Abstract
The blistering skin disorder Epidermolysis bullosa simplex (EBS) results from dominant mutations in K5 or K14 genes, encoding the intermediate filament network of basal epidermal keratinocytes. The mechanisms governing keratin network formation and collapse due to EBS mutations remain incompletely understood. Drosophila lacks cytoplasmic intermediate filaments, providing a ‚null’ environment to examine the formation of keratin networks and determine mechanisms by which mutant keratins cause pathology. Here, we report that ubiquitous co-expression of transgenes encoding wild-type human K14 and K5 resulted in the formation of extensive keratin networks in Drosophila epithelial and non-epithelial tissues, causing no overt phenotype. Similar to mammalian cells, treatment of transgenic fly tissues with phosphatase inhibitors caused keratin network collapse, validating Drosophila as a genetic model system to investigate keratin dynamics. Co-expression of K5 and a K14R125C mutant that causes the most severe form of EBS resulted in widespread formation of EBS-like cytoplasmic keratin aggregates in epithelial and non-epithelial fly tissues. Expression of K14R125C/K5 caused semi-lethality; adult survivors developed wing blisters and were flightless due to lack of intercellular adhesion during wing heart development. This Drosophila model of EBS is valuable for the identification of pathways altered by mutant keratins and for development of EBS therapies.
INTRODUCTION
The keratin cytoskeleton protects epithelia against mechanical and other stresses and contributes to strong intercellular adhesion by interaction with desmosomes and hemidesmosomes (Homberg and Magin, 2014). Among the 54 type I and type II keratins, which form cell-specific cytoskeletal networks in all epithelial tissues, mutations in ≥20 keratin genes cause a large variety of disorders in stratified epithelia and modify complex disorders in simple epithelia [www.interfil.org (Szeverenyi et al., 2008)]. Keratins form heterodimers consisting of type I and type II keratins whereby K14 is the obligate K5 binding partner. Dominant mutations in keratin genes K5 or K14 composing the keratin network of basal keratinocytes, lead to the blistering skin disorder Epidermolysis bullosa simplex (EBS), characterized by collapse of the keratin network into cytoplasmic protein aggregates and tissue fragility (Coulombe et al., 2009). The mechanisms governing assembly of keratin subunits into filaments and bundles are partially characterized, however, the requirements for three-dimensional network organization and dynamics in vivo remain to be elucidated. It is proposed that keratin-intrinsic determinants and associated proteins such as plakin cytolinkers and 14-3-3 proteins are required for network formation (Lee and Coulombe, 2009; Windoffer et al., 2011). The occurrence of keratin arrays in epithelial and non-epithelial tissues suggests that these determinants lack tissue- and keratin-isotype specificity (Bader et al., 1988; Bader et al., 1991; Capetanaki et al., 2007; Traweek et al., 1993).
The missense mutation Arg125Cys is the most frequent mutation in K14 and gives rise to the most severe form of EBS (Dowling-Meara subtype), characterized by extensive cytoplasmic keratin aggregates. The molecular mechanisms by which these and additional mutations in keratin genes cause EBS and other keratinopathies are not well understood. Furthermore, it is unclear whether these disease phenotypes result from a loss or gain of function (Coulombe and Lee, 2012). Thus, there is a need for genetic models. To address this need, we developed a Drosophila model of EBS. We show that ectopic expression of human keratins K5 and K14 form a keratin network in Drosophila that caused no overt detrimental phenotype. In contrast, expression of mutant K14 and wild type K5, a combination that causes EBS in humans, resulted in semi-lethality at the pupal stage. Adult ‚escapers’ had blistered wings due to cell-cell adhesion defects during wing heart development. Our findings imply a gain of toxic function for keratin aggregates and provide a genetic model that will allow for rapid identification of conditions that ameliorate the pathological phenotypes.
RESULTS AND DISCUSSION
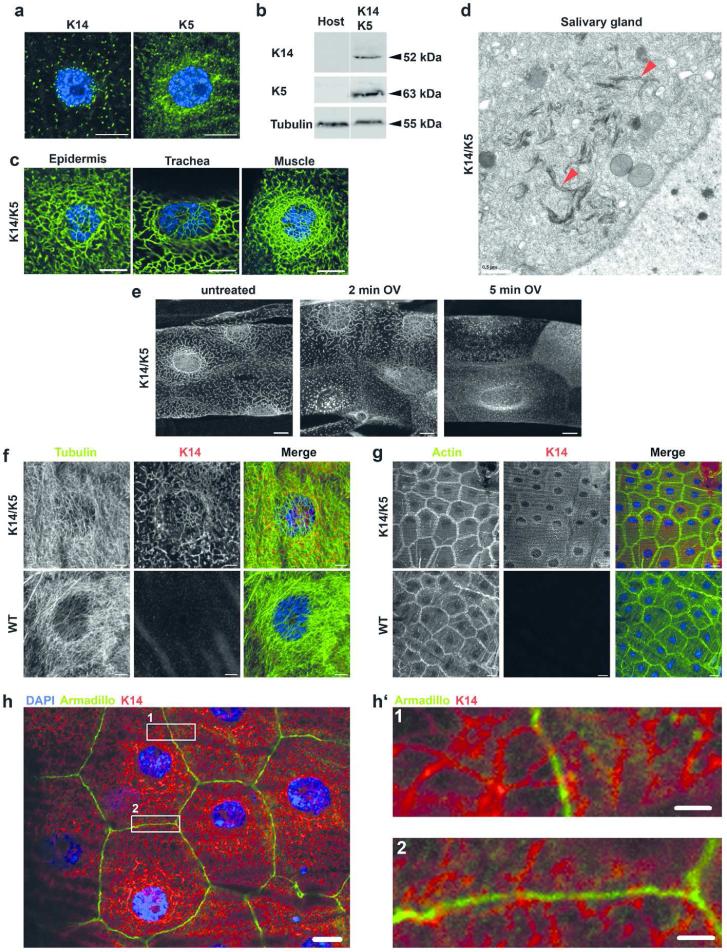
Drosophila lacks keratins and other cytoplasmic intermediate filament (IF) proteins (Goldstein and Gunawardena, 2000), providing a ‚null’ in vivo system to investigate mechanisms underlying keratin network formation and network collapse resulting from keratin gene mutations. Given the heterodimeric nature of keratin IF building blocks, we determined the consequences of expressing human K5 and K14 alone and in combination, in Drosophila. This was accomplished using the GAL4/UAS expression system, which allowed these keratins to be expressed globally or in specific tissues depending on the GAL4 driver (Brand and Perrimon, 1993; Duffy, 2002). Using multiple GAL4 drivers to express single keratins, only small cytoplasmic keratin aggregates resulted, but no filaments formed (Figure 1a). In contrast, co-expression of human K14 and K5 from a variety of GAL4 drivers resulted in the accumulation of extensive cytoplasmic keratin networks extending from the nuclear surface to the plasma membrane (Figure 1b/c, S1). Such keratin networks formed in all tissues tested. Electron microscopy demonstrated that K5 and K14 assembled into fine bundles, remarkably similar to those that form naturally in the vertebrate epidermis. One noticeable difference was the apparent shorter length of filament bundles, possibly due to low concentrations of keratins in the fly relative to human tissue (Figure 1d).
Figure 1. Keratin network organization in Drosophila melanogaster is similar to that observed in mammals.
a) Confocal immunofluorescence images of muscle tissue from third instar larvae expressing either K14 or K5 using Mef2-GAL4 muscle-specific driver. K14 and K5 are indicated by green and DAPI-stained DNA is indicated by blue. b) Western analysis of total protein extracted of adult flies expressing wt K14/K5 using Act5C-GAL4 and extracts from the host stock, which does not express keratins. Images are from the same membrane (see Figure S1). c) Confocal immunofluorescence images from third instar larvae. K14 (green) and K5 were expressed in epidermis and trachea using Act5C-GAL4 and in muscle using Mef2-GAL4 driver. d) Electron microscopy images of salivary glands of third instar larvae expressing K14 and K5 using the salivary gland-specific driver Sgs3-GAL4. Arrowheads indicate keratin bundles. e) Maximum intensity projections of entire confocal stacks of tracheal cells from third instar larvae expressing K14 (white) and K5 using the ubiquitous Act5C-GAL4 driver. Dissected tracheas were treated 2 or 5 min with 5 mM sodium orthovanadate (OV) dissolved in PBS. f-h) Confocal immunofluorescence images of third instar larvae epidermis expressing K14/K5 using the Act5C-GAL4 driver. Double labeling of the epidermis with a K14 antibody and either tubulin a), actin b) or armadillo c). DAPI-stained nuclei are blue. h’) Two magnified images from panel h showing contact sites between of keratin and the cell membrane. Scale bars: a/c/e/h 10 μm; d 0.5 μm; g 20 μm; f 5 μm; h’ 2 μm
To further substantiate that Drosophila is a suitable model system to study mechanisms underlying keratin organization and keratin-associated diseases, we tested if human keratin filaments behave in a similar way in fly tissues as in mammalian cells. Treatment of interphase keratin networks with tyrosine phosphatase inhibitors is well known to cause rapid filament breakdown and accumulation of cytoplasmic keratin granules in mammalian cells (Strnad et al., 2002). We treated freshly dissected trachea of third instar larvae with sodium orthovanadate, a tyrosine phosphatase inhibitor known to disassemble keratin filaments (Strnad et al., 2002). In analogy to mammalian cells, treatment with orthovanadate led to rapid breakdown of K14/K5 filaments and formation of keratin granules in tracheal cells (Figure 1e), highlighting the similarities between both systems. Remarkably similar results were obtained upon treatment with the serine/threonine phosphatase inhibitor okadaic acid (Strnad et al., 2001) (Figure S2). These findings suggest that phosphorylation, a major posttranslational modification that regulates keratin filament organization and turnover (Snider and Omary, 2014) is conserved in the fly. Thus, our model possesses many features that will be useful for a genetic analysis of keratin network formation and the identification of pathological mechanisms.
The fact that the wt K14/K5 combination could be expressed in a broad range of different tissues (Table S1) without deleterious effects suggests that the presence of keratin IF is well tolerated in Drosophila. In support of this, microtubules and actin filaments, adherens and septate junctions appeared unaltered in the presence of extensive keratin networks (Figure 1f/g/h, S3c/d, S4). Thus, we concluded that human keratin networks in Drosophila tissues cause no obvious alteration in the endogenous cytoskeleton and the main junctional complexes.
We noted that keratin IF appeared in very close approximation to Drosophila cell membranes in an orthogonal arrangement (Figure 1h’) reminiscent of mammalian epithelia. Although Drosophila lacks typical desmosomes, this could indicate the existence of yet unknown proteins able to mediate keratin interactions with the plasma membrane. Alternatively, amphiphilic sequences predicted in the aminoterminal head domain of type II keratins might be involved in such interactions (Ouellet et al., 1988). Future experiments are required to elucidate the nature of such interactions and a potential role in intercellular adhesion.
Non-mutually exclusive mechanisms might explain the network-forming ability of the K5 and K14 partners in Drosophila. Intrinsic keratin properties could be sufficient to form a network in an ectopic environment. It is possible that evolutionary conserved determinants in Drosophila such as Hsp70 (Boorstein et al., 1994), 14-3-3 proteins (Rosenquist et al., 2000) and plakins (Sonnenberg and Liem, 2007) facilitate network formation, despite the absence of endogenous keratins. Regardless of the mechanism, the network forms and causes no overt deleterious phenotype.
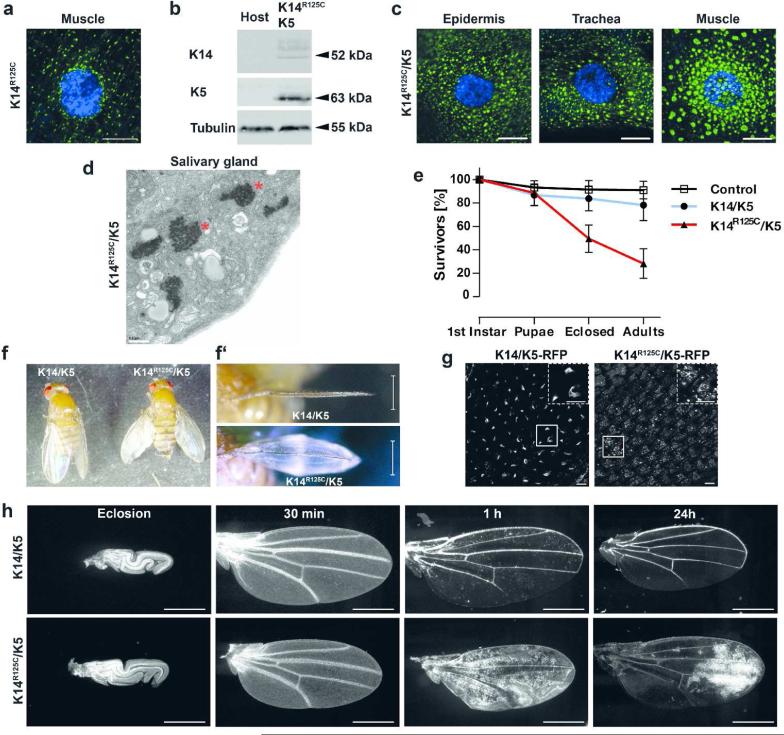
Having established commonalities between keratin networks in Drosophila and vertebrates, we investigated the effects of expressing the EBS K14R125C mutant alone and with a wt K5 partner using a variety of GAL4 drivers. Expression of K14R125C alone yielded only small aggregates, similar to those observed with K14 alone, (Figure 2a) and did not result in any overt phenotypes. In contrast, expression of K14R125C in combination with K5 (Figure 2b) resulted in large cytoplasmic keratin aggregates (Figure 2c). Electron microscopy (Figure 2d) revealed that these aggregates appeared highly similar to keratin aggregates detected in EBS patients (Anton-Lamprecht and Schnyder, 1982). These keratin aggregates caused no recognizable phenotype when expressed with a variety of tissue-specific GAL4 drivers (Table S1). In contrast, expression of the K14R125C/K5 combination by strong ubiquitous GAL4 drivers such as Act5C and Ubi (Table S1) resulted in lethality and semi-lethality, depending on the driver. Most remarkably, when expressed with Act5C-GAL4, the adult “escapers” that survived the pupal lethality stage (Figure 2e) formed wing blisters and were flightless (Figure 2f/f’). These wing blisters were filled with hemolymph (the “blood” of the fly), reminiscent of fluid-filled EBS blisters (Coulombe and Lee, 2012).
Figure 2. The human disease-causing mutant K14R125C causes wing blistering.
a) Confocal immunofluorescence image of muscle tissue from third instar larvae expressing K14R125C (green) using Mef2-GAL4 muscle-specific driver. DAPI-stained DNA is indicated by blue. b) Western analysis of total protein extracts of adult flies expressing K14R125C/K5 using Act5C-GAL4 and extracts from the host stock, which does not express keratins. Full size image of the western is shown in Figure S1. c) Confocal immunofluorescence images of third instar larval tissues expressing K14R125C (green) and K5. K14R125C/K5 were expressed in epidermis and trachea by Act5C-GAL4 or muscle specific by Mef2-GAL4. DAPI-stained DNA is indicated by blue. d) Electron microscopy images of salivary glands of third instar larvae expressing K14R125C/K5 by the salivary gland specific by Sgs3-GAL4 driver. Asterisks indicate keratin aggregates. e) Survival curves of flies expressing wt K14/K5 and K14R125C/K5 keratins by Act5C-GAL4. Adults were analyzed 3 to 4 days after eclosion from the pupal case. Mean ± SD, n≥6. f) Image of adult flies expressing wt or mutant K14 in combination with K5 using Act5C-GAL4. Frontal views of the wings are shown in f’). g) Expression of K14/K5-RFP or K14R125C/K5-RFP in wings immediately after unfolding, using Act5C-GAL4. RFP epifluorescence signal was recorded 5 to 10 min after unfolding. Dashed boxes are magnification of smaller boxes. h) Intervein cell clearance in K14/K5 or K14R125C/K5 expressing flies using Act5C-GAL4,UAS-GFP. GFP epifluorescence signal in wings were recorded at eclosion and at indicated time points after wing unfolding. Scale bars: a/c/g 10 μm; d 0.5 μm; f’ and h 500 μm
To investigate the generality of the observed phenotypes beyond the K14R125C allele, we tested a second K14 EBS patient mutation. Ubiquitous expression of K14R125P/K5 with Act5C-GAL4 resulted in effects similar to those observed with the K14R125C/K5 combination (Figure S5). Importantly, control flies expressing wt K14/K5 using strong, ubiquitous drivers did not affect viability and resulted in no such phenotypes. Western analyses of total fly protein extracts showed that K14R125C/K5 protein levels were lower than wt K14/K5 (Figure S1a). As quantitative real-time PCR analysis of total fly RNA demonstrated very similar mRNA levels in K14R125C/K5 and wt K14/K5 flies (Figure S6), this indicated elevated turnover of mutant K14 as potential mechanism for the reduced protein levels (Loffek et al., 2010). Thus, blistering was not caused by excessive IF protein levels, known to induce tissue defects (e.g. vimentin (Capetanaki et al., 1989), K8 (Casanova et al., 2004) and K1/K10 (Blessing et al., 1993)), and can be attributed to the mutant version of K14.
To investigate the apparent dominant nature of the K14R125C mutation, we expressed one or two copies of K5 along with one copy of wt K14 plus K14R125C by Act5C-GAL4. Both genetic backgrounds resulted in formation of extensive aggregates throughout the cytoplasm with occasional residual filaments close to the nucleus (Figure S7a/b), in full agreement with observations in primary EBS keratinocytes (Kitajima et al., 1989). In contrast, flies expressing one or two copies of K5 with two wt K14 copies showed extensive filaments (Figure S7a/b). The dominant nature of K14R125C was further confirmed by the appearance of wing blisters in K14/K14R125C;K5 expressing flies. In contrast, K14/K14;K5 flies showed no wing blisters. We note that Act5C-GAL4 driven K14/K14R125C;K5/K5 and K14/K14;K5/K5 expression (two copies of each transgenes) resulted in larval lethality by unknown mechanisms.
Since the Act5C-GAL4 driver is active in wing epithelia (Figure 2g), we suspected defects in wing maturation, a process that requires delamination of intervein cells from the cuticle and subsequent clearance from the wing. This developmental process is a prerequisite for tight attachment of dorsal and ventral cuticle layers necessary for formation of a flexible wing blade (Kiger et al., 2007; Kimura et al., 2004) (Figure S8a). Expression of wt keratins had no effect on wing maturation. In contrast, K14R125C/K5 caused retention of the intervein cells, preventing stable attachment of dorsal and ventral wing surfaces (Figure 2h). As delamination of wing epithelial cells from the cuticle involves an EMT-like process (Kiger et al., 2007), we examined whether this process was disturbed. Video imaging of GFP-labeled wing epithelial cells showed free-floating delaminated cells, supporting an unimpaired EMT-like process (Video S1). Given that wing blistering can also result from altered integrin expression (Brown et al., 2000), dysregulation of extracellular matrix components (Godenschwege et al., 2000) and altered signal transduction (Kimura et al., 2004), we addressed this by wing-restricted expression of mutant keratins using appropriate drivers (Table S1). Unexpectedly, these drivers failed to cause blistering. Although we cannot fully exclude the possibility that the wing-restricted drivers produced insufficient levels of keratins in wing precursors cells, wing discs and adult wings, the lack of an effect with the wing drivers suggested that the blistering might be not wing cell-autonomous (Figure S8b).
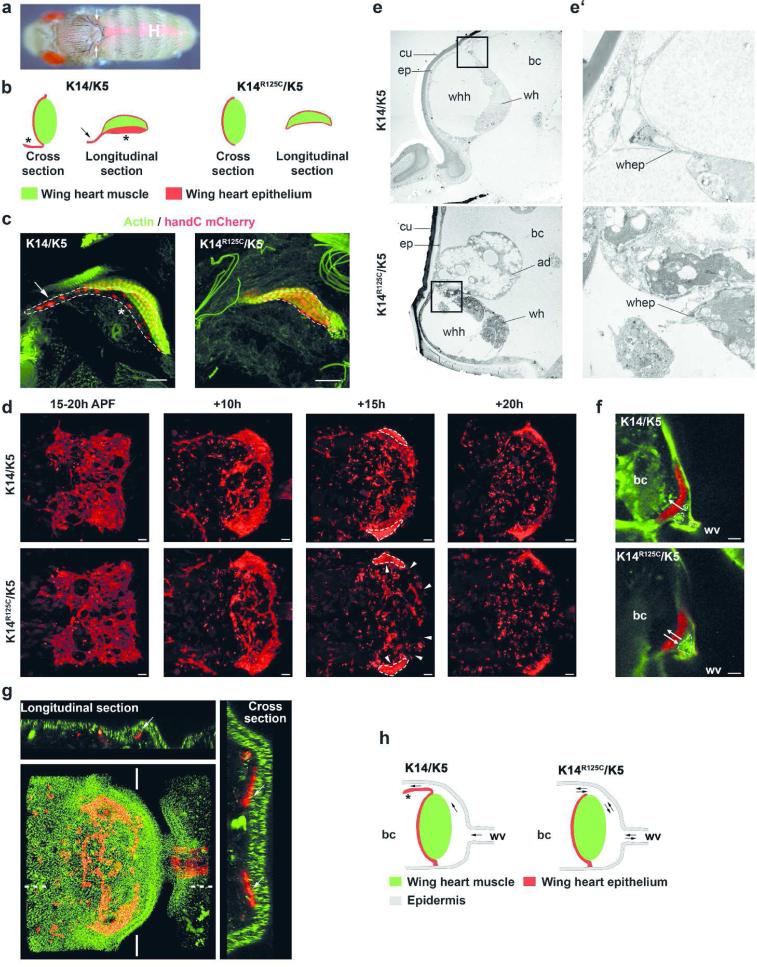
We considered the possibility that wing blistering resulted from defects in the wing heart (Togel et al., 2013a). Wing hearts (Figure 3a) are essential for wing maturation, maintenance of hemolymph flow in the wings and for the ability to fly (Togel et al., 2008). Wing hearts are positioned in the thorax near the wing attachment point and consist of muscle covered by a thin epithelium that is connected to the body wall epidermis. The wing hearts act as suction pumps that draw hemolymph from the wings into the body cavity. During wing maturation the pumping action is used to clear the intervein cells that have undergone EMT from the wing. To examine wing heart integrity and function we used the handC mCherry reporter, which is expressed in the wing heart, to visualize wing heart muscle and epithelium (Paululat and Heinisch, 2012; Togel et al., 2013b). Flies expressing wt K14/K5 showed no obvious wing heart defects. In contrast, expression of K14R125C/K5 caused structural defects in the wing heart epithelium (Figure 3b/c) leading to improper wing heart function (Figure 3f/h, Video S2). In wt and K14/K5 expressing flies, the wing heart epithelium functions normally and prevents the back-flow of hemolymph and cells during cardiac cycles (Figure 3e/e’/f/h, S9a, Video S3). In contrast, the K14R125C/K5 mutant lacked the essential parts of the epithelium that control flow of cells out of the wing. Video imaging confirmed that over extended periods of time, cells moved back and forth without net movement out of the wing as observed for wt (Video S2 and S3). This finding suggested that the epithelial defect caused a poor seal between the wing heart and the body cavity, leading to substantially reduced wing heart suction power. This resulted in a severe delay of intervein cell clearance (Figure 2h) and clotting of cells that block the hemolymph flow across the hemocoel. Ultimately, hemolymph accumulated and inflated the wing.
Figure 3. Mutant K14R125C causes a wing heart epithelial defect.
a) Epifluorescence image of an adult fly expressing the handC mCherry reporter, demonstrating the bilateral location of wing hearts (arrows) in the scutellum. The heart is marked by H. b) Diagram of cross and longitudinal sections through wing hearts expressing wt and mutant keratins. Asterisk indicates the epithelial back-flow valve; Arrow indicates epithelial elongation (sheet) of unknown function. c) Maximum intensity projections of total confocal stacks through wing hearts. Act5C-GAL4,handC mCherry was used to visualize wing heart epithelial cells (red nuclei) and actin stained by phalloidin (green). The dashed line indicates the extent of the wing heart epithelium. Arrow and asterisk indicate the same as in panel b. d) In vivo epifluorescence images of developing wing hearts and wing heart epithelium during pupal development using Act5C-GAL4,handC mCherry. Shown are maximum intensity projections of total confocal stacks of the developing wing hearts. Still images are from video S4 and S5. Dashed line indicates the muscle of the wing heart; arrowheads indicate areas of tissue disintegration. e-e’) Transmission electron micrographs of ultra-thin sections of wing hearts of adult flies (e at 3000 × and e’ at 12.000 × magnification). K14/K5 or K14R125C/K5 were expressed by Act5C-GAL4. f) Epifluorescence images of wing hearts of flies expressing K14/K5 or K14R125C/K5 with the Act5C-GAL4,handC mCherry/CyO; GFP reporter line. Arrows indicate the movements of delaminated intervein cells. Still images were taken from video S2 and S3. Act5C expressing cells in green, dashed lines indicate delaminated wing intervein cells in the wing heart hemocoel. g) Maximum intensity projection of an entire confocal stack of developing wing hearts. Flies expressing K14R125P-YFP;K5 were crossed with flies expressing the Act5C-GAL4,handC mCherry/CyO transgenes to visualize wing hearts and wing heart epithelial cells (red), and K14R125P-YFP expressing cells (green). Images were taken at ~35 hours after puparium formation. The white lines indicate origin of cross section; dotted lines indicate origin of longitudinal section. Arrows indicate tight contact of developing wing heart epidermal cells with the overlaying epidermis. h) Scheme of wing heart epithelial defect in mutant keratin flies. Arrows indicate possible direction of cell movement. Asterisk indicates epithelial back-flow valve. ad: adipocyte, bc: body cavity, cu: cuticle, ep: epidermis, wh: wing heart, whh: wing heart hemocoel, whep: wing heart epithelial cells, wv: wing vein, Scale bars: 20 μm
To further support the notion that suction defects resulted from the epithelial defect, electron microscopy of the wing heart was performed. The images provided evidence of retrograde flow of adipocytes into the hemocoel of mutant flies, due to impaired epidermal contact (Figure 3e/e’, S9a). The phenotype caused by expression of K14R125C/K5 is remarkably similar to the phenotype produced by the loss of the Drosophila hand gene (Togel et al., 2013a). These flies develop a wing heart epithelial defect, characterized by a blister wing phenotype and flightlessness. To further characterize wing heart epithelium defects, its formation during development was analyzed. Until 30-35 h after puparium formation (APF), wing heart formation proceeded undisturbed in wt and mutant keratin flies. Immediately afterwards, wing heart epithelial cell structures not directly attached to the muscle were found to disintegrate by a yet unknown mechanism (Figure 3d, Videos S4 and S5).
To examine whether this defect originated in the wing heart, Act5C-GAL4 activity in this organ was examined. In congruence with our finding that wing heart specific expression (handCGAL4) of mutant keratins led to no obvious phenotype, we noted that the Act5C-GAL4 driver was inactive in the wing heart. The notion that during development the wing heart progenitors are tightly attached to the pupal epidermis (Togel et al., 2013b) (Figure 3g, S9b) led us to hypothesize that epidermal cells instruct the underlying developing wing heart epithelial cells directly. In fact, Act5C-driven co-expression of the YFP tagged EBS mutant K14R125P and K5 caused formation of extensive keratin aggregates in the pupal epidermis (Figure 3g). To examine their significance to wing blistering, K14R125C/K5 were specifically expressed in the pupal epidermis, using the pnr-GAL4 driver, known to be specific for this tissue (Calleja et al., 2000; Togel et al., 2013b). Consistent with our hypothesis, expression of K14R125C/K5 caused wing blistering and flightlessness. Given that intercellular adhesion defects occur in keratin-deficient mice and keratinocytes (Bar et al., 2013; Kroger et al., 2013), these findings suggest that presence of mutant keratins in the Drosophila pupal epidermis causes wing blistering by compromising intercellular adhesion (Figure 3e/e’). Recent studies in mammalian systems have demonstrated a major role of keratins in the maintenance of intercellular adhesion. Both the loss of keratins and overexpression of mutant keratins caused diminished cell adhesion due to destabilization of desmosomes (Bar et al., 2013; Kroger et al., 2013; Liovic et al., 2009; Seltmann et al., 2013b). In addition, mutant keratins cause cell fragility (Coulombe and Lee, 2012; Ramms et al., 2013; Seltmann et al., 2013a), inflammation (Depianto et al., 2010; Lessard et al., 2013; Lu et al., 2007; Roth et al., 2012; Roth et al., 2009) and elevated protein stress (Loffek et al., 2010). Collectively, these findings suggest that epidermal keratin aggregates induced wing heart defects by mechanisms known to be targeted by mutant keratins in mammals (Togel et al., 2013a).
In summary, our data show that the expression of human keratins in Drosophila, an organism that does not require cytoplasmic intermediate networks due to its exoskeleton, is sufficient to form keratin networks that share properties with those in mammalian cells. In mammals, it is unclear whether disease-causing keratin mutations act as dominant gain of function or loss of function (Coulombe and Lee, 2012). The observation that widespread expression of the dominant K14R125C/K5 causes mutant phenotypes in Drosophila supports a gain of toxic function mechanism. This dominance is further supported by the fact that expression of K14R125C in the presence of wt K14/K5 caused mutant phenotypes. Our analysis also revealed that keratin aggregates disrupt intercellular adhesion, possibly by physically disrupting cellular networks or signaling involved in adhesion. Collectively, these findings support the notion that severe EBS mutations act by gain of toxic function to disrupt cell and tissue pathology through multiple mechanisms, including compromising cytoskeletal integrity. The semi-lethality and wing blisters that occur in Drosophila provided a unique opportunity for rapid genetic screens and whole organism compound screens to identify keratin network organizers and cellular pathways dysregulated by mutant keratins.
MATERIALS AND METHODS
Drosophila stocks
All Drosophila stocks were obtained from the Bloomington Stock Center (Bloomington, IN) unless otherwise noted. To generate stocks expressing human keratins, sequences encoding K14, K14R125C, K14R125P and K5 were PCR-amplified from full length cDNA and either cloned directly or in frame with either RFP or YFP into the pUAST transformation vector (Brand and Perrimon, 1993). Transgenic stocks were generated using standard P-element transformation techniques (Rubin and Spradling, 1982). The injection host stock y, 1w67c23 was used as a control. All stocks were cultured at 25°C. The GAL4-UAS system (Brand and Perrimon, 1993; Duffy, 2002) was used to drive expression of the keratins in specific tissues. GAL4-driver lines used in this study are listed in Table S1. A58-GAL4 and e22c-GAL4 were kindly provided by M. J. Galko (M.D. Anderson Cancer Center). Act5C-GAL4,handC-mCherry/CyO-GFP flies were generated by recombination of Act5C-GAL4 onto the handC-mCherry containing chromosome. K14,Act5C-GAL4;K5/T(2;3)SM6a-TM6B flies were generated by recombination of Act5C-GAL4 onto the K14 containing chromosome. Act5C-GAL4,handC-mCherry/CyO; UAS-GFP was generated by crossing Act5C-GAL4,handC-mCherry/CyO-GFP flies with a third chromosome UAS-GFP fly stock, gift of Y. Li (University of Iowa). Animal care and experimental procedures were in accordance with the institutional and governmental guidelines.
Immunohistochemistry, histology and electron microscopy
Indicated tissues were dissected fixed and stained as stated. For details see Supplementary Methods.
Phosphatase inhibitor treatment
Tracheal branches from third instar larvae were dissected and treated with okadaic acid (Santa Cruz Biotechnology, Heidelberg, Germany) or sodium orthovanadate (Sigma-Aldrich, Munich, Germany) dissolved in PBS at 25°C.
Documentation of intervein cell clearance
Stocks possessing the K14;K5 and K14R125C;K5 transgenes were crossed to Act5C-GAL4,UAS-GFP/ CyO flies and cultured at 25°C. Wings of adult progeny were cut off at different time points and GFP epifluorescence was recorded using a Nikon SMZ1500 stereomicroscope.
Recording of beating wing hearts
Stocks possessing the K14;K5 and K14R125C;K5 transgenes were crossed to Act5CGAL4,handC mCherry/ CyO; UAS-GFP flies and cultured at 25°C. Approximately 50 min after wing unfolding, mCherry positive progeny were anesthetized using carbon dioxide and attached with dorsal side down to cover slips with a small drop of Voltalef 10S oil (VWR, Dresden, Germany). Beating wing hearts were analyzed using a fluorescence laser-scanning confocal microscope (LSM 780, Carl Zeiss, Jena, Germany). GFP and mCherry epifluorescence was recorded simultaneously to allow higher frame rates. The videos were recorded with 5 frames per second. Analysis and processing of acquired images were carried out using Zen software (Carl Zeiss, Jena, Germany).
Time lapse analyses of wing heart development
Stocks possessing K14;K5 and K14R125C;K5 transgenes were crossed with Act5C-GAL4,handC mCherry/ CyO-GFP flies at 25°C. Pupae positive for mCherry were dissected from their pupal case 10 to 15 hours APF and attached with dorsal side down to cover slips with a small drop of Voltalef 10S oil (VWR, Dresden, Germany). To prevent dehydration, pupae were covered with the lid of a 2 ml Eppendorf tube inlaid with a moist filter paper.
Western analysis
SDS-PAGE was performed as previously described (Vijayaraj et al., 2009) In brief, total proteins of adult flies were extracted in SDS-PAGE sample buffer under repeated heating (95°C) and sonication cycles. Separation of total protein extracts was performed by standard procedures (10% SDS-PAGE). Western analysis was performed as described earlier (Vijayaraj et al., 2009). Primary antibodies used were anti-K5 1:10.000, anti-K14 1:62 (Abcam, Cambridge, UK) and anti-tubulin 1:12.000 (Sigma-Aldrich, Munich, Germany).
Flight test
The flight test was carried out with 100 individuals per genotype between 3 and 24 h after wing unfolding as previously described (Togel et al., 2008). In brief, flies were placed individually in a transparent container and flight ability was scored positive when flies flew in the container or landed on the walls. Flies that fell directly to the bottom and were unable to fly even upon mechanical stimulation were counted as being flightless.
Lethal phase analysis
Stocks expressing the K14;K5 and K14R125C;K5 transgenes were crossed with Act5C-GAL4/ CyO-GFP flies and cultured at 25°C. Adults were allowed to lay eggs for 3 h on apple juice agar plates (4.5 g agar, 2.5 g sucrose in 200 ml apple juice). After 24 hours, the first instar larvae expressing keratins or the balancer chromosome CyO-GFP were selected and transferred to new vials (30 per vial). Individuals were counted at the indicated time points. Adults were 3 to 4 days old. The CyO-GFP balancer chromosome flies were used as controls.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for stocks from the Bloomington Stock Center (Bloomington, IN), M. J. Galko (M.D. Anderson Cancer Center) and Y. Li (University of Iowa) for stocks. We thank A. Weinhold, M. Richter, K. Etzold, C. Lehmacher and O. Jones for excellent technical assistance. This work was partially supported by the Translational Center for Regenerative Medicine, TRM, Leipzig, PtJ-Bio, 0315883, and a University of Iowa Helen C. Levitt Endowed Annual Visiting Professorship to T. M. Magin; R4119 to J. Bohnekamp, by funding from the German Federal Ministry of Education and Research (BMBF 1315883), by the DFG (SFB 944) and the State of Lower-Saxony, Hannover to A. Paululat and by a Travel Fellowship to J. Bohnekamp from The Company of Biologists Ltd. Electron microscopy was partially supported by National Institutes of Health grants S10 RR018998 and P30CA086862 to the University of Iowa.
ABBREVATIONS
- APF
after puparium formation
- EBS
Epidermolysis bullosa simplex
- IF
intermediate filament
- K5
keratin 5
- K14
keratin 14
- wt
wild-type
Footnotes
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interest.
REFERENCES
- Anton-Lamprecht I, Schnyder UW. Epidermolysis bullosa herpetiformis Dowling-Meara. Report of a case and pathomorphogenesis. Dermatologica. 1982;164:221–35. [PubMed] [Google Scholar]
- Bader BL, Jahn L, Franke WW. Low level expression of cytokeratins 8, 18 and 19 in vascular smooth muscle cells of human umbilical cord and in cultured cells derived therefrom, with an analysis of the chromosomal locus containing the cytokeratin 19 gene. Eur J Cell Biol. 1988;47:300–19. [PubMed] [Google Scholar]
- Bader BL, Magin TM, Freudenmann M, et al. Intermediate filaments formed de novo from tail-less cytokeratins in the cytoplasm and in the nucleus. J Cell Biol. 1991;115:1293–307. doi: 10.1083/jcb.115.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar J, Kumar V, Roth W, et al. Skin Fragility and Impaired Desmosomal Adhesion in Mice Lacking All Keratins. J Invest Dermatol. 2013;134:1012–22. doi: 10.1038/jid.2013.416. [DOI] [PubMed] [Google Scholar]
- Blessing M, Ruther U, Franke WW. Ectopic synthesis of epidermal cytokeratins in pancreatic islet cells of transgenic mice interferes with cytoskeletal order and insulin production. J Cell Biol. 1993;120:743–55. doi: 10.1083/jcb.120.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown NH, Gregory SL, Martin-Bermudo MD. Integrins as mediators of morphogenesis in Drosophila. Dev Biol. 2000;223:1–16. doi: 10.1006/dbio.2000.9711. [DOI] [PubMed] [Google Scholar]
- Calleja M, Herranz H, Estella C, et al. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development. 2000;127:3971–80. doi: 10.1242/dev.127.18.3971. [DOI] [PubMed] [Google Scholar]
- Capetanaki Y, Bloch RJ, Kouloumenta A, et al. Muscle intermediate filaments and their links to membranes and membranous organelles. Exp Cell Res. 2007;313:2063–76. doi: 10.1016/j.yexcr.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Capetanaki Y, Smith S, Heath JP. Overexpression of the vimentin gene in transgenic mice inhibits normal lens cell differentiation. J Cell Biol. 1989;109:1653–64. doi: 10.1083/jcb.109.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova ML, Bravo A, Martinez-Palacio J, et al. Epidermal abnormalities and increased malignancy of skin tumors in human epidermal keratin 8-expressing transgenic mice. FASEB J. 2004;18:1556–8. doi: 10.1096/fj.04-1683fje. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Kerns ML, Fuchs E. Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J Clin Invest. 2009;119:1784–93. doi: 10.1172/JCI38177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Lee CH. Defining keratin protein function in skin epithelia: epidermolysis bullosa simplex and its aftermath. J Invest Dermatol. 2012;132:763–75. doi: 10.1038/jid.2011.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depianto D, Kerns ML, Dlugosz AA, et al. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010;42:910–4. doi: 10.1038/ng.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Godenschwege TA, Pohar N, Buchner S, et al. Inflated wings, tissue autolysis and early death in tissue inhibitor of metalloproteinases mutants of Drosophila. Eur J Cell Biol. 2000;79:495–501. doi: 10.1078/0171-9335-00072. [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Gunawardena S. Flying through the drosophila cytoskeletal genome. J Cell Biol. 2000;150:F63–8. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg M, Magin TM. Beyond expectations: novel insights into epidermal keratin function and regulation. International review of cell and molecular biology. 2014;311:265–306. doi: 10.1016/B978-0-12-800179-0.00007-6. [DOI] [PubMed] [Google Scholar]
- Kiger JA, Jr., Natzle JE, Kimbrell DA, et al. Tissue remodeling during maturation of the Drosophila wing. Dev Biol. 2007;301:178–91. doi: 10.1016/j.ydbio.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Kodama A, Hayasaka Y, et al. Activation of the cAMP/PKA signaling pathway is required for post-ecdysial cell death in wing epidermal cells of Drosophila melanogaster. Development. 2004;131:1597–606. doi: 10.1242/dev.01049. [DOI] [PubMed] [Google Scholar]
- Kitajima Y, Inoue S, Yaoita H. Abnormal organization of keratin intermediate filaments in cultured keratinocytes of epidermolysis bullosa simplex. Arch Dermatol Res. 1989;281:5–10. doi: 10.1007/BF00424265. [DOI] [PubMed] [Google Scholar]
- Kroger C, Loschke F, Schwarz N, et al. Keratins control intercellular adhesion involving PKC-alpha-mediated desmoplakin phosphorylation. J Cell Biol. 2013;201:681–92. doi: 10.1083/jcb.201208162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Coulombe PA. Self-organization of keratin intermediate filaments into cross-linked networks. J Cell Biol. 2009;186:409–21. doi: 10.1083/jcb.200810196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JC, Pina-Paz S, Rotty JD, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A. 2013;110:19537–42. doi: 10.1073/pnas.1309576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liovic M, D'Alessandro M, Tomic-Canic M, et al. Severe keratin 5 and 14 mutations induce down-regulation of junction proteins in keratinocytes. Exp Cell Res. 2009;315:2995–3003. doi: 10.1016/j.yexcr.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Loffek S, Woll S, Hohfeld J, et al. The ubiquitin ligase CHIP/STUB1 targets mutant keratins for degradation. Hum Mutat. 2010;31:466–76. doi: 10.1002/humu.21222. [DOI] [PubMed] [Google Scholar]
- Lu H, Chen J, Planko L, et al. Induction of inflammatory cytokines by a keratin mutation and their repression by a small molecule in a mouse model for EBS. J Invest Dermatol. 2007;127:2781–9. doi: 10.1038/sj.jid.5700918. [DOI] [PubMed] [Google Scholar]
- Ouellet T, Levac P, Royal A. Complete sequence of the mouse type-II keratin EndoA: its amino-terminal region resembles mitochondrial signal peptides. Gene. 1988;70:75–84. doi: 10.1016/0378-1119(88)90106-0. [DOI] [PubMed] [Google Scholar]
- Paululat A, Heinisch JJ. New yeast/E. coli/Drosophila triple shuttle vectors for efficient generation of Drosophila P element transformation constructs. Gene. 2012;511:300–5. doi: 10.1016/j.gene.2012.09.058. [DOI] [PubMed] [Google Scholar]
- Ramms L, Fabris G, Windoffer R, et al. Keratins as the main component for the mechanical integrity of keratinocytes. Proc Natl Acad Sci U S A. 2013;110:18513–8. doi: 10.1073/pnas.1313491110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, et al. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J Mol Evol. 2000;51:446–58. doi: 10.1007/s002390010107. [DOI] [PubMed] [Google Scholar]
- Roth W, Kumar V, Beer HD, et al. Keratin 1 maintains skin integrity and participates in an inflammatory network in skin through interleukin-18. J Cell Sci. 2012;125:5269–79. doi: 10.1242/jcs.116574. [DOI] [PubMed] [Google Scholar]
- Roth W, Reuter U, Wohlenberg C, et al. Cytokines as genetic modifiers in K5−/−mice and in human epidermolysis bullosa simplex. Hum Mutat. 2009;30:832–41. doi: 10.1002/humu.20981. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–53. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Seltmann K, Fritsch AW, Kas JA, et al. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc Natl Acad Sci U S A. 2013a;110:18507–12. doi: 10.1073/pnas.1310493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K, Roth W, Kroger C, et al. Keratins mediate localization of hemidesmosomes and repress cell motility. J Invest Dermatol. 2013b;133:181–90. doi: 10.1038/jid.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider NT, Omary MB. Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nature reviews Molecular cell biology. 2014;15:163–77. doi: 10.1038/nrm3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Strnad P, Windoffer R, Leube RE. In vivo detection of cytokeratin filament network breakdown in cells treated with the phosphatase inhibitor okadaic acid. Cell Tissue Res. 2001;306:277–93. doi: 10.1007/s004410100455. [DOI] [PubMed] [Google Scholar]
- Strnad P, Windoffer R, Leube RE. Induction of rapid and reversible cytokeratin filament network remodeling by inhibition of tyrosine phosphatases. J Cell Sci. 2002;115:4133–48. doi: 10.1242/jcs.00096. [DOI] [PubMed] [Google Scholar]
- Szeverenyi I, Cassidy AJ, Chung CW, et al. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum Mutat. 2008;29:351–60. doi: 10.1002/humu.20652. [DOI] [PubMed] [Google Scholar]
- Togel M, Meyer H, Lehmacher C, et al. The bHLH transcription factor hand is required for proper wing heart formation in Drosophila. Dev Biol. 2013a;381:446–59. doi: 10.1016/j.ydbio.2013.05.027. [DOI] [PubMed] [Google Scholar]
- Togel M, Pass G, Paululat A. The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev Biol. 2008;318:29–37. doi: 10.1016/j.ydbio.2008.02.043. [DOI] [PubMed] [Google Scholar]
- Togel M, Pass G, Paululat A. In vivo imaging of Drosophila wing heart development during pupal stages. Int J Dev Biol. 2013b;57:13–24. doi: 10.1387/ijdb.120111ap. [DOI] [PubMed] [Google Scholar]
- Traweek ST, Liu J, Battifora H. Keratin gene expression in non-epithelial tissues. Detection with polymerase chain reaction. Am J Pathol. 1993;142:1111–8. [PMC free article] [PubMed] [Google Scholar]
- Vijayaraj P, Kroger C, Reuter U, et al. Keratins regulate protein biosynthesis through localization of GLUT1 and-3 upstream of AMP kinase and Raptor. J Cell Biol. 2009;187:175–84. doi: 10.1083/jcb.200906094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, et al. Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol. 2011;194:669–78. doi: 10.1083/jcb.201008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.