Abstract
Compressed sensing (CS) aims to recover images from fewer measurements than that governed by the Nyquist sampling theorem. Most CS methods use analytical predefined sparsifying domains such as Total variation (TV), wavelets, curvelets, and finite transforms to perform this task. In this study, we evaluated the use of dictionary learning (DL) as a sparsifying domain to reconstruct PET images from partially sampled data, and compared the results to the partially and fully sampled image (baseline).
A CS model based on learning an adaptive dictionary over image patches was developed to recover missing observations in PET data acquisition. The recovery was done iteratively in two steps: a dictionary learning step and an image reconstruction step. Two experiments were performed to evaluate the proposed CS recovery algorithm: an IEC phantom study and five patient studies. In each case, 11% of the detectors of a GE PET/CT system were removed and the acquired sinogram data were recovered using the proposed DL algorithm. The recovered images (DL) as well as the partially sampled images (with detector gaps) for both experiments were then compared to the baseline. Comparisons were done by calculating RMSE, contrast recovery and SNR in ROIs drawn in the background, and spheres of the phantom as well as patient lesions.
For the phantom experiment, the RMSE for the DL recovered images were 5.8% when compared with the baseline images while it was 17.5% for the partially sampled images. In the patients’ studies, RMSE for the DL recovered images were 3.8%, while it was 11.3% for the partially sampled images. Our proposed CS with DL is a good approach to recover partially sampled PET data. This approach has implications towards reducing scanner cost while maintaining accurate PET image quantification.
Keywords: Compressive sensing, positron emission tomography (PET), Image recovery, Image reconstruction, gap filling
1. Introduction
Positron emission tomography (PET) imaging has gained wide acceptance primarily for its use in cancer staging, therapy response monitoring, and drug discovery (Mankoff et al 2006). PET scanners however are relatively expensive imaging systems ranging between 1–3 million dollars and hence are less accessible to patients and clinicians in regional and community centers (Saif 2010). One approach to decrease the scanner cost is to reduce the number of detectors since these components are the most expensive in PET systems. However, image reconstruction from fewer observations (lines of response), while maintaining image quality, is a challenging task.
Several approaches have been proposed to estimate missing samples from tomographic data. One approach relies on various forms of interpolation (De Jong et al 2003, Karp et al 1988, Tuna et al 2010, Zhang 2008), while a second approach utilizes a statistical framework such as Maximum Likelihood Expectation Maximization (MLEM) (Nguyen 2010, Kinahan 1997, Raheja 1999). The first approach is highly sensitive to local variation, which results in substantial error in the data-fitting process while the second approach suffers from error augmentation as the number of iterations increase especially when the MLEM algorithms is used. A third approach that is also worthy of mentioning is texture synthesis which has been used for metal artifact reduction in CT imaging (Chen et al 2011, Effros et al 1999). These methods assume a model such as the Markov Random Field to fill in missing voxels, based on neighboring pixels.
An alternative approach to overcome this challenge in signal processing is the use of compressive sensing (CS) techniques (Sidky et al 2008, Donoho 2006, Otazo et al 2010, Ahn et al 2012, Valiollahzadeh et al 2012, 2013, 2015). CS enables the recovery of images/signals from fewer measurements than that governed by the traditional Nyquist sampling theorem, because the image/signal can be represented by sparse coefficients in an alternative domain (Donoho 2006). The sparser the coefficients the better the image recovery will be (Donoho 2006). Most CS methods use analytical predefined sparsifying domains (transforms) such as wavelets, curvelets, and finite transforms (Otaza et al 2010). For medical images however, one of the very commonly used sparsifying domains is the gradient magnitude domain (GMD) (Sidky et al 2008, Pan et al 2009, Ahn et al 2012, Valiollahzadeh et al 2012, 2015) and its associated minimization approach known as total variation (TV). The underlying assumption for using this domain is that medical images can often be described as piecewise constant (Ritschl et al 2011) such that when the gradient operator of GMD is applied, the majority of the resultant image coefficients become zero. Using GMD however has some immediate drawbacks: First, the TV constraint is a global requirement that cannot adequately represent structures within an object; and second, the gradient operator cannot distinguish true structures from image noise (Xu et al 2012). Consequently, images reconstructed using the TV constraint may lose some fine features and generate a blocky appearance particularly for undersampled and noisy cases. Hence, it is necessary to investigate superior sparsifying methods for CS image reconstruction. Previously, we showed a CS model based on a combination of gradient magnitude and wavelet domains to recover missing observations in PET data acquisition (Valiollahzadeh et al 2015). The model also incorporated a Poisson noise term that modeled the observed noise while suppressing its contribution by penalizing the Poisson log likelihood function. The results from this approach showed similar quantitative measurements compared to those with no missing samples suggesting good image recovery. However, the final reconstructed image suffered from some structured residual errors that were particularly visible on the difference image between the original and recovered images suggesting the potential for additional improvements.
Recently, the approach of sparsifying images using a redundant dictionary, known as dictionary learning (DL), has attracted interest in image processing, particularly in magnetic resonance imaging (Saiprasad et al 2011), ultrasound (Tosic et al 2010) and CT (Xu et al 2012, Yang et al 2012) primarily due to its ability to recover missing data. A dictionary is an overdetermined basis (the matrix has more columns than rows) whereby the elements of the basis, called atoms, are learned from training images obtained from decomposing an image into small overlapped patches. Because the dictionary is learned from these patches, and the patches are derived from the image itself, it is expected that DL will have a better sparsifying capability than any generic sparse transform and hence a superior image recovery. Furthermore, the redundancy of the atoms further facilitates a sparser representation (Xu et al 2012, Yang et al 2012). More importantly, the dictionary tends to effectively capture local image features as well as structural similarities without sacrificing resolution (Aharon et al 2006).
From an algorithmic point of view, there are two general classes of DL: optimization techniques and Bayesian techniques. In the optimization class, a dictionary is found by optimizing a cost function that include parameters such as the dictionary size, patch size, and patch sparsity (Xu et al 2012, Yang et al 2012). In the Bayesian class on the other hand, a statistical model is assumed to represent the data while all parameters of this model are directly inferred from the data (Ding et al 2013, Huang et al 2013). In general, optimization techniques have the advantage of faster convergence, but require several parameters that need to be justified. Bayesian techniques on the other hand have the advantage of not requiring any predetermined parameter but at the cost of slower convergence.
Dictionary learning has been utilized in medical imaging (CT, MR, Ultrasound) primarily for denoising, classification, and image restoration/reconstruction (Xu et al 2012, Saiprasad et al 2011,Chen et al 2010, Ma et al 2013, Li et al 2012, Zhao et al 2012). However, to the best of our knowledge, the use of DL to recover missing data while reconstructing PET images has not been studied before. In this work, we use a patch-based DL optimization t echnique to recover missing data in PET image reconstruction. The model we propose, simultaneously learns a dictionary and reconstructs PET images in an iterative manner from undersampled and noisy sinogram data. In this regard this approach can be termed as adaptive dictionary learning. Once the dictionary is learned, it is applied to reconstruct the partially observed data and in the next iteration, the dictionary is adaptively updated based on patches that are derived from the updated reconstructed image and the process continues until convergence.
In the following sections, we first develop the model for image recovery using DL. The model is then verified using phantom and patient studies by comparing its performance to fully sampled, and partially sampled reconstructions.
2. Material and Methods
2.1 Dictionary learning image reconstruction
Our proposed DL technique requires two steps; the first step is to determine the dictionary (Dictionary learning) and its associated coefficients while the second step is to recover and reconstruct the PET image.
2.1.1 Dictionary learning
The determination of a dictionary and its associated coefficients requires the knowledge of an image (m0) which when decomposed into overlapping patches and vectorized will represent the columns of a matrix X.
Let X=Dα describe the equation relating a sample training matrix (X) to the dictionary (D) and its corresponding coefficient matrix (α). Each patch of image m0, defined as a square subimage of size , is vectorized and placed as a column in the training matrix X while each column in matrix α is the corresponding coefficient of columns of X. The columns of D and α are learned simultaneously from X and the goal of DL is to find the sparsest representation of α (most zero elements of α) such that X=Dα; while m0 is the initial reconstruction of the undersampled data using ordered subset expectation maximization (OSEM with 2 iterations and 21 subsets). Mathematically, learning the dictionary and the corresponding coefficients can be represented as:
| (1) |
which means we want to express the entire image as a linear transform of Dα̲j in such a way that the corresponding coefficient α̲j is sparse and the highest number of nonzero elements is L. P is the number of elements in the vectorized image. The typical approach to solve this constrained optimization is as follows:
| (2) |
where λ is a regularization constant that balances the reconstruction noise level (first term) and the sparsity level of the image (the second term) to be recovered.
The optimization problem in Equation 2 is hard to solve in one step, so we estimate the solution with two optimization steps:
| (3) |
| (4) |
In other words, the algorithm alternates between finding the sparse coefficients when the dictionary (D) is fixed (equation 3) and then finding the best dictionary when the coefficients (α) are fixed (equation 4). To accomplish this two step optimization, we use the singular value decomposition (K-SVD) algorithm (Aharon et al 2006) initialized with left singular vector values for faster performance to find D and then use the Orthogonal Matching pursuit (OMP) algorithm (Tropp et al 2004) to derive the sparse coefficients (α) of each image patch. Once the coefficients are found, each column of the dictionary is updated for only those data that lead to nonzero coefficients. This makes the learning process faster and the next update either sparser or maintain the same sparsity level.
2.1.2 Image recovery and reconstruction
Following the determination of the dictionary and its associated coefficients, the next step is the image recovery and reconstruction process.
Let m be the 2-dimensional PET image that we want to reconstruct; for the fully sampled measurements we have y = Gm in which y is the measurement in sinogram space and G is the system matrix. For the undersampled case, yu =Gum where yu is the (undersampled) measurements and Gu is the modified undersampled system matrix. In this section we make use of the dictionary that is learned from the image patches from the previous section.
Given an undersampled vectorized sinogram yu, the data recovery and image reconstruction comes from the following optimization:
| (5) |
where Rij is the matrix that extracts the patches from the image with the top left corner of the patch located at (i,j), m is the reconstructed image, and D and α are the previously described dictionary and its corresponding coefficients respectively.
The first term of equation 5 enforces the sparsity of image p atches, while the second term (fidelity term) ensures that the solution (reconstructed image m) when multiplied by the system matrix matches the acquired measurement. The parameter μ is a regularization term that affects the weighting of the sparsity factor and data fidelity.
In order to solve Equation 5, the problem is again split into two steps, as discussed in the previous section:
| (6) |
| (7) |
Equation 6 is the same as equation 1 and is discussed in the previous section for determining D and α. Given the D and α, the objective now is to reconstruct the image (equation 7).
Equation 7 is a least-squares type problem. Since , equation (7) can be written as:
| (8) |
The solution can be found by setting the derivative of the cost function equal to zero.
| (9) |
which is further simplified to:
| (10) |
The first term in Equation 10 is of size 16384×16384 (corresponding to a typical image size of 128×128), so it is impractical to invert the matrix due to its large size.
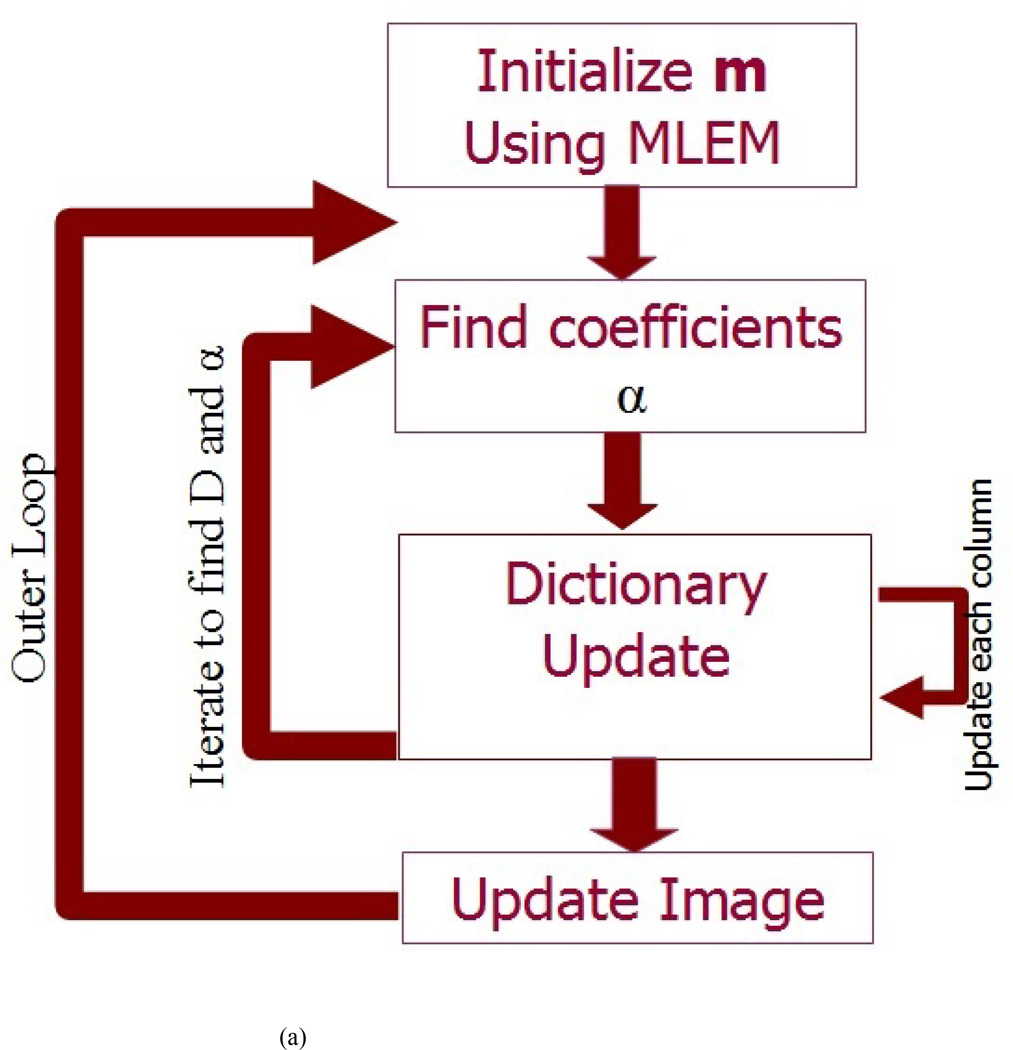
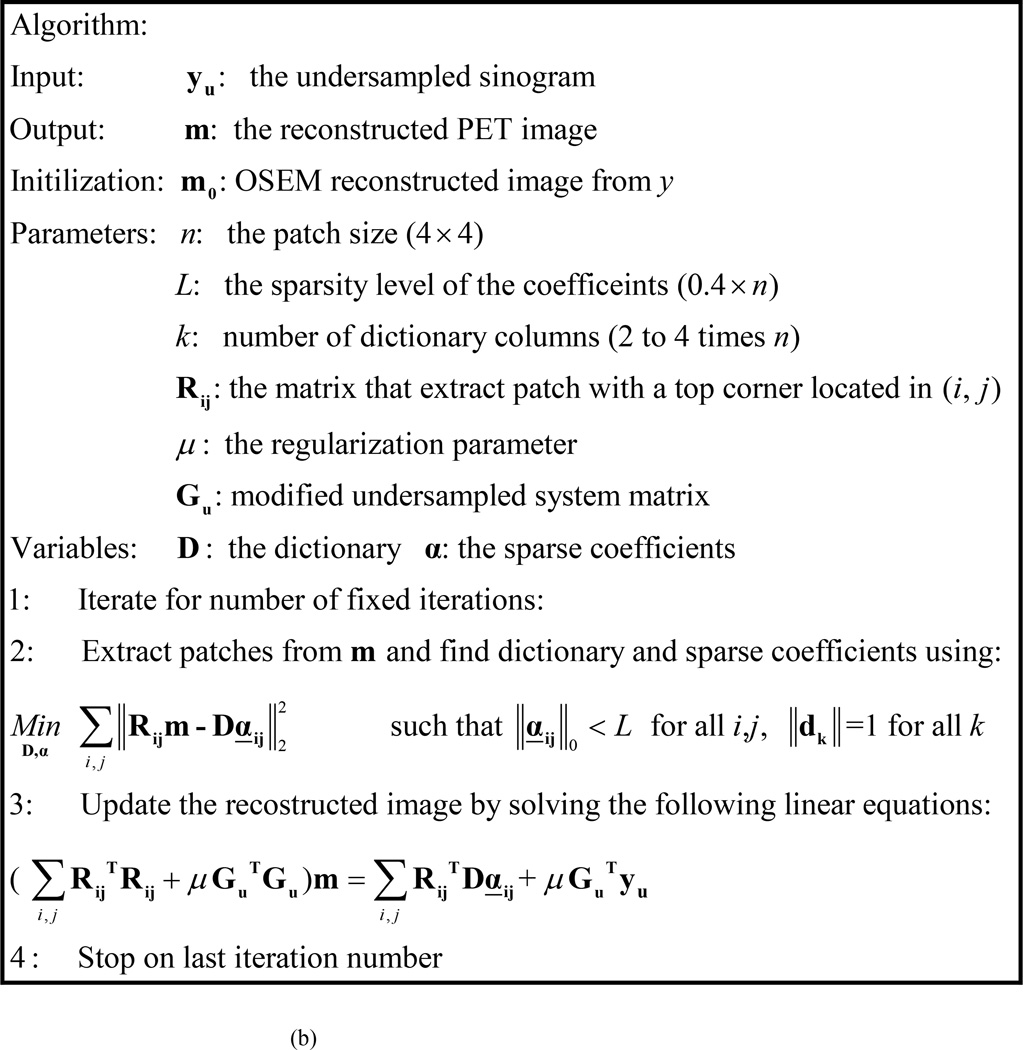
However, since the first term is a diagonal matrix and the second term GuTGu is a matrix that has nonzero elements only along the diagonal and its vicinity, we can solve equation 10 as a set of linear equations for the nonzero indices only. The solution of Equation 10, provides an updated reconstruction for image (m). The updated image is then used as a new image to extract patches, and an updated dictionary (D) and coefficients (α) are learned using Equation 6. This process is repeated for a total of 15 iterations in our implementation. A flow chart of the algorithm is shown in Figure1a. The summary of the algorithm is shown in Figure1b.
FIG. 1.
(A) A Flow chart of proposed DL PET data recovery (B) The proposed DL algorithm for PET data recovery
In general, the number of iterations required for the algorithm to converge varies depending on the complexity of the image and noise. To achieve convergence, we usually evaluate whether a change between the image and its update is below a preset threshold (which in this case is calculated based on . However, for practical purposes we also set a maximum number of iterations as a stopping criterion as well. This is useful where the optimization does not reach the desired convergence criteria (preset threshold) quickly. Our choice of using the partially sampled image as the initial image helped achieve convergence since the partially sampled image is believed to be in the local neighborhood of the true solution.
The recovery algorithm was written in Matlab ver. 7.9 (R2009b) and applied in 2D for all experiments - phantom and patient studies.
2.2. Experimental Setup
Two studies were performed to assess the performance of the proposed algorithm. A phantom study and 5 patient studies. All data recovery was performed in 2D.
2.2.1 Phantom study
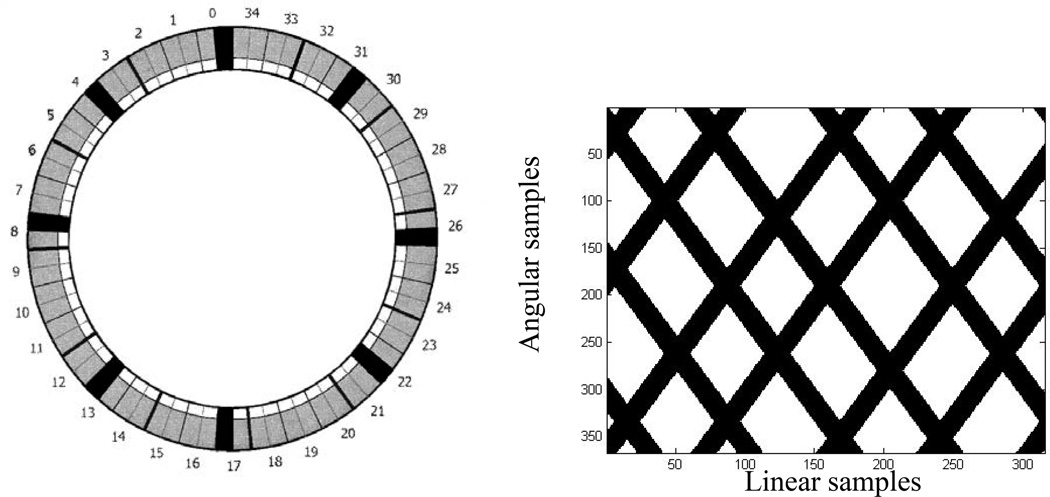
For the phantom evaluation, we used an International Electrotechnical Commission (IEC) image-quality phantom consisting of a lung insert filled with Styrofoam beads with no radioactivity and six internal spheres that were filled with Fluorine-18 water with a sphere-to-background ratio of 10:1. The spheres had an activity concentration of 37 kBq/cc (1 µCi/cc), and the background had an activity concentration of 3.7 kBq/cc (0.1 µCi/cc). The phantom was imaged twice for 3 min on a GE DRX PET/CT scanner: once with all detectors operational (baseline), and once with four detector blocks (11%) turned off at 0, 90, 180, and 270 degrees corresponding to the first block in module numbers 0, 4, 8, 13, 17, 22, 26, and 31 as shown in Figure 2. To turn off detector blocks, the corresponding photomultiplier tube (PMT) gains were set to zero prior to data acquisition. The GE DRX PET/CT system design and its performance characteristics can be found in Kemp et. al. (Kemp et al 2006). Figure 2B shows the mask corresponding to detector removal in the sinogram space.
FIG. 2.
(A) The PET scanner configuration. showing 35 detector modules (2 Blocks/module). The first block in modules 0, 4, 8, 13, 17, 22, 26, 31 was turned off (Black) to simulate missing (removed) detectors. (B) The corresponding mask in the sinogram space; black corresponds to the removed detectors in the sinogram.
The undersampled sinograms were used to extract the mask that captures the exact geometrical effect of detector removal in the sinogram space. The fully sampled sinograms were then reconstructed using OSEM (2 iterations, 21 subsets), with all data corrections (random, scatter, attenuation, normalization, decay, etc.), performed within the reconstruction loop to maintain the integrity of the Poisson statistics. These images were then forward- projected and masked. Although this approach does not truly simulate the imaging conditions of a scanner with detector gaps (see Discussion), it eliminates the need to apply data correction (for attenuation, scatter, random, decay, etc.) because the resulting sinograms were generated from images that had included these corrections. The corresponding partially sampled sinograms were then reconstructed using OSEM with 2 iterations and 21 subsets and an undersampled system matrix. The resultant image then served as the initialization point of the proposed DL algorithm as well as the partially sampled image with no data recovery.
The reconstructed images for both the DL and the partially sampled image were then compared with the fully sampled image (no gaps) by evaluating the root-mean-square-error (RMSE) in regions of interest (ROIs) between corresponding images for each of the six spheres, lung insert, and background. Background in this case was defined as the whole area of the phantom excluding the six spheres and lung insert. RMSE was calculated as follows:
| (13) |
In addition difference images between the recovered (using DL) and the fully sampled images were generated to identify any reconstruction irregularities. A plot showing the convergence of the cost function value versus iteration number was also generated to illustrate the convergence behavior of the algorithm. Furthermore RMSE, and contrast recovery (CR) of the 6 spheres and lung insert were calculated for each reconstruction and compared to the fully sampled image. SNR and CR for hot (spheres) and cold (lung insert) areas were defined as (NEMA 2001):
| (14) |
and
| (15) |
Where H stands for the mean of the measured sphere activity, B is the mean of the measured background, C stands for the mean of the measured lung insert activity, and true refers to the known values in the spheres and the background. In both cases, the mean was based on manual drawing of ROIs around the spheres. The standard deviation in the SNR calculation was based on 5 ROIs drawn in the background.
In this experiment, the nominal values of the algorithm parameters were Niter=15 (outer loop iteration number), k=32 (number of dictionary columns), patch size=4×4, L=6 (sparsity of the coefficients), K-SVD-iteration=30 which was initialized with the left singular vector values, and OMP with a threshold set to 0.02.
2.2.2 Patient study
Five patients (3 men and 2 women; mean age ± SD, 60 ± 15 years) referred for PET/CT imaging of various lesions were also evaluated to test the performance of the proposed DL approach. Following 4 hours of fasting, four of the five patients were intravenously injected with 296–444 MBq (8–12 mCi) of FDG while one patient was injected with 18-NaF with 317MBq (8.57 mCi). No specific criteria were used to select the patients for this study other than no adverse imaging artifacts during the image acquisition process. Images of NaF and FDG were selected to show the applicability of the proposed approach over different PET images. In all patients, imaging started 60–90 min after injection. The imaging process consisted of a scout followed by a whole-body CT scan and a whole-body PET scan covering the area from the patients’ orbits to thighs for the FDG patients and head to toe for the NaF study. The duration of the PET scan was 3 min per bed position. During the imaging process, the patients were requested to breathe regularly. The acquired sinograms were then reconstructed in a similar manner to the phantom data and the resultant images Linear samples (baseline, partially sampled and DL recovered) were compared by evaluating the RMSE in manual ROIs drawn around patient lesions on the corresponding images. Furthermore SNR and CR of the ROIs were calculated for each reconstruction and compared to the fully sampled image. In addition difference images between the recovered (using DL) and the fully sampled images were generated to identify any reconstruction irregularities and a plot showing the convergence of the cost function value versus iteration number was also generated to illustrate the convergence behavior of the algorithm. The DL recovery algorithm parameters for the patients studies were similar to those of the phantom study.
2.2.3 Algorithm Parameter assessment
We also evaluated the impact of four different parameters on the outcome of our proposed reconstruction algorithm using two of our patient studies (P1 and P4). The parameters evaluated were: 1) the noise level (γ), 2) percentage of detector removed, 3) patch size (n), and 4) the sparsity of coefficient elements (L).
We evaluated the effect of noise by adding ten different Poisson noise levels (γ =0, 1, 2, …, 10) to the undersampled patients’ study (P1 and P4) sinogram and reconstructed the resulting data using the DL algorithm. In order to simulate the noise, an image was forward projected to generate its corresponding sinogram (this represented a sinogram with γ =0). The “poissrnd” matlab command was then applied to this sinogram on pixel by pixel to generate a noisy sinogram equal to γ =1. We then divided the sinogram by 2 and repeated the process to generate noisy sinograms (γ =2). This process was repeated from γ =2 to γ =10. The corresponding images were then reconstructed and compared to the original fully sampled image divided by γ, by calculating the RMSE between the original and DL recovered image for increasing noise levels and the results were plotted.
For percent of detectors removed, we forward-projected the same fully sampled patient images (P1 and P4), and then reconstructed the resulting sinograms using DL with increasing undersampling ratios ranging from 5 to 55% detector removal. All other parameters were kept constant. In each case, the undersampling was done by removing detectors in an equidistant manner along the circumference of the scanner. The resultant images were then compared to the original fully sampled image by calculating the RMSE between the original and recovered image. A plot of RMSE recovery for increasing detector removal was generated. The procedure was repeated for different noise levels (γ =0, 1, 2, 3, 4, 5, 10).
To evaluate the effect of patch size effect on the proposed algorithm, we tested patch sizes ranging from 2× 2 to 10×10 while keeping all other parameters constant (inherent noise γ =0, 11% detector removal) and then reconstructed the resulting data using DL. The RMSE between the resultant images and original fully sampled image was plotted for increasing patch size.
Finally, the effect of sparsity of the coefficients level (L) on the proposed algorithm was tested by varying L from 10% to 90% of the patch size. In this case, a patch size of 4×4 was used with inherent noise and 11% detector removal. Here, also the RMSE between the resultant images and original fully sampled image was plotted for increasing sparsity of the coefficients level.
3. Results
3.1 Phantom Study
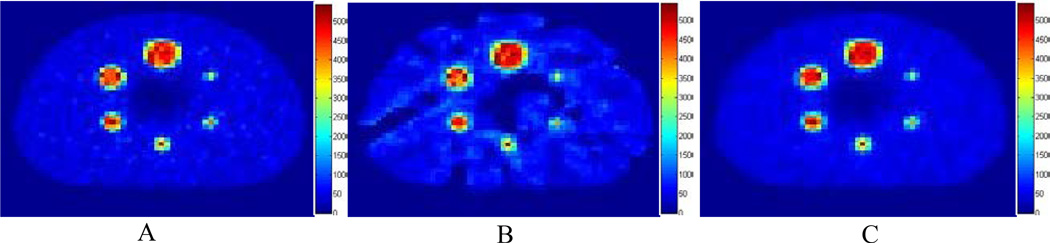
Figure 3 shows the results of the phantom study. The partially sampled image exhibits streak artifacts owing to the missing observations (samples), whereas the DL image shows decreased streak artifacts. The RMSE between activity concentration values of the spheres among the fully sampled, partially sampled, and recovered images for the DL approach are shown in Table 1. The Table also shows the CR and SNR for these cases. The results in Table 1 as well as Figure 3 show that the DL approach has better overall performance (RMSE, CR, SNR) compared to the partially sampled image. All images are displayed using the same scale.
FIG. 3.
The phantom study comparing 2 methods of recovering partially sampled PET data. (A) The baseline image, (B) the partially sampled image, and (C) the DL-recovered image.
Table 1.
RMSE, SNR and CR of the phantom study: C1 (largest sphere) t o C6 (smallest sphere).
| Baseline | Partially sampled | DL | ||||||
|---|---|---|---|---|---|---|---|---|
| SNR | CR | RMSE% | SNR | CR | RMSE% | SNR | CR | |
| C1 | 25.8 | 1 | 9.4 | 23.9 | 0.86 | 3.7 | 25.3 | 0.97 |
| C2 | 25.4 | 0.96 | 12.3 | 22.9 | 0.86 | 5.6 | 24.5 | 0.96 |
| C3 | 24.8 | 0.94 | 11.5 | 22.6 | 0.83 | 1.5 | 23.8 | 0.95 |
| C4 | 21.2 | 0.71 | 13.6 | 18.4 | 0.73 | 7.0 | 17.8 | 0.81 |
| C5 | 14.0 | 0.67 | 23.1 | 10.6 | 0.56 | 1.3 | 10.5 | 0.61 |
| C6 | 7.9 | 0.49 | 21.2 | 3.2 | 0.10 | 4.8 | 7.4 | 0.19 |
| Bkg | 31.6 | 17.1 | ||||||
| Lung insert | 0.73 | 0.62 | 0.70 | |||||
Abbreviations: RMSE, root-mean-square error; Bkg, background; TV, total variation; DL, dictionary learning
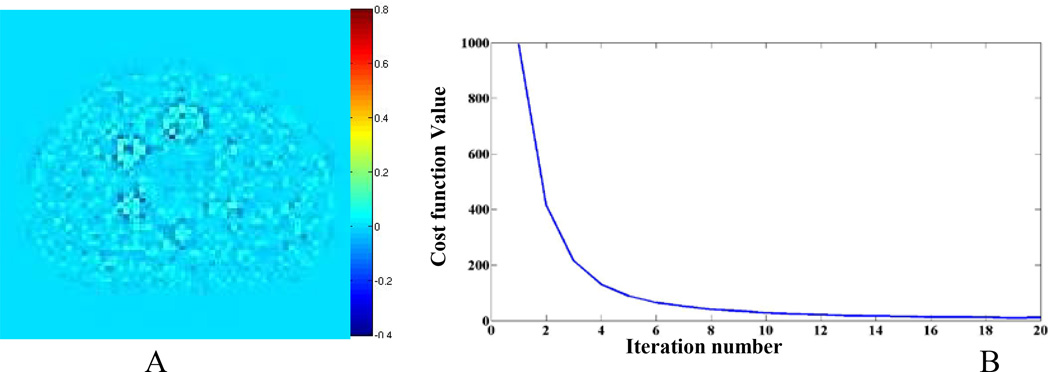
Figure 4A shows a difference image between the DL-recovered image and the fully sampled image. The image shows there is little difference between the recovered DL and fully sampled images. Figure 4B shows the convergence of the cost function versus iteration numbers. The plot indicates that with 15 iterations, the algorithm reaches convergence.
FIG. 4.
Phantom results, (A) The difference between the baseline image and the DL-recovered image (B) the cost function versus iteration number for the DL recovery algorithm
3.2 Patient Study
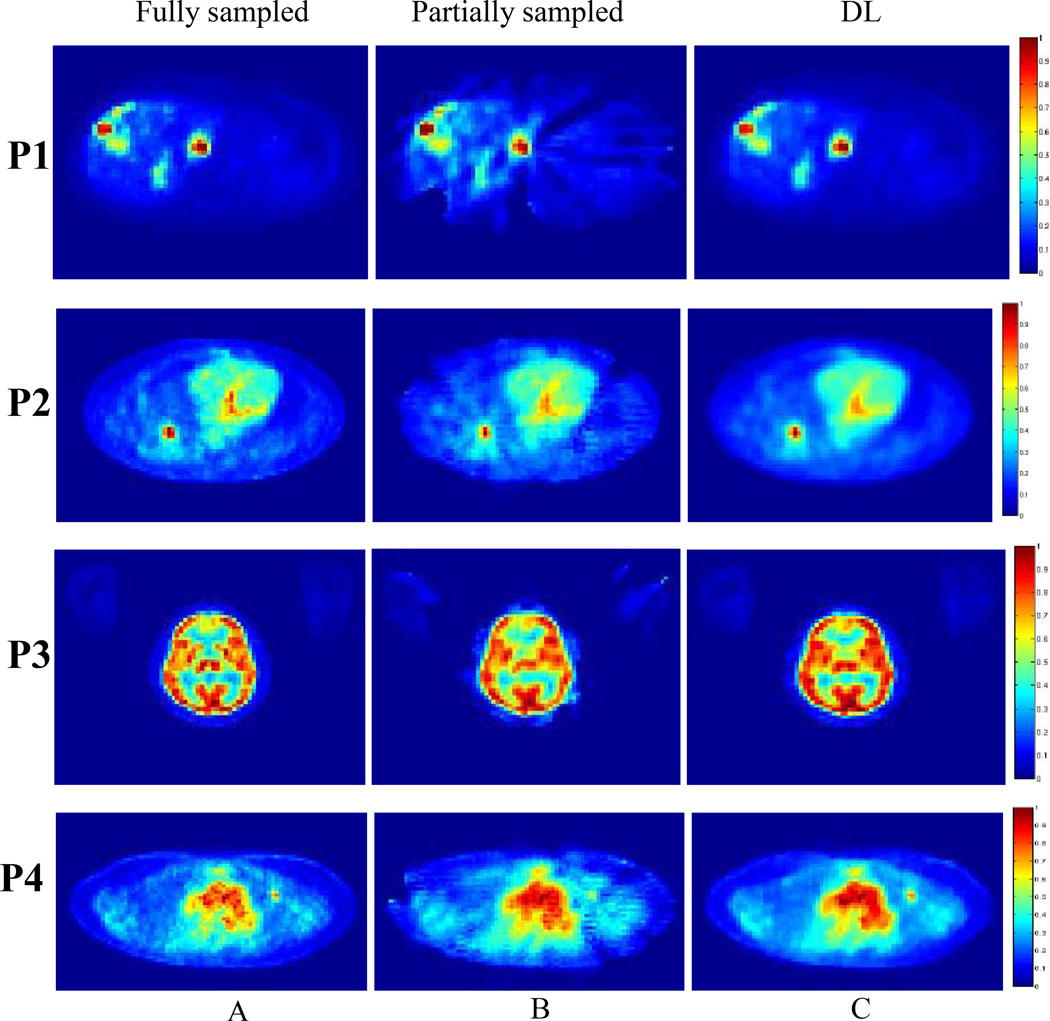
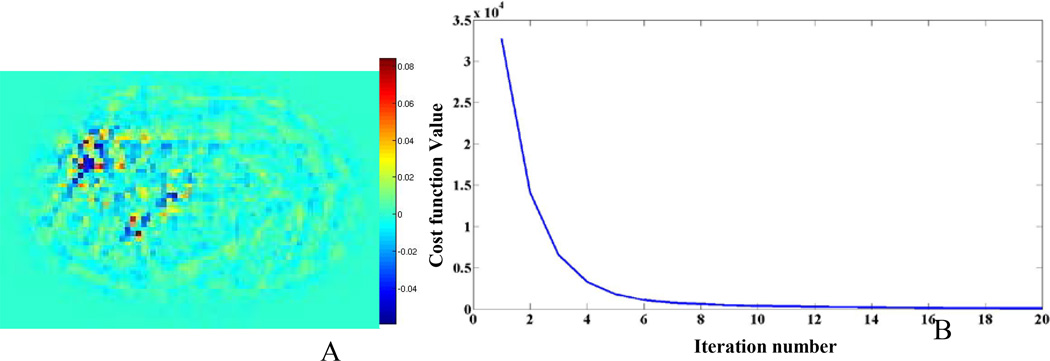
Figure 5 shows various axial images from 4 different patient studies (P1–P4) using the fully sampled, partially sampled, and DL-recovered images. The RMSE, SNR, CR for all lesions in the 5 patient studies are shown in Table 2. The results in this table indicate that the DL approach (Fig. 5A) has better overall performance than the partially sampled image (Fig. 5B). Furthermore, the SNR and CR results of the DL approach are very similar to baseline. Figure 6A shows a difference image between the DL-recovered and fully sampled images of one of the patients (P1). The difference image clearly shows that the DL approach produces an image that is very similar to the fully sampled image. Figure 6B shows the convergence of the cost function versus number of iterations for patient (P1). The plot indicates that with 15 iterations, the algorithm has convergence.
Table 2.
The RMSE, SNR, and CR of 10 lesions from five patient studies
| Patient # |
Tumor # |
Baseline SNR |
Partially sampled | DL | ||||
|---|---|---|---|---|---|---|---|---|
| RMSE% | SNR | CR | RMSE% | SNR | CR | |||
| 1 | 1 | 7.6 | 8.4 | 5.8 | 0.67 | 2.9 | 6.4 | 0.78 |
| 2 | 7.2 | 7.6 | 6.7 | 0.80 | 4.4 | 6.7 | 0.89 | |
| 2 | 3 | 11.6 | 12.9 | 9.4 | 0.70 | 2.8 | 9.7 | 0.74 |
| 4 | 9.5 | 15.3 | 7.0 | 0.66 | 3.3 | 8.3 | 0.83 | |
| 3 | 5 | 10.0 | 7.5 | 9.3 | 0.89 | 2.9 | 9.7 | 0.95 |
| 6 | 9.5 | 15.1 | 6.1 | 0.53 | 5.2 | 7.2 | 0.68 | |
| 4 | 7 | 5.5 | 7.2 | 5.2 | 0.90 | 3.9 | 5.3 | 0.92 |
| 8 | 10.5 | 15.6 | 8.6 | 0.78 | 5.4 | 10.0 | 0.94 | |
| 5 | 9 | 9.4 | 16.1 | 7.1 | 0.68 | 4.1 | 8.3 | 0.84 |
| 10 | 5.9 | 6.9 | 5.6 | 0.80 | 3.1 | 5.7 | 0.90 | |
Abbreviations: RMSE, root-mean-square error; Bkg, background; TV, total variation; DL, dictionary learning
FIG. 5.
Images from four patient studies showing (A) The baseline image, (B) the partially sampled image, and (C) the DL-recovered image. All the images are displayed with the same scale.
FIG. 6.
Results from patient (P1) (A) The difference between the baseline image and the DL721 recovered image; (B) the cost function versus iteration number for the DL recovery algorithm
3.3 Algorithm parameter assessment
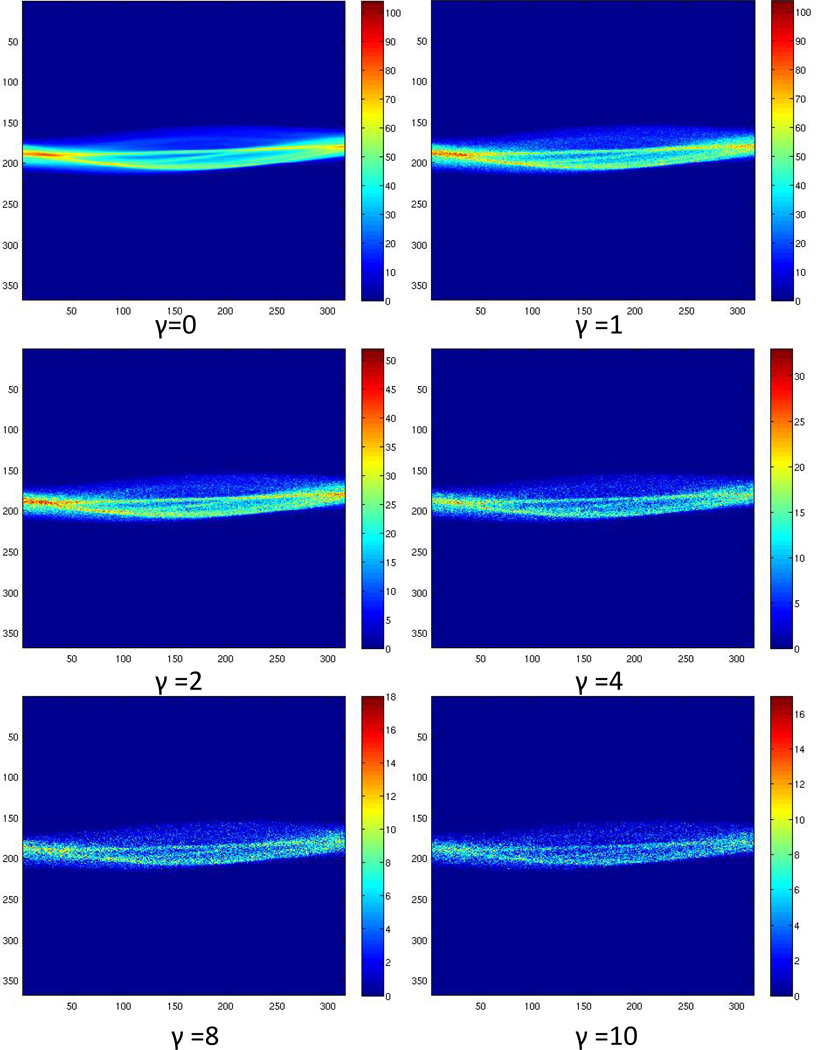
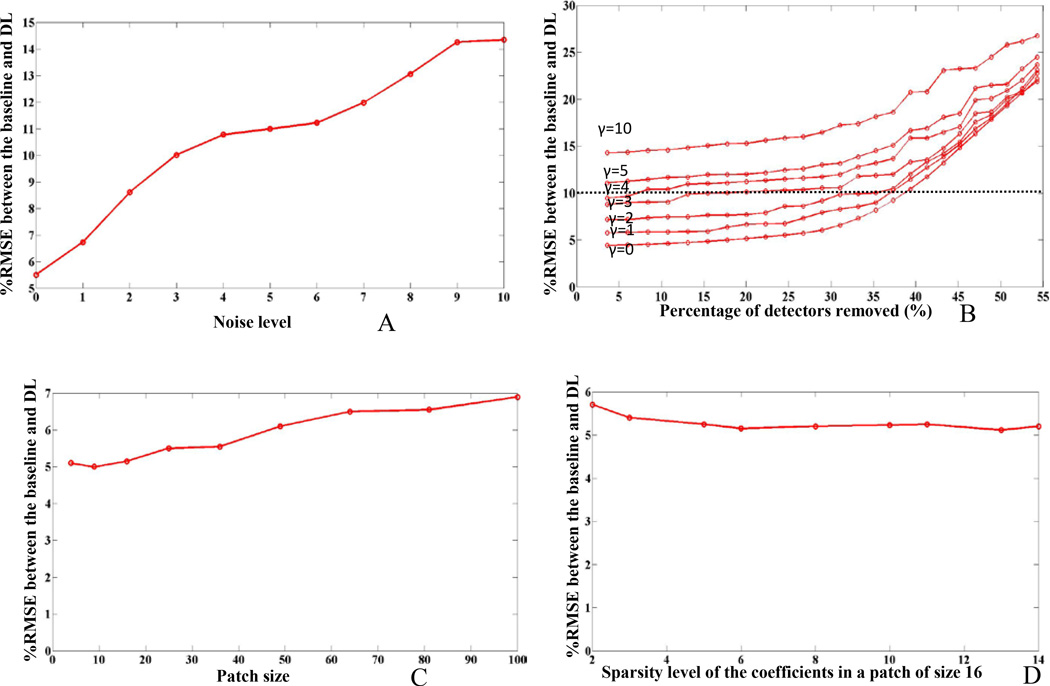
The algorithm parameter assessment was performed on two patient studies (P1 and P4). The results showed similar trends; however, data from only one patient study (P4) are shown. Figure 7 shows the noisy sinograms for γ =0, 1, 2, 4, 8, 10 levels of Poisson noise. Figure 8A shows a plot of %RMSE between the DL and baseline image for increasing noise levels.
FIG. 7.
Example of sinograms with varying Poisson noise levels (γ) 0, 1, 2, 4, 8, 10
FIG. 8.
Algorithm parameter assessment for patient study (P1). % RMSE between the recovered image and the baseline image with varying: (A) Poisson noise level, (B) the percent detectors removed, (C) the patch size (γ=0), and (D) the sparsity of coefficients (γ=0).
The results show that the proposed algorithm recovers undersampled images with RMSE of 5% to 15% with increasing noise levels. The result of DL performance with increasing number of detectors removed is shown in figure 8B. The figure shows that the proposed DL approach can restore the image with upto 10% RMSE (horizontal dotted line) with respect to baseline even when 35% of detectors are equidistantly removed and a noise level of γ ≤2.
Figure 8C shows the impact of patch size on DL recovery. Reconstructions for patch sizes of 2× 2 to 10×10 are shown in the figure. The results suggest that the best RMSE correspond to a patch size of 3×3 while increasing the patch size results in an increase in RMSE.
Figure 8D shows the change in RMSE with the sparsity level of the patch of size 16 (4×4). There is a slight decrease in the percent RMSE (image quality improves) when the sparsity level increases from 2 to 6 but remains relatively constant afterwards.
4. Discussion and Conclusions
We developed an adaptive signal recovery model for PET images from partially sampled data based on CS with DL using image patches. The model was evaluated using phantom studies and 5 patient studies. In addition, we also evaluated the impact of four different parameters on the outcome of the proposed algorithm using two of the patient studies. The parameters evaluated were the noise level (γ), percentage of detector removed, patch size (n), and the sparsity of coefficient elements (L). Our results showed that the DL recovery algorithm is robust with regard to the investigated parameters and produces images that are comparable to fully sampled (no detector gaps) data. To the best of our knowledge this work represents the first time that DL has been used for PET image recovery and reconstruction.
Conventional CS reconstruction techniques often use a TV regularization method that produces blocky results in practical applications owing to the piecewise constant image model. In contrast, DL is aimed at learning local patterns and structures on image patches and simultaneously suppressing noise, resulting in reduced artifacts. In addition, DL adapts from the acquired data and thus provides sparser representation and better recovery quality.
Our results in Tables 1 and 2 show that the DL image recovery results in only 5.8% and 3.8% average differences in RMSE when compared with the fully sampled images for phantom and patient studies, respectively. On average the SNR and CR for DL were 10.29 and 0.81 respectively.
The DL algorithm is initiated with the partially sampled reconstructed image using OSEM which is considered a good initial condition since it is believed to be in the local neighborhood of the true solution, and makes the convergence to a global minimum closer to the real solution. In general, the number of iterations required for the algorithm to converge varies depending on the complexity of the image and noise. However, since equation (5) is split into two steps (equation 6 and 7) and in each step, there is a monotonic decrease in the corresponding cost functions (which is nonnegative), we are guaranteed convergence. Nonetheless, for practical purposes, we also set a preset maximum number of iterations (in this case 15) as a stopping criterion as well, to constraint the convergence to a relatively short processing time. An alternative initialization point is a FBP-reconstructed image which is characterized by faster reconstruction, however at the cost of lower image quality compared to OSEM which might impact the final DL image recovery. In this study, we have not investigated the advantages and/or shortcomes of FBP or OSEM as an initial reconstruction step.
The minimization step for image reconstruction (equation 10), requires a least square optimization which was simplified by leveraging the pseudo diagonal nature of T GuTGu to reduce the computational cost. In this regard, the algorithm had an average run time of 2 min/iteration and 30 min in total for one image when implemented in Matlab ver. 7.9 (R2009b), running on a Dual 6c Xeon E5-2667 CPU machine using 2.9 GHz and 64 GB memory with Linux 2.6.32 as an operating system using 15 iterations for image recovery. The algorithm speed however can be further improved by employing fast and more efficient optimization techniques for both learning the dictionary (instead of K-SVD) and finding the sparse coefficients (instead of OMP). Also implementing the algorithm in C++ and GPUs is expected to further decrease the reconstruction times.
The robustness of the proposed algorithm was evaluated by varying several model parameters (noise level (γ), the percentage of detector removed, patch size (n), and the sparsity of coefficient elements (L)). For noise level, the RMSE between the DL recovered images and the baseline is shown in figure 8 for increasing noise levels (γ =0, 1, 2, 3, 4, 5, 10). The reason the recovery works well with DL even in the presence of high noise content is that DL locally sifts noise while learning the structures of small subimages. In addition, the resultant image pixel values are derived from the average of all the overlapping patches that correspond to that pixel thereby further suppressing the noise content. This process is also applicable even when the noise is nonstationary owing to the local patch-based nature of recovery (Aharon et al 2006).
For percentage of detectors removed, our results (Figure 8b) suggest that even with 35% of detectors removed, the DL recovered image had only 10% error with γ ≤2. Further removal of detectors, up to 50% shows 20% in RMSE with γ ≤3 between the DL recovered and the baseline images with γ =0. This is mainly due to the dictionary’s learning ability to enforce sparsity owing to the inherent representation of the data. For comparison, the partially sampled OSEM showed a 10% RMSE when 35% of the detectors were removed only under noiseless conditions (γ =0); moreover, for 50% detector removal, the partially sampled OSEM resulted in 27 and 41% RMSE when γ =0 and 10 respectively (partially sampled OSEM data not shown).
Our patch size evaluation suggested that the best performance was achieved when the size ranged between 2×2 and 4×4 primarily because a large patch size reduces the ability to learn localized details. However, in general the optimal patch size might vary with different images depending on their complexity. Furthermore, computational complexity increases with increasing the patch size due to the dependence of the majority of the other parameters on this metric. The patch size in our experiments was 4×4 while the size of the dictionary was 16×32. In this regard, the number of rows in the dictionary was equal to the patch size, while the number of columns was 2 to 4 times larger than the number of rows. In this work we did not study the effect of varying the number of dictionary columns as an independent parameter on the performance of the proposed DL algorithm.
Finally, the sparsity of coefficient elements in each patch showed that a value of 6 or larger, results in the smallest RMSE between the DL recovered image and baseline. However, since the larger this value is the longer the recovery will be, we set the number to the smallest value in this range (L=6). However as can be seen in Figure 8d, changing the sparsity level has relatively little impact on the RMSE results. Furthermore, the evaluation did not include an evaluation of the effects of using a different recovery algorithm for the initial condition, nor studied the effect of varying the number of dictionary columns, on the performance of the algorithm
The collective effect of these findings suggests that the algorithm is robust to all its internal parameters, since the RMSE between the recovered image and baseline is rather small (<10%). However, it is important to note that the evaluation of the proposed algorithm parameters was done in only two patient studies and hence further verification is needed before a conclusive assessment can be made.
The parameter μ in Equation 5, and 10 requires careful weighting between the data fidelity and the DL terms such that each term is well represented in the final results. The data fidelity errors are mainly determined by the noise levels of the scan, whereas the optimal DL term values depend on the spatial variation of the true object. In our implementation we empirically set this parameter to 200/γ however a generalized cross-validation method can be used to determine its optimal value (Golub et al 1979). It is important to note that the parameter μ is inversely proportional to the noise level (γ); therefore in a noiseless scenario, it will have a very large number thereby making the results more dependent on the term corresponding to the fidelity term. In noisy situations on the other hand, the μ value balances the effects of , respectively.
We recovered the missing PET data by first reconstructing the images with no gap, forward-projecting these images with the system matrix, and then applying a mask corresponding to preset detector voids/gaps. In doing so, the resultant sinograms included all data corrections, albeit with detector gaps. Although this approach does not represent an actual measured sinogram with missing data, we believe that our approach does not affect the objective of our investigation since it affects the evaluation in a similar manner when DL-recovered or OSEM-recovered images are compared with the fully sampled data. In practice, to correct the sinogram that is obtained from the scanner with the missing samples, correction factors for attenuation, decay, random, scatter, and normalization should also be included as inputs to our recovery algorithm.
Owing to computational complexity, the DL framework in this investigation was developed in two dimensions (2D), and the reconstruction was performed slice by slice. Each slice was recovered independently with no information obtained from adjacent slices. Since the input to the algorithm was a 2D-corrected sinogram, the signal recovery and image reconstruction performed in this evaluation was conducted in 2D mode as well. Extending the approach to 3D is straightforward but requires more computational time.
One limitation of this work is that we only evaluated partial detector removal along the cardinal axes of one axial slice for the phantom and patient studies. An optimal detector removal pattern depends on many factors including individual patient geometry, individual patient uptake distribution, scanner geometry, the sparsifying domain, and measurement space. Future work will investigate optimal detector removal configurations for site-specific scanning.
Another limitation of this work is the use of a Gaussian noise model for the fidelity term rather than a Poisson noise model which more adequately describes the acquired data. Our choice was primarily to simplify the solution of equation 7 (quadratic term) which was presented in equation 10. A more accurate noise modeling would incorporate Poisson or weighted least square type fidelity term (Bouman et al) which was not studied in this work.
Decreasing the number of detectors per ring while employing CS recovery techniques can be beneficial in several ways. This technique could lower the cost of the scanner because the scintillator and the associated photomultiplier tube are among the most expensive components of PET scanners. Furthermore, less expensive scanners with fewer detectors would make such systems more accessible to patients and physicians. Another potential advantage of the CS techniques is to decrease the scan time by increasing scanner sensitivity through exchanging the removed detectors between the transverse FOV and the axial FOV (adding gapped PET detector rings in the axial direction). Increasing the axial FOV, increases the scanner sensitivity as well as the extent of body coverage, both of which lead to shorter scan times while maintaining image quality. Furthermore, decreasing the scan time could increase patient throughput and decrease the likelihood of patient movement during the scan. Finally, increasing scanner sensitivity could also be traded for lower injected activity and hence lower patient radiation dose, while maintaining similar image quality.
In addition to cost reduction and/or improving scanner performance, our proposed approach might also have potential applications in open PET geometries, such as some designs used in Positron Emission Mammography (Shkumat 2011), in-beam PET for in-vivo dosimery in hadron therapy (Cabello et al 2013, Vecchio 2009) or when novel PET prototypes or demonstrators are developed and consist of few detector heads (Sakellios 2006, Yamaya 2008). These systems however, have large detector gaps which might not be adequately recovered using our proposed approach. In this regard, an in depth investigation about the value of our proposed approach to such systems is warranted.
In conclusion, CS techniques using DL is a promising approach to recover relatively accurate activity concentrations even in the presence of partially sampled data.
Acknowledgments
The authors thank Jill Delsigne and Diane Hackett in the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center. This research was supported in part by the National Institutes of Health through the MD Anderson Cancer Center Support Grant (CA016672).
References
- Aharon M, Elad M, Bruckstein A. K-SVD: An algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans. in Signal Process. 2006;54(11):4311–4322. [Google Scholar]
- Ahn S, Kim S, Son J, Lee D, Lee J. Gap compensation during PET image reconstruction by constrained, total variation minimization. Med Phys. 2012;39:589–602. doi: 10.1118/1.3673775. [DOI] [PubMed] [Google Scholar]
- Bouman C, Sauer k. A unified approach to statistical tomography using coordinate descent optimization. IEEE Transactions on Image Processing. 1996;5(3):480–492. doi: 10.1109/83.491321. [DOI] [PubMed] [Google Scholar]
- Cabello J, Torres-Espallardo I, Gillam J, Rafecas M. PET reconstruction from truncated projections using total-variation regularization for hadron therapy monitoring. IEEE Trans Nucl Sci. 2013;60:3364–3372. [Google Scholar]
- Chen Y, Ye X, Huang F. A novel method and fast algorithm for MR image reconstruction with significantly under-sampled data. Inverse Problems Imaging. 2010;4:223–240. [Google Scholar]
- Chen YL, Hsu C. Time-variant modeling for general surface appearance. 18th IEEE International Conference on Image Processing (ICIP; Brussels. 2011. pp. 1077–1080. [Google Scholar]
- De Jong H, Boellaard R, Knoess C, Lenox M, Michel C, Casey M, Lammertsma A. Correction methods for missing data in sinograms of the HRRT PET scanner. IEEE Trans. Nucl. Sci. 2003;50:1452–1456. [Google Scholar]
- Ding X, Paisley J, Huang Y, Chen X, Huang F, Zhang X. Compressed Sensing MRI With Bayesian Dictionary Learning; 20th IEEE International Conference on Image Processing (ICIP); 2013. pp. 2319–2323. [Google Scholar]
- Donoho D. Compressed sensing. IEEE Trans. Info. Theory. 2006;52(4):1289–1306. [Google Scholar]
- Efros A, Leung T. Texture synthesis by non-parametric sampling. The Proceedings of the Seventh IEEE International Conference on Computer Vision; Kerkyra. 1999. pp. 1033–1038. [Google Scholar]
- Golub G, Heath M, Wahba G. Generalized cross-validation as a method for choosing a good ridge parameter. Technometrics. 1979;21:215–223. [Google Scholar]
- Huang Y, Paisley J, Lin Q, Ding X, Fu X, Zhang X. Bayesian Nonparametric Dictionary Learning for Compressed Sensing MRI. ArXiv. 2013 doi: 10.1109/TIP.2014.2360122. e-prints 1302.2712. [DOI] [PubMed] [Google Scholar]
- Karp J, Muehllehner G, Lewitt R. Constrained Fourier space method for compensation of missing data in emission computed tomography. IEEE Trans. Med. Imaging. 1988;7:21–25. doi: 10.1109/42.3925. [DOI] [PubMed] [Google Scholar]
- Kemp B, Kim C, Williams J, Ganin A, Lowe V. NEMA NU 2-2001 performance measurements of an LYSO-based PET CT system in 2D and 3D acquisition modes. J. Nucl. Med. 2006;47:1960–1967. [PubMed] [Google Scholar]
- Kinahan P, Fessler J, Karp J. Statistical image reconstruction in PET with compensation for missing data. IEEE Trans. Nucl. Sci. 1997;44:1552–1557. [Google Scholar]
- Li S, Yin H, Fang L. Group-sparse representation with dictionary learning for medical image denoising and fusion. IEEE Trans. Biomed. Eng. 2012;59(12):3450–3459. doi: 10.1109/TBME.2012.2217493. [DOI] [PubMed] [Google Scholar]
- Ma D, Gulani V, Seiberlich N, Liu K, Sunshine J, Duerk J, Griswold M. Magnetic resonance fingerprinting. Nature. 2013;145:187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoff D, Krohn K. PET imaging of response and resistance to cancer therapy. In: Teicher, editor. Cancer Drug Resistance. Chapter 5B. Humana Press; 2006. pp. 105–122. [Google Scholar]
- National Electrical Manufacturers Association (NEMA) NEMA Standards Publication NU 2-2001. Rosslyn, VA: National Electrical Manufacturers Association; 2001. Performance Measurements of Positron Emission Tomographs. [Google Scholar]
- Nguyen VG, Lee SJ. Image reconstruction from limited-view projections by convex nonquadratic spline regularization. Opt. Eng. 2010;49(3):37001–37001. [Google Scholar]
- Otazo R, Sodickson DK. Adaptive compressed sensing MRI. Proceedings of the 18th Scientific Meeting of ISMRM; Stockholm. 2010. pp. 4867–4867. [Google Scholar]
- Pan X, Sidky E, Vannier M. Why do commercial CT scanners still employ traditional, filtered back-projection for image reconstruction? Inverse Probl. 2009;25:1–36. doi: 10.1088/0266-5611/25/12/123009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheja A, Doniere TF, Dhawan AP. Multiresolution Expectation Maximization Reconstruction Algorithm For Positron Emission Tomography Using Wavelet Processing. IEEE Trans Nuclear Science. 1999;46:594–602. [Google Scholar]
- Ritschl L, Bergner F, Fleischmann C, Kachelrieß M. Improved total variation-based CT image reconstruction applied to clinical data. Physics in medicine and biology. 2011;56(6):1545–1561. doi: 10.1088/0031-9155/56/6/003. [DOI] [PubMed] [Google Scholar]
- Saif MW, Tzannou I, Makrilia, Makrilia N, Syrigos K. Role and cost effectiveness of PET/CT in management of patients with cancer. Yale J Biol Med. 2010;83(2):53–65. [PMC free article] [PubMed] [Google Scholar]
- Saiprasad R, Bresler Y. MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Transactions on Medical Imaging. 2011;30(5):1028–1041. doi: 10.1109/TMI.2010.2090538. [DOI] [PubMed] [Google Scholar]
- Sakellios N, Rubio J, Karakatsanis N, Kontaxakis G, Loudos G, Santos A, Nikita K, Majewski S. GATE simulations for small animal SPECT/PET using voxelized phantoms and rotating-head detectors; Nuclear Science Symposium Conference Record; 2006. pp. 2000–2003. [Google Scholar]
- Shkumat N, Springer A, Walker C, Rohren E, Yang W, Adrada B, Arribas E, Carkaci S, Chuang H, Santiago L, Mawlawi O. Investigating the limit of detectability of a positron emission mammography device: a phantom study. Medical physics. 2011;38(9):5176–5185. doi: 10.1118/1.3627149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidky E, Pan X. Image reconstruction in circular cone-beam computed tomography by constrained, total-variation minimization. Phys. Med. Biol. 2008;53:4777–4807. doi: 10.1088/0031-9155/53/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosic I, Jovanovic I, Frossard P, Vetterli M, Duric N. Ultrasound tomography with learned dictionaries; IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP); 2010. pp. 5502–5505. [Google Scholar]
- Tropp J, Gilbert A. Signal recovery from random measurements via orthogonal matching pursuit. IEEE Trans on Information Theory. 2007;53(12):4655–4666. [Google Scholar]
- Tuna U, Peltonen S, Ruotsalainen U. Gap-filling for the highresolution PET sinograms with a dedicated DCT-domain filter. IEEE Trans. Med. Imaging. 2010;29:830–839. doi: 10.1109/TMI.2009.2037957. [DOI] [PubMed] [Google Scholar]
- Valiollahzadeh SM, Chang T, Clark J, Mawlawi O. Image recovery in PET scanners with partial detector rings using compressive sensing; Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC); 2012. pp. 3036–3039. [Google Scholar]
- Valiollahzadeh SM, Clark J, Mawlawi O. Dictionary Learning in Compressed Sensing Using Undersampled Data in PET Imaging. Medical Physics. 2013;40(6):400–401. [Google Scholar]
- Valiollahzadeh SM, Clark J, Mawlawi O. Using compressive sensing to recover images from positron emission tomography (PET) scanners with partial detector rings. Medical Physics. 2015;42(1):121–133. doi: 10.1118/1.4903291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio S, Attanasi F, Belcari N, Camarda M, Cirrone G, Cuttone G, Rosa F. A PET prototype for “in-beam” monitoring of proton therapy. IEEE Transactions on Nuclear Science. 2009;56(1):51–56. [Google Scholar]
- Xu Q, Yu H, Mou X, Zhang L, Hsieh J, Wang G. Low-dose X-ray CT reconstruction via dictionary learning. IEEE Transactions on Medical Imaging. 2012;31(9):1682–1697. doi: 10.1109/TMI.2012.2195669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya T, Inaniwa T, Minohara S, Yoshida E, Inadama N, Nishikido F, Shibuya K, Lam C, Murayama H. A proposal of an open PET geometry. Physics in medicine and biology. 2008;53(3):757–773. doi: 10.1088/0031-9155/53/3/015. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhao J, Wang G. Few-view image reconstruction with dual dictionaries. Physics in medicine and biology. 2012;57(1):173–189. doi: 10.1088/0031-9155/57/1/173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu X. Image Interpolation by Adaptive 2-D Autoregressive Modeling and Soft-Decision Estimation. IEEE Trans. Image Process. 2008;17:887–896. doi: 10.1109/TIP.2008.924279. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ding H, Lu Y, Wang G, Zhao J, Molloi S. Dual-dictionary learning-based iterative image reconstruction for spectral computed tomography application. Phys Med Biol. 2012;57(24):8217–8229. doi: 10.1088/0031-9155/57/24/8217. [DOI] [PMC free article] [PubMed] [Google Scholar]