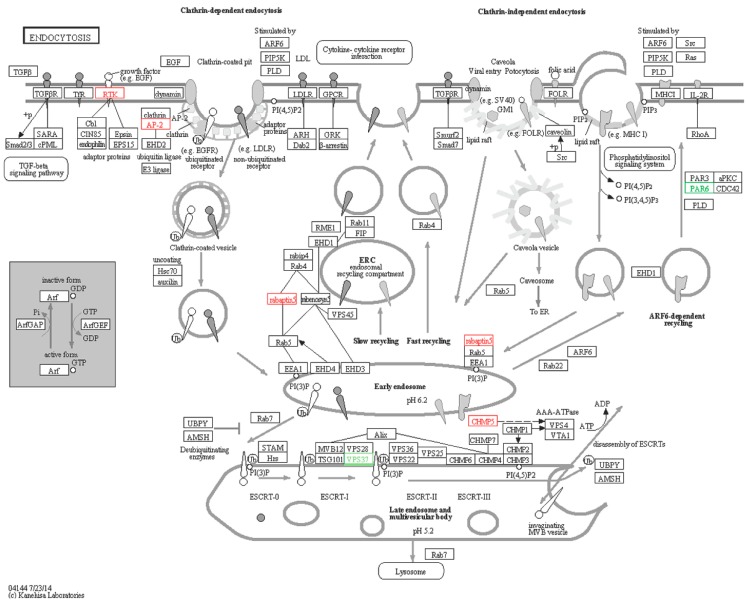
2.6.4. Endocytosis
Endocytosis is a process which brings extracellular large molecules or other cells (like bacteria) into the cell interior by cytomorphosis. There are three types of endocytosis: phagocytosis, pinocytosis and receptor mediated endocytosis. RTK/EGFR-like, AP-2-like, PAR6-like, rabaptin5-like, CHMP5-like, and VPS37-like (Figure 13) were the DEGs enriched in endocytosis, among which VPS37-like and PAR6-like were down-regulated. AP-2 is localized to the plasma membrane and mediates receptor endocytosis [56,57,58]. CHMP5 participated in biosynthesis of multivesicular bodies in endocytosis [59], therefore its expression will be up-regulated after Vibrio splendidus infection. Depletion of HCRP1/hVps37A in mammals hinders degradation of EGFR [60]. Rabaptin 5 is a major component of clathrin-coated vesicles in the Golgi network, taking part in endosomal recycling compartment with Rab4 [61]. PAR6 is also a key adaptor that links Cdc42 and atypical PKCs to Par3 in endocytosis [62].
Figure 13.
Endocytosis signaling pathway. Red boxes represent up-regulated genes, and green boxes represent down-regulated genes.
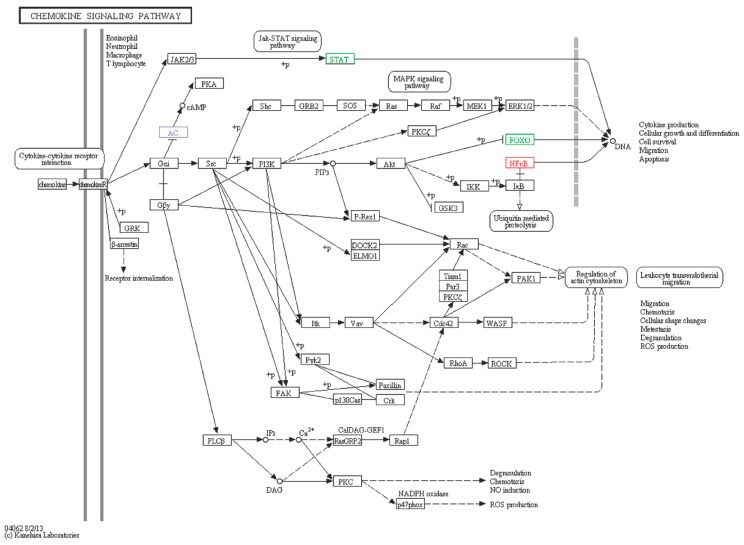
2.6.5. Chemokine Signaling Pathway
The chemokine receptor is one kind of small protein polypeptide of the cytokines superfamily and contains specific Cysteine motifs in their amino acid sequence [63]. Chemokines and their receptors are also important factors in B and T cell development [64,65], infections, angiogenesis, and tumor growth as well as metastasis. AC-like, NFkB-like, FOXO-like and STAT5B-like (Figure 14) participated in chemokine signaling pathways, of which FOXO-like and STAT-like were down-regulated. We predicted that the pathways including FOXO and STAT were inhibited, and immune response was depended mainly on the pathways involving in NFkB. Although above-mentioned genes are involved in immune signaling pathways, it is very necessary to validate their functions further.
Figure 14.
Chemokine signaling pathway. Red boxes represent up-regulated genes, green boxes represent down-regulated genes, and blue box includes two genes of which one is an up-regulated gene and the other is a down-regulated gene.
2.7. Differential Expression Verification of Putative Genes
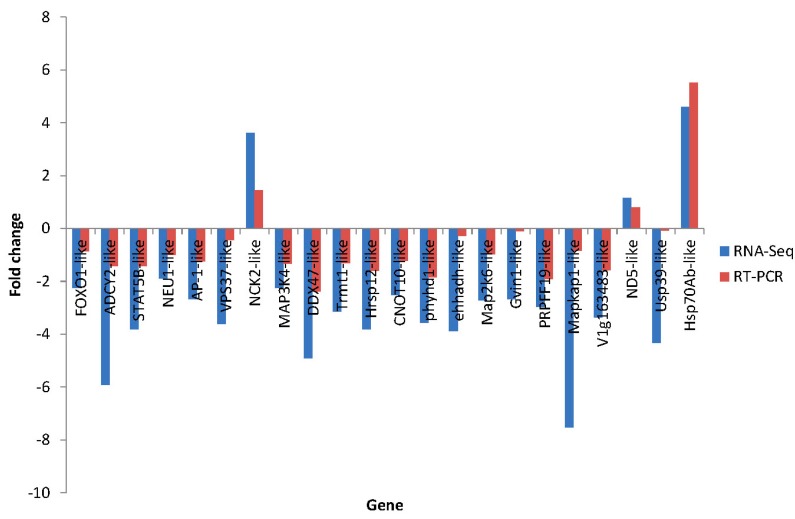
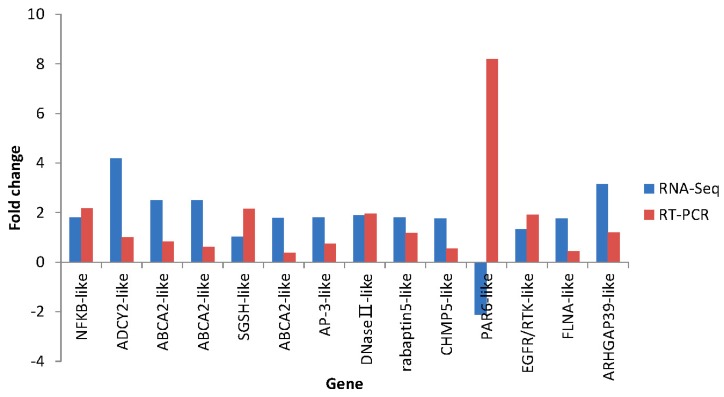
The primers of 36 genes were designed and all primer sequences are listed in Table S1. The candidate immune genes were used for qRT-PCR validation. The results showed the differential expression level of these genes between the control group and the disease-resistant group or susceptibility group (Figure 15 and Figure 16). Among them, up-regulation or down-regulation of 35 genes are consistent with the results of RNA-Seq. In conclusion, validation results (qRT-PCR vs. RNA-Seq) are overall reliable with the clear exception of PAR6-like, but further studies are still needed to verify the functions of these genes.
Figure 15.
Comparison of 22 putative disease-resistant gene expression levels between RNA-Seq (blue bar) and RT-PCR (red bar). “−”indicates down-regulation.
Figure 16.
Comparison of 14 putative susceptibility gene expression levels between RNA-Seq (blue bar) and RT-PCR (red bar). “−”indicates down-regulation.
3. Experimental Section
3.1. Sea Cucumber and Microbial Challenge
Two hundred healthy sea cucumber juveniles from a culture corporation in Qingdao with average weight of 10.3 ± 2.1 g were randomly selected from a full-sib family which is medium-sensitive to Vibrio splendidus and acclimatized in 4 plastic tanks (83 cm × 64 cm × 60 cm) with filtered ozone sterilized seawater for a week. The temperature was maintained at 15 ± 1.5 °C during the whole period of the experiment. The water was changed every three days and the sea cucumbers of treatment group were fed with formula every three days after changing the water.
Vibrio splendidus strain used in the experiment was initially isolated from a skin ulceration syndrome sea cucumber and identified as the pathogen of this syndrome in our laboratory [6]. Cryopreserved Vibrio splendidus strain were revived on tryptone soy broth medium (TSB) medium, and then inoculated into liquid TSB medium at 28 °C for 12 h with shaking at 200 rpm. The cultured bacteria were collected by centrifuging at 4000 rpm for 2 min, and then re-suspended in sterilized seawater.
For microbial challenge, 1 tank with 50 samples was served as control (without any treatment), and the other 3 tanks with 150 samples were immersed with high density of Vibrio splendidus at the final pathogen concentration of 108 cfu·mL−1. The challenge experiment lasted for 30 days and the water was changed every three days. The onset of skin ulceration syndrome was observed from 3 to 25 days in succession after the challenge. The skin ulceration syndrome appeared in 96 sea cucumbers (64% of the challenged sea cucumbers) and the residual 54 sea cucumbers (36%) were not infected by the disease. The first 25% infected sea cucumbers (24 sea cucumbers) were considered as bacterial-susceptible group (S) and the 36 uninfected sea cucumbers were considered as bacterial-resistant group (A). In all, three groups: control group (K), bacterial-resistant group (A) and bacterial-susceptible group (S) were used for the next steps.
3.2. Total RNA Extraction and cDNA Library Construction
Three sea cucumber individuals from each group were collected (S1, S2, and S3 from bacterial-susceptible group with obvious symptoms, A1, A2 and A3from bacterial-resistant group, K1 K2 and K3 from the control group). Coelomic fluid weas extracted and centrifuged at 3000 rpm for 2 min to harvest the coelomocytes. Total RNA was extracted using TaKaRa Mini BEST Universal RNA Extract Kit (Takara, Dalian, China). Total RNA quantity and purity were analyzed using Bioanalyzer 2100 and RNA 6000 Nano Lab Chip Kit (Agilent, Palo Alto, CA, USA) with RIN number >7.0 [66,67]. Nine RNA samples were collected from the 9 samples. RNA-Seq of every sample was performed respectively for gene expression profile analysis. Approximately 10 μg of total coelomocyte RNA was subjected to isolate Poly (A) mRNA with poly-T oligo attached magnetic beads (Invitrogen, Carlsbad, CA, USA). Following purification, the mRNA is fragmented into small pieces using divalent cations under elevated temperature. Then the cleaved RNA fragments were reverse-transcribed to create the final cDNA library in accordance with the protocol of the mRNA-Seq sample preparation kit (Illumina, San Diego, CA, USA), and the average insert size for the paired-end libraries was 300 bp (±50 bp). Subsequently, we performed the paired-end sequencing on an Illumina Hiseq2500 (LC Sciences, Hongzhou, China), following the recommended protocol from the vendor.
3.3. Sequencing and Assembly
Transcriptome sequence of the pooled nine samples was conducted using the Illumina paired-end RNA-Seq approach with Illumina 2500 sequence platform. Prior to assembly, the raw reads were first filtered by removing the adapter sequences, primer sequences and potential contaminations, which are the reads with unknown base greater than 5 and also with low-quality (<Q20) with existing tools: CutAdapt, NGS QC Toolkit and Trimmomatic. After that, paired-end trimmed reads were produced. The raw sequence data were then submitted to the NCBI Short Read Archive with accession number of SRP 057956. Also, trimmed reads were assembled using Trinity (Available online: http://trinityrnaseq.sourceforge.net/) and to remove the effect of different isoforms or alternative splicing, the longest contig of each isoform set was selected as the representative contig in the downstream analysis.
3.4. Annotation of Representative Contig
Representative contigs were first annotated to protein databases (download date: 7 March 2014) Nr, Swiss-prot, Pfam, KEGG and COG separately. Gene names were assigned to each assembled sequence based on the best BLAST hit (highest score). For homologous annotation, sample representative contigs were compared with NCBI non-redundant protein (Nr) (Available online: ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz), Swiss-Prot (Available online: ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz), Cluster of Orthologous Groups (COG) (Available online: http://www.ncbi.nlm.nih.gov/COG/grace/shokog.cgi), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Available online: http://www.kegg.jp/kegg/download/) and Pfam (Available online: ftp://ftp.sanger.ac.uk/pub/databases/Pfam/releases/Pfam27.0/Pfam-A.fasta.gz) database using algorithm blastx with E-value cut-off of 10−5. Gene ontology (GO) categories [68] were used for gene annotation using the BLAST 2 GO software [69,70]. The top 20 hits extracted from the blastx results were used for gene annotation and GO analysis (level 2), illustrating general functional categories. KEGG pathways were assigned to the assembled sequences using the online KEGG Automatic Annotation Server (KAAS, available online: http://www.genome. jp/kegg/kaas/). The bi-directional best hit (BBH) method was used to obtain KEGG Orthology (KO) assignment [71].
3.5. SNPs and SSRs Detection
The SNPs in the transcriptome level were analyzed based on the massively parallel Illumina technology. The Bowtie (Available online: http://bowtie-bio.sourceforge.net/) and Samtools (Available online: http://samtools.sourceforge.net/) software with default parameters (cDNA mode) were used to identify the SNPs. The SNP identification was limited to the transcripts (≥200 bp) containing at least 100 reads for each allele. The sample data were mapped to the contig via the Bowtie software after pretreatment, based on the library of transcription. Further SNP analysis was done according to the mapping results, and then variable sites with higher possibility were further filtered using the software of Samtools.
The MISA (Microsatellite) Perl script (Available online: http://pgrc.ipkgatersleben.de/misa) was used for the identification of SSRs. The BatchPrimer3 V1.0 program was used to design primers pairs for amplification of the SSR motifs [72]. Monomers, Dimers, Trimers, Quadmers, Pentamers and Hexamers were all considered as the searching criteria for SSRs in MISA script.
3.6. Identification of Differentially Expressed Genes (DEGs)
To investigate the expression level of each representative contig in different groups, digital gene expression profiles of the three groups were constructed and transcripts expression levels were calculated using RPKM (Reads per kilobase of exon model per million mapped reads) [73]. p-Value was used to identify the DEGs between two groups using chi-square test (2 × 2), and the significance threshold of the p-value in multiple tests was set based on the FDR (FDR ≤ 0.05). The fold changes (log2 (RERPKM/PERPKM)) were also estimated according to the normalized gene expression levels. Considering the above researches, “p-value < 0.01 and |log2fold change| ≥ 1 (FDR ≤ 0.05)” were set as the threshold. Putative Vibrio splendidus-resistant genes were screened through comparing the expression level of genes between disease-resistant group (A) and control group (K), and susceptibility genes were screened through comparing the expression difference between bacterial-susceptible group (S) and control group (K).
3.7. Identifying Potential Immune Genes and Pathway Analysis
The gene ontology (GO) was conducted for Functional classification of the putative disease resistant and susceptibility genes, and the pathway analysis was carried out by using KEGG.
3.8. Validation of Illumina Sequencing Results by qRT-PCR
Quantitative RT-PCR (qRT-PCR) was used to verify the expression level of putative immune genes that were identified in RNA-Seq analysis. Primers were designed using the Primer5 software and β-actin gene was used as the reference gene [74]. The qRT-PCR reactions were performed in a 20 mL volume composed of 2 μL of cDNA, 4 μM of each primer, and 10 mL Master mix, 2× conc (Roche, Penzberg, Germany) in the Eppendorf Real time PCR System. The thermal cycling program was 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 57 °C for 20 s and 72 °C for 30 s. Melting curve analysis was performed by the end of each PCR to confirm the PCR specificity. Three replications were used for each qRT-PCR validation. The relative expression of target genes was calculated using the 2−∆∆Ct method [75]. Differential expression level between control group and experimental group was determined using Log2 (A/K) or Log2 (S/K).
4. Conclusions
We conducted transcriptome sequencing and gene expression profile analysis of coelomocytes RNA in sea cucumber; 30 potential disease-resistant genes and 19 potential susceptibility genes were obtained, respectively, according to GO, KEGG, NCBI annotation and relevant published references. Furthermore, the genes were involved in immune signaling pathways, such as Endocytosis, Lysosome, MAPK, ERBB, and Chemokine, playing key roles in the interactive network of genes. Our study might provide useful information for future investigation of defense mechanism for Vibrio splendidus challenge.
Acknowledgments
This study was supported by the National High Technology Research and Development Program, China (2012AA10A412), National Natural Science Foundation of China (No. 31202016), Agriculture Seed Improvement Project of Shandong Province, Special Funds for Technology R&D Program in Research Institutes (2011EG134219), and the Science and Technology Program of Qingdao (13-4-1-65-hy).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/07/16347/s1.
Author Contributions
Qiong Gao and Meijie Liao conducted the major part of this study including sample collection, bioinformatic analysis and manuscript preparation; Yingeng Wang conceived and designed the experiment, supervised the entire study and revised the manuscript; Bin Li, Zheng Zhang and Guiping Chen were involved in the sample preparation and microbial challenge experiment; Xiaojun Rong and Lan Wang were involved in data analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sloan N.A. Echinoderm fisheries of the world: A review. In: Keegan B.F., O’Connor B.D.S., editors. Proceedings of the 5th International Echinoderm Conference on Echinodermata; Galway, Ireland. 24–29 September 1984; Rotterdam, The Netherlands: A.A. Balkema; 1984. pp. 109–124. [Google Scholar]
- 2.Huang H.W., Wang Y.G. Current situation, questions and prospect in the sea cucumber industry. China Fish. 2007;10:50–53. [Google Scholar]
- 3.Liu H.Z., Zheng F.R., Sun X.Q., Hong X.G., Dong S.L., Wang B., Tang X.X., Wang Y.Q. Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka) J. Invertebr. Pathol. 2010;105:236–242. doi: 10.1016/j.jip.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y.G., Zhang C.Y., Rong X.J., Chen J.J., Shi C.Y. Diseases of cultured sea cucumber Apostichopus japonicus in China. FAO Fish. Tech. Paper. 2005;463:297–310. [Google Scholar]
- 5.Deng H., Zhou Z.C., Wang N.B., Liu C. The syndrome of sea cucumber (Apostichopus) japonicas infected by virus and bacteria. Virol. Sin. 2008;23:63–67. doi: 10.1007/s12250-008-2863-9. [DOI] [Google Scholar]
- 6.Zhang C.Y., Wang Y.G., Rong X.J. Isolation and identification of causative pathogen for skin ulcerative syndrome in Apostichopus japonicus. J. Fish. China. 2006;30:118–123. [Google Scholar]
- 7.Li X.H., Cui Z.X., Liu Y., Song C.W., Shi G.H. Transcriptome analysis and discovery of genes involved in immune pathways from Hepatopancreas of microbial challenged mitten crab Eriocheir sinensis. PLoS ONE. 2013;8:e68233. doi: 10.1371/journal.pone.0068233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glin’ski Z., Jarosz J. Immune phenomena in echinoderms. Arch. Immunol. Ther. Exp. 2000;48:189–193. [PubMed] [Google Scholar]
- 9.Eliseikina M.G., Magarlamov T.Y. Coelomocyte morphology in the Holothurians Apostichopus japonicus (Aspidochirota: Stichopodidae) and Cucumaria japonica (Dendrochirota: Cucumariidae) Russ. J. Mar. Biol. 2002;28:197–202. doi: 10.1023/A:1016801521216. [DOI] [Google Scholar]
- 10.Dolmatova L.S., Eliseikina M.G., Romashina V.V. Antioxidant enzymatic activity of coelomocytes of the Far East sea cucumber Eupentacta fraudatrix. J. Evol. Biochem. Physiol. 2004;40:126–135. doi: 10.1023/B:JOEY.0000033803.35634.46. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z.M., Ma Y.X., Yang Z.P., Li M., Liu J., Bao P.Y. Immune responses and disease resistance of the juvenile sea cucumber Apostichopus japonicus induced by Metschnikowia sp. C14. Aquaculture. 2012;368:10–18. doi: 10.1016/j.aquaculture.2012.09.009. [DOI] [Google Scholar]
- 12.Ma Y.X., Liu Z.M., Yang Z.P., Li M., Liu J., Song J. Effects of dietary live yeast Hanseniaspora opuntiae C21 on the immune and disease resistance against Vibrio splendidus infection in juvenile sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2012;34:66–73. doi: 10.1016/j.fsi.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Gowda N.M., Goswami U., Khan M.I. T-antigen binding lectin with antibacterial activity from marine invertebrate, sea cucumber (Holothuria scabra): Possible involvement in differential recognition of bacteria. J. Invertebr. Pathol. 2008;99:141–145. doi: 10.1016/j.jip.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z.C., Sun D.P., Yang A.F., Dong Y., Chen Z., Wang X.Y., Guan X.Y., Jiang B., Wang B. Molecular characterization and expression analysis of a complement component 3 in the sea cucumber (Apostichopus japonicus) Fish Shellfish Immunol. 2011;31:540–547. doi: 10.1016/j.fsi.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y., Sun H.J., Zhou Z.C., Yang A.F., Chen Z., Guan X.Y., Gao S., Wang B., Jiang B., Jiang J.W. Expression analysis of immune related genes identified from the coelomocytes of sea cucumber (Apostichopus japonicus) in response to LPS challenge. Int. J. Mol. Sci. 2014;15:19472–19486. doi: 10.3390/ijms151119472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A.F., Zhou Z.C., Dong Y., Jiang B., Wang X.Y., Chen Z., Guan X.Y., Wang B., Sun D.P. Expression of immune-related genes in embryos and larvae of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2010;29:839–845. doi: 10.1016/j.fsi.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Cong L., Yang X., Wang X., Tada M., Lu M., Liu H., Zhu B. Characterization of an i-type lysozyme gene from the sea cucumber Stichopus japonicus, and enzymatic and nonenzymatic antimicrobial activities of its recombinant protein. J. Biosci. Bioeng. 2009;107:583–588. doi: 10.1016/j.jbiosc.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Yang A.F., Zhou Z.C., He C.B., Hu J.J., Chen Z., Gao X.G., Dong Y., Jiang H., Liu W.D., Guan X.Y., et al. Analysis of expressed sequence tags from body wall, intestine and respiratory tree of sea cucumber (Apostichopus japonicus) Aquaculture. 2009;296:193–199. doi: 10.1016/j.aquaculture.2009.08.016. [DOI] [Google Scholar]
- 19.Li C.H., Feng W.D., Qiu L.H., Xia C.G., Su X.R., Jin C.H., Zhou T.T., Zeng Y., Li T.W. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 2012;33:436–441. doi: 10.1016/j.fsi.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P., Li C.H., Li Y., Zhang P.J., Shao Y.N., Jin C.H., Li T.W. Proteomic identification of differentially expressed proteins in sea cucumber Apostichopus japonicus coelomocytes after Vibrio splendidus infection. Dev. Comp. Immunol. 2014;44:370–377. doi: 10.1016/j.dci.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P.J., Li C.H., Zhang P., Jin C.H., Pan D.D., Bao Y.B. iTRAQ-based proteomics reveals novel members involved in pathogen challenge in sea cucumber Apostichopus japonicus. PLoS ONE. 2014;9:e100492. doi: 10.1371/journal.pone.0100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-Seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du H.X., Bao Z.M., Hou R., Wang S., Su H.L., Yan J.J., Ti M.L., Li Y., Wei W., Lu W., et al. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867) PLoS ONE. 2012;7:e33311. doi: 10.1371/journal.pone.0033311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velculescu V.E., Kinzler K.W. Gene expression analysis goes digital. Nat. Biotechnol. 2007;25:878–880. doi: 10.1038/nbt0807-878. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Gerstein M., Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallin R.P., Lundqvist A., Moré S.H., von Bonin A., Kiessling R., Ljunggren H.G. Heat-shock proteins as activators of the innate immune system. Trends Immunol. 2002;23:130–135. doi: 10.1016/S1471-4906(01)02168-8. [DOI] [PubMed] [Google Scholar]
- 29.Zügel U., Kaufmann S.H.E. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong Y.W., Dong S.L., Meng X.L. Effects of thermal and osmotic stress on growth, osmoregulation and Hsp70 in sea cucumber (Apostichopus japonicus Selenka) Aquaculture. 2008;276:179–186. doi: 10.1016/j.aquaculture.2008.01.028. [DOI] [Google Scholar]
- 31.Meng X.L., Ji T.T., Dong Y.W., Wang Q.L. Thermal resistance in sea cucumbers (Apostichopus japonicus) with differing thermal history: The role of Hsp70. Aquaculture. 2009;294:314–318. doi: 10.1016/j.aquaculture.2009.06.015. [DOI] [Google Scholar]
- 32.Wang X.Y., Zhou Z.C., Yang A.F., Dong Y., Chen Z., Guan X.Y., Jiang B., Wang B. Molecular characterization and expression analysis of heat shock cognate 70 after heat stress and lipopolysaccharide challenge in sea cucumber (Apostichopus japonicus) Biochem. Genet. 2013;51:443–457. doi: 10.1007/s10528-013-9576-y. [DOI] [PubMed] [Google Scholar]
- 33.Samuel S.J., Tzung S.P., Cohen S.A. Hrp12, a novel heat-responsive, tissue-specific, phosphorylated protein isolated from mouse liver. Hepatology. 1997;25:1213–1222. doi: 10.1002/hep.510250525. [DOI] [PubMed] [Google Scholar]
- 34.Feng J., de Jesus P.D., Su V., Han S., Gong D., Wu N.C., Tian Y., Li X., Wu T.T., Chanda S.K., et al. RIOK3 is an adaptor protein required for IRF3-mediated antiviral type I interfereon production. J. Virol. 2014;88:7987–7997. doi: 10.1128/JVI.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y.Z., Batra S., Sassano A., Majchrzak B., Levy D.E., Gaestel M., Fish E.N., Davis R.J., Platanias L.C. Activation of mitogen-activated protein kinase kinase (MKK) 3 and MKK6 by Type I interferons. J. Biol. Chem. 2005;280:10001–10010. doi: 10.1074/jbc.M410972200. [DOI] [PubMed] [Google Scholar]
- 36.Hicks S.D., Parmele K.T., DeFranco D.B., Klann E., Callaway C.W. Hypothermia differentially increases extracellular signal-regulated kinase and stress-activated protein kinase/c-Jun terminal kinase activation in the hippocampus during reperfusion after asphyxial cardiac arrest. Neuroscience. 2000;98:677–685. doi: 10.1016/S0306-4522(00)00169-X. [DOI] [PubMed] [Google Scholar]
- 37.Klamp T., Boehm U., Schenk D., Pfeffer K., Howard J.C. A giant GTPase, very large inducible GTPase-1, is inducible by IFNs. J. Immunol. 2003;171:1255–1265. doi: 10.4049/jimmunol.171.3.1255. [DOI] [PubMed] [Google Scholar]
- 38.Moon S.Y., Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/S0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 39.Van Aelst L., D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 40.Qiu R., Sun B.G., Li J., Liu X., Sun L. Identification and characterization of a cell surface scavenger receptor cysteine-rich protein of Sciaenops ocellatus: Bacterial interaction and its dependence on the conserved structural features of the SRCR domain. Fish Shellfish Immunol. 2013;34:810–818. doi: 10.1016/j.fsi.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 41.Mu Y.N., Ding F., Cui P., Ao J.Q., Hu S.N., Chen X.H. Transcriptome and expression profiling analysis revealed changes of multiple signaling pathways involved in immunity in the large yellow croaker during Aeromonas hydrophila infection. BMC Genomics. 2010;11:506. doi: 10.1186/1471-2164-11-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oda K., Matsuoka Y., Funahashi A., Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lian G., Lu J., Hu J., Zhang J., Cross S.H., Ferland R.J., Sheen V.L. Filamin a regulates neural progenitor proliferation and cortical size through wee1-dependent Cdk1 phosphorylation. J. Neurosci. 2012;32:7672–7684. doi: 10.1523/JNEUROSCI.0894-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craig E.A., Stevens M.V., Vaillancourt R.R., Camenisch T.D. MAP3Ks as central regulators of cell fate during development. Dev. Dyn. 2008;237:3102–3114. doi: 10.1002/dvdy.21750. [DOI] [PubMed] [Google Scholar]
- 45.Bogoyevitch M.A., Kobe B. Uses for JNK: The many and varied substrates of the c-Jun N-terminal kinases. Microbiol. Mol. Biol. Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beg A.A., Baldwin A.S., Jr. Activation of multiple NF-κB/Rel DNA-binding complexes by tumor necrosis factor. Oncogene. 1994;9:1487–1492. [PubMed] [Google Scholar]
- 47.Beinke S., Ley S.C. Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem. J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J.X., Leonard W.J. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 49.Farrar M.A., Harris L.M. Turning transcription on or off with STAT5: When more is less. Nat. Immunol. 2011;12:1139–1140. doi: 10.1038/ni.2163. [DOI] [PubMed] [Google Scholar]
- 50.Kremer B.E., Adang L.A., Macara I.G. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mack J.T., Beljanski V., Tew K.D., Townsend D.M. The ATP-binding cassette transporter ABCA2 as a mediator of intracellular trafficking. Biomed. Pharmacother. 2006;60:587–592. doi: 10.1016/j.biopha.2006.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonifacino J.S., Lippincott-Schwartz J. Coat proteins: Shaping membrane transport. Nat. Rev. Mol. Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- 53.Mullins C., Hartnell L.M., Wassarman D.A., Bonifacino J.S. Defective expression of the mu3 subunit of the AP-3 adaptor complex in the Drosophila pig mentation mutant carmine. Mol. Gen. Genet. 1999;262:401–412. doi: 10.1007/s004380051099. [DOI] [PubMed] [Google Scholar]
- 54.Le Borgne R., Alconada A., Bauer U., Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Biol. Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- 55.Peden A.A., Oorschot V., Hesse B.A., Austin C.D., Scheller R.H., Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirchhausen T., Bonifacino J.S., Riezman H. Linking cargo to Vesicle formation: Receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 1997;9:488–495. doi: 10.1016/S0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 57.Lewin D.A., Mellman I. Sorting out adaptors. BBA-Mol. Cell. Res. 1998;1401:129–145. doi: 10.1016/S0167-4889(97)00130-4. [DOI] [PubMed] [Google Scholar]
- 58.Hirst J., Robinson M.S. Clathrin and adaptors. Biochim. Biophys. Acta. 1998;1404:173–193. doi: 10.1016/S0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 59.Bache K.G., Slagsvold T., Cabezas A., Rosendal K.R., Raiborg C., Stenmark H. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol. Biol. Cell. 2004;15:4337–4346. doi: 10.1091/mbc.E04-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang H.T., Connell J.W., Brown S.E., Thompson A., Reid E., Sanderson C.M. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Vitale G., Rybin V., Christoforidis S., Thornqvist P., McCaffre M., Stenmark H., Zerial M. Distinct Rab-binding domains mediate the interaction of Rabaptin5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joberty G., Petersen C., Gao L., Macara I.G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 63.Rossi D., Zlotnik A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 64.Forster R., Emrich T., Kremmer E., Lipp M. Expression of the G-protein-oupled receptor BLR1 defines mature, recirculating B cells and a subset of Thelper memory cells. Blood. 1994;84:830–840. [PubMed] [Google Scholar]
- 65.Vicari A.P., Figueroa D.J., Hedrick J.A., Foster J.S., Singh K.P., Menon S., Copeland N.G., Gilbert D.J., Jenkins N.A., Bacon K.B., et al. TECK: A novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity. 1997;7:291–301. doi: 10.1016/S1074-7613(00)80531-2. [DOI] [PubMed] [Google Scholar]
- 66.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7 doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mueller O., Lightfoot S., Schroeder A. RNA integrity number (RIN)–standardization of RNA quality control. Agilent Appl. Note Publ. 2004:1–8. [Google Scholar]
- 68.Balakrishnan R., Harris M.A., Huntley R., van Auken K., Cherry J.M. A guide to best practices for Gene Ontology (GO) manual annotation. Database (Oxf.) 2013 doi: 10.1093/database/bat054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 70.Götz S., García-Gómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talón M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.You F.M., Huo N., Gu Y.Q., Luo M.C., Ma Y., Hane D., Lazo G.R., Dvorak J., Anderson O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008;9 doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X., Wang X.W., Wang L.K., Feng Z.X., Zhang X.G. A review on the processing and analysis of next-generation RNA-Seq data. Prog. Biochem. Biophys. 2010;37:834–846. doi: 10.3724/SP.J.1206.2009.00151. [DOI] [Google Scholar]
- 74.Yang A.F., Zhou Z.C., Dong Y., Jiang B., Wang X.Y., Chen Z., Guan X.Y., Wang B., Sun D.P. Stability comparison of cytb and β-actin genes expression in sea cucumber (Apostichopus japonicus) J. Agric. Sci. Tech-Iran. 2010;12:79–84. [Google Scholar]
- 75.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−∆∆Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.