Abstract
Microtubule plus‐end dynamics are regulated by a family of proteins called plus‐end tracking proteins (+TIPs). We recently demonstrated that the transforming acidic coiled‐coil (TACC) domain family member, TACC3, can function as a +TIP to regulate microtubule dynamics in Xenopus laevis embryonic cells. Although it has been previously reported that TACC3 is the only TACC family member that exists in Xenopus, our examination of its genome determined that Xenopus, like all other vertebrates, contains three TACC family members. Here, we investigate the localization and function of Xenopus TACC1, the founding member of the TACC family. We demonstrate that it can act as a +TIP to regulate microtubule dynamics, and that the conserved C‐terminal TACC domain is required for its localization to plus‐ends. We also show that, in Xenopus embryonic mesenchymal cells, TACC1 and TACC3 are each required for maintaining normal microtubule growth speed but exhibit some functional redundancy in the regulation of microtubule growth lifetime. Given the conservation of TACC1 in Xenopus and other vertebrates, we propose that Xenopus laevis is a useful system to investigate unexplored cell biological functions of TACC1 and other TACC family members in the regulation of microtubule dynamics. © 2015 The Authors. Cytoskeleton, Published by Wiley Periodicals, Inc.
Keywords: transforming acid coiled coil domain, microtubule dynamics, Xenopus, live imaging, plusTipTracker
Abbreviations used
- EB1
end‐binding protein 1
- GFP
green fluorescent protein
- KD
knockdown
- MO
antisense morpholino oligonucleotide
- MT
microtubule
- OE
overexpression
- TACC
transforming acidic coiled coil domain
- +TIP
plus‐end tracking protein
Introduction
Microtubule (MT) plus‐end dynamics are regulated by a diverse family of proteins called plus‐end tracking proteins (+TIPs) [Akhmanova and Steinmetz, 2008]. Previously, we reported that the centrosome‐associated protein TACC3, a member of the transforming acidic coiled‐coil (TACC) domain family, can act as a +TIP in multiple Xenopus embryonic cell types [Nwagbara et al., 2014]. This +TIP function of TACC3 was recently confirmed to be conserved in human cells as well [Gutierrez‐Caballero et al., 2015]. However, while manipulation of TACC3 resulted in abnormal MT dynamics in Xenopus cells [Nwagbara et al., 2014], it did not significantly alter MT dynamics in human RPE1 cells [Gutierrez‐Caballero et al., 2015]. One possible explanation for this difference is that it has been reported that Xenopus only contains a single member, TACC3/Maskin [Ha et al., 2013; Peset and Vernos, 2008], while humans possess three members [Gergely et al., 2000]. Thus, there might be TACC functional redundancy in human cells that is not present within organisms possessing only a single family member. Yet, it has not been demonstrated that TACC family members, TACC1 and TACC2, can function as +TIPs in vertebrates. Moreover, it is unclear whether Xenopus truly contains only a single TACC family member, as this notion was postulated prior to the sequencing of the Xenopus laevis and tropicalis genomes [Karpinka et al., 2015]. Given that all other examined vertebrates contain three members of the TACC family, we sought to discern whether Xenopus laevis also possesses the remaining TACC orthologs. Indeed, we have determined that both TACC1 and TACC2 are present in Xenopus genomic sequence (GenBank numbers NW_004668234.1 for TACC1 and NW_004668240.1 for TACC2 in Xenopus tropicalis). Thus, it appears that Xenopus, like other vertebrates, possess all three members of the TACC family. Here, we focus on further investigation of Xenopus laevis TACC1, which we cloned from a cDNA library of stage 22 Xenopus laevis embryos.
TACC1, the founding member of the TACC family, was first cloned from human breast carcinoma tissue as a potential gene that promotes tumorigenicity [Still et al., 1999]. In mammals, TACC1 appears to be more widely‐expressed than TACC3, with TACC1 showing almost ubiquitous expression in adult murine and human tissues [Sadek et al., 2003; Still et al., 1999]. However, TACC1 is also spatiotemporally regulated during murine development [Lauffart et al., 2006; Still et al., 1999]. Further characterization of the human TACC family proteins by immunostaining demonstrated that all three family members interact with MTs and specifically concentrate at centrosomes and mitotic spindles [Gergely et al., 2000]. TACC3 has been the most heavily studied of the TACC family members, but a number of reports in the last decade have also begun to unravel some aspects of TACC1 functionality. For example, TACC1 is similar to TACC3 in that it interacts with the C‐terminus of the MT polymerase, ch‐TOG [Booth et al., 2011; Lauffart et al., 2002]. This begs the question of whether TACC1 localizes to the MT plus‐end, as do both TACC3 and ch‐TOG/XMAP215.
In this study, we began to explore the function of Xenopus laevis TACC1. We sought to determine whether TACC1 could function as a +TIP, and whether it can have an effect on modulating MT plus‐end dynamics. Here, we demonstrate that TACC1 is expressed throughout Xenopus laevis embryonic development. Moreover, we discover that TACC1 can track MT plus‐ends in vertebrate embryonic cells, and we show that the conserved C‐terminal TACC domain is both necessary and sufficient for MT plus‐end localization. We also demonstrate that TACC1 can positively modulate MT dynamics by increasing average MT growth track velocity, as well as average MT growth track length, in a similar manner to that of TACC3. While knockdown of either TACC1 or TACC3 results in the same microtubule growth velocity defect, with no effect on growth lifetime, we find that combined knockdown of both +TIPs reveals a reduction in microtubule growth lifetime. Thus, we have identified new functions for TACC1 in the regulation of MT dynamics, in concert with TACC3.
Results and Discussion
Xenopus Laevis TACC1 is Orthologous to Other TACC1 Proteins
In order to examine and characterize Xenopus laevis TACC1, we first cloned the TACC1 cDNA from stage 22 embryos, after designing primers based on the annotated Xenopus laevis genome (see “Methods” for details on primer sequence design). Examination of the cloned Xenopus laevis TACC1 sequence in comparison to human and mouse shows that there is moderate conservation between the species (50% identity of Xenopus laevis TACC1 to the long TACC1 isoforms of both mammals) (Fig. S1A in Supporting Information). We noted that the C‐terminal domain was particularly well‐conserved, with 80% identity among the three species. This domain, called the TACC domain (which contains two coiled‐coil domains), is known to be well‐conserved throughout the TACC family [Gergely et al., 2000; Peset and Vernos, 2008]. We also observed that while Xenopus laevis TACC1 and TACC3 are only 35% identical, the TACC domains are 53% identical and 71% similar between these family members (Fig. S1B in Supporting Information). We further confirmed that this Xenopus gene is the correct TACC1 ortholog, based on reciprocal BLAST hits with TACC1 from other species, as well as conservation of gene synteny comparing Xenopus laevis and human (not shown). Given the conservation between TACC1 in Xenopus and other vertebrates, we propose that Xenopus laevis is a useful system to investigate the unexplored cell biological functions of TACC1.
TACC1 is Expressed in Multiple Tissue Types During Embryonic Development
In the murine system, comparison of TACC1 and TACC3 expression levels have shown that TACC1 is regulated in a dynamic manner during embryogenesis, with ubiquitous expression of at least one TACC1 isoform [Lauffart et al., 2002]. Moreover, TACC3 is highly upregulated in specific tissues during development, while TACC1 levels are more widespread [Sadek et al., 2003]. We sought to examine whether Xenopus TACC1 was spatiotemporally regulated during embryonic development, in comparison to TACC3. RT‐PCR of Xenopus laevis cDNA libraries from various developmental time points and tissues showed that TACC1 was expressed at all examined embryonic stages, in neural, epidermal, and mesoendodermal tissues (Fig. S1C in Supporting Information). We observed that TACC1 and TACC3 have the highest expression in early stage embryos, whereas levels decline throughout development. Note that the particularly high expression of TACC3 may reflect the prominent mitotic role for this protein [Gergely et al., 2000; Kinoshita et al., 2005; Yao et al., 2007], but its constant presence in later stages would indicate an additional continued role throughout embryonic development. TACC1 appears to be expressed similarly to TACC3, although its expression is not as strong in the early embryo. Both TACC1 and TACC3 are expressed in neural, epidermal, and mesoendodermal tissues in the stage 22 neurula embryo (Fig. S1C in Supporting Information). Thus, TACC1 may function in multiple tissue types throughout development.
TACC1 can act as a Plus‐End Tracking Protein in Embryonic Cells
We previously demonstrated that the C‐terminal domain of TACC3 is required for MT plus‐end tracking in Xenopus [Nwagbara et al., 2014]. Since this domain is well‐conserved between TACC1 and TACC3 (Fig. S1B in Supporting Information), we questioned whether TACC1 might also behave as a +TIP. Thus, we examined the subcellular dynamics of GFP‐tagged TACC1 in cultured embryonic Xenopus cells. GFP‐TACC1 localized to the growing plus‐ends of MTs in mesenchymal cells derived from the neural tube (Figs. 1A–1E, Movie S1 in Supporting Information). This localization was in addition to its expected presence at the centrosome [Fig. S2 and Movie S2 in Supporting Information; Gergely et al., 2000]. Co‐localization analysis of GFP‐TACC1 with mKate2‐tagged end‐binding protein 1 (EB1), which binds all growing MT plus‐ends [Stepanova et al., 2003], showed that TACC1 partially overlaps with, but is distal to, EB1 (Figs. 1F–1J and Movie S3 in Supporting Information). Co‐localization analysis of GFP‐TACC1 with mKate2‐TACC3 showed that they mostly overlap; however, at many time points, mKate2‐TACC3 appears to be just distal to GFP‐TACC1 localization along the plus‐end (Figs. 1K–1O and Movie S4 in Supporting Information). Note that while the channels were imaged sequentially, we corrected for the acquisition difference (for Figs. 1N and 1O) by translating the green channel based on measured plus‐end velocity (see Fig. 1 legend for details). However, even when we imaged the red channel prior to the green channel, GFP‐TACC1 signal was still proximal to the peak of mKate2‐TACC3 (not shown). This further supports the finding that GFP‐TACC1 overlaps with but is slightly proximal to mKate2‐TACC3 on the MT plus‐end (Fig. 1O). Note that the peak of GFP‐TACC1 is approximately 0.4 microns distal to mKate2‐EB1 (Fig. 1J) and approximately 0.1 μm proximal to mKate2‐TACC3 (Fig. 1O). While these are imprecise measurements based on experimental limitations, it is notable that they are consistent with the previous measurement of 0.5 μm between mKate2‐EB1 and GFP‐TACC3 [Nwagbara et al., 2014]. Additionally, we observed that the GFP‐TACC1 plus‐end accumulations were considerably lengthened upon overexpression of mKate2‐TACC3 (Fig. 1O, compare green peak in 1J and 1O), compared to the punctate‐like accumulations when expressing GFP‐TACC1 alone (compare green localization in Figs. 1I and 1N). Thus, this data confirm that TACC1, like TACC3, can act as a +TIP during embryonic development, that TACC1 appears to be slightly proximal to TACC3 on the MT plus‐end, and that there may be an interaction between TACC1 and TACC3 that impacts TACC1 localization to the MT plus‐end.
Figure 1.
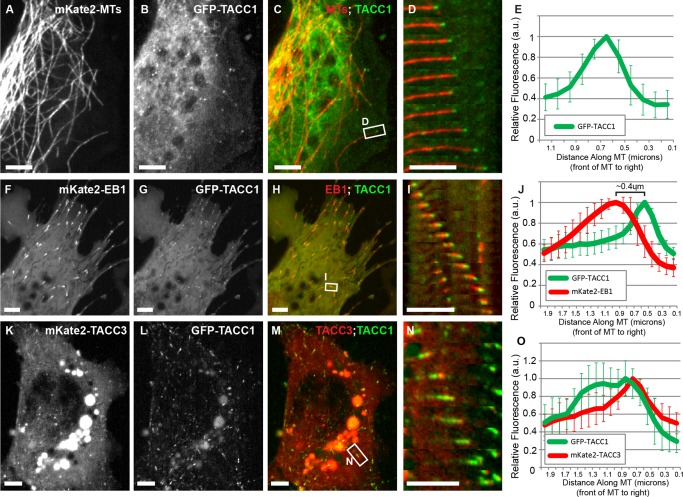
TACC1 can act as a plus‐end tracking protein in embryonic cells. Expression of mKate2‐tubulin (A), GFP‐TACC1 (B), and merge (C) in cultured mesenchymal cells derived from embryonic neural tube; see also Movies S1 in Supporting Information. (D) Magnified time‐lapse montages of the boxed region in panel C. The time interval between frames is 2.9 s. (E) Fluorescence intensity profile of GFP‐TACC1. Signals from 20 individual MTs were quantified by intensity line scans to present the relative fluorescence intensity profiles, with the plus‐end of the MT toward the right. Expression of mKate2‐EB1 (F), GFP‐TACC1 (G), and merge (H) in cultured embryonic mesenchymal cells; see also Movie S3 in Supporting Information. (I) Magnified time‐lapse montage of the boxed region in panel H. The time interval between frames is 2.7 s. For this panel, the green channel has been translated to the left by 0.27 µm to account for the different acquisition times of the red and green channels (the velocity of the plus‐end was consistently moving at 0.27 µm/s in this time series, and the green channel was imaged 1 s after the red channel). This allows for a more accurate visualization of colocalization between the two +TIPs. This demonstrates that, even after correcting for the time displacement, GFP‐TACC1 overlaps with and is distal to mKate2‐EB1. (J) Fluorescence intensity profiles of GFP‐TACC1 and mKate2‐EB1. Signals from 18 MTs were quantified by intensity line scans to present the relative fluorescence intensity profiles, with the plus‐end of the MT toward the right. For each measurement, the green channel has been translated to the left (approximately 0.2 µm) based on each individual MTs velocity, to account for the time displacement between channels. After the correction, the highest peak intensity of GFP‐TACC1 is ∼0.4 μm distal to the peak of mKate2‐EB1. Expression of mKate2‐TACC3 (K), GFP‐TACC1 (L), and merge (M) in cultured embryonic mesenchymal cells; see also Movie S4 in Supporting Information. (N) Magnified time‐lapse montage of the boxed region in panel M. The time interval between frames is 3.9 s. For this panel, the green channel has been translated to the left by 0.25 μm to account for the different acquisition times of the red and green channels (the velocity of the plus‐end was consistently moving at 0.21 µm/s in this time series, and the green channel was imaged 1.2 s after the red channel). (O) Fluorescence intensity profiles of GFP‐TACC1 and mKate2‐TACC3. Signals from 12 MTs were quantified by intensity line scans to present the relative fluorescence intensity profiles, with the plus‐end of the MT toward the right. For each measurement, the green channel has been translated to the left (approximately 0.1 µm) based on each individual MTs velocity, to account for the time displacement between channels. While mKate2‐TACC3 and GFP‐TACC1 mostly overlap, the peak of mKate2‐TACC3 is just slightly distal to GFP‐TACC1. Also, we found that GFP‐TACC1 comets were consistently longer when mKate2‐TACC3 was expressed. Note that mKate2‐TACC3 is less effective at tracking MT plus‐ends than GFP‐TACC3 [observed in Nwagbara et al., 2014]. One reason for this is that the presence of the mKate2 tag leads to more efficient cleavage of the C‐terminal TACC domain (not shown), which is required for MT plus‐end tracking. Moreover, note that in images K–M, yolk granules are strongly autofluorescing. Bar 5 µM.
The C‐Terminal TACC Domain is Necessary and Sufficient for MT Plus‐End Tracking
To further examine the mechanism by which TACC1 tracks the MT plus‐end, we constructed two structural domain mutants of TACC1 (Figs. 2A and 2B). An N‐terminal construct, which lacks the highly‐conserved C‐terminal TACC domain, was unable to track MT plus‐ends and instead remains cytoplasmic (Figs. 2C–2G and Movie S5 in Supporting Information). However, a C‐terminal construct that contains the entire TACC domain was able to efficiently track MT plus‐ends (Figs. 2H–2L and Movie S6 in Supporting Information). Thus, the C‐terminal TACC domain appears to be both necessary and sufficient for promoting MT plus‐end tracking of TACC1. As this domain is 80% identical between Xenopus and mammals (Fig. S1A in Supporting Information), we predict that the plus‐end tracking ability of TACC1 is likely conserved throughout vertebrates. Additionally, we note that while the C‐terminal domain allows for plus‐end tracking, we also observed some MT lattice‐binding with this construct (Fig. 2K, arrowhead), and fluorescence intensity profiles of the tagged C‐terminal domain did not show the sharp peak distal to mKate2‐EB1 which occurred with full‐length GFP‐TACC1 (compare Figs. 2L to 1J). This suggests that the N‐terminal domain might function to restrict plus‐end tracking to the more distal MT tip. Interestingly, when we performed a Western blot to confirm protein expression of the TACC1 constructs, we noticed that the full‐length GFP‐TACC1 showed a weak band at around 68 kD, in addition to a stronger band at around 96 kDa (Fig. 2B), which is the expected approximate size for full‐length GFP‐TACC1. The weaker band size was indistinguishable from that of GFP‐NtermTACC1, suggesting in a small subset of protein, the C‐terminal TACC domain was cleaved off. It has been observed in mouse that the C‐terminal TACC domain could be cleaved from the rest of TACC3 using a thrombin cleavage site [Thakur et al., 2014]. This particular cleavage site is not present in Xenopus laevis TACC3 or TACC1, and it was noted by Thakur et al. 2014 authors that it is unclear whether the thrombin cleavage site in mouse has any physiological relevance. However, given that we observed a protein fragment that appears to result from the cleavage of the TACC domain in Xenopus TACC1 (Fig. 2B), we predict that cleavage of the TACC domain may represent a mode of regulation of TACC family function. This possibility will need to be more thoroughly addressed in the future.
Figure 2.
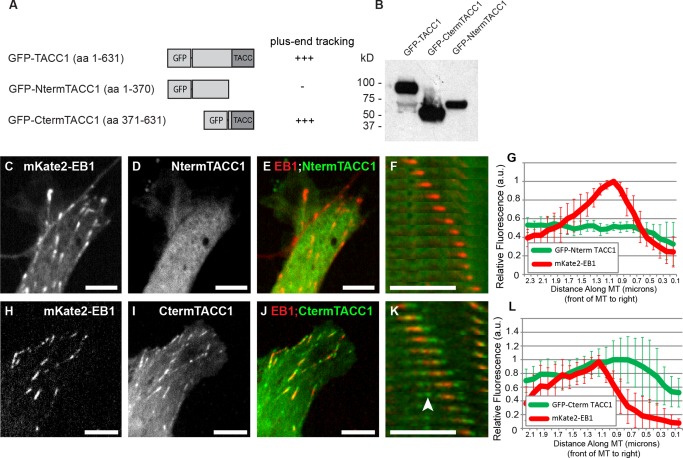
The C‐terminal TACC domain is necessary and sufficient for MT plus‐end tracking. (A) Schematic representation of GFP‐tagged TACC1 protein and deletion constructs. The amino acid residue numbers refer to those listed in Fig S1A in Supporting Information. (B) Western blot of embryonic lysates following expression of constructs, blotted for GFP. This blot is intentionally overexposed in order to reveal the smaller band in the full‐length construct lane. Expression of mKate2‐EB1 (to identify MT plus‐ends; C, H), GFP‐tagged TACC1 constructs (D, I), and merged images of both channels (E, J). Plus‐end accumulation is apparent in I (with GFP‐CtermTACC1) but not in D (with GFP‐NtermTACC1); see also Movies S5 and S6 in Supporting Information. (F, K) Magnified time‐lapse montages of a representative EB1 comet from cell in (E, J) confirms that CtermTACC1 localizes to growing MT plus‐ends, while NtermTACC1 does not. Arrowhead in (K) points to MT lattice binding. The time interval between frames in panel F is 3.3 s and panel K is 2.1 s. (G, L) Fluorescence intensity profiles of GFP‐TACC1 constructs and mKate2‐EB1. For each plot, signals from 10 MTs were quantified by intensity line scans to present the relative fluorescence intensity profiles, with the plus‐end of the MT toward the right. For K, the green channel has been translated to the left (approximately 0.1 µm) based on each individual MTs velocity, to account for the time displacement between channels. GFP‐Cterm TACC1 comets were consistently longer than full‐length GFP‐TACC1 (compare Figs. 2L to 1J). Bar 5 µM.
TACC1 can Regulate MT Plus‐End Dynamics
Given the localization of TACC1 at the plus‐ends of MTs, we next sought to examine whether TACC1 plays a role to regulate MT plus‐end dynamics. To test this, we overexpressed TACC1 levels and then acquired high‐resolution live images of fluorescently‐tagged EB1, which is used as a marker for MT polymerization. We then quantified parameters of MT polymerization dynamics using the Matlab‐based open source software, plusTipTracker [Applegate et al., 2011]. This software program has been validated for accurate tracking of fluorescently tagged EB1 comets in Xenopus laevis embryonic fibroblasts and other cell types, using similar imaging conditions to those used in the present study [Nwagbara et al., 2014; Stout et al., 2014]. mKate2‐EB1 comet velocity was on average 24% faster upon TACC1 overexpression (OE) (Fig. 3A). Average MT growth‐track lifetime (which measures the number of seconds of MT polymerization in a given growth track before pausing or undergoing catastrophe) was not significantly different (Fig. 3B). Average MT growth‐track length (which measures the distance of persistent MT polymerization before pausing or undergoing catastrophe) was 25% longer (Fig. 3C). These results were similar to previous MT dynamics effects by TACC3 OE [Nwagbara et al, 2014] (and also shown in Figs. 3A–3C), suggesting that TACC1 may modulate MT plus‐end dynamics through a mechanism related to that of TACC3.
Figure 3.
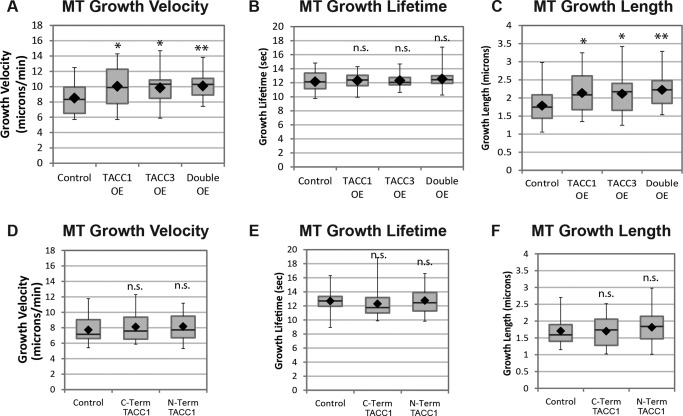
TACC1 overexpression (OE) increases MT growth velocities. Quantification of MT growth track parameters after TACC1 and TACC3 overexpression in cultured embryonic mesenchymal cells derived from neural tube. TACC proteins are expressed approximately twice as much as in controls. mKate2‐EB1 localizes to the ends of growing MTs and is thus a marker for MT polymerization. Automated tracking of mKate2‐EB1 comets is used to calculate MT growth track velocity (A), MT growth track lifetime (B), and MT growth track length (C). Mean values for all the experiments combined (three were performed in total): MT growth velocity, control, 8.5 µm/min, TACC1 OE 10.1 µm/min, TACC3 OE 9.84 µm/min, double OE 10.1 µm/min; MT growth lifetime, control, 12.1 s, TACC1 OE 12.3 s, TACC3 OE 12.3 s, double OE 12.5 s; MT growth length, control, 1.8 µm, TACC1 OE 2.1 µm, TACC3 OE 2.1 µm, double OE 2.2 µm. Control data are the means of 26 cells, representing a total of 4292 analyzed MT growth tracks; TACC1 OE are from 35 cells and 7606 analyzed MT growth tracks; TACC3 OE are from 38 cells and 9138 MT growth analyzed tracks; double OE are from 30 cells and 7214 MT growth analyzed tracks. (D–F) Quantification of MT parameters after overexpression of TACC1 deletion constructs. Mean values: MT growth velocity, control, 7.7 µm/min, C‐term TACC1 8.1 µm/min, N‐term TACC1 8.2 µm/min; MT growth lifetime, control, 12.7 s, C‐term TACC1 12.3 s, N‐term TACC1 12.8 s; MT growth length, control, 1.7 µm, C‐term TACC1 1.7 µm, N‐term TACC1 1.8 µm. Control data are the means of 40 cells (from 5 individual experiments), representing a total of 9425 analyzed MT growth tracks; C‐term TACC1 data are from 33 cells and 6960 analyzed MT growth tracks; N‐term TACC1 data are from 46 cells and 9701 analyzed MT growth tracks. Box‐and‐whisker plots indicate the mean (diamond), median, extrema, and quartiles. An unpaired t test was performed to assess significance of overexpression conditions compared to control. **P < 0.01, *P < 0.05; n.s., not significant.
As there is similarity between TACC1 and TACC3 with respect to MT dynamics modulation, we next tested whether TACC1 and TACC3 could display any functional synergy in driving MT polymerization by performing combinatorial manipulations of protein levels. When we overexpressed both TACC1 and TACC3 at the same time, the parameters of MT dynamics were not significantly different than in each of the single overexpression conditions (Figs. 3A–3C). Thus, TACC1 and TACC3 do not show a synergistic interaction and instead appear to be functioning within the same mechanistic pathway in promoting MT dynamics. As previously noted, we did observe that when both GFP‐TACC1 and mKate2‐TACC3 were expressed, there was qualitatively more GFP‐TACC1 accumulating on MT plus‐ends compared to when GFP‐TACC1 was expressed alone (compare Fig. 1N, where GFP‐TACC1 appears as a comet, with Fig. 1D, where it appears as a puncta). Thus, TACC3 may affect the localization of TACC1, but expressing both TACC members simultaneously cannot increase MT polymerization rates any more than expressing only one TACC member.
We also examined whether overexpression of either the N‐terminal or C‐terminal TACC1 constructs could have any effect on MT dynamics. However, we found that overexpression of each of these constructs did not result in any changes in MT dynamics compared to control conditions (Figs. 3D–3F). This suggests that while the C‐terminal domain is sufficient for plus‐end localization, its effect in modulating MT dynamics requires at least part of the N‐terminal domain.
Knockdown Analysis Reveals an Effect of TACC1 Function on MT Growth Velocity and Lifetime
We next determined whether knockdown of TACC1 might have an effect on MT dynamics. An antisense morpholino oligonucleotide (MO) was designed against TACC1 which prevented correct splicing of the first intron and led to over 90% knockdown of normal mRNA levels (Fig. 4A). Compared to controls, MT growth velocity was reduced by 10% upon TACC1 KD (Fig. 4B). MT growth track lifetime was not affected by TACC1 KD (Fig. 4C), while MT growth track length was reduced 11% (Fig. 4D). Thus, inhibiting TACC1 levels led to a small, but significant effect on MT growth velocity and track length. These results are almost identical to what we previously observed after TACC3 KD [Nwagbara et al., 2014].
Figure 4.
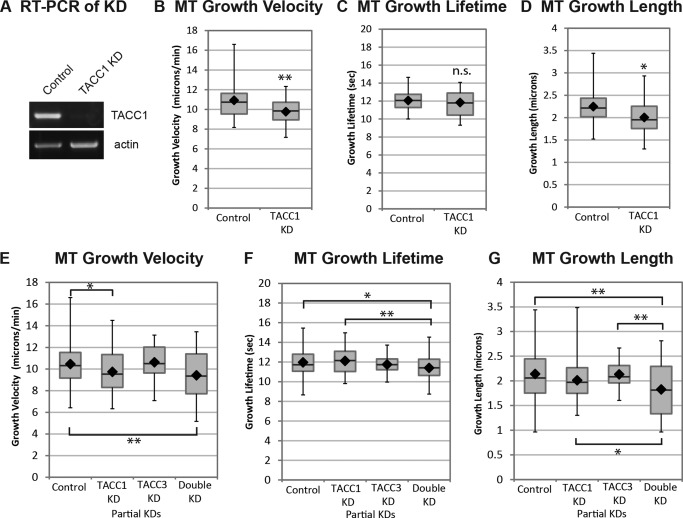
TACC1 KD reduces MT growth velocities, while double KD of both TACC1 and TACC3 also reduces MT growth lifetime. Quantification of MT growth track parameters after TACC1 and TACC3 knockdown in cultured embryonic mesenchymal cells derived from neural tube. (A) RT‐PCR demonstrates knockdown of normal TACC1. Automated tracking of mKate2‐EB1 comets after TACC1 knockdown is used to calculate MT growth track velocity (B), MT growth track lifetime (C), and MT growth track length (D). Mean values: MT growth velocity, control, 10.9 µm/min, TACC1 KD 9.8 µm/min; MT growth lifetime, control, 12.1 s, TACC1 KD 11.8 s; MT growth length, control, 2.2 µm, TACC1 KD 2.0 µm. Control data are the means of 52 cells, representing a total of 12,947 analyzed MT growth tracks; TACC1 KD are from 24 cells and 9435 analyzed MT growth tracks. (E–G) Quantification of MT parameters after double TACC1/TACC3 knockdowns (TACC1 mRNA levels were reduced approximately 25% while TACC3 was reduced approximately 40%, not shown). Mean values: MT growth velocity, control, 10.5 µm/min, TACC1 partial KD 9.7 µm/min, TACC3 partial KD 10.6 µm/min, TACC1/TACC3 double partial KD 9.4 µm/min; MT growth lifetime, control, 12.0 s, TACC1 partial KD 12.1 s, TACC3 partial KD 11.8 s, TACC1/TACC3 double partial KD 11.4 s; MT growth length, control, 2.1 µm, TACC1 partial KD 2.0 µm, TACC3 partial KD 2.1 µm, TACC1/TACC3 double partial KD 1.8 µm. Control data are the means of 92 cells (from 5 individual experiments), representing a total of 25,266 analyzed MT growth tracks; TACC1 partial KD are from 80 cells and 22,637 analyzed MT growth tracks; TACC3 partial KD are from 31 cells and 8,143 analyzed MT growth tracks; TACC1/TACC3 double partial KD are from 44 cells and 21,531 analyzed MT growth tracks. Box‐and‐whisker plots indicate the mean (diamond), median, extrema, and quartiles. To determine statistical significance for panels E–G, an ANOVA test was performed first (which revealed that the means were not the same for any condition), followed by an unpaired t‐test to assess significance between conditions. **P < 0.01, *P < 0.05; n.s., not significant.
Figure 5.
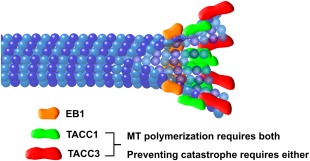
Cartoon schematic of TACC localization at the MT plus‐end. Our data suggest a model in which TACC3 (red) is at the distal‐most end, with TACC1 (green) overlapping with and slightly proximal to TACC3. EB1 (orange) lies further proximal to TACC1. Both TACC1 and TACC3 are required for maintaining normal rates of MT polymerization, while normal levels of either of TACC1 or TACC3 appears to be sufficient for preventing MT catastrophe. Note that it is still unclear whether TACC3 and TACC1 can bind to the plus‐end independently of other factors, or whether other +TIPs are required (such as XMAP215, which interacts directly with TACC3). Not drawn to scale.
Given the conservation between TACC1 and TACC3 (particularly within their highly conserved C‐terminal domains responsible for plus‐end tracking), as well as their similar knockdown phenotypes, we wondered if TACC1 and TACC3 might display any functional synergy. Namely, if we partially knockdown each of them individually or together, do any phenotypes increase in severity? When we used a low dose of MO to only partially knock down TACC1 and TACC3 (TACC1 mRNA levels were reduced approximately 25% while TACC3 was reduced approximately 40%, not shown), we observed no effect on MT growth velocity in the TACC3 KD but we still saw a statistically‐significant 8% reduction in MT growth velocity with TACC1 KD (Fig. 4E). This effect was not significantly enhanced in the double KD (Fig. 4E), suggesting that partially inhibiting TACC3 does not worsen the phenotype. However, with the double KD, we uncovered a MT growth lifetime defect which was not present in either partial KD alone (Fig. 4F), nor was it present in the stronger TACC1 KD (Fig. 4C) or in a stronger TACC3 KD [Nwagbara et al., 2014]. As MT growth lifetime is a reflection of catastrophe rate (the longer the lifetime, the longer the time between catastrophes), this suggests that TACC family members do play a role in regulating MT catastrophe, but that either TACC1 or TACC3 alone can function in this capacity. When both proteins are compromised in their function, then catastrophe rates increase. Consistent with the MT growth velocity and lifetime results, MT growth length was also significantly reduced (by 14%) with the double KD, compared to partial KD of each TACC member alone (Fig. 4G). To summarize, wild‐type levels of TACC1 and TACC3 are each required for maintaining normal microtubule growth speed, but they exhibit some functional redundancy in the regulation of microtubule growth lifetime. An effect on MT growth lifetime (i.e., catastrophe rate) is only uncovered when both TACC family members are partially inhibited.
In conclusion, we provide the first definitive evidence that Xenopus laevis possesses more than a single TACC family member, and that TACC1 can function as a +TIP to regulate MT dynamics. This work further illustrates the utility of using Xenopus embryonic cell culture as a system for studying the cell biological functions of the TACC family members, as well as other known and suspected +TIPs. Future studies will show whether this plus‐end regulatory function of TACC1 is conserved throughout vertebrates, whether Xenopus TACC1 and TACC3 are differentially regulated to affect MT dynamics, and whether the final member of the TACC family, TACC2, can also regulate MT plus‐end dynamics.
Materials and Methods
Embryos
Eggs obtained from female Xenopus laevis frogs (NASCO, Fort Atkinson, WI) were fertilized in vitro, dejellied, and cultured at 13–24°C in 0.1× Marc's modified Ringer's (MMR) using standard methods [Sive et al., 2010]. Embryos were staged according to Nieuwkoop and Faber [Nieuwkoop and Faber, 1994]. All experiments were approved by the Boston College Institutional Animal Care and Use Committee (IACUC) and were performed according to national regulatory standards.
Culture of Xenopus Embryonic Explants
Embryos were cultured in 0.1× MMR at 22°C to stages 22–24, and embryonic explants were dissected and cultured on poly‐l‐lysine (100 µg/mL) and laminin‐coated (20 µg/mL) coverslips, as described previously [Lowery et al., 2012; Nwagbara et al., 2014]. Cells were imaged at room temperature 18–24 h after plating.
Constructs and RNA
Capped mRNA was transcribed in vitro using SP6 or T7 mMessage mMachine Kit (Life Technologies, Grand Island, NY). RNA was purified with LiCl precipitation and re‐suspended in nuclease free water. Constructs used: GFP‐TACC1, GFP‐TACC1‐Cterm, GFP‐TACC1‐Nterm, GFP‐TACC3 [TACC3 pET30a was gift from Richter lab (University of Massachusetts Medical, Worcester, MA)], mKate2‐TACC3 (all TACC constructs subcloned into pCS2+ vector), mKate2‐tubulin [Shcherbo et al., 2009] in pT7TS, EB1‐GFP in pCS107 [gift from Danilchik lab (Oregon Health Sciences University, Portland, OR)], mKate2‐EB1 in pCS2+. The dorsal blastomeres of embryos were injected four times at the two‐to‐four cell stage (in 0.1× MMR containing 5% Ficoll) with total mRNA amount per embryo: 1000–2000 pg GFP‐TACC1, 100 to 300 pg EB1‐GFP or mKate2‐EB1, 2000 pg mKate2‐tubulin, 1000–3000 pg mKate2‐TACC3. For overexpression analysis (Fig. 3), 1000 pg of each construct was used. This leads to approximately twice as much protein as in controls.
RT‐PCR Time Course and Cloning of TACC1
Total RNA was extracted from staged wild‐type embryos using Trizol reagent (Life Technologies, Grand Island, NY), followed by chloroform extraction and isopropanol precipitation. For isolation of neural tissues, neural tubes were isolated as previously described [Lowery et al., 2012], and 40 neural tubes were used for isolating RNA. For isolation of epidermal and mesoendodermal tissues, embryos were cut in half as in the neural prep and soaked in collagenase, the neural tube was completely removed, and then the epidermis was isolated by peeling it off with fine forceps. Epidermis from 40 embryos was used for isolating RNA. The remaining tissue, after neural and epidermal removal, was designated as mesodendodermal (40 embryos) and used for isolating RNA. Total RNA was further purified with a phenol chloroform extraction and an ethanol precipitation. cDNA synthesis was performed with Super Script II Reverse Transcriptase (Life Technologies, Grand Island, NY) and random hexamers. PCR was then performed, with the following primers: For cloning TACC1 (forward 5′‐TTTTCTCGAGATGTCGTTCAGCCCGTGG‐3′, reverse 5′‐TTTTCTCGAGTCACTGCGTCCCATCTTTGC‐3′), for testing for expression (tacc1F 5′‐CTCGAGGGCTCATCACTCGAACTGGATG‐3′, tacc1R 5′‐TTTTCTCGAGTCACTGCGTCCCATCTTTGC‐3′), TACC3 expression (tacc3F 5′‐AGCTTCAGAACTCACCAGCA‐3′, tacc3R 5′‐GCAGCACCAGAATCCTGGG‐3′), and actin as a loading control (actin F 5′‐AAGGAGACAGTCTGTGTGCGTCCA‐3′, actin R, 5′‐CAACATGATTTCTGCAAGAGCTCC‐3′). For TACC1 cloning, the above primers were designed based upon the TACC1 annotated sequence from the laevis genome v6.0 gene model (http://www.xenbase.org). Specifically, we used the UTexas Oktoberfest transcript model of Scaffold27535:373746.413683, which was predicted to contain the Xenopus laevis TACC1 genomic sequence. The TACC1 mRNA sequence was submitted to Genbank, accession number KP866215.
Morpholinos
MOs targeted to an early splice site of X. laevis TACC1 (5′‐ACTGCCCGTGTCACTCACCCAGACT‐3′), the translation start site of X. laevis TACC3 (5′‐AGTTGTAGGCTCATTCTAAACAGGA‐3′) [used in Nwagbara et al., 2014], or standard control MO (5′‐cctcttacctcagttacaatttata‐3′; purchased from Gene Tools (Philomath, OR) were injected into two to four‐cell‐stage embryos (28 ng/embryo for full (>90%) TACC1 knockdown, 7 ng/embryo for partial TACC1 knockdown, 20 ng/embryo for partial TACC3 knockdown and control MOs). TACC3 knockdown was assessed by Western blot of embryos at stages 35–36. TACC1 knockdown was assessed by RT‐PCR using primers that were designed to the correct splicing product (tacc1F 5′‐CTCGAGGGCTCATCACTCGAACTGGATG‐3′, tacc1R_Nterm 5′‐GCGCCTCGAGTTCATAATTTTGCACTTTACAGC‐3′).
Western Blotting
Embryos were lysed in buffer (50 mM Tris pH 7.5, 5% glycerol, 0.2% IGEPAL, 1.5 mM MgCl2, 125 mM NaCl, 25 mM NaF, 1 mM Na3V04, 1 mM DTT), supplemented with complete protease inhibitor cocktail with or without EDTA (Roche, Indianapolis, IN). Blotting was carried out using rabbit anti‐GFP [1:2500, ab290, Abcam (Cambridge, MA)]. For blocking membrane, 2% nonfat milk was used. Detection was done by chemiluminescence using Amersham ECL Western blot reagent (GE Healthcare, Pittsburgh, PA).
Confocal Microscopy
Live images were collected with a Yokogawa CSU‐X1M 5000 spinning disk confocal on a Zeiss Axio Observer inverted motorized microscope with a Zeiss 63X Plan Apo 1.4 NA lens. Images were acquired with a Hamamatsu ORCA R2 CCD camera controlled with Zen software (Zeiss, Thornwood, MY). For time‐lapse, images were collected every 2–4 s for 1–3 min, depending on the experiment. Laser power for 488 nm was 30–40%, with exposure time 1000–1500 ms. Laser power of 561 nm was 25–50%, with exposure time 600–1500 ms. For two‐color co‐localizations in Figures 1 and 2, red channel was imaged 600–800 ms before the green channel. Thus, the true extent of potential overlap between plus‐end accumulations could not be examined from these image time‐series without correcting for plus‐end movement. Figures 1A–1H, 1K–1M and 2 show the original acquisitions without correction. However, for the single MT montages (Figs. 1I and 1N), we performed x axis translation on the green channel to correct for the time displacement. For this analysis, the green channel was translated in the x axis, after calculating the frame‐to‐frame velocity of the growing MT plus‐end, in order to account for the time delay between channels, for each examined MT (using ImageJ Translate function). Details of displacement measurements are in the figure legend. To further confirm correct translation, time‐lapse colocalizations were examined with both combinations of imaging—red channel first, green channel second; then green channel first, red channel second. Fluorescently tagged TACC1, TACC3, and EB1 MT plus‐end accumulations were measured with the line tool in ImageJ along their longest axis, which was defined from their trajectory in the previous and subsequent frames. Line intensities were gathered using Plot Profile.
PlusTipTracker Software Analysis
MT dynamics were analyzed from EB1‐GFP or mKate2‐EB1 movies using plusTipTracker [Applegate et al., 2011; Lowery et al., 2013; Stout et al., 2014]. The same parameters were used for all movies: maximum gap length, 8 frames; minimum track length, 3 frames; search radius range, 5–12 pixels; maximum forward angle, 50°, maximum backward angle, 10°; maximum shrinkage factor, 0.8; fluctuation radius, 2.5 pixels; time interval 2 s. Only cells with a minimum number of 10 MT tracks in a one‐minute time‐lapse were included for analysis. The Peltier Tech Box and Whisker Chart Utility for Excel was used to make box plots.
Image Analysis and Statistics
Phenotypic quantification was performed from multiple experiments to ensure reproducibility. Graphs were made in Microsoft Excel, and histograms were generated using the Analysis Tool Pak. To determine statistical differences, unpaired two‐tailed t tests were used for comparing two conditions, while ANOVA tests were used to compare multiple conditions (GraphPad, La Jolla, CA).
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Movie 1.
Supporting Information Movie 2.
Supporting Information Movie 3.
Supporting Information Movie 4.
Supporting Information Movie 5.
Supporting Information Movie 6.
Supporting Information
Acknowledgments
We thank members of the Lowery lab for helpful critiques of the manuscript. We also thank Aleks Ostojic, Leslie Carandang, Claire Stauffer, Paul Paris, Alexandra Mills, and Jackson Bowers for technical assistance with experiments and Nancy McGilloway and Todd Gaines for excellent Xenopus husbandry. L.A.L. is funded by National Institutes of Health R00 MH095768.
Author Contributions
CL, ME and LAL designed the study. CL, ME, BN, PE, CB, JV, AF, SR performed the experiments and analyzed the data. LAL wrote the manuscript, and all authors edited the manuscript.
Monitoring Editor: Bruce Goode
References
- Akhmanova A, Steinmetz, MO . 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9:309–322. [DOI] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. 2011. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol 176:168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Hood FE, Prior IA, Royle SJ. 2011. A tacc3/ch‐TOG/clathrin complex stabilises kinetochore fibres by inter‐microtubule bridging. EMBO J 30:906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. 2000. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA 97:14352–14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez‐Caballero C, Burgess SG, Bayliss R, Royle SJ. 2015. TACC3‐ch‐TOG track the growing tips of microtubules independently of clathrin and Aurora—a phosphorylation. Biol Open 4:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha GH, Kim JL, Breuer EK. 2013. Transforming acidic coiled‐coil proteins (TACCs) in human cancer. Cancer Lett 336:24–33. [DOI] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James‐Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. 2015. Xenbase, the xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res 43:D756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. 2005. Aurora a phosphorylation of tacc3/maskin is required for centrosome‐dependent microtubule assembly in mitosis. J Cell Biol 170:1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffart B, Dimatteo A, Vaughan MM, Cincotta MA, Black JD, Still IH. 2006. Temporal and spatial expression of tacc1 in the mouse and human. Develop Dyn 235:1638–1647. [DOI] [PubMed] [Google Scholar]
- Lauffart B, Howell SJ, Tasch JE, Cowell JK, Still IH. 2002. Interaction of the transforming acidic coiled‐coil 1 (tacc1) protein with ch‐TOG and gas41/NuBI1 suggests multiple TACC1‐containing protein complexes in human cells. Biochem J 363:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Faris AE, Stout A, Van Vactor D. 2012. Neural explant cultures from Xenopus laevis J Vis Exp 68:e4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Stout A, Faris AE, Ding L, Baird MA, Davidson MW, Danuser G, Van Vactor D. 2013. Growth cone‐specific functions of xmap215 in restricting microtubule dynamics and promoting axonal outgrowth. Neural Develop 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. 1994. Normal Table of Xenopus Laevis (Daudin). New York: Garland Publishing Inc. [Google Scholar]
- Nwagbara BU, Faris AE, Bearce EA, Erdogan B, Ebbert PT, Evans MF, Rutherford EL, Enzenbacher TB, Lowery LA. 2014. Tacc3 is a microtubule plus‐end tracking protein that promotes axon elongation and also regulates microtubule plus‐end dynamics in multiple embryonic cell types. Mol Biol Cell 25:3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I. 2008. The TACC proteins: TACC‐ling microtubule dynamics and centrosome function. Trends Cell Biol 18:379–388. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Pelto‐Huikko M, Tujague M, Steffensen KR, Wennerholm M, Gustafsson JA. 2003. Tacc3 expression is tightly regulated during early differentiation. Gene Expr Patterns 3:203–211. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. 2009. Far‐red fluorescent tags for protein imaging in living tissues. Biochem J 418:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. 2010. Microinjection of xenopus embryos. Cold Spring Harbor Protoc 2010:pdb.ip81. [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3‐GFP (end‐binding protein 3‐green fluorescent protein). J Neurosci 23:2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still IH, Hamilton M, Vince P, Wolfman A, Cowell JK. 1999. Cloning of tacc1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene 18:4032–4038. [DOI] [PubMed] [Google Scholar]
- Stout A, D'Amico S, Enzenbacher T, Ebbert P, Lowery LA. 2014. Using plusTipTracker software to measure microtubule dynamics in Xenopus laevis growth cones. J Vis Exp 91:e52138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur HC, Singh M, Nagel‐Steger L, Kremer J, Prumbaum D, Fansa EK, Ezzahoini H, Nouri K, Gremer L, Abts A, Schmitt L, Raunser S, Ahmadian MR, Piekorz RP. 2014. The centrosomal adaptor tacc3 and the microtubule polymerase chTOG interact via defined C‐terminal subdomains in an Aurora‐a kinase‐independent manner. J Biol Chem 289:74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Natsume Y, Noda T. 2007. Tacc3 is required for the proper mitosis of sclerotome mesenchymal cells during formation of the axial skeleton. Cancer Sci 98:555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Movie 1.
Supporting Information Movie 2.
Supporting Information Movie 3.
Supporting Information Movie 4.
Supporting Information Movie 5.
Supporting Information Movie 6.
Supporting Information


