Abstract
Optical coherence tomography (OCT) has great potential for deciphering the role of mechanics in normal and abnormal heart development. OCT images tissue microstructure and blood flow deep into the tissue (1–2mm) at high spatiotemporal resolutions allowing unprecedented images of the developing heart. Here, we review the advancement of OCT technology to image heart development and report some of our recent findings utilizing OCT imaging under environmental control for longitudinal imaging. Precise control of the environment is absolutely required in longitudinal studies that follow the growth of the embryo or studies comparing normal versus perturbed heart development to obtain meaningful in vivo results. These types of studies are essential to tease out the influence of cardiac dynamics on molecular expression and their role in the progression of congenital heart defects.
Index Terms: Biomedical imaging, cardiography, in vivo imaging, optical imaging
I. Introduction
The embryonic cardiovascular system develops at an extraordinary pace. For example, in the avian model, the heart progresses from a straight heart tube to a c-shaped loop and then an s-shaped loop on its way to becoming a septated four-chambered heart all in the span of several days [1]–[3]. Precise control orchestrating this transformation is imperative for normal development. An understanding of the etiology of congenital heart defects (CHDs) that result from abnormal development is largely incomplete at this time. It is the consensus that a complex interplay between genetic and environmental factors is responsible for many of the defects. For example, researchers have recently shown that the prevalence of heart defects in CITED2 knockout mice (C57BL6/J) goes up over twofold after the mothers are fed a high-fat diet [4]. Several groups have presented studies demonstrating that altered hemo-dynamics can lead to heart defects [5]–[10]. It is thought that biomechanical forces lead to altered gene expression that can lead to altered cardiac structure and function that can in turn lead back to altered biomechanical forces. Unfortunately, this feedback loop is poorly understood because of the lack of proper tools to assess forces on the early looping heart and their resultant effects in cardiogenesis.
Optical coherence tomography (OCT) has the potential to be a robust tool to help decipher and understand some of the feedback pathways influenced by the cardiac function in early heart development. The rapid beating of the minute heart organ and the complex ultrafine vasculature has severely limited the options for assessing dynamics. OCT is based upon low-coherence interferometry and was first demonstrated in 1991 [11]. OCT specifications that enable characterization of structure and dynamics of the cardiovascular system during cardiac looping include the following.
High spatial resolution (2–20 µm)—allows visualization and measurement of cardiac layers (myocardium, cardiac jelly, and endocardium) and ultrafine structures (e.g., tethers connecting the endocardium to the myocardium).
High temporal resolution (10 to >100 kHz line rates)—enables assessment of cardiac dynamics in one, two, and three dimensions.
Appropriate imaging depth (1–2 mm)—permits imaging of four-chamber heart development under physiological conditions.
Doppler OCT—allows measurement of hemodynamics (e.g., blood velocity, blood flow, etc.) and biomechanical forces induced by blood flow (e.g., shear stress).
Vascular mapping—enables the visualization and measurement of the vasculature.
Early OCT reports have shown exciting results, but many studies have not used precise environmental control during imaging. In our experience, the heart rate of an embryo can drop drastically within the first minute of being removed from environmental control (data not shown). Maintaining physiological conditions while imaging allows one to perform longitudinal imaging (follow the growth of an embryo) and compare the results to embryos with applied stressors. In order to determine the influence of biomechanics and the time course of its effects on normal and abnormal heart development, it is imperative to investigate heart development longitudinally under physiological conditions.
Here, we demonstrate longitudinal four-dimensional (4-D) imaging of a developing embryonic chick heart during cardiac looping. We use the term longitudinal 4-D imaging to mean 3-D volumetric image sets recorded throughout a short time scale (one heartbeat, 4-D imaging) and also in time lapse over a long time scale (hours or days of development). The 4-D image sets contain 90 volumes/heartbeat and allow us to observe systolic dynamics within the heart tube. Also, we present the first images of the developing circulatory system in the embryonic chick in vivo at these early stages. Distinguishing cardiovascular abnormalities can be crucial in pinpointing the genesis and determining the mechanisms of abnormal growth. Finally, we apply a stressor (hyaluronidase) to the developing embryo and compare morphology and dynamics with an embryo under normal development. These experimental methods will enable studies to determine how cardiac morphology and dynamics play a role in cardiac development and how environmental modifications to the heart affect development. Thus, OCT has the potential to help connect congenital heart diseases with the early events that set the heart on an abnormal course.
First, we review OCT imaging in heart development in Section II. In Sections III and IV, we present our current methods and results for longitudinal imaging of the developing heart. In Section V, we discuss and state our conclusions.
II. OCT Imaging in Developmental Cardiology
In 1996, Boppart et al. applied OCT technology to image developing embryonic morphology for the first time [12]. Subsequently, Boppart et al. concentrated on studying the structure and dynamics of the embryonic Xenopus heart [13]. From these early experiments, it became evident that OCT had enormous potential for investigating embryonic heart development. Not only did OCT have the spatial resolution to distinguish the structures in the diminutive embryo heart, but the technology was also noninvasive and had the temporal resolution to assess cardiac dynamics. Within 15 years since the first paper, embryonic and larval hearts of many animal models have been examined including the chicken [14]–[29], quail [15], [30]–[34], mouse [14], [15], [35]–[42], Xenopus [12], [13], [43]–[46], drosophila [47]–[49], and zebrafish [50]–[52]. The capabilities of OCT imaging has been expanded over the years and has allowed one to image the heart in four dimensions, assess hemodynamics using Doppler OCT, gauge heart mechanics using modeling, perform live imaging by incorporating culturing and environmental chambers along with the OCT system. Also, investigators have begun to use this technology to answer questions about the mechanisms of normal and abnormal development. The following sections give a more detailed account of the developments of OCT technology and its application in the field of embryonic cardiology.
A. Four-Dimensional OCT Imaging
Initially, OCT systems were too slow to capture 2-D cross-sectional images (B-scans) of the beating heart without significant motion artifacts. Yazdanfar et al. used a retrospective cardiac gating scheme to produce 2-D cross-sectional Doppler images of the beating Xenopus heart with a time-domain OCT system capable of imaging 8 lines (A-scans) per second [43]. Because of the periodic nature of the beating heart, one can image over multiple heartbeats and either trigger data acquisition to start at a certain phase in the cardiac cycle (prospective gating) or reorganize the data in postprocessing using a physiological signal or image similarity (retrospective gating) to capture an image set with greatly reduced motion artifact. As OCT developed video-rate capabilities (2–8 kHz line rates) [53], [54], the speed enabled real-time cross-sectional imaging of the beating heart [22], [44], [53]. Unfortunately, the increased speed was not capable of capturing the beating heart in three dimensions. Jenkins et al. employed a prospective gating method to record the first 4-D datasets from both chicken and mouse embryo hearts [14]. The hearts were excised and paced electrically. The electrical signal was also used to trigger data acquisition. Next, groups found ways to the image live embryonic hearts in four dimensions by measuring Doppler signals from the blood. Jenkins et al. employed a prospective gating scheme using a laser Doppler velocimeter signal [31], while Mariampillai utilized a signal-based retrospective gating method, where a second OCT system recorded a Doppler optical cardiogram signal [45].
Around this time, OCT was making a fundamental change from time domain (temporal interference patterns) to frequency-domain (spectral interference patterns) OCT [55]. Frequency domain’s signal-to-noise ratio (SNR) was superior to time domain and enabled the construction of high-speed systems (orders of magnitude faster than time-domain OCT) utilizing either a spectrometer or a swept laser source [56]–[61]. The first examples of these systems demonstrated that one could image the 3-D beating heart in real time (≥10 volumes/s) [30], [46]. Jenkins et al. also demonstrated that the high speed allowed one to accurately visualize the heart movement during systole, but only with cross-sectional images. Therefore, gating would again be needed to accurately assess systolic motion in three dimensions. This time gating was accomplished in postprocessing by looking for correlations in the images. Gargesha et al. adapted an algorithm from confocal imaging [62] to combine gated imaging with ultrahigh speed OCT (>100 kHz line rate) to capture 90 volumes/heart beat (quail embryo) with a temporal accuracy of 5 ms [33]. Other groups have since applied 4-D image-based algorithms to image the murine embryo heart in four dimensions [38] and the outflow tract (OFT) in a stage-18 chick embryo [18]. Recently, imaging speeds increased dramatically again [63], and as data acquisition instrumentation improves, it will be feasible to image at >100 volumes/s. However, new gating algorithms may still be needed for tissue and blood cell tracking.
B. Doppler OCT
Like ultrasound, OCT can use the phase information in the signal to measure a Doppler shift imposed by a moving particle and compute the velocity component in the direction of the illuminating beam of light. Several groups have employed Doppler OCT for imaging embryonic hearts [17], [20], [25]–[27], [34], [36], [39], [43]–[45], [64]. Doppler OCT was applied to the embryonic Xenopus heart in 1997 [43]. At this time point OCT imaging was still extremely slow (8-Hz line rate) and Doppler signals were processed along the individual A-scans using a short-time Fourier transform. Several years later as imaging speeds increased (2–8 kHz line rates), Yang et al. measured phase differences between adjacent A-scans to determine the Doppler shift and demonstrated Doppler imaging at 4–32 images per second in the Xenopus heart [44]. This work clearly showed that Doppler OCT could noninvasively image velocity distribution during heart cycles and established the potential of Doppler OCT in developmental studies.
Like standard structural OCT imaging, Doppler OCT benefited from the switch to Fourier domain OCT. The faster imaging speeds allow one to image faster flows and the static reference arm reduces phase noise in the signal [65]. Davis et al. first applied Doppler OCT in the Fourier domain to examine embryonic chick hearts [26].When the flow of fluids induces a Doppler phase shift greater than 2π between subsequent A-scans, the velocity is not uniquely determined from the phase. To correct for phase wrapping, Davis et al. employed a cellular-automata method for phase unwrapping [66]. The investigators were then able to make measurements of blood velocity in the looping heart [26] and measurements of blood velocity, blood flow, and shear rate in the vitelline vessels [27]. Because OCT is sensitive only to the velocity component parallel with the incident beam, many groups measure the direction of the vessel of interest to determine absolute velocity [26], [27]. Iftimia et al. worked to overcome this limitation by constructing a dual-beam OCT system to image zebrafish [52]. If the vessel of interest lied within a single plane, absolute velocity could be obtained. Determining absolute velocity in the beating heart is difficult because of the constant change in blood flow direction. Jenkins et al. overcame this limitation by utilizing a 4-D Doppler OCT dataset to measure the relative blood flow in the plane perpendicular to the illuminating beam [34]. Using the perpendicular plane, a blood flow measurement only relies on the velocity component acquired by OCT and measurements of stroke volume and cardiac output can be made without obtaining the absolute velocity. Ma et al. used a snake algorithm to determine the centerline to the OFT of the heart tube [20]. By assuming that blood flow is in the direction of the centerline and that the centerline was constant for all phases of the cardiac cycle, absolute velocity was estimated. In the future, simpler and more robust techniques are warranted for calculating absolute velocity.
Blood flow asserts forces upon the endocardial wall of the cardiovascular system and it is assumed that these forces affect gene transcription and heart development. As our data and others indicate that the architecture of even the earliest tubular heart is not a simple tube, the complexity of measuring these forces on different sections of the endocardium is a challenge. Jenkins et al. first employed OCT to measure shear stress in the developing quail embryo heart [34]. Instead of measuring the Doppler signal directly, one can also use modeling to determine the range of these forces [16], [19], [21]. Recently, Liu et al. developed a finite-element model that utilized structural OCT images to determine the location of the walls and pressure measurements at both the inlet and the outlet of the OFT. The model predicted shear stresses consistent with past OCT [34] and µPIV measurements [67], [68].
The growth of the embryonic vasculature can give an indication of normal versus abnormal development. Because Doppler OCT is insensitive to measuring blood flow transverse to the illumination beam, vascular mapping is incomplete. Fingler et al. used phase variance contrast to create vascular maps in a zebrafish and were able to pick out transverse vessels and vessels with no flow [50]. By looking at the temporal variance in the structural image, Sudheendran et al. imaged vessels in the yolk sac of the mouse. We have used the same technique to image the vasculature of the developing quail embryo longitudinally (see Fig. 5). Although these techniques are incapable of measuring the velocity of the blood, they will be extremely useful to determine how the morphology of the blood vessels varies in normal and abnormal development.
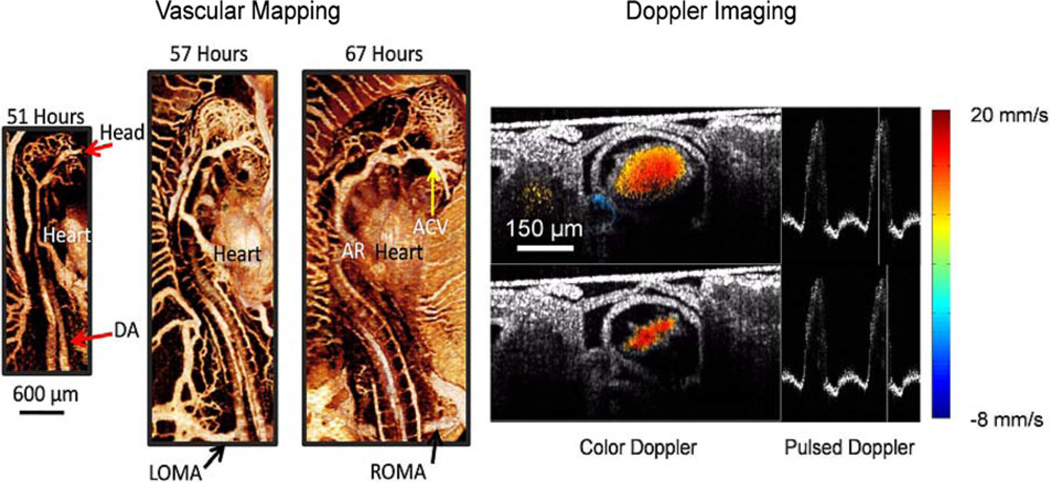
Fig. 5.
Vascular maps and Doppler imaging. (Left) Vascular imaging is combined with longitudinal imaging. Vascular maps are shown of a quail embryo at 51, 57, and 67 h. Vascular maps allow investigations into the entire cardiovascular system. (Right) Gated Doppler imaging through the middle (transverse) of a stage 14 embryonic quail heart. Two time points in the cardiac cycle are shown. Gated imaging allows us to capture 60 frames/heartbeat. Pulsed Doppler profiles adjacent to the color Doppler images show the shape of the heartbeat and the position of the frame in the cardiac cycle. The color Doppler images display the relative blood velocity as shown in the color bar (blue—toward the OCT beam; red—away from the OCT beam). Here, we set velocities resulting from forward flow to be positive. DA—dorsal aorta; AR—aortic root; LOMA—left omphalomesenteric artery; ROMA—right omphalomesenteric artery; and ACV—anterior cardinal vein.
C. Methods for Live Imaging
Many early OCT experiments on the developing heart were designed to demonstrate the capability of OCT imaging and did not adequately control the environment for the embryo to develop under physiological conditions. Not only is it important to control the environment to accurately assess normal development, but also when comparing control embryos with genetically modified or experimentally perturbed embryos. Also, with the proper environment, one can follow the growth of the embryos over several stages of development. Some initial studies demonstrated the ability of OCT to follow the growth of the avian embryo over several stages of development without robust environmental control (heating pads) [26], [30]. Unfortunately, even brief periods away from the incubator leads to sharp changes in the heart rate and better control is needed. Filas et al. employed a culture dish that controlled temperature, humidity, nutrients, and gas levels for investigating avian embryos in a new culture [28], which places the embryo ventral side up. We have constructed an OCT scanner within an environmental chamber that can precisely control temperature, humidity, and oxygen and has successfully investigated avian embryos in shell-less cultures [15], [33], [34].
Although the mouse is well suited for genetic manipulations, assaying murine embryonic heart dynamics under physiological conditions is challenging because of the protective maternal intrauterine environment that is hard to penetrate or duplicate. Echocardiography has been used extensively for the study of these embryos but limitations in resolution have confined its use to later stage embryos. Larina et al. demonstrated the first structural and Doppler OCT images of the live murine embryo by combining an OCT system with a static culture [36]. Briefly, embryos were dissected with the yolk sac intact and cultured in a medium (89% DMEM/F12, 10% FBS, and 1% 100× Pen-strep solution), placed in a 37 °C, 5% CO2 incubator, and imaged with OCT. Subsequent papers demonstrated OCT imaging of the developing murine embryo heart at several stages of development (E 7.5–E 10.5) [38], imaging of the embryonic rat heart [64] and velocity determination of individual blood cells in an E 8.5 mouse embryo using Doppler OCT [39]. More recently, Syed et al. externalized the uterine horn in survival surgeries and imaged embryos at E 13.5, E 15.5 and E 17.5 [69]. Unfortunately, the decidua, which degrades at E 12.5 and highly scatters light, prevents OCT imaging of the embryo before E 12.5 when the heart is accessible with OCT imaging. OCT clearly has the capability to image murine heart development from the earliest stages. In the future, it will be important to assess if and how the physiology of the embryo may be altered by culturing the embryo outside the womb. As the technology develops, OCT should become a robust tool to rapidly assess phenotypic differences in murine heart morphology and function in four dimensions.
D. Experiments and Descriptive Studies
The morphology of the looping heart and nearby structures influences both function and development. Filas et al. employed OCT to demonstrate that the splanchnopleure (a membrane pressing against the ventral side of the heart tube) exerts forces on the heart that drives c-looping and alters cardiac jelly distribution [28], [29]. When the splanchnopleure was removed rightward torsion of the heart was delayed proving the influence of the membrane on the shape of the heart. Jenkins et al. first described tether connections (thin processes connecting the endocardium to the myocardium) seen with OCT in 2007 [30]. Garita et al. expanded these studies by collocating known mechanotransducing molecules (fibronectin, tenascin C, α-tubulin, and nonmuscle myosin II) at myocardial–endocardial attachment areas [15]. Manner et al. utilized OCT to demonstrate that the endocardium undergoes eccentric narrowing and widening during the cardiac cycle of stages 10–12 chicken embryos rather than concentric narrowing and widening [23]. It was surmised that this eccentric pattern may allow the tubular heart to pump more efficiently. At later stages (15–17), they showed that the endocardium becomes bell shaped and hypothesized that the shape may represent an optimization between pumping efficiency and supplying energy to the myocardium [24]. Finally, Garita et al. demonstrated in a stage-13 quail that the cardiac jelly shifts to prevalve areas to prevent regurgitation [15]. From these studies, we are just beginning to determine how the heart forms and pumps blood in the early stages. Manner et al. discuss current theories regarding the mechanisms of unidirectional flow in the valveless heart and discuss the need for more studies to determine the mechanisms [70].
Genetic manipulations and physical perturbations play an important role in deciphering mechanisms in biology. Rugonyi et al. demonstrated the ability of OCT to measure blood velocity and wall motion after OFT banding and vitelline vessel ligation [17]. Jenkins et al. utilized OCT to morphologically phenotype-excised murine hearts discerning hexamethylene-bisacetamide-inducible protein 1 (HEXIM1) mutants from their wild-type littermates. As of yet, OCT has not been used to phenotype live murine embryos, but with the progress in live murine cultures, this should come in the near future. Drosophila is another model system that can easily be genetically altered. Several groups have demonstrated, in larval- to adult-stage Drosophila, the ability of OCT to phenotype alterations in heart dynamics in mutant animals [47]–[49], [71]–[74]. Although Drosophila does not have a four-chambered heart, and among other differences from humans, there are still much that can be learned by studying this model organism. Choma et al. have pointed out the similarities in Drosophila and humans [47].
III. Materials and Methods
A. Embryo Preparation
Fertilized quail (Coturnix Coturnix) eggs were incubated in a humidified forced draft incubator at 38 °C. Embryos were removed from the egg between 40 and 50 h postfertilization and cultured on the yolk in a sterilized 3.5-cm-diameter Petri dish. Next, the embryos were placed within the environmental imaging chamber and repeatedly imaged over the next 24–36 h. For hyaluronidase experiments, embryos were injected with either 25 µL of hyaluronidase or 25 µL of phosphate-buffered saline (control) at 18-h postincubation before being cultured in a Petri dish for 40 h of development and imaged with OCT in the environmental chamber.
B. Environmental Control
Embryos require incubation, where temperature and humidity are precisely regulated. For environmental control, we constructed an environmental chamber (4 × 3 × 2 feet) with Plexiglas walls housing the scanner of our OCT system as shown in Fig. 1(b) [15], [33], [34]. Two bare-hand entry iris ports allow interaction with specimens without affecting the environment. The temperature control system consists of a temperature sensor, a PID controller, and wire heating elements. A fan blows the heated air over distilled water, which creates humidity. By manually controlling the surface area of the water, one can control the level of relative humidity in the chamber. The temperature and humidity levels stabilize at 102.1 ± 0.35oF and 60.0 ± 0.31% RH. The OCT scanner is attached to an XYZ positioning gantry (Velmex, Inc., Bloomfield, NY) capable of spanning 0.61 ft3 with accuracy on the scale of 10 µm. The stage can fit up to 25 embryos at once, which will allow for high-throughput longitudinal studies.
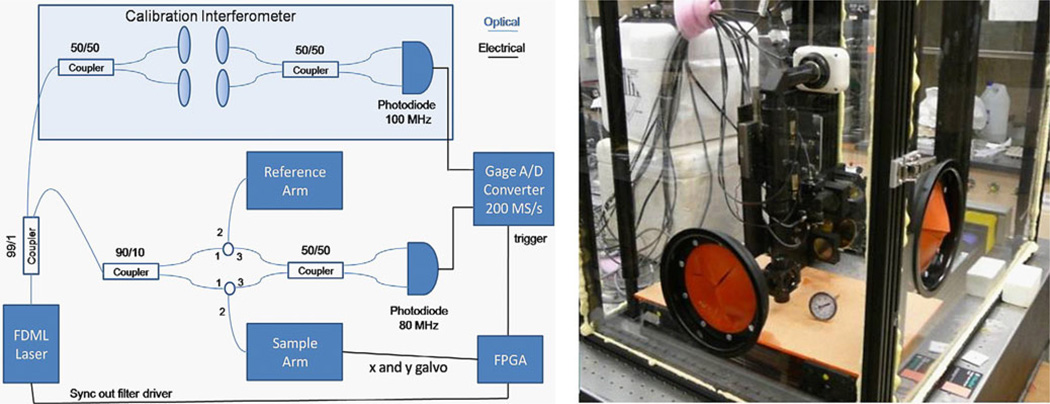
Fig. 1.
OCT setup and environmental chamber. (Right) Diagram of the OCT setup. The system employs an FDML laser operating at 117-kHz line rate. The scanner in the sample arm consists of quasitelecentric optics with an attached microscope. A calibration interferometer is utilized to resample the data evenly in wave number. (Left) Photograph of our environmental imaging chamber. The chamber precisely controls temperature and humidity, while allowing OCT imaging.
C. OCT Imaging
We performed ultrahigh-speed OCT imaging by employing a buffered Fourier Domain Mode Locked (FDML) laser [30], [61] operating at 117-kHz line rate. Fig. 1(a) shows a diagram of the OCT setup. The laser outputs 13mW of power and sweeps a range of wavelengths centered at 1296 nm. The full sweep range of the laser is 120 nm and the full-width at half-maximum (FWHM) of the spectrum is approximately 75 nm, which resulted in an axial resolution of 8 µm in tissue. The sensitivity falloff is 3 dB at 3.5mm of mismatch. A Mach–Zehnder interferometer (the splitting ratio 90/10) is used to collect the OCT data. The scanner in the sample arm consists of quasi-telecentric optics with an attached microscope [75].With a beam analyzer, we measured the spot size of the system to be 10 µm FWHM in air. We designed the lateral resolution of the OCT system to be able to easily distinguish the layers of the embryonic heart, while still preserving an imaging depth capable of capturing the heart tube in vivo. A field programmable gate array (FPGA) sends signals to control the scanning mirrors and data acquisition. The ultrahigh-speed system enables real-time 4-D imaging at ~5 volumes/heartbeat, and combined with cardiac gating, the ultrahigh speed allows for highly accurate gating schemes to capture systolic dynamics in three dimensions. All image processing and visualization of the OCT images (image-based retrospective gating, Doppler OCT, and vascular mapping) were realized in MATLAB (The MathWorks, Inc.) and Amira (Visage Imaging GmbH).
D. Image-Based Retrospective Gating
We developed a signal-free retrospective gating algorithm to accurately reconstruct 4-D image sets of the embryonic heart during looping at 90 volumes/heartbeat [33]. Briefly, the cardiac gating algorithm consisted of period determination, temporal interpolation, determination of temporal offsets between adjacent slices, and reordering of all images in absolute time. Through validation experiments, we found that the retrospective gating algorithm could accurately reassemble 4-D datasets with a temporal displacement error of less than 5 ms. To save time, the image data were decimated by 4 for the first three steps of the algorithm.
E. Doppler OCT
To accommodate Doppler imaging, OCT A-scans were recorded at 1.3 µm steps in the B-scan direction. A calibration interferometer was used both to resample the data evenly in wave number and to improve the Doppler signal by correcting for small fluctuations in sweep progression. A Goldstein algorithm was utilized to phase unwrap the Doppler data [76]. To decrease noise in the color Doppler images, a rolling five-line average was employed. We obtained pulsed Doppler waveforms from the center of the lumen by selecting a small window (5 × 5 pixels). The pixels in the window were utilized to compute the pulsed Doppler waveform in the same way as using a line segment in pulsed Doppler ultrasound.
F. Vascular Mapping
We created maps of the developing vasculature in the embryo by assessing the variance of the speckle in the OCT images. At each slice position approximately ten cross-sectional images (B-scans) were taken. Three to four images were used to compute the variance as described in [77] and [78] and the additional images allowed for averaging to reduce noise. A median filter further reduced the noise in the images.
IV. Results
A. Longitudinal Imaging
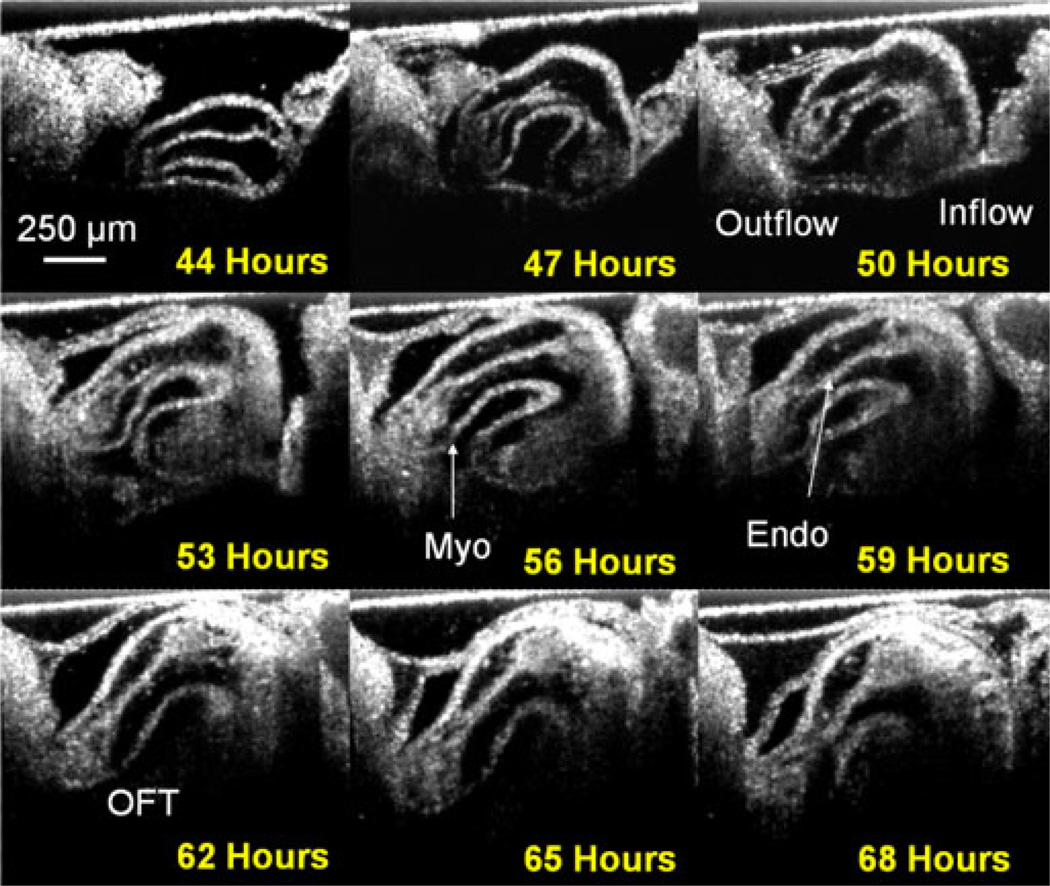
Fig. 2 shows the heart of a growing embryo every 3 h fora24-h period. The displayed images were selected from the contracted phase of the cycle. At 44 h, the embryo is between stages 11 and 12 of development and the embryonic heart appears as a straight tube. By 68 h, (stages 18/19) the heart has become an s-shaped tube. After 60 h of development, it becomes difficult to visualize the entire inflow of the heart with this choice of lenses in the scanner, but the OFT is still easily visualized. The sequence of images illustrates the rapid growth of the heart in a short time period. The resolution is high enough to reveal details of endocardial cushion (cardiac jelly) and myocardial morphology and blood flow. The B-scans in Fig. 2 were processed with an optimized nonorthogonal wavelet-based denoising algorithm [32].
Fig. 2.
Development in 24 h. The panel shows the heart of a growing embryo every 3 h for a 24-h period. The sequence of images illustrates the enormous growth of the embryos over a short period of time. The heart starts as a straight tube (stages 11/12) and progresses to a c-shaped tube and finally an s-shaped tube (stages 18/19). Myo—myocardium; Endo—endocardium; and OFT—OFT.
Fig. 3 displays surface renderings of the embryonic quail heart (endocardium—red; myocardium—blue) as it develops over a 24-h period. At each time point, a 4-D dataset was collected and processed. A total of ten 4-D datasets were collected and comprise the first longitudinal 4-D OCT dataset of an embryonic quail heart during looping. The entire longitudinal 4-D dataset (10 4-D sets of 90 volumes each) produced large amounts of data (~150 GB) and took considerable processing time. The progression of looping is clearly evident. Each rendering was manually segmented and is displayed at the phase when the heart is most compressed at the top of the loop. The endocardium of the 40-h heart is smooth and the heart has just begun to loop, whereas the endocardium of the 64-h heart has many ridges and folds and has grown significantly over a short period of time. Being able to monitor the rapid heart growth in the looping stages of development will enable many experiments not previously possible.
Fig. 3.
Three-dimensional reconstructions of the heart tube growing over a 24-h period. Transparent blue is the myocardium and red is the endocardium. The endocardium starts out smooth and develops more folds and ridges as time passes. Myo—myocardium; Endo—endocardium; CJ—cardiac jelly; A— anterior; P—posterior; D—dorsal; V—ventral; R—right; and L—left.
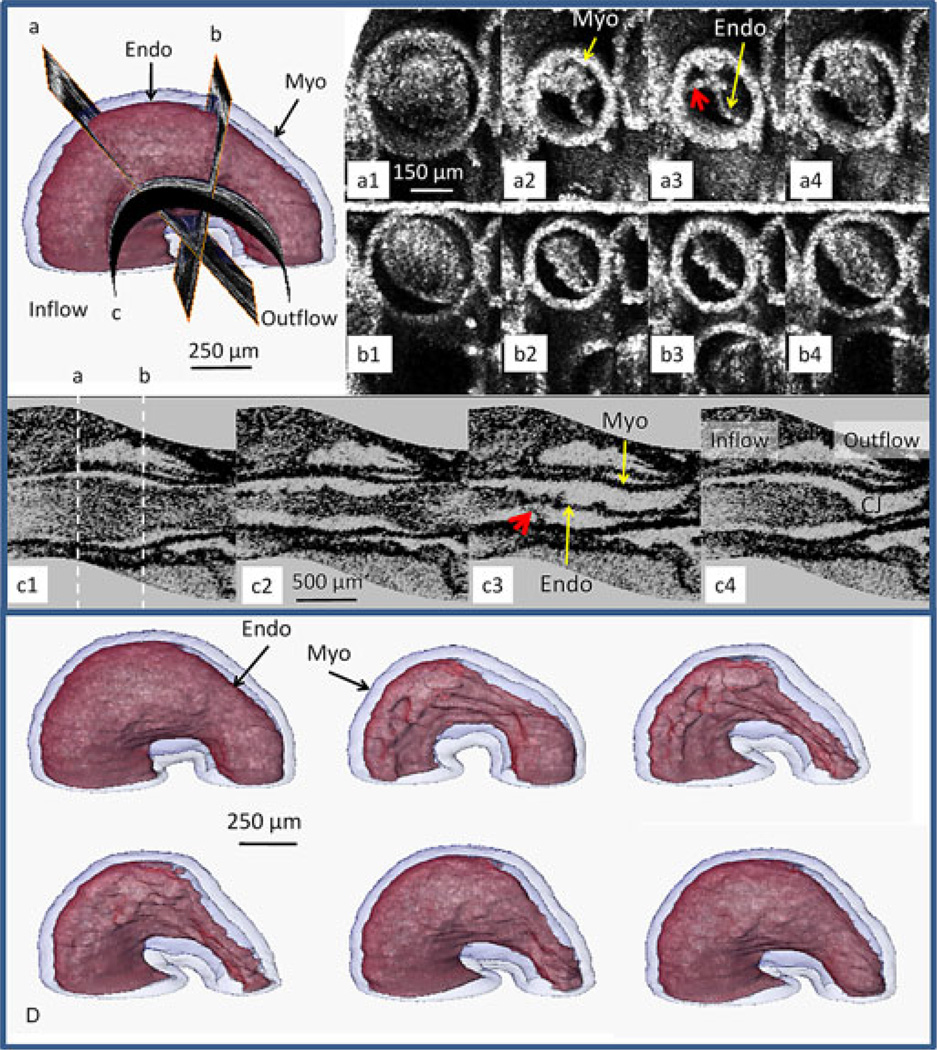
Fig. 4 exhibits one 4-D dataset (stage 13) taken from a longitudinal 4-D dataset. Surface renderings of segmented endocardium (red) and myocardium (transparent blue) as the heart progresses through one cardiac cycle reveal that the endocardium folds into longitudinal ridges as the myocardium contracts indicating a complex interplay between the three cardiac layers (panel D). Endocardial connections to the myocardium may explain the endocardial folding. Visualization of two orthogonal and one curved slice (see Fig. 4) from the 4-D image set demonstrates an eccentric (B1–4), and complex deformation pattern (A1–A4) that suggest tether-like connections (red arrows) between the endocardium and myocardium. The curved surface view (C1–C4) provides an informative orientation not possible with standard 2-D OCT imaging or with histological sectioning. Four-dimensional OCT will enable us to better understand the intricate morphogenesis of the beating heart and integrate the role of mechanics with morphogenesis during heart development.
Fig. 4.
Slices through a stage-13 quail heart. (Upper left) Position of the three slices a, b, and c on the 3-D heart. A1–A4 and B1–B4 time sequences of the beating heart at slice locations a and b oriented orthogonal to the heart tube. By capturing the full 4-D dataset, it was possible to reconstruct true orthogonal slices even when the orientation of the slice (e.g., slice a) was not possible to obtain directly. B1–B4 demonstrates eccentric deformation, while A1-A4 displays a more complex deformation pattern and suggests tether connections (red arrows) between the endocardium and myocardium. C1–C4 illustrates a curved surface through the center of the length of the heart tube. This orientation illustrates the nonuniform morphology of the three layers throughout the heart tube. The endocardium has many outpocketings and twists. The white dashed lines indicate the locations of slices a and b. Panel D displays surface renderings of segmented endocardium (red) and myocardium (transparent blue) as the heart progresses through one cardiac cycle and reveals that the endocardium folds into longitudinal ridges as the myocardium contracts indicating a complex interplay between the three cardiac layers. Myo—myocardium and Endo—endocardium.
Fig. 5 illustrates the ability of OCT to visualize the vasculature and measure blood velocity. Here, we have combined vascular mapping with longitudinal imaging to follow the growth of a quail embryo from 51–67 h of development. Clearly, the vascular system develops extremely rapidly. Vascular maps can give us insights into how the cardiovascular system develops and how nutrients are delivered to the entire embryo. The right panel shows a transverse section through the middle of a stage-14 embryonic quail heart (51 h of development) at two time points in the cardiac cycle. The images to the left are color Doppler overlays on top of the structure images. Pulsed Doppler profiles adjacent to the color Doppler images show the shape of the heartbeat and the position of the frame in the cardiac cycle. The images are taken from a gated OCT dataset consisting of 60 frames/heartbeat. Learning how the vasculature and blood flow are influenced in development and how they influence development will be important for future studies utilizing OCT.
B. Hyaluronidase Experiments
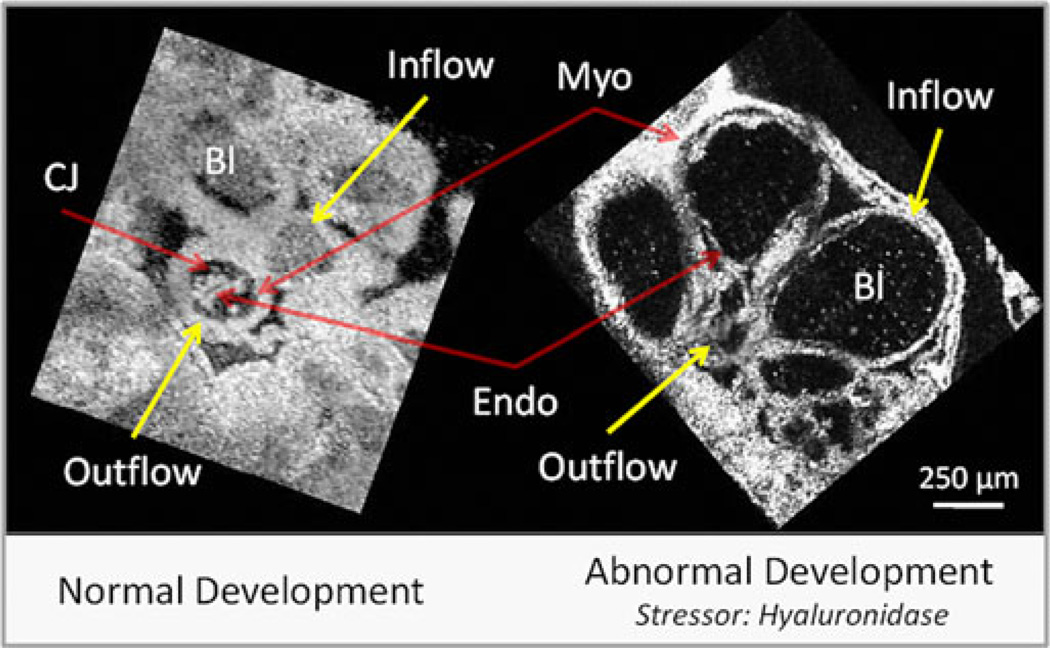
Fig. 6 presents multiplanar reformatted OCT images from two 4-D image sets of embryonic quail hearts. The left panel displays a stage-18 embryonic quail heart undergoing normal development, while the right panel shows a stage-19 embryonic quail heart that was exposed to hyaluronidase at 18 h of development. From Fig. 6, it is clear that there is a reduction of blood cells and the myocardial walls are thinner in the perturbed heart. The normal heart in Fig. 6 exhibits a fourfold increase in peak wall velocity at the inflow (2.4 mm/s) compared to the OFT (0.6 mm/s), which corresponds to the initiation of the beat at the inflow. The hyaluronidase heart does not display a significant difference in wall velocity between the inflow and OFT. The decreased inflow wall velocity suggests that the cardiac jelly may play an important role in regulating cardiac conduction. Either signaling between the cardiac jelly and the myocardium has been altered or the missing jelly has modified the stress on the myocardium.
Fig. 6.
Normal versus abnormal heart development. En face slices from 4-D image sets (90 volumes/heartbeat). Hyaluronidase degrades the proteoglycan component of the cardiac jelly found between the myocardium and the endocardium. From the images, it is clear that there is a reduction of blood cells and the myocardial walls are thinner in the perturbed heart. Also, the maximum wall velocity between the inflow (2.4 mm/s) and outflow (0.6 mm/s) tract varies significantly in normal development, while the perturbed heart did not have a higher maximum velocity in the inflow. The decreased inflow wall velocity suggests that the cardiac jelly may play an important role in regulating cardiac conduction. CJ—cardiac jelly; Myo—myocardium; Endo—endocardium; and Bl—blood.
V. Discussion and Conclusion
The following results demonstrate the feasibility of longitudinal imaging of the avian embryo using the several described OCT imaging modes. These techniques will enable measurements of various morphologic and hemodynamics parameters (wall width, wall velocity, blood velocity, shear stress, etc.) at different stages of development. We have demonstrated the first longitudinal 4-D images of the developing heart. Following the developmental progression of the heart tube can greatly aid in determining the time points when heart development is altered. We are able to collect datasets from stages 10 to 18. By stage 18, the depth penetration of our system was not able to capture the entire depth of the heart tube. By switching objectives, one can choose a lower numerical aperture lens that will image deeper at the expense of lowered resolution. The longitudinal 4-D dataset is extremely large and cumbersome. In the future, algorithms for handling and displaying the data more rapidly will greatly enhance the utility of longitudinal 4-D imaging. For example, it is not practical to capture longitudinal 4-D datasets of large groups of embryos at the current time.
We also present the first longitudinal vascular maps of the developing embryo. The vascular maps show tremendous growth over a very short time period. Selecting the image time between B-scans determines the range of flows that the variance masking algorithm can pick up. Unfortunately, increased image time also increases motion artifact noise; so, a balance between detecting low flow vessels and image time must be optimized.
Finally, we used our longitudinal imaging system to show a baseline study comparing the development of normal quail embryo hearts with hearts injected with hyaluronidase. Hyaluronidase caused significant abnormalities in the heart (e.g., altered wall thickness, blood cell development, wall velocity, etc.). Longitudinal imaging greatly enhances comparisons between normal hearts and hearts with applied stressors. For example, by imaging various stages of development, we were able to see that hyaluronidase alters cardiac jelly from the beginning of development and the heart is never able to compensate for its loss. Future studies following the growth of the embryo are needed to fully characterize normal parameters of development and begin deciphering how cardiac dynamics play a role in the development of congenital defects.
Acknowledgment
The authors would like to thank M. Gargesha for assistance with processing the datasets, L. Barwick for helping with manual segmentation, and A. Raina for helping with the hyaluronidase experiments.
This work was supported in part by National Institutes of Health (NIH) under Grant RO1-HL083048 and Grant RO1 HL095717. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program under Grant C06 RR12463-01 from the National Center of Research Resources, NIH.
Biographies

Michael W. Jenkins received the Ph.D. degree in biomedical engineering from Case Western Reserve University, Cleveland, OH, in 2008. He was a Research Associate and Postdoctoral Fellow in both Dr. A. Rollins’ and M. Watanabe’s laboratory from 2008–2010. Since 2011, he has been the Research Assistant Professor in the Department of Biomedical Engineering, Case Western Reserve University. His research interest includes the development and application of OCT technology for studying the developing heart.

Michiko Watanabe received the Ph.D. degree from Wesleyan University, Middletown, CT, in 1981. She was a Postdoctoral Researcher in developmental biology at Indiana University, Bloomington, Indiana, with Dr. Mahowald’s group and at the School of Medicine, Case Western Reserve University, Cleveland, OH, in Dr. Rutishauser’s laboratory. She is currently a Full Professor in the Department of Pediatrics and has secondary appointments in the Departments of Genetics and Anatomy. She is also a trainer in various programs including the Developmental Biology, Physiology, and Biomedical Engineering Programs and is the Director of Research for the Pediatric Cardiology Fellowship Program. Her current research interests include the understanding of the mechanism of coronary vessel development and growth in the embryo and adult heart. She collaborates closely with Dr. Rollins, Dr. Jenkins, and other biomedical engineering colleagues to apply the latest technology to the study of the heart development and disease.

Andrew M. Rollins received the Ph.D. degree in biomedical engineering from Case Western Reserve University, Cleveland, OH, in 2000. He is currently an Associate Professor in the Department of Biomedical Engineering and Medicine, Case Western Reserve University. His current research interests include the development and application of advanced optics and photonics technologies for imaging and characterization of biological samples, with particular emphasis on detection of early disease and monitoring of therapy in human tissues and investigating embryonic development. His primary research interest includes the technique of OCT.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Michael W. Jenkins, Email: mwj5@case.edu, Department of Biomedical Engineering and Medicine, Case Western Reserve University, Cleveland, OH 44106 USA.
Michiko Watanabe, Email: mxw13@case.edu, Department of Pediatrics, Case Western Reserve University, Cleveland, OH 44106 USA.
Andrew M. Rollins, Email: amr9@case.edu, Department of Biomedical Engineering and Medicine, Case Western Reserve University, Cleveland, OH 44106 USA.
References
- 1.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick-embryo, (Reprinted from Journal of Morphology, Vol 88, 1951) Develop. Dyn. 1992 Dec;195:231. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 2.Manner J. Cardiac looping in the chick embryo: A morphological review with special reference to terminological and biomechanical aspects of the looping process. Anatom. Rec. Adv. Integr. Anatomy Evol. Biol. 2000 Jul.259:248–262. doi: 10.1002/1097-0185(20000701)259:3<248::AID-AR30>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 3.Manner J. The anatomy of cardiac looping: A step towards the understanding of the morphogenesis of several forms of congenital cardiac malformations. Clin. Anatomy. 2009 Jan.22:21–35. doi: 10.1002/ca.20652. [DOI] [PubMed] [Google Scholar]
- 4.Bentham J, et al. Maternal high-fat diet interacts with embryonic Cited2 genotype to reduce Pitx2c expression and enhance penetrance of left-right patterning defects. Hum. Mol. Genetics. 2010 Sep.19:3394–3401. doi: 10.1093/hmg/ddq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogers B, et al. Unilateral vitelline vein ligation alters intracardiac blood flow patterns and morphogenesis in the chick embryo. Circ. Res. 1997 Apr.80:473–481. doi: 10.1161/01.res.80.4.473. [DOI] [PubMed] [Google Scholar]
- 6.Hogers B, et al. Extraembryonic venous obstructions lead to cardiovascular malformations and can be embryolethal. Cardiovascular Res. 1999 Jan.41:87–99. doi: 10.1016/s0008-6363(98)00218-1. [DOI] [PubMed] [Google Scholar]
- 7.Hove JR, et al. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003 Jan.421:172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 8.Reckova M, et al. Hemodynamics is a key epigenetic factor in development of the cardiac conduction system. Circ. Res. 2003 Jul.93:77–85. doi: 10.1161/01.RES.0000079488.91342.B7. [DOI] [PubMed] [Google Scholar]
- 9.Lucitti JL, et al. Increased arterial load alters aortic structural and functional properties during embryogenesis. Amer. J. Physiol. Heart Circ. Physiol. 2006 Oct.291:H1919–H1926. doi: 10.1152/ajpheart.01061.2005. [DOI] [PubMed] [Google Scholar]
- 10.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009 Nov.7 doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang D, et al. Optical coherence tomography. Science. 1991 Nov.254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boppart SA, et al. Investigation of developing embryonic morphology using optical coherence tomography. Dev. Biol. 1996 Jul.177:54–63. doi: 10.1006/dbio.1996.0144. [DOI] [PubMed] [Google Scholar]
- 13.Boppart SA, et al. Noninvasive assessment of the developing Xenopus cardiovascular system using optical coherence tomography. Proc. Natl. Acad. Sci. USA. 1997 Apr.94:4256–4261. doi: 10.1073/pnas.94.9.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins MW, et al. 4D embryonic cardiography using gated optical coherence tomography. Opt. Exp. 2006 Jan.14:736–748. doi: 10.1364/opex.14.000736. [DOI] [PubMed] [Google Scholar]
- 15.Garita B, et al. Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping. Amer. J. Physiol. Heart Circ. Physiol. 2011 Mar.300:H879–H891. doi: 10.1152/ajpheart.00433.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu A, et al. Finite element modeling of blood flow-induced mechanical forces in the outflow tract of chick embryonic hearts. Comput. Struct. 2007 Jun.85:727–738. [Google Scholar]
- 17.Rugonyi S, et al. Changes in wall motion and blood flow in the outflow tract of chick embryonic hearts observed with optical coherence tomography after outflow tract banding and vitelline-vein ligation. Phys. Med. Biol. 2008 Sep.53:5077–5091. doi: 10.1088/0031-9155/53/18/015. [DOI] [PubMed] [Google Scholar]
- 18.Liu A, et al. Efficient postacquisition synchronization of 4-D nongated cardiac images obtained from optical coherence tomography: Application to 4-D reconstruction of the chick embryonic heart. J. Biomed Opt. 2009 Jul-Aug;14:044020. doi: 10.1117/1.3184462. [DOI] [PubMed] [Google Scholar]
- 19.Liu A, et al. Dynamic variation of hemodynamic shear stress on the walls of developing chick hearts: Computational models of the heart outflow tract. Eng. Comput. 2009;25:73–86. [Google Scholar]
- 20.Ma Z, et al. Measurement of absolute blood flow velocity in outflow tract of HH18 chicken embryo based on 4D reconstruction using spectral domain optical coherence tomography. Biomed. Opt. Exp. 2010;1:798–811. doi: 10.1364/BOE.1.000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu A, et al. Quantifying blood flow and wall shear stresses in the outflow tract of chick embryonic hearts. Comput. Struct. 2011 Jun.89:855–867. doi: 10.1016/j.compstruc.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yelbuz TM, et al. Optical coherence tomography: A new high-resolution imaging technology to study cardiac development in chick embryos. Circulation. 2002 Nov.106:2771–2774. doi: 10.1161/01.cir.0000042672.51054.7b. [DOI] [PubMed] [Google Scholar]
- 23.Manner J, et al. High-resolution in vivo Imaging of the cross-sectional deformations of contracting embryonic heart loops using optical coherence tomography. Dev. Dyn. 2008 Apr.237:953–961. doi: 10.1002/dvdy.21483. [DOI] [PubMed] [Google Scholar]
- 24.Manner J, et al. In vivo imaging of the cyclic changes in cross-sectional shape of the ventricular segment of pulsating embryonic chick hearts at stages 14 to 17: A contribution to the understanding of the ontogenesis of cardiac pumping function. Dev. Dyn. 2009 Dec.238:3273–3284. doi: 10.1002/dvdy.22159. [DOI] [PubMed] [Google Scholar]
- 25.Thrane L, et al. Field programmable gate-array-based real-time optical Doppler tomography system for in vivo imaging of cardiac dynamics in the chick embryo. Opt. Eng. 2009 Feb.48 [Google Scholar]
- 26.Davis AM, et al. In vivo spectral domain optical coherence tomography volumetric imaging and spectral Doppler velocimetry of early stage embryonic chicken heart development. J. Opt. Soc. Amer. A Opt. Image Sci. Vis. 2008 Dec.25:3134–3143. doi: 10.1364/josaa.25.003134. [DOI] [PubMed] [Google Scholar]
- 27.Davis A, et al. Quantitative measurement of blood flow dynamics in embryonic vasculature using spectral doppler velocimetry. Anatom. Rec. Adv. Integr. Anatomy Evol. Biol. 2009 Mar.292:311–319. doi: 10.1002/ar.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filas BA, et al. Optical coherence tomography as a tool for measuring morphogenetic deformation of the looping heart. Anatom. Rec. Adv. Integr. Anatomy Evol. Biol. 2007;290:1057–1068. doi: 10.1002/ar.20575. [DOI] [PubMed] [Google Scholar]
- 29.Ramasubramanian A, et al. On modeling morphogenesis of the looping heart following mechanical perturbations. J. Biomech. Eng. 2008 Dec.130:061018. doi: 10.1115/1.2978990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins MW, et al. Ultrahigh-speed optical coherence tomography imaging and visualization of the embryonic avian heart using a buffered Fourier Domain Mode Locked laser. Opt. Exp. 2007 May 14;15:6251–6267. doi: 10.1364/oe.15.006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins MW, et al. In vivo gated 4D imaging of the embryonic heart using optical coherence tomography. J. Biomed. Opt. 2007 May-Jun;12 doi: 10.1117/1.2747208. [DOI] [PubMed] [Google Scholar]
- 32.Gargesha M, et al. Denoising and 4D visualization of OCT images. Opt. Exp. 2008 Aug.16:12313–12333. doi: 10.1364/oe.16.012313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gargesha M, et al. High temporal resolution OCT using image-based retrospective gating. Opt. Exp. 2009 Jun.17:10786–10799. doi: 10.1364/oe.17.010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins MW, et al. Measuring hemodynamics in the developing heart tube with four-dimensional gated Doppler optical coherence tomography. J. Biomed. Opt. 2010 Nov-Dec;15:066022. doi: 10.1117/1.3509382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larina IV, et al. Enhanced OCT imaging of embryonic tissue with optical clearing. Laser Phys. Lett. 2008 Jun.5:476–479. [Google Scholar]
- 36.Larina IV, et al. Live imaging of blood flow in mammalian embryos using Doppler swept-source optical coherence tomography. J. Biomed. Opt. 2008;13 doi: 10.1117/1.3046716. 060506-3. [DOI] [PubMed] [Google Scholar]
- 37.Bhat S, et al. Multiple-cardiac-cycle noise reduction in dynamic optical coherence tomography of the embryonic heart and vasculature. Opt. Lett. 2009 Dec.34:3704–3706. doi: 10.1364/OL.34.003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larin KV, et al. Live imaging of early developmental processes in mammalian embryos with optical coherence tomography. J. Innov. Opt. Health Sci. 2009 Jan.2:253–259. doi: 10.1142/S1793545809000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larina IV, et al. Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT. Opt. Lett. 2009 Apr.34:986–988. doi: 10.1364/ol.34.000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudheendran N, et al. Speckle variance OCT imaging of the vasculature in live mammalian embryos. Laser Phys. Lett. 2011;8:247–252. [Google Scholar]
- 41.Jenkins MW, et al. Phenotyping transgenic embryonic murine hearts using optical coherence tomography. Appl. Opt. 2007 Apr.46:1776–1781. doi: 10.1364/ao.46.001776. [DOI] [PubMed] [Google Scholar]
- 42.Luo W, et al. Three-dimensional optical coherence tomography of the embryonic murine cardiovascular system. J. Biomed. Opt. 2006 Mar-Apr;11 doi: 10.1117/1.2193465. [DOI] [PubMed] [Google Scholar]
- 43.Yazdanfar S, et al. High resolution imaging of in vivo cardiac dynamics using color Doppler optical coherence tomography. Opt. Exp. 1997;1:424–431. doi: 10.1364/oe.1.000424. [DOI] [PubMed] [Google Scholar]
- 44.Yang VX, et al. High speed, wide velocity dynamic range Doppler optical coherence tomography—Part II: Imaging in vivo cardiac dynamics of Xenopus laevis. Opt. Exp. 2003 Jul.11:1650–1658. doi: 10.1364/oe.11.001650. [DOI] [PubMed] [Google Scholar]
- 45.Mariampillai A, et al. Doppler optical cardiogram gated 2D color flow imaging at 1000 fps and 4D in vivo visualization of embryonic heart at 45 fps on a swept source OCT system. Opt. Exp. 2007 Feb.15:1627–1638. doi: 10.1364/oe.15.001627. [DOI] [PubMed] [Google Scholar]
- 46.Yelin R, et al. Multimodality optical imaging of embryonic heart microstructure. J. Biomed. Opt. 2007 Nov-Dec;12 doi: 10.1117/1.2822904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choma MA, et al. Physiological homology between Drosophila melanogaster and vertebrate cardiovascular systems. Disease Models Mech. 2011 May 1;4:411–420. doi: 10.1242/dmm.005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradu A, et al. Dual optical coherence tomography/fluorescence microscopy for monitoring of Drosophila melanogaster larval heart. J. Biophoton. 2009 Jul.2:380–388. doi: 10.1002/jbio.200910021. [DOI] [PubMed] [Google Scholar]
- 49.Ma L, et al. Arrhythmia caused by a Drosophila tropomyosin mutation is revealed using a novel optical coherence tomography instrument. PLoS One. 2010;5:e14348. doi: 10.1371/journal.pone.0014348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fingler J, et al. Mobility and transverse flow visualization using phase variance contrast with spectral domain optical coherence tomography. Opt. Exp. 2007 Oct.15:12636–12653. doi: 10.1364/oe.15.012636. [DOI] [PubMed] [Google Scholar]
- 51.Kagemann L, et al. Repeated, noninvasive, high resolution spectral domain optical coherence tomography imaging of zebrafish embryos. Mol. Vis. 2008;14:2157–2170. [PMC free article] [PubMed] [Google Scholar]
- 52.Iftimia NV, et al. Dual-beam Fourier domain optical Doppler tomography of zebrafish. Opt. Exp. 2008 Sep.16:13624–13636. doi: 10.1364/oe.16.013624. [DOI] [PubMed] [Google Scholar]
- 53.Rollins A, et al. In vivo video rate optical coherence tomography. Opt. Exp. 1998 Sep.3:219–229. doi: 10.1364/oe.3.000219. [DOI] [PubMed] [Google Scholar]
- 54.Tearney GJ, et al. In vivo endoscopic optical biopsy with optical coherence tomography. Science. 1997 Jun.276:2037–2039. doi: 10.1126/science.276.5321.2037. [DOI] [PubMed] [Google Scholar]
- 55.Fercher AF, et al. Measurement of intraocular distances by backscattering spectral interferometry. Opt. Commun. 1995 May 15;117:43–48. [Google Scholar]
- 56.Choma MA, et al. Sensitivity advantage of swept source and Fourier domain optical coherence tomography. Opt. Exp. 2003 Sep.11:2183–2189. doi: 10.1364/oe.11.002183. [DOI] [PubMed] [Google Scholar]
- 57.de Boer JF, et al. Improved signal-to-noise ratio in spectral-domain compared with time-domain optical coherence tomography. Opt. Lett. 2003 Nov.28:2067–2069. doi: 10.1364/ol.28.002067. [DOI] [PubMed] [Google Scholar]
- 58.Leitgeb R, et al. Performance of fourier domain vs. time domain optical coherence tomography. Opt. Exp. 2003 Apr.11:889–894. doi: 10.1364/oe.11.000889. [DOI] [PubMed] [Google Scholar]
- 59.Yun SH, et al. High-speed optical frequency-domain imaging. Opt. Exp. 2003 Nov.11:2953–2963. doi: 10.1364/oe.11.002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nassif N, et al. In vivo human retinal imaging by ultrahigh-speed spectral domain optical coherence tomography. Opt. Lett. 2004 Mar.29:480–482. doi: 10.1364/ol.29.000480. [DOI] [PubMed] [Google Scholar]
- 61.Huber R, et al. Fourier Domain Mode Locking (FDML): A new laser operating regime and applications for optical coherence tomography. Opt. Exp. 2006 Apr.14:3225–3237. doi: 10.1364/oe.14.003225. [DOI] [PubMed] [Google Scholar]
- 62.Liebling M, et al. Four-dimensional cardiac imaging in living embryos via postacquisition synchronization of nongated slice sequences. J. Biomed. Opt. 2005 Sep-Oct;10 doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- 63.Wieser W, et al. Multi-megahertz OCT: High quality 3D imaging at 20 million A-scans and 4.5 GVoxels per second. Opt. Exp. 2010 Jul.18:14685–14704. doi: 10.1364/OE.18.014685. [DOI] [PubMed] [Google Scholar]
- 64.Larina IV, et al. Live imaging of rat embryos with Doppler swept-source optical coherence tomography. J. Biomed. Opt. 2009 Sep-Oct;14:050506. doi: 10.1117/1.3241044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White B, et al. In vivo dynamic human retinal blood flow imaging using ultra-high-speed spectral domain optical coherence tomography. Opt. Exp. 2003 Dec.11:3490–3497. doi: 10.1364/oe.11.003490. [DOI] [PubMed] [Google Scholar]
- 66.Ghiglia DC, et al. Cellular-automata method for phase unwrapping. J. Opt. Soc. Amer. A. Opt. Image Sci. Vis. 1987 Jan.4:267–280. [Google Scholar]
- 67.Vennemann P, et al. In vivo micro particle image velocimetry measurements of blood-plasma in the embryonic avian heart. J. Biomech. 2006;39:1191–1200. doi: 10.1016/j.jbiomech.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 68.Poelma C, et al. Measurements of the wall shear stress distribution in the outflow tract of an embryonic chicken heart. J. R. Soc. Interface. 2010 Jan.7:91–103. doi: 10.1098/rsif.2009.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Syed SH, et al. Optical coherence tomography for high-resolution imaging of mouse development in utero. J. Biomed. Opt. 2011 Apr.16:046004. doi: 10.1117/1.3560300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manner J, et al. How does the tubular embryonic heart work? Looking for the physical mechanism generating unidirectional blood flow in the valveless embryonic heart tube. Dev. Dyn. 2010 Apr.239:1035–1046. doi: 10.1002/dvdy.22265. [DOI] [PubMed] [Google Scholar]
- 71.Choma MA, et al. Images in cardiovascular medicine: In vivo imaging of the adult Drosophila melanogaster heart with real-time optical coherence tomography. Circulation. 2006 Jul.114:e35–e36. doi: 10.1161/CIRCULATIONAHA.105.593541. [DOI] [PubMed] [Google Scholar]
- 72.Choma MA, et al. Heart wall velocimetry and exogenous contrast-based cardiac flow imaging in Drosophila melanogaster using Doppler optical coherence tomography. J. Biomed. Opt. 2010 Sep-Oct;15 doi: 10.1117/1.3503418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsai M-T, et al. Observations of cardiac beating behaviors of wild-type and mutant Drosophilae with optical coherence tomography. J. Biophoton. 2011 doi: 10.1002/jbio.201100009. [DOI] [PubMed] [Google Scholar]
- 74.Li A, et al. Changes in the expression of the alzheimer’s disease-associated presenilin gene in drosophila heart leads to cardiac dysfunction. Curr. Alzheimer Res. 2011 Apr. doi: 10.2174/156720511795563746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu Z, Rollins A. Quasi-telecentric optical design of a microscope-compatible OCT scanner. Opt. Exp. 2005 Aug.13:6407–6415. doi: 10.1364/opex.13.006407. [DOI] [PubMed] [Google Scholar]
- 76.Goldstein RM, et al. Satellite radar interferometry: Two-dimensional phase unwrapping. Radio Sci. 1988 Jul-Aug;23:713–720. [Google Scholar]
- 77.Mariampillai A, et al. Optimized speckle variance OCT imaging of microvasculature. Opt. Lett. 2010 Apr.35:1257–1259. doi: 10.1364/OL.35.001257. [DOI] [PubMed] [Google Scholar]
- 78.Mariampillai A, et al. Speckle variance detection of microvasculature using swept-source optical coherence tomography. Opt. Lett. 2008 Jul.33:1530–1532. doi: 10.1364/ol.33.001530. [DOI] [PubMed] [Google Scholar]