Abstract
Plant genomes encode large numbers of F-box proteins (FBPs), the substrate recognition subunit of SKP1–CULLIN–F-box (SCF) ubiquitin ligases. There are ~700 FBPs in Arabidopsis, most of which are uncharacterized. TIR1 is among the best-studied plant FBPs and functions as a receptor for the plant hormone auxin. Here we use a yeast two-hybrid system to identify novel TIR1 mutants with altered properties. The analysis of these mutants reveals that TIR1 associates with the CULLIN1 (CUL1) subunit of the SCF through the N-terminal H1 helix of the F-box domain. Mutations that untether TIR1 from CUL1 stabilize the FBP and cause auxin resistance and associated growth defects, probably by protecting TIR1 substrates from degradation. Based on these results we propose that TIR1 is subject to autocatalytic degradation when assembled into an SCF. Further, our results suggest a general method for determining the physiological function of uncharacterized FBPs. Finally, we show that a key amino acid variation in the F-box domain of auxin signalling F-box (AFB1), a closely related FBP, reduces its ability to form an SCF, resulting in an increase in AFB1 levels.
The SCF ubiquitin protein ligases (E3s) are composed of CUL1, the RING protein RBX1, SKP1 (Arabidopsis SKP1-like protein (ASK) in plants), and an FBP1,2. Most eukaryotes have a large number of FBPs, ranging from 20 in budding yeast to approximately 700 in Arabidopsis1–3. Although a growing number of Arabidopsis FBPs have been characterized to various extents, the function of the vast majority of these proteins is unknown2. Even for those FBPs with a clear physiological or developmental role, it can be very challenging to identify the relevant substrates.
The abundance of FBPs can be regulated at a number of levels2,4–7. In particular, studies in mammals and yeast have shown that FBPs can be degraded by an autocatalytic mechanism once they are assembled into the SCF, particularly in the absence of substrate8–13. This mechanism has not been demonstrated in plants, but may contribute to remodelling of SCF complexes in response to changing cellular conditions.
In Arabidopsis, SCFTIR1 functions in auxin perception and promotes the degradation of the Aux/IAA (indole-3-acetic acid) transcriptional repressors14. TIR1 consists of the F-box domain at the N-terminus, and a large leucine-rich-repeat domain that forms the auxin-binding pocket15. It is one of a small family of FBPs, called the AFB proteins, all of which function as auxin receptors16,17. Here we describe genetic studies that identify residues in the H1 helix of the F-box domain of TIR1 that are required for binding to CUL1 and assembly into an SCF complex. When not assembled into an SCF, TIR1 is relatively stable and alters auxin responses. Our results suggest that expression of mutant forms of uncharacterized FBPs may provide information on their physiological function.
Results
Isolation of novel TIR1 mutants in yeast
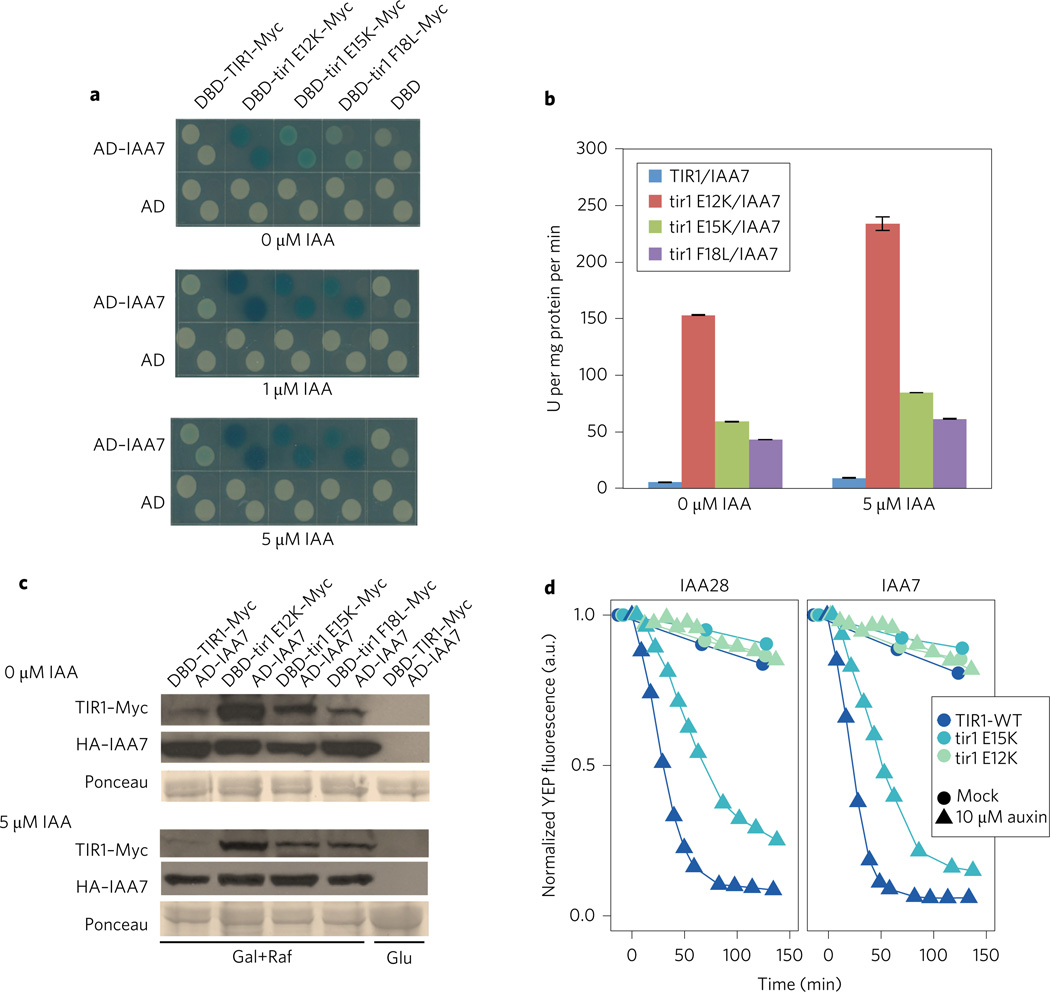
In previous studies we have demonstrated that the yeast two-hybrid system can be used to screen for TIR1 mutants that exhibit an altered interaction with their substrates, the Aux/IAA proteins18–20. We introduced the library of mutagenized TIR1 cDNAs fused to the LexA DNA binding domain described in these studies, into yeast cells expressing the AD–IAA7 fusion protein and grown on selective medium containing 1 µM IAA. The fastest growing colonies were isolated and tested on X-gal plates. A total of eight mutants were recovered. Three TIR1 mutants dramatically increased the level of β-galactosidase activity in the absence and presence of auxin compared with control yeast colonies (Fig. 1a,b). The mutations result in the amino acid substitutions E12K, E15K and F18L, all in the first helix (H1) of the F-box domain of TIR1 (Supplementary Fig. 1a). To determine if this effect is specific for IAA7, we also examined the interaction between the mutant TIR1 proteins and six additional Aux/IAA proteins. Except for IAA31, the combination of each Aux/IAA protein with the three TIR1 mutant proteins resulted in increased β-galactosidase activity compared with wild-type TIR1 (Supplementary Fig. 1b). The IAA31 protein has a diverged DII domain and interacts weakly with TIR1 (ref. 18). We also asked if analogous mutations in other members of the TIR1/AFB family have the same effect. The results in Supplementary Fig. 1c show that afb2E7K, afb2E10K and afb2F13L all produced a stronger two-hybrid response than wild-type AFB2, indicating that the residues in question have a conserved function in the family. (Note: herein lowercase denotes the mutated forms of genes and proteins.)
Figure 1. Isolation of TIR1 mutants in yeast.
a, The TIR1 proteins E12K, E15K and F18L dramatically increase X-Gal staining when in combination with IAA7. b, Activity of β-galactosidase in extracts from yeast strains in a. Error bars are s.e.m. c, Levels of TIR1 and IAA7 proteins in yeast. TIR–Myc levels in extracts prepared from yeast strains in a determined by protein blot. Protein samples (20 µg) were loaded in each lane. d, Auxin-induced degradation of YFP–IAA fusion proteins in yeast in the presence of wild-type and mutant TIR1 proteins. Fluorescence levels were captured using time-lapse flow cytometry. Degradation curves were normalized to the starting fluorescence. a.u., absorbance units; U, units.
To determine if the mutant proteins are expressed at a similar level as the wild type, we performed a protein blot using anti-Myc antibody. Surprisingly, the mutant proteins accumulate to a higher level in yeast cells than in the wild-type control, strongly suggesting that increased β-galactosidase activity is related to higher levels of protein rather than increased affinity between TIR1 and the Aux/IAA (Fig. 1c).
The tir1E12K and E15K proteins are deficient in Aux/IAA degradation in yeast
Since the E12K and E15K mutations had the most dramatic effect on protein level in yeast, we concentrated our further studies on these two proteins. To determine if the E12K and E15K proteins are functional, we tested their activity in Saccharomyces cerevisiae20,21. Previous studies have shown that YFP–Aux/IAA fusion proteins are rapidly degraded by SCFTIR1 upon auxin treatment in yeast20–22. We expressed the E12K and E15K proteins in yeast and measured the YFP–IAA7 or YFP–IAA28 protein level by flow cytometry (Fig. 1d). The Aux/IAA degradation rate k5 was calculated as previously described (Supplementary Table 1)21. Faster degrading Aux/IAAs have larger k5 values. Our results show that neither the E12K nor the E15K proteins are as effective as the wild type in degrading the Aux/IAAs. The E12K protein has very low activity whereas E15K protein exhibits substantial, but clearly reduced, activity.
The tir1E12K and E15K substitutions stabilize TIR1 in Arabidopsis plants
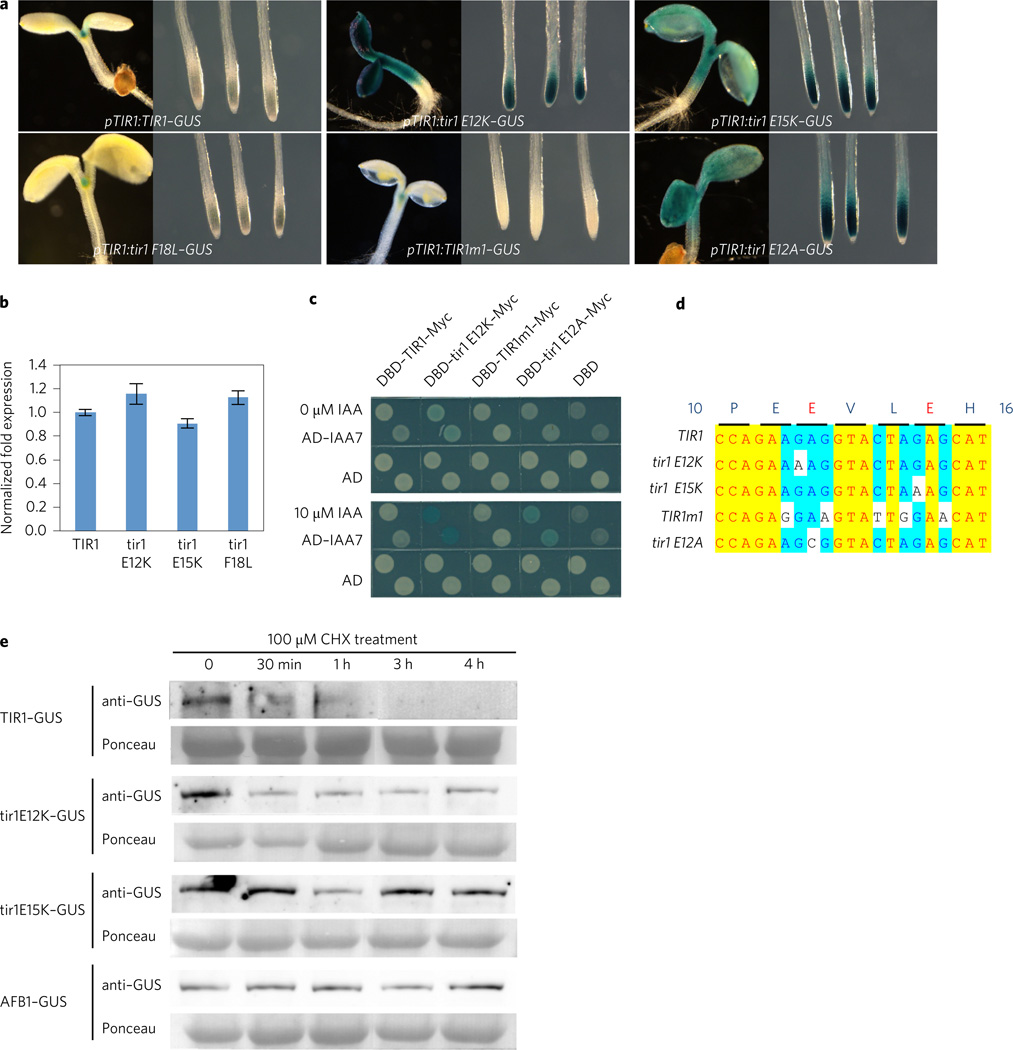
To determine the effects of the TIR1 mutations in Arabidopsis plants, we generated TIR1–GUS (β-glucuronidase) constructs regulated by the TIR1 promoter, and introduced these into the tir1-1 recessive mutant. Between eight and ten lines were recovered and analysed, and a representative line selected for further characterization. The pTIR1:TIR1–GUS construct was shown previously to complement the phenotype of the tir1-1 mutant23. Representative lines are shown in Fig. 2a. GUS staining revealed that the tir1E12K–GUS and tir1E15K–GUS proteins accumulate to much higher levels than TIR1–GUS (Fig. 2a). To assess the level of transgene transcript, we performed real-time quantitative polymerase chain reaction (qPCR) using primers for the GUS portion of the transcript. The results show that each transcript is present at a similar level, indicating that the effect of the mutations is post-transcriptional (Fig. 2b).
Figure 2. The tir1E12K and E15K proteins are stable in plants.
a, β-Glucuronidase (GUS) histochemical staining of transgenic lines expressing wild-type and mutant TIR1. b, GUS transcript levels in 7-day-old seedlings determined by qPCR. Data shown are from three biological replicates. Error bars are s.e. c, The TIR1m1 protein behaves like TIR1 in a Y2H assay. d, Summary of base substitutions in the TIR1 mutants. Yellow indicates 100% identity, turquoise indicates 80% identity. e, tir1E12K–GUS, tir1E15K–GUS, and AFB1–GUS are more stable than TIR1 in 7-day-old seedlings treated with cycloheximide (CHX). Total proteins were extracted from seedlings after treatment with 100 µM CHX for times indicated.
To establish that the effects of the E12K and E15K mutations are related to the respective amino acid substitutions rather than changes in nucleic acid sequence, we introduced five synonymous base substitutions into the pTIR1:TIR1–GUS construct within the region encoding TIR1 E12 to E15 (Fig. 2c,d). The mutant gene (pTIR1:TIR1m1–GUS) was introduced into the tir1-1 mutant and transgenic lines were stained for GUS. The results show that changes in the RNA sequence within this region of the transcript do not affect TIR1 levels. To determine if an increased TIR1 protein level is specifically associated with the substituted lysine residue, we transformed the pTIR1:tir1E12A–GUS construct into tir1-1 plants (Fig. 2d). The results show that the E to A substitution also increases the TIR1–GUS level compared with the wild type (Fig. 2). Similar results can be observed in the yeast two-hybrid system (Fig. 2c). The tir1E12A protein dramatically increases β-galactosidase activity with IAA7 whereas TIR1m1 exhibits similar β-galactosidase activity to wild-type TIR1 on auxin medium.
Our results suggest that E12K and E15K affect stability of the TIR1 protein. To confirm this we performed a cycloheximide (CHX) experiment. Total proteins were extracted from pTIR1: TIR1–GUS, pTIR1:tir1E12K–GUS, and pTIR1:tir1E15K–GUS seedlings after treatment with CHX for 4h and TIR1–GUS levels were assessed by protein blot. The results indicate that TIR1 is an unstable protein and that both mutant proteins are relatively stable (Fig. 2e).
A number of fungal and animal FBPs have been shown to be unstable due to auto-ubiquitination when assembled into an active SCF complex9–13. Thus one possible explanation for the increased stability of the tir1E12K and tir1E15K proteins is that they fail to assemble into an active SCF. A previous study demonstrated that TIR1 is degraded by the proteasome24. To confirm this and to determine if the mutant proteins are also degraded by the proteasome, we treated the pTIR1:TIR1–GUS, pTIR1:tirE12K–GUS, and pTIR1:tirE15K–GUS lines with the proteasome inhibitors MG132 and bortezomib. The results in Supplementary Fig. 2 show that both inhibitors stabilize TIR1–GUS. In contrast, the inhibitors have little effect on tir1E12K levels. The effect on tir1E15K levels is intermediate.
Because the mutations affect residues within the F-box domain of TIR1, we first tested their ability to interact with ASK1 using the yeast two-hybrid system. The results show that both mutant proteins interact strongly with ASK1 (Supplementary Fig. 3). X-gal staining is stronger for the mutants, presumably because they accumulate to higher levels than the wild-type protein.
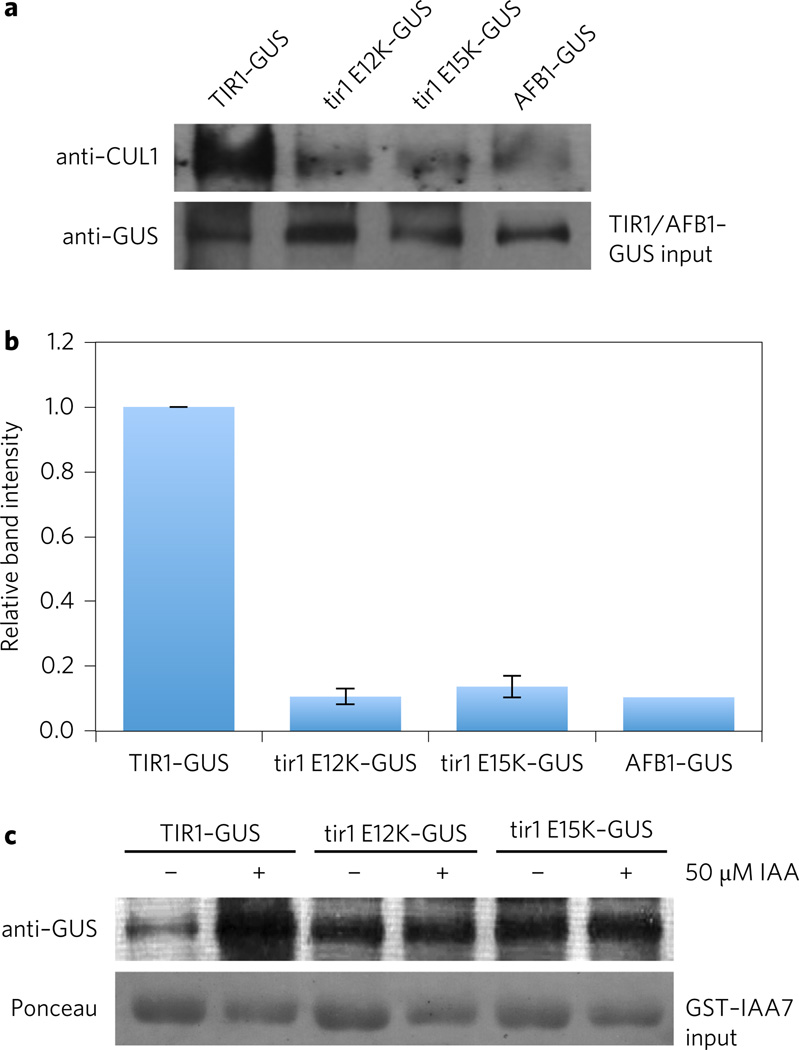
Structural studies of human SCFSKP2 indicate that residues in the first helix (H1) of the F-box domain of SKP2 interact with residues in CUL1 (ref. 25). In particular, conserved residues P113, D114 and E115 contact CUL1. Since TIR1E12 is homologous to SKP2E115, it is possible that the E12K mutation affects interaction with CUL1. To test this possibility, GUS fusions were immunoprecipitated from extracts prepared from pTIR1:TIR1–GUS, pTIR1:tir1E12K, and pTIR1:tir1E15K plants using anti-GUS antibody and the immunoprecipitates were analysed for the presence of CUL1 after SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Because the tir1E12K and tir1E15K proteins are so much more abundant than the wild-type protein, we elected to dilute the mutant extracts so that they contained an equivalent amount of GUS fusion protein as the wild-type extract. The amount of fusion protein in each extract was first assessed by anti-GUS western blot. The results in Fig. 3a,b demonstrate a dramatic reduction in the level of CUL1 bound to tir1E12K or tir1E15K. Based on these data we propose that the E12K and E15K mutations disrupt the interaction between the TIR1–ASK1 complex and CUL1. Thus as in human SKP2, the H1 region of TIR1 contributes to CUL1 binding. It is likely that the relative stability of the mutant proteins is due to reduced assembly into an SCF and as a consequence, decreased auto-ubiquitination and degradation. Consistent with this, we found that MG132 and bortezomib have little effect on the levels of the mutant proteins (Supplementary Fig. 2). Alternatively, it is possible that TIR1 is a substrate for a different E3 ligase.
Figure 3. The tir1E12K and E15K proteins are deficient in SCF assembly.
a, Co-immunoprecipitation (IP) assays performed with wild-type and mutant GUS fusion proteins. CUL1 was detected with an anti-CUL1 antibody. The tir1E12K, tir1E15K and AFB1 extracts were diluted relative to the wild-type protein so that approximately equal amounts of fusion protein was present during the IP. b, Quantification of band intensities shown in a using a chemiluminescence imager. Values are expressed relative to the wild type. Error bars are s.d. c, In vivo pull-down of GST–IAA7 from extracts prepared from lines expressing wild-type and mutant GUS fusion protein. As in a approximately equal amount of GUS fusion protein were present in each pull-down reaction.
To further assess the function of the mutant TIR1 proteins in plants, we tested their ability to interact with the Aux/IAA protein IAA7 in a GST pull-down assay. Again equivalent amounts of GUS fusion protein were present in each assay. As expected, recovery of wild-type TIR1 in the pull-down increased with the addition of IAA (Fig. 3c). In contrast, tir1E12K and tir1E15K interacted better than wild type in the absence of IAA, but recovery of the FBP was not enhanced by the addition of IAA.
Expression of stable TIR1 proteins has a dramatic effect on auxin response and plant growth
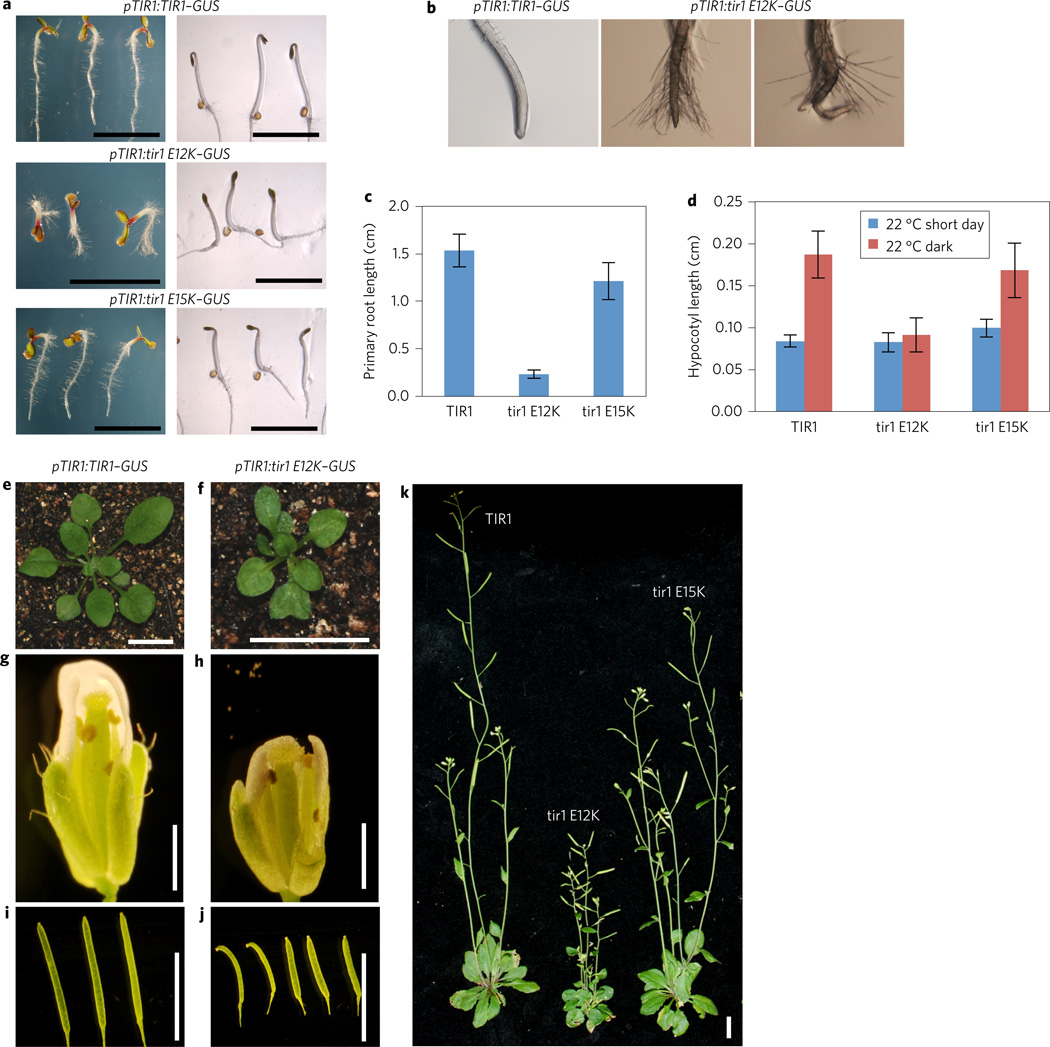
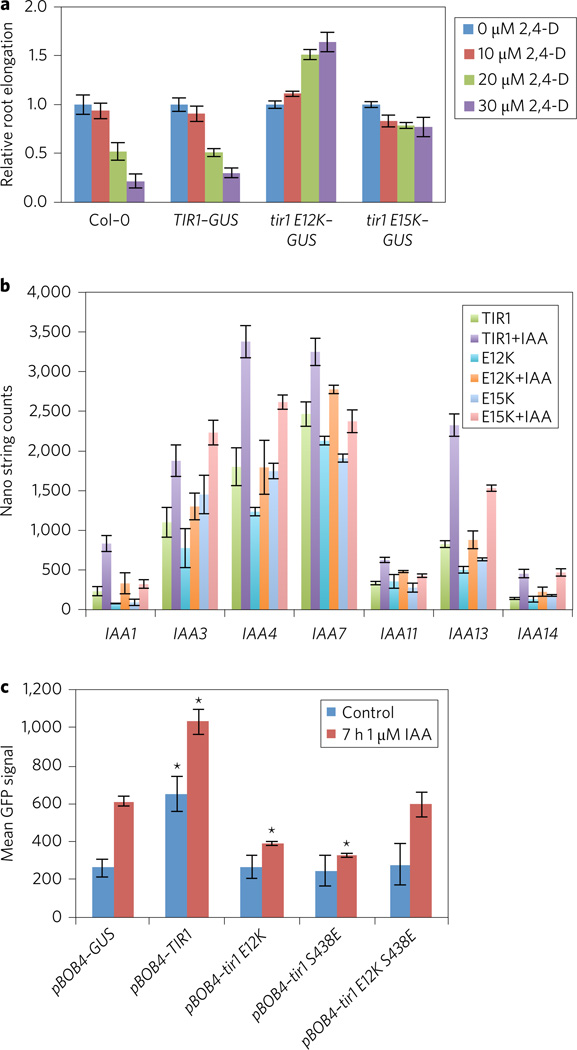
The pTIR1:tir1E12K–GUS and pTIR1: tir1E15K–GUS lines exhibited numerous defects in development compared with either wild-type, pTIR1:TIR1–GUS or tir1 plants (Fig. 4)16,26. In general, the pTIR1:tir1E12K–GUS lines were the most severely affected, consistent with higher levels of TIR1 accumulation in yeast and plants. Five-day-old pTIR1:tir1E12K–GUS seedlings grown in constant light display severe defects in development, including much shorter primary roots, pronounced root hair elongation and defects in both root and shoot gravitropism (Fig. 5a–c and Supplementary Fig. 4).
Figure 4. The pTIR1:TIR1–GUS transgenic lines display severe defects throughout plant growth.
a, Five-day-old transgenic plants expressing wild-type and mutant TIR1-GUS protein grown in constant light (left) and dark (right). b, Close-up of root tips of seedlings shown in a. c, Root length of 5-day-old seedlings. d, Hypocotyl length of 5-day-old light- and dark-grown seedlings. For c and d, error bars represent s.e.m. e–j, Rosette (e,f), flower (g,h), and fruit (i,j) growth are all reduced in the pTIR1:tir1E12K-GUS transgenic line compared to pTIR1:TIR1–GUS. k, Four-week-old pTIR1:TIR1-GUS (left), pTIR1:tir1E12K-GUS (centre) and pTIR1:tir1E15K-GUS (right) plants. Scale bar, 1 cm except in g,h (1 mm).
Figure 5. Effects of TIR1 mutations on auxin response.
a, Effect of auxin on root growth of Col-0 and transgenic seedlings expressing TIR1–GUS, tir1E12K–GUS and tir1E15K–GUS. Error bars are s.e. b, Abundance of Aux/IAA transcripts in transgenic plants expressing TIR1 and TIR1 mutants with or without 1 µM IAA for 2 h. Transcript levels were determined with the Nanostring nCounter Gene Expression assay. Error bars are s.d. c, DR5-GFP expression was measured in protoplasts transiently transformed with mCherry-tagged TIR1 proteins or an mCherry–GUS control. Protoplasts were treated for 7 h with 1 µM IAA prior to cytometric quantification of the mean GFP fluorescence intensity (arbitrary units) in mCherry-positive events. The average of three independent treatments is presented in a histogram ± s.d., n = 3. Asterisks indicate a significant difference (t-test P < 0.01) with the GUS control under the same conditions.
In the dark, expression of the mutant proteins appears to enhance the de-etiolation response. Compared with the control line, dark-grown pTIR1:tir1E12K–GUS seedlings have shorter hypocotyls and do not form the typical apical hook (Fig. 4a–d). pTIR1: tir1E15K–GUS seedlings have an intermediate phenotype. Later in development, the leaves of pTIR1:tir1E12K–GUS plants are smaller, rounder and often display hyponastic growth. After flowering, pTIR1:tir1E12K–GUS plants exhibit highly branched dwarf inflorescences with smaller flowers and fruit (Fig. 4e–k).
Although the pTIR1:tir1E12K–GUS and pTIR1:tir1E15K–GUS plants accumulate high levels of TIR1, in many respects they resemble auxin-resistant mutants such as axr1, tir/afb and various gain-of-function aux/iaa mutants16,27–31. The one exception to this is the increased root hair elongation, which has not been observed in auxin-resistant mutants. To learn more about the effects of the mutant proteins on auxin response, we examined root growth on increasing concentrations of the synthetic auxin 2,4-D. As expected, root growth in both Col-0 and pTIR1:TIR1–GUS seedlings was strongly inhibited by 2,4-D. In contrast, both lines expressing the mutant proteins were highly resistant to the auxin (Fig. 5a). In fact, 2,4-D actually promoted root elongation of the pTIR1: tir1E12K–GUS line compared with the untreated control.
To examine auxin response in these mutants at the level of transcription, we used NanoString technology to assess the transcript level of auxin-responsive Aux/IAA genes. The results show that both pTIR1:tir1E12K–GUS and pTIR1:tir1E15K–GUS lines are deficient in auxin-induction of these genes (Fig. 5b). For most genes, transcript levels are also reduced without auxin treatment in the mutants. Aux/IAA genes that are not auxin regulated are not affected by expression of E12K or E15K (Supplementary Fig. 5).
Our results show that tir1E12K and E15K do not assemble normally into an SCF complex, but nevertheless bind the Aux/IAA proteins in the absence and presence of auxin. Thus we hypothesize that the mutant proteins act to reduce auxin response by binding to and stabilizing the Aux/IAAs. To test this hypothesis, we introduced a second mutation in the auxin-binding pocket of TIR1 and tir1E12K (S438E)15. This mutation has been shown to disrupt the in vivo interaction between TIR1 and Aux/IAAs18. If tir1E12K affects auxin response by directly interacting with the Aux/IAAs, introduction of S438E should reduce this effect. The double- and single-mutant proteins, along with the wild-type TIR1 and a GUS control, were fused to a red fluorescent protein (mCherry) and transiently expressed in root protoplasts carrying the auxin-responsive DR5:GFP (green fluorescent protein) reporter (Fig. 5c). Compared to the mCherry–GUS control, protoplasts expressing TIR1–mCherry showed significantly increased GFP levels both with and without auxin treatment (Fig. 5c). In accordance with the auxin resistance observed in pTIR1:tir1E12K–GUS plants, tir1E12K–mCherry-expressing protoplasts exhibited significantly reduced activation of DR5:GFP after auxin treatment. Surprisingly, expression of tir1S438E–mCherry also reduced DR5 activity, perhaps because accumulation of the mutant protein depletes ASK levels. However, the combination of E12K and S438E partially restored activity, indicating that Aux/IAA binding is necessary for the negative effect of tir1E12K on DR5 activity (Fig. 5c). Importantly, TIR1–mCherry and tir1E12K–mCherry were both localized to the nucleus (Supplementary Fig. 6).
Accumulation of the AFB1 protein is associated with a defect in SCF assembly
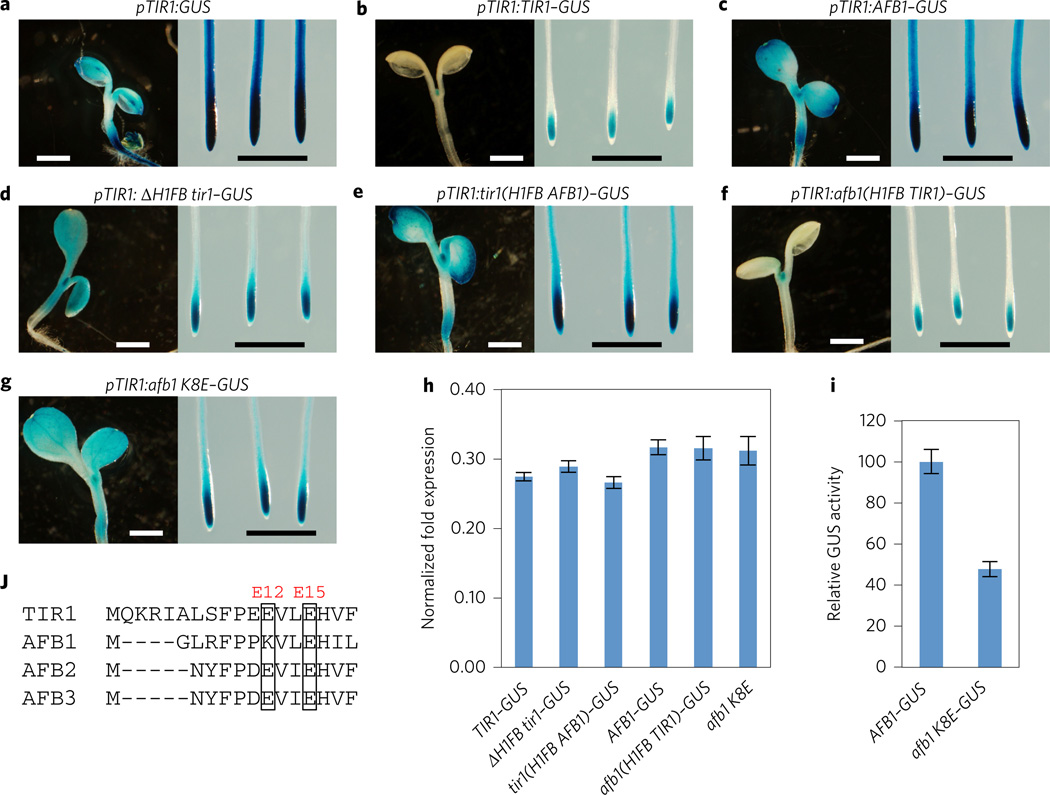
In a previous study we showed that the AFB1 protein accumulates to a higher level than the TIR1 protein and that this difference is conferred by the coding region of these proteins23. In Fig. 2e we confirm that AFB1 is much more stable than TIR1. Examination of the protein sequences revealed that the E12 position in TIR1 is represented by K8 in AFB1 (Fig. 6j). Since the E to K substitution in TIR1 is responsible for stabilizing TIR1, it is possible that accumulation of AFB1 may be related to a natural E to K variation at this position. To address this possibility, we constructed a series of deletion and domain swap TIR1/AFB1 genes and introduced them into the tir1-1 mutant. Deletion of helix 1 of the TIR1 F-box domain (residues 2–18aa) in pTIR1: ΔH1FB TIR1–GUS results in a strong increase in GUS staining compared with the wild-type protein, an effect that is not associated with a change in transcript level (Fig. 6a,b,d). When the first helix of the F-box domain from TIR1 is replaced by this region from AFB1 in the pTIR1:tir1(H1FBAFB)–GUS line, the level of GUS staining increased compared with the pTIR1:TIR1–GUS line (Fig. 6b,e). In contrast, GUS staining in the pTIR1:afb1 (H1FB TIR1)–GUS line, strongly decreases GUS expression compared with the pTIR1:AFB1–GUS line (Fig. 6c,f). Real-time qPCR results indicate that these transgenic lines have a similar GUS RNA level (Fig. 6h). These findings, together with other results reported here, suggest that differences in the level of GUS fusion protein are related to protein stability. It is important to note, however, that other types of post-transcriptional regulation may also be involved. Similar results were obtained using the yeast two-hybrid system (Supplementary Fig. 7). β-Galactosidase activity in lines expressing the DNA binding domain (DBD)–tir1 (H1FB AFB1)–Myc and AD–IAA7 was much higher than with DBD–TIR1–Myc, whereas enzyme activity in lines expressing DBD–afb1 (H1FB TIR1)–Myc is less than DBD–AFB1–Myc on auxin medium. Protein blot shows that β-galactosidase levels are related to the abundance of the DBD fusion proteins.
Figure 6. The H1 region of the F-box domain stabilizes AFB1.
a–g, GUS staining of TIR1/AFB transgenic lines. Scale bar, 1 mm. h, The pTIR1:tir1/afb1–GUS transgenic seedlings have similar levels of GUS transcript. Seven-day-old seedlings were analysed by qPCR. Data shown are from three biological replicates. i, GUS enzyme activity in the pTIR1:AFB1–GUS and pTIR1:afb1 K8E–GUS transgenic lines. Error bars in h and i are s.e.m. j, Sequence of the H1 F-box domain among TIR1/AFB1-3.
To confirm that the differences we observe are related to residue TIR1E12/AFB1K8, we mutated AFB1K8 to a glutamic acid and introduced this gene into tir1-1 plants. We found that this line exhibited much lower GUS levels than the pTIR1:AFB1–GUS line consistent with the proposed role of TIR1E12 in SCF assembly (Fig. 6g,i).
If we are correct in our explanation for the high level of AFB1 accumulation, the protein should be deficient in SCF assembly, similar to tir1E12K. A coimmunoprecipitation experiment shows that, as predicted, the level of CUL1 recovered with AFB1–GUS is dramatically reduced compared with TIR1–GUS (Fig. 3a,b). Together these results indicate that AFB1 accumulates to a much higher level than TIR1 because it does not efficiently assemble into an SCF and is therefore not subjected to autocatalytic degradation.
Discussion
During SCF assembly, the SKP1 subunit is generally thought to be responsible for binding of the SKP1–FBP subcomplex to CUL1 (ref. 32). However, the structure of SCFSKP2 shows that residues in the first helix (H1) of the F-box domain of SKP2 also interact with residues in CUL1 (ref. 25). In particular, conserved amino acids P113, D114, and E115 contact CUL1. In this study we show that residues in helix H1 of TIR1 and AFB1 are required for assembly of SCFTIR1. One of these residues, TIR1E12 (or AFB1K8) occupies the same position as SKP2E115. A second TIR1 residue required for CUL1 binding, E15, is conserved among the TIR1/AFB proteins but not in SKP2. In fact, this residue is not broadly conserved among Arabidopsis FBPs.
Autocatalytic degradation of FBPs has been demonstrated in fungal and animal systems7–12. This occurs when, in the absence of substrate, the ubiquitin moiety is transferred from the E2 to an available lysine on nearby proteins such as the FBP. In this study we show that mutant forms of TIR1 that do not assemble into an SCF are stable relative to the wild-type protein. Similarly the AFB1 protein is deficient in SCF assembly and relatively stable. Thus in Arabidopsis, as in yeast and animals, assembly of FBPs into an SCF is associated with degradation, possibly by an autocatalytic mechanism. It is also formally possible that TIR1 is a substrate for a different E3 ligase. However, this E3 would also have to be present in yeast cells, a possibility that seems unlikely. Interestingly, the related FBP COI1 is also degraded by the proteasome, but in this case, assembly into an SCF complex stabilizes COI1 (ref. 33). The reasons for the difference between TIR1 and COI1 in this regard are unknown, but may be related to substrate levels or the highly dynamic nature of auxin signalling. In another interesting example, auxin was shown to promote proteasome-dependent degradation of the FBP SKP2A (ref. 34). The mechanism in this case is unknown.
Expression of the tir1E12K and E15K proteins results in a reduction in auxin response and dramatic changes in plant growth and development, indicating that accumulation of mutant protein represses auxin signalling, possibly by protecting the Aux/IAAs from degradation. This is consistent with the observation that a mutation in the auxin-binding pocket of tir1E12K acts to partially suppress the negative effect in protoplasts. These results suggest a method to study the large number of uncharacterized FBPs in Arabidopsis and other species. Notably, the TIR1E12 position is occupied by a glutamate or aspartate in a large number of FBPs in plants and animals suggesting that it may be possible to easily generate an interfering form of uncharacterized FBPs (Supplementary Fig. 8). It is also possible that the mutant FBPs could be used to trap substrates for analysis by mass spectrometry.
By analogy with tir1E12K, one might expect AFB1 to have a negative effect on auxin response. However, the available genetic evidence indicates that AFB1 has a positive, but relatively minor effect on auxin response in the seedling16. Although the reasons for this are not clear, AFB1 genes, encoding a protein with K at the AFB1K8 position, are found in the Brassicaceae but not in other plant lineages. Since this family diverged from the Capparaceae ~70 million years ago, it is likely that AFB1 and orthologues retain a unique function (Supplementary Fig. 9). Future detailed genetic studies in Arabidopsis or related species may shed light on this question.
Methods
Plant materials and conditions
All Arabidopsis thaliana mutants used in this study were generated in the Columbia-0 (Col-0) ecotype. Seeds were surface sterilized for 20 min in 30% commercial bleach, plated on ATS medium (Arabidopsis thaliana solution) supplemented with 0.8% agar, and stratified for 2–4 days at 4 °C. ATS medium consists of 1% sucrose, 5 mM KNO3, 2.5 mM KPO4, 2 mM MgSO4, 2 mM Ca(NO3)2, 50 µM Fe-EDTA and 1 mL/L micronutrients. All seedling experiments were performed under long-day conditions (16 h light and 8 h dark) in a growth chamber (80 µmol m−2 s−1, 22 °C) unless otherwise stated. Plants in soil were grown in long-day conditions at 22 °C.
RNA extraction and real-time RT-PCR
Total RNA was extracted from 7-day-old seedlings using the RNeasy plant mini kit (Qiagen, no. 74904) and the RNA yield was quantified using the Thermo Scientific NanoDrop 2,000. RNA (1 µg), pre-treated with DNase, was used for cDNA synthesis through the Superscript III First-Strand Synthesis kit (Invitrogen, no. 18080-0444). Quantitative RT-PCR was performed as described17.
Protein extraction and western blotting
Total yeast protein was prepared by Y-PER Reagent (Thermo, no. 78990) and the yield was quantified by Bradford assay. A 20 µg protein sample was eluted in SDS-PAGE sample buffer, heated for 8 min at 90 °C, cooled on ice for 2 min and fractionated by SDS–PAGE. The membrane was stained with Ponceau (0.1% (w/v) Ponceau S in 5% (v/v) acetic acid) for loading control. Target proteins were detected by immunoblotting with anti-c-Myc-peroxidase antibody (Roche no. 11814150001) and anti-HA-peroxidase antibody (Roche no. 12013819001), and visualized using the ECL Plus Western Blotting Detection System (Amersham no. RPN2232).
For pull-down assays from plant extracts, tissue was harvested by grinding to a powder in liquid nitrogen and vortexing vigorously in extraction buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% NP-40, complete protease inhibitor (Roche no. 11836170001), 20 µM MG-132). Cellular debris was removed by centrifugation and total protein concentration was quantified by the Bradford assay. The complex was pulled down with 20 µl of anti-c-Myc agarose beads (Clontech no. 631208) and washed with extraction buffer five times.
For immunoprecipitation experiments, protein samples were extracted as described above. The level of GUS-fused protein was estimated by anti-GUS western blotting to allow equivalent amounts of GUS-fused proteins. The protein extracts were incubated with 4 µg of anti-GUS antibody (Invitrogen no. A5790) crosslinked to Protein G Dynabeads (Invitrogen no. 10004D), for 4 h at 4 °C. The protein–antibody–bead complexes were washed with protein extraction buffer and then eluted by 2× SDS sample buffer. Protein samples were applied to SDS-PAGE. CUL1 was detected using an anti-CUL1 antibody (a gift from William Gray, University of Minnesota).
Transient transformation and cytometry of root protoplasts
Protoplasts derived from the roots of 1-week-old DR5:GFP seedlings were generated, polyethylene glycol-transformed and cytometrically analysed as previously described35. Transient expression vectors with an N- or C-terminal mCherry tag were constructed by ligation of an mCherry PCR product (pmCherry-C1 template (Clontech) up- or downstream, respectively, of the Gateway cassette in p2GW7 (ref. 36) using the SpeI and SacII restriction sites, respectively, to generate p2ChGW7 and p2GWCh7. Primers are described in Supplementary Table 2. These vectors will be made available through the VIB/Ghent website (http://gateway.psb.ugent.be). Protoplast solutions were treated with 1 µM IAA (Sigma no. 15148) for 7 h prior to cytometric analysis of the GFP expression levels in mCherry-positive cells using an LSRII cytometer (BD Biosciences). The mean GFP intensity of three independent treatments was measured.
NanoString analysis
Seven-day-old seedlings were treated with or without 1 µM IAA for 2 h at 22 °C. Five seedlings from each treatment were collected for RNA isolation and three biological replicates were performed. RNA samples (1 µg) were sent to Nanostring (NanoString Technologies, Inc.) for analysis.
Aux/IAA degradation assays in yeast
Yeast degradation assays were carried out as in ref. 21. Briefly, yeast strains coexpressing stably integrated TIR1 mutants and YFP-tagged IAA proteins were diluted in Synthetic Complete (SC) media such that cultures were in log phase 16 h later and for the duration of the experiment. All cultures were grown at 30 °C with shaking. Pre-auxin measurements were taken to determine baseline YFP–IAA expression, followed by addition of auxin (10 µM IAA) or mock treatment (95% (v/v) ethanol). Fluorescence measurements were captured over the course of 150 min following auxin treatment, with intervals ranging from 10 min early in the YFP–IAA degradation phase to 15 min later in the degradation phase. Controls were measured every hour for the duration of the experiment. Quantitative analysis of IAA degradation profiles and calculation of Aux/IAA degradation rate (k5) was conducted as in20,21.
Supplementary Material
Acknowledgements
We gratefully acknowledge Eric Klavins for use of the cytometer and Shelly Jang for assistance with calculation of k5 values. This work was supported by the National Institutes of Health (grant no. GM43644, M.E.), the Howard Hughes Medical Institute (M.E.), the Gordon and Betty Moore Foundation (M.E.), and the Paul G. Allen Family Foundation (J.N.).
Footnotes
Author contributions
H.Y., Y.Z., B.M., B.O.R.B., R.W., J.N. and M.E. conceived and designed experiments. H.Y., Y.Z., B.M., B.O.R.B. and R.W. performed the experiments. M.P. did the phylogenetic analysis. H.Y., Y.Z., B.M., M.P. and M.E. wrote the manuscript.
Additional information
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 3.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl Acad. Sci. USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olmedo G, et al. ETHYLENE-INSENSITIVE5 encodes a 5′–>3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl Acad. Sci. USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potuschak T, et al. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 7.Si-Ammour A, et al. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011;157:683–691. doi: 10.1104/pp.111.180083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl Acad. Sci. USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luke-Glaser S, et al. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol. Cell. Biol. 2007;27:4526–4540. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol. Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirbelauer C, et al. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014;21C:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 16.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Greenham K, et al. The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr. Biol. 2011;21:520–525. doi: 10.1016/j.cub.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Calderon Villalobos LI, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nature Chem. Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prigge MJ, Lavy M, Ashton NW, Estelle M, et al. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 2010;20:1907–1912. doi: 10.1016/j.cub.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, et al. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 23.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl Acad. Sci. USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuttmann J, Parker JE, Noel LD. Novel aspects of COP9 signalosome functions revealed through analysis of hypomorphic csn mutants. Plant Signal. Behav. 2009;4:896–898. doi: 10.4161/psb.4.9.9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 26.Ruegger M, et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1108. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- 29.Wilson AK, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- 30.Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 32.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nature Rev. Mol. Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, et al. The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell. 2013;25:486–498. doi: 10.1105/tpc.112.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurado S, et al. The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell. 2010;22:3891–3904. doi: 10.1105/tpc.110.078972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bargmann BO, Birnbaum KD. Positive fluorescent selection permits precise, rapid, and in-depth overexpression analysis in plant protoplasts. Plant Physiology. 2009;149:1231–1239. doi: 10.1104/pp.108.133975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.