Abstract
Purpose
A distinct subset of genes, so-called “late fiber genes,” is expressed in cells bordering the central, organelle-free zone (OFZ) of the lens. The purpose of this study was to identify additional members of this group.
Methods
Fiber cells were harvested from various layers of the lens by laser micro-dissection and subjected to microarray, in situ hybridization, and Western blot analysis.
Results
Expression of Livin, a member of the inhibitor of apoptosis protein (IAP) family encoded by Birc7, was strongly upregulated in deep cortical fiber cells. The depth-dependent distribution of Livin mRNA was confirmed by quantitative PCR and in situ hybridization. The onset of Livin expression coincided with loss of organelles from primary fiber cells. Livin expression peaked at 1 month but was sustained even in aged lenses. Antibodies raised against mouse Livin labeled multiple bands on immunoblots, reflecting progressive proteolysis of the parent molecule during differentiation. Mice harboring a floxed Birc7 allele were generated and used to conditionally delete Birc7 in lens. Lenses from knockout mice grew normally and retained their transparency, suggesting that Livin does not have an indispensable role in fiber cell differentiation.
Conclusions
Birc7 is a late fiber gene of the mouse lens. In tumor cells, Livin acts as an antiapoptotic protein, but its function in the lens is enigmatic. Livin is a RING domain protein with putative E3 ubiquitin ligase activity. Its expression in cells bordering the OFZ is consistent with a role in organelle degradation, a process in which the ubiquitin proteasome pathway has been implicated previously.
Keywords: fiber cell differentiation, organelle degradation, apoptosis, hypoxia, Livin
Livin, an antiapoptotic protein, is expressed immediately before organelle degradation begins in fiber cells located in the deep lens cortex.
Lens fiber cells are formed continuously by differentiation of cells at the edge of the lens epithelium. Newly formed cells are aligned on top of existing fiber cells, stacked like planks of wood in a lumber yard. The cell stacks (also referred to as radial cell columns1) arise deep in the tissue and extend to the surface of the lens. There is no cell turnover in the fiber cell compartment. Consequently, each stack contains a complete cellular record, with the oldest fibers at the base and the youngest on the top.
All fiber cells are transparent and share an elongated prismatic shape. This has sometimes led to the assumption that they are a homogeneous population. In fact, depending on the relative positions of fibers in the cell stack, differences in biochemical or cell biological properties can be stark.2 For example, the basal tips of superficial fiber cells make direct contact with the posterior lens capsule. In contrast, older cells, located farther down the stack, have detached from the capsule and instead form connections with other fiber cells, in a region called the lens suture. Deep in the stack, fiber cells are deprived of oxygen, due to consumption by overlying cells.3 Consequently, the proportion of cellular ATP derived from oxidative phosphorylation differs markedly between the hypoxic inner cell layers and the relatively oxygenated outer fibers.4 Finally, a few hundred micrometers below the lens surface, fibers undergo programmed organelle destruction, whereby the nucleus and other membranous organelles are rapidly and synchronously degraded.5 The dissolution of the organelles ensures the removal of light-scattering structures and results in the formation of the central, organelle-free zone (OFZ) of the lens.6 Unlike the synthetically active surface cells, fiber cells within the OFZ lack the capacity for de novo protein synthesis.7 Based on such considerations it is likely that the biochemical differences between fibers located at different depths in the cell stack are as profound as those that exist between fiber cells and epithelial cells.
Once fiber cells have begun the process of terminal differentiation, gene expression is adjusted continuously, so that the cell biology of a fiber is appropriate to its position in the stack. Of particular interest are genes expressed in cells bordering the OFZ. This set of “late fiber genes” is likely to include genes associated with the organelle degradation process or stabilizing the fiber cell phenotype. Founder members of this group of late fiber genes include Hopx,8 Capn3,9 Dnase2b (DLAD),10,11 and lengsin (Lgsn).12 Hopx is expressed in fiber cells that have lost their direct connection to the lens capsule.8 In keratinocytes, Hopx is expressed in the suprabasal layer, where it may control expression of differentiation-dependent genes. Capn3 encodes a calcium-activated endopeptidase, involved in remodeling of the spectrin cytoskeleton.13 Dnase2b is a lysosomal nuclease.11 It has been shown to mediate chromatin breakdown during organelle loss and its absence results in cortical cataract due to incomplete DNA removal.10 Lengsin is expressed in cells during the membrane remodeling phase and interacts directly with the intermediate filament proteins Cp49 and vimentin, perhaps facilitating cytoskeletal reorganization.12
To identify other late fiber genes, we used laser microdissection to harvest cells from various layers of the lens for comparative transcriptional analysis. We sought to identify genes that were expressed immediately prior to organelle breakdown. In this report, we characterized the expression of one such transcript, Livin (encoded by Birc7), which was strongly expressed in cells bordering the OFZ. Livin is an antiapoptotic protein (IAP) with suspected roles in progression of melanoma14 and other malignancies.15
Methods
Mice
Wild-type (C57BL/6J) mice were obtained from a commercial supplier (Jackson Lab, Bar Harbor, ME, USA). Mice harboring a floxed Birc7 allele were generated by homologous recombination (Supplementary Fig. S1). Two independent transgenic lines, LeCre16 and MLR10,17 expressing Cre recombinase in lens, were used for tissue-specific inactivation of Birc7. A germline knockout of Birc718 (generated while these studies were in progress) was obtained and included in the analysis. To examine the effect of Birc7 inactivation on lens cell architecture, Birc7-null mice were crossed with mT/mG reporter mice ([B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB–tdTomato,–EGFP)Luo/J]tdTomato). The mT/mG mice (Jackson Lab) express tandem dimer Tomato (tdTomato) in cell membranes throughout the body,19 allowing cell morphology to be imaged in living intact lenses. All procedures were approved by the Washington University Animal Studies Committee and performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Laser Microdissection and Microarray Analysis
Three tissue samples were isolated from 1-month-old lenses: an epithelial sample (EP; obtained by manually dissecting the epithelium from the lens), nucleated outer cortical fiber cells (OC; comprising fiber cells located 0–100 μm beneath the lens surface), and nucleated inner cortical cells (IC; comprising cells located 100–200 μm beneath the surface). The inner cortical cell sample directly abutted the OFZ, the border of which, in 1-month-old mice, is located approximately 200 μm beneath the equatorial lens surface. The samples of OC and IC were obtained by laser microdissection. For that purpose, enucleated globes were embedded in optimal cutting temperature (OCT) medium (TissueTek; Miles, Elkhart, IN, USA) and frozen in 2-methylbutane over dry ice. Sections (20-μm thick) were cut on a cryostat and mounted on glass RNase-free PEN-Membrane Slides (Leica Microsystems, Buffalo Grove, IL, USA). Sections were washed in RNase-free water for 30 seconds, fixed in 70% ethanol for 1 minute, and air dried for 10 minutes. The samples of OC and IC were laser-microdissected (LMD6000, Leica Microsystems) and collected directly into the lysis buffer supplied with the RNA isolation kit (RNeasy Micro Kit; Qiagen, Valencia, CA, USA). We quantified RNA using a commercial assay (Quant-iT RNA; Invitrogen, Grand Island, NY, USA). Synthesis of cDNA was accomplished using an amplification system (NuGEN Ovation Pico WTA System; NuGEN Technologies, Inc., San Carlos, CA, USA). Independent quadruplicate replicates were applied to bead microarrays according to manufacturer instructions (Illumina Mouse 6; Illumina, San Diego, CA, USA). These experiments were modeled on published methodology.20
Endpoint PCR
Two splice variants (α and β) of Livin have been described, differing by a stretch of 18 amino acids located between the BIR and RING domains.21 Transcripts corresponding to the α and β forms were differentiated by endpoint PCR using the following primers:
Forward: 5′ TCCTCTGGGGCTGCCTTCCTT 3′ (349-369)
Reverse: 5′ AGGCACCCTGCAGATAGGGCA 3′ (930-949)
The predicted amplicon included exon 6, which contains the alternatively spliced region.
RT-qPCR
For real-time quantitative PCR (RT-qPCR), cDNA was synthesized with 1-μg lens RNA as a template using a reverse transcription supermix for RT-qPCR (iScript; BioRad, Hercules, CA, USA). The following primers were used:
Gapdh-F 5′ CCTCGTCCCGTAGACAAAATG 3′
Gapdh-R 5′ TGAAGGGGTCGTTGATGGC 3′
Ppia-F 5′ GCGGCAGGTCCATCTACG 3′
Ppia-R 5′ GCCATCCAGCCATTCAGTC 3′
Birc7-F 5′ GGGGAGCCTGCCTTCCCTGA 3′
Birc7-R 5′ AGGGGTCATCCCCACGCTCC 3′
Experiments with RT-qPCR were designed according to the MIQE guidelines,22 using a supermix according to manufacturer instructions (SsoFast EvaGreen; BioRad). Each biological sample was obtained from a pool of five different animals, unless otherwise specified, and three technical replicates were performed per sample. Reference genes (Gapdh and Ppia) were selected using Normfinder23 software and verified by determining the M value. Efficiency of RT-qPCR was determined for all primers and was within 93% to 102% range.
Northern Blot
The size and distribution of Livin mRNA transcripts were evaluated by Northern blotting, as described.7 Total RNA (1 μg) was separated on denaturing gels and transferred to nitrocellulose. Blots were probed with digoxigen-labeled sense and antisense riboprobes (spanning region 579–811 of the Livin transcript) prepared using Dig RNA labeling mix (Roche Diagnostics, Indianapolis, IN, USA).
In Situ Hybridization
In situ hybridization was performed as described.24 Briefly, 4-μm thick, midsagittal sections of paraffin-embedded lens tissue were prepared. A target probe set raised against the Livin transcript was obtained from Advanced Cell Diagnostics (ACD, Hayward, CA, USA). The probe set consisted of 20 pairs of oligonucleotides spanning a ≈1-kb contiguous region of the target mRNA transcript (NM_001163247.1: 2-1141). In situ hybridization was performed using a multiplex assay (RNAscope 2.0; ACD). Following proprietary preamplification and amplification steps, target probes were detected using an alkaline-phosphatase–conjugated label probe with Fast Red as substrate and Gill's hematoxylin as counterstain. As negative controls, some sections were hybridized with target probe against DapB, a bacterial gene. Alternatively, the Livin probe set was hybridized to lens sections from Birc7-null mice.
Antibodies and Immunoblotting
Two independently generated antibodies against mouse Livin were used for immunofluorescence and Western blotting applications. A recombinant protein corresponding to amino acid positions 22 through 285 of mouse Livin and containing an N-terminal histidine tag was generated in Escherichia coli and used to immunize rabbits (PrimmBiotech, West Roxbury, MA, USA). Anti-Livin was purified from the antiserum using CNBr-sepharose affinity column chromatography. The second antibody (clone 7H5.1.1-IgG2a) was raised against mouse Livin and characterized in an earlier study.18 Both antibodies recognized recombinant Livin and endogenous Livin on Western blot and exhibited little or no immunoreactivity on lens samples from Birc7-null mice. Western blotting was performed as described previously.25 Livin antibodies were used at 1:500 dilution. To examine Livin expression in various strata of the tissue, lenses were progressively solubilized and fractions collected for analysis, as described.9
Immunofluorescence
To visualize the expression of Livin protein, P1 mouse lenses were fixed for 2 hours in 4% PBS, washed and sectioned at 150 μm on a tissue slicer (Vibratome series 1000; TPI, Inc., St. Louis, MO, USA). Lens slices were permeabilized in 0.1% Triton X-100/PBS for 30 minutes and blocked with 1% bovine serum albumin/10% normal goat serum in PBS. Slices were incubated overnight in anti-Livin, washed in PBS, and incubated for 2 hours in secondary antibody. Draq5 (Invitrogen) was included in the washing solution as a nuclear counterstain. Slices were coverslipped and examined using a confocal microscope (LSM510, Carl Zeiss, Inc., Thornwood, NY, USA). To visualize the fate of cytoplasmic organelles, paraffin sections were incubated with an antibody against protein disulfide isomerase (PDI; an ER resident protein), as described.26
Multiphoton Imaging of Fiber Cell Structure
Wild-type or Birc7-null mice were crossed with a reporter strain (mT/mG) expressing membrane targeted TdTomato. Lenses were dissected from the eye and incubated in warm artificial aqueous humor solution.3 Image stacks were collected using a multiphoton microscope (Fluoview1000 MPE; Olympus, Center Valley, PA, USA) using an excitation wavelength of 785 nm and an XLPlan N ×25 water immersion objective lens (1.05 NA).
Results
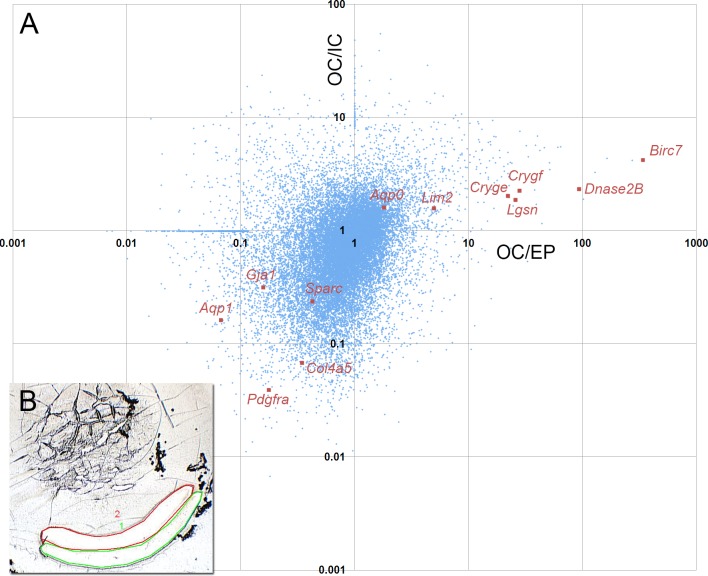
Tissue samples from the lens epithelium (EP), outer cortex (OC), and inner cortex (IC) were harvested manually (EP) or by laser microdissection (OC, IC) and relative levels of gene expression in the three samples were determined by microarray analysis (Fig. 1). On this analysis, mRNA products of known late fiber genes (such as Dnase2B and Lgsn) segregated with transcripts (Cryge and Crygf) characterized by enhanced expression in the OC and IC samples. Surprisingly, the most upregulated of this group was Livin, which was undetectable in the EP sample, but expressed strongly in the OC sample and even more so in the IC sample. Livin (encoded by Birc7) is a member of the IAP family.27 Previous studies have shown Livin to be expressed in certain types of melanoma cells as well as some fetal tissues.28
Figure 1.
Microarray analysis of depth-dependent gene expression in 1-month-old mouse lenses. (A) Scatterplot of microarray data showing relative levels of gene expression in EP cells, OC and IC fiber cells. The quadrants define four distinct patterns of lens gene expression. The lower left quadrant, for example, contains transcripts that are highly expressed in the epithelium (with respect to either of the fiber samples). Epithelial specific transcripts, Aqp1, Pdgfra, Gja1, Sparc, and Col4a5 are located in this quadrant. Transcripts that show upregulated expression in both the early and late stages of fiber differentiation are shown in the upper right quadrant. Members of this group include genes known to be expressed predominantly in fiber cells, such as Aqp0, Lim2, and the gamma crystallins Cryge and Crygf. Among strongly upregulated genes in this quadrant are the late fiber genes Dnase2B and Lgsn. The most upregulated transcript, with expression levels several hundred–fold higher in the inner cortical fibers than in the epithelium, is Livin. The other two quadrants contain transcripts that are transiently upregulated (lower right quadrant) or downregulated (upper left quadrant) during early fiber cell differentiation. (B) The inset shows an example of a frozen lens section from which the OC (green) and IC (red) samples were collected by laser microdissection.
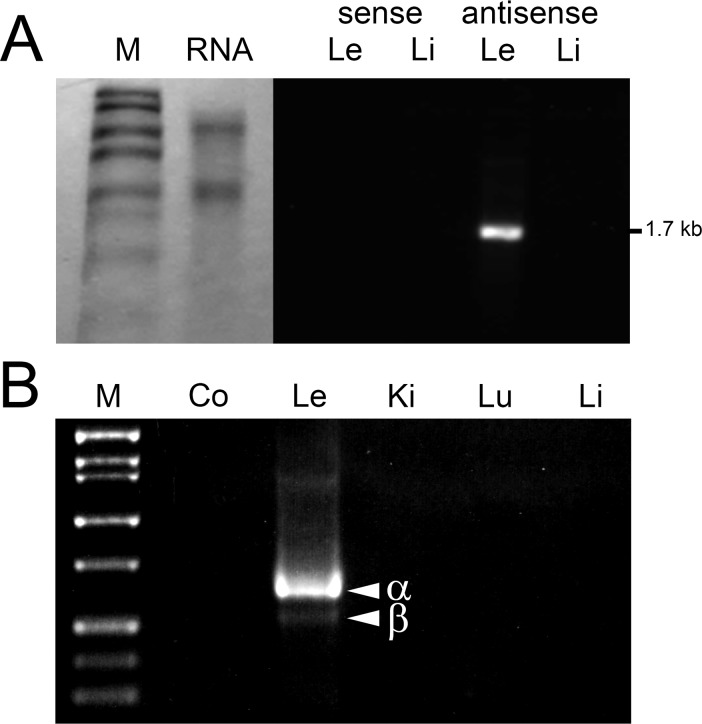
Expression of Livin mRNA in lenses from 1-month-old mice was evaluated by Northern blot (Fig. 2). A prominent 1.7-kb RNA transcript was detected in RNA extracted from lens but not liver (Fig. 2A). Two alternative transcripts (α and β) of Livin have been described21 and, by endpoint PCR, both were detected in lens (Fig. 2B). Livin α, the larger of the transcripts, was the more abundant. Endpoint PCR failed to detect Livin expression in kidney, lung, or liver samples.
Figure 2.
Expression of Livin mRNA in mouse tissues. (A) Northern blot analysis identifies a 1.7-kb transcript in total RNA extracted from lens (Le) but not liver (Li). Sense control probe does not hybridize to RNA from either tissue. (B) Two amplicons (α and β) are detected following endpoint PCR amplification of lens cDNA with Livin-specific primers. The larger and more abundant amplicon corresponds to Livin α and the smaller amplicon to Livin β. The identity of the amplicons was confirmed by sequencing. Co, negative control (lacking template DNA); Ki, kidney; Lu, lung; M, marker.
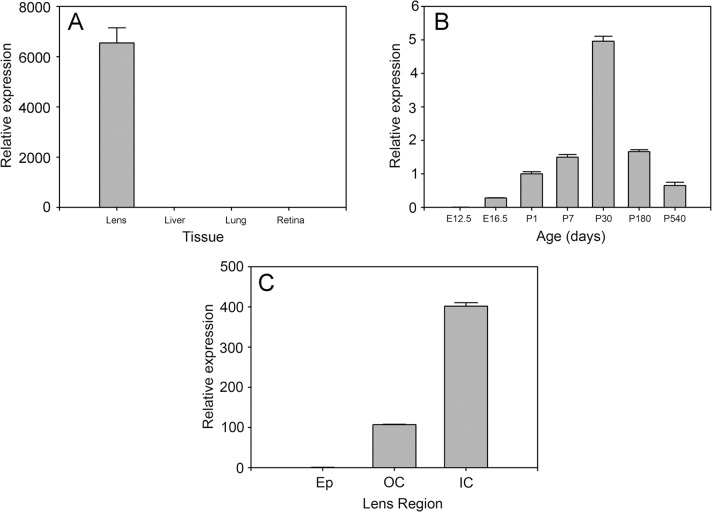
We used RT-qPCR to quantify Livin expression in lens and other tissues (Fig. 3). Livin was strongly expressed in lens but not liver, lung, or retina (Fig. 3A). The mouse lens forms at ≈day 11 of embryonic development (E11), but Livin expression was first detected at E16.5 (Fig. 3B), the stage at which organelle degradation commences in the innermost fiber cells.29 Livin expression increased in the lens during the early postnatal period, peaking in 1-month-old lenses (Fig. 3B). Expression was sustained in the maturing lens and the Livin transcript was readily detected even in lenses from 18-month-old mice (the oldest examined). In laser microdissected samples from 1-month-old lenses, Livin expression was undetectable in epithelial samples (Fig. 3C) and ≈4-fold greater in inner cortical cells than outer cortical cells, substantiating the microarray results (see Fig. 1).
Figure 3.
Quantification by RT-qPCR of Livin expression in mouse tissue. (A) In 1-month-old mice, Livin is strongly expressed in lens compared with other tissues (liver, lung, retina) examined. (B) Livin expression in the lens commences between E12.5 and E16.5. Expression increases throughout the early postnatal period, peaking at postnatal (P) day 30. Although expression levels are somewhat reduced in older animals, Livin is an abundant transcript at all ages. (C) Within the lens, Livin expression is undetectable in the epithelium, and strongest in the inner cortical fiber cells. Data represent mean and standard deviation of at least three determinations in each case.
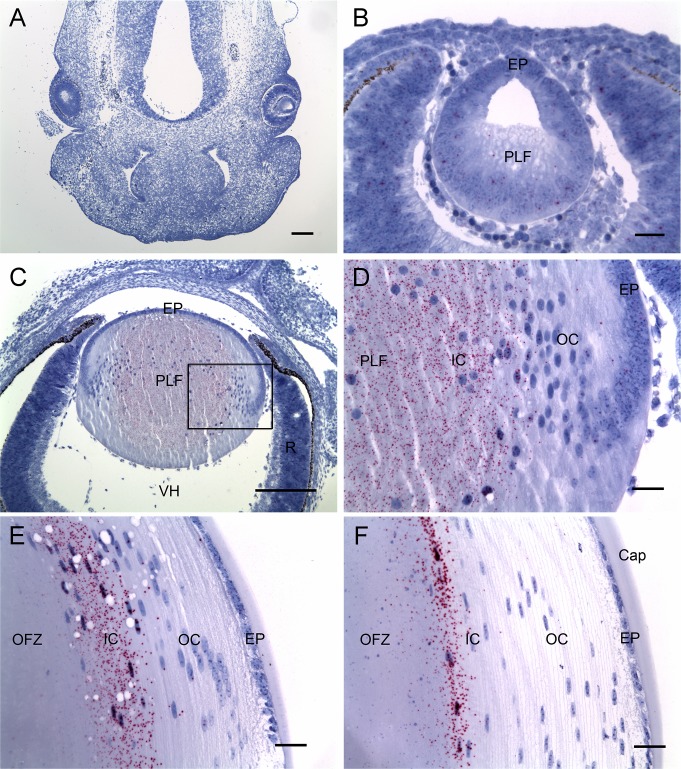
To visualize the expression of Livin mRNA at the single cell level within the developing mouse lens, paraffin sections were hybridized with a target probe set raised against a 1-kb region of the Livin mRNA transcript (Fig. 4). At the first stage examined, E12.5, the lens consisted of a hollow ball of cells (the lens vesicle). The posterior wall of the vesicle comprised the primary lens fibers, which at that stage were in the process of elongating. At E12.5, Livin expression was low throughout the embryonic head, including the developing eye and lens (Figs. 4A, 4B). However, by E16.5, Livin mRNA was readily detected in nucleated primary fiber cells located in the center of the lens (Figs. 4C, 4D). Thus, expression of Livin immediately preceded organelle breakdown, which commences in the central fiber cells during the late embryonic period.29 Elimination of organelles from the innermost fiber cells leads to the formation of the OFZ.5 The process of organelle degradation continues postnatally. At age 1 or 6 months, Livin was expressed by a contingent of cortical fiber located adjacent to the OFZ (Figs. 4E, 4F). Thus, at all ages, Livin was expressed by fiber cells shortly before organelle degradation began. Transcripts were not detected within the anucleated cells of the OFZ nor in the anterior lens epithelium.
Figure 4.
In situ hybridization analysis of Livin mRNA expression in the developing mouse eye. (A) At day 12.5 of embryonic development, Livin expression (red dots) is undetectable in the embryonic head. (B) The lens, at this stage a hollow vesicle, is composed of epithelial cells (EP) and primary lens fibers (PLF) in the process of elongation. Livin expression in the E12.5 lens is barely above background. (C, D) At day 16.5 of embryonic development, Livin is expressed strongly by primary fiber cells located in the center of the lens. The boxed region in (C) is shown at higher magnification in (D). Livin expression is undetectable elsewhere in the eye. (E) At postnatal day 30, Livin expression is restricted to nucleated IC fiber cells bordering the central OFZ. (F) At age 6 months, Livin expression is restricted to a thin layer of fiber cells adjacent to the OFZ. Cap, capsule; R, retina; VH, vitreous humor. Scale bars: (A, C) 200 μm, (B, D–F) 25 μm.
Livin was originally cloned from a melanoma cell line.28 To compare the pattern of Livin expression in lens and melanoma, histopathology samples of human uveal melanoma were examined. Livin mRNA distribution within the tumor was visualized by in situ hybridization, using a probe set raised against human Livin (Fig. 5). Livin was detected in four of six melanoma specimens examined. Significantly, in Livin-positive tumors, expression was restricted to the core of the tumor. Melanoma cells in the superficial region of the tumor (Fig. 5A) or regions adjacent to vascular elements (Fig. 5B) did not express Livin. Thus, expression of Livin in melanoma samples recapitulated its expression pattern in lens, where the Livin transcript was restricted to the inner cell layers.
Figure 5.
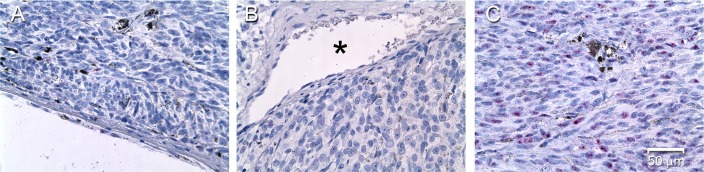
In situ hybridization analysis of Livin expression in human uveal melanoma. Expression is undetectable in the superficial (A) region of the tumor (at the vitreous surface) or around vascular (*) elements (B). Marked Livin expression (red dots) is detected only in the core of the tumor (C). Scale bar: 50 μm.
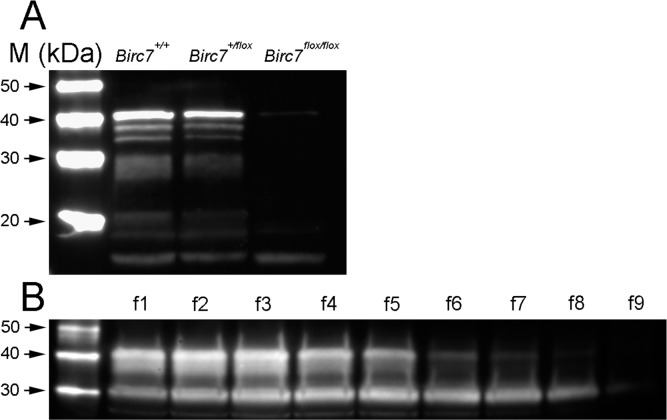
To visualize the expression of Livin protein, antibodies were raised against recombinant mouse Livin and used to probe Western blots of whole lens samples prepared from wild-type or MLR10tg/−;Birc7flox/flox mice (the latter serving as a negative control). Several immunopositive bands were detected in lens samples. The largest and most prominent band had an apparent molecular mass of ≈ 41 kDa (Fig. 6A) and likely corresponds to full-length Livin. However, the presence of additional immunopositive bands with apparent masses of ≈37, ≈34, and ≈22 kDa was noted and a diffuse band of ≈30 kDa was also present. Immunopositive bands were not detected in lens samples prepared from MLR10tg/−;Birc7flox/flox mice, implying that antibody labeling was specific and that the 22- to 41-kDa bands corresponded to authentic Livin splice variants or posttranslationally modified species. To follow the fate of Livin protein during fiber cell differentiation and aging, lenses were progressively solubilized (Fig. 6B). This semiquantitative technique allows fractions to be collected from progressively deeper layers of the lens, although the precise spatial relationships between the fractions are uncertain (it is unlikely, e.g., that fractions are derived from strata of equal thickness). The multiple immunopositive bands observed in whole-lens lysates (Fig. 6A), were evident even in the outermost fractions (containing the youngest fiber cells), suggesting that the lower molecular weight bands were not simply proteolytic fragments generated during cellular aging. However, depth-dependent changes in Livin expression were noted. For example, the putative full length (41 kDa) form was not detected in the deeper fractions and no immunopositive bands were detected in the innermost fiber cells.
Figure 6.
Western blot analysis of Livin protein expression in the mouse lens. (A) Several immunopositive bands are present in lens samples from wild-type mice, or mice heterozygous for the floxed allele. To conditionally delete Birc7 in the lens, Birc7flox/flox animals were crossed with MLR10 mice. No immunopositive bands were detected in the conditional null lenses. (B) Progressive solubilization of lenses allowed the collection of nine fractions, corresponding with various strata of the lens. In the outmost fractions (f1–f5) two immunopositive bands (at ≈40 and ≈30 kDa) are present. The larger band is absent in fractions from the inner fiber cells (f6–f8). Neither band is detected in the innermost fraction (f9).
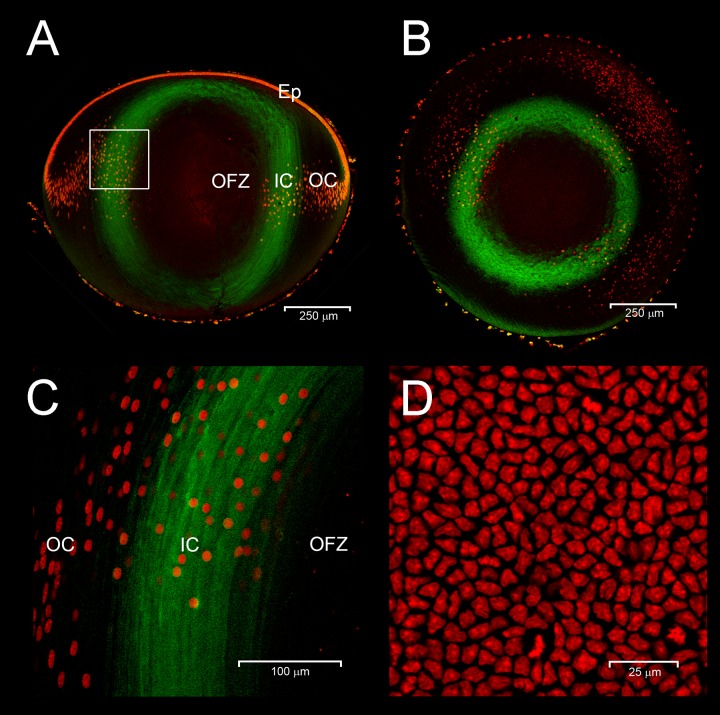
To better localize Livin protein expression, we used confocal immunofluorescence microscopy of vibratome sections prepared from early postnatal lenses (Fig. 7). Analysis of sagittal sections from P1 lenses revealed that Livin expression was first evident in fiber cells that had recently detached from the lens epithelium (Fig. 7A). In equatorial sections, Livin immunofluorescence formed an annular pattern (Fig. 7B). Livin protein was detected in the deeper layers of nucleated IC fiber cells, located just outside the central OFZ (Fig. 7C). In expressing cells, Livin protein was diffusely distributed throughout the cytoplasm (Fig. 7C). Expression was undetectable in the overlying epithelium (Fig. 7D) consistent with the results of RT-qPCR analysis (Fig. 3C).
Figure 7.
Livin expression in P1 mouse lens. Sagittal (A) and equatorial sections (B) show Livin expression (green) in fiber cells in the IC region. The boxed region in (A) is shown at higher magnification in (C). The disappearance of fiber cell nuclei (red) marks the border of the OFZ. Note that Livin is expressed by fiber cells immediately prior to denucleation. In expressing fiber cells, Livin immunofluorescence is distributed throughout the cytoplasm. En face views of the lens epithelium (D) demonstrate the absence of Livin expression in this cell population. Scale bars: (A) 250 μm, (B) 250 μm, (C) 100 μm, (D) 25 μm.
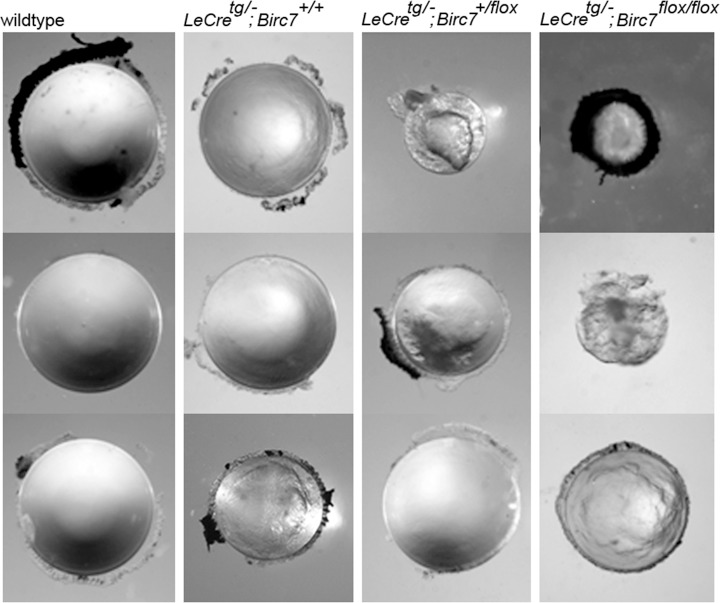
Fiber cells are among the longest lived cells in the body. Based on its expression pattern and putative antiapoptotic role in other systems, we hypothesized that Livin might contribute to lens cell longevity by suppressing cell death in the hypoxic core of the tissue. Alternatively, Livin, which is expressed by cells as they approach the OFZ (Fig. 7C), might have a role in organelle degradation. To test these hypotheses directly, we generated mice carrying a floxed Birc7 allele (see Supplementary Fig. S1), Birc7flox/flox mice were crossed with either LeCre16 or MLR1017 mice to conditionally delete Birc7 in the lens. The LeCre and MLR10 strains express Cre recombinase in lens and have been widely used to conditionally delete genes in this tissue. The two strains differ primarily in the timing of the onset of lens Cre expression (E8.75 for LeCre and E10.5 for MLR10). Since our data suggested that Livin expression does not commence until after E12.5 (Fig. 3B), we anticipated that the efficacy of the two lines (and, therefore, any resulting phenotypes) should be broadly similar. Unexpectedly, there was a striking qualitative difference in the lens phenotypes between LeCretg/−;Birc7flox/flox and MLR10tg/−;Birc7flox/flox animals. The eyes of LeCretg/−;Birc7flox/flox mice were invariably (16/16) smaller than wild type (Fig. 8). Histologic analysis revealed that the microphthalmic phenotype was associated with the presence of a small and severely disrupted lens (Supplementary Fig. S2). Fiber cells in the LeCretg/−;Birc7flox/flox mice were heavily vacuolated and cell death assays revealed the presence of numerous apoptotic cells in the central lens fibers, as early as E16 (data not shown). These data were consistent with the hypothesis that Livin expression actively suppressed cell death in the hypoxic lens interior. The majority (12/16 or 75%) of lenses from mice heterozygous for the floxed allele exhibited a similar lens phenotype. Surprisingly, however, analysis of lenses from LeCretg/−;Birc7+/+ littermate controls revealed a relatively high incidence (12/32 or 37%) of microphthalmia. In affected animals, the lens phenotype was often indistinguishable from that observed in the conditional knockouts. Thus, at least on the mixed genetic background used here, expression of the Cre transgene in the LeCre strain produced a phenotype independent of the presence of the floxed Birc7 allele. In contrast, no overt lens phenotype was observed in MLR10tg/−;Birc7flox/flox animals (Fig. 9). We confirmed that Birc7 was efficiently disrupted (as evidenced by absence of Livin protein) in both LeCretg/−;Birc7flox/flox and MLR10tg/−;Birc7flox/flox mice (Supplementary Fig. S3).
Figure 8.
Conditional deletion of Livin in the lens using the LeCre transgene results in small and disrupted lenses. Compared with wild-type lenses from 1-month old animals (left column), lenses from mice homozygous for the floxed Birc7 allele (right column) were severely disrupted, with 100% penetrance. However, a similar lens phenotype was also observed in 37% of control (LeCretg/−;Birc7+/+) littermates. A phenotype was observed in 75% of lenses from mice that were heterozygous for the floxed allele and hemizygous for the LeCre transgene.
Figure 9.
Lens clarity and refractive properties are unaffected in Livin knockout lenses. We used MLR10 to conditionally delete Birc7 in the lens. Lenses from 1-month-old wild-type or conditional knockout animals were photographed against a grid pattern. The optical properties of control animals (wild-type or mice hemizygous for the MLR10 transgene) did not differ noticeably from mice in which the Birc7 locus was partially (Birc7+/flox) or completely (Birc7flox/flox) inactivated.
We examined lenses from MLR10tg/−;Birc7flox/flox mice for the presence of cataracts or growth defects. Histological analysis indicated that the lens cellular architecture was indistinguishable from wild type (Supplementary Fig. S4). Lenses retained their clarity at all ages examined (up to 12 months; data not shown) and careful measurements of lens size failed to detect any significant growth delays (Fig. 10). To substantiate findings obtained with the MLR10 mice, we also examined lenses from germline Birc7-null mice18 that became available in the course of our studies. Lenses from Birc7-null animals were indistinguishable from MLR10tg/−;Birc7flox/flox lenses (data not shown).
Figure 10.
Lens growth is not affected by the absence of Livin. No significant differences in the equatorial diameter of the lens were noted among the various genotypes. Data represent mean values ± SD (n = 4 in each case).
In wild-type lenses, the observation that Livin was expressed in cells near the border of the OFZ (Fig. 7), suggested a potential role for Livin in organelle degradation. To test this hypothesis, lenses from MLR10tg/−;Birc7flox/flox mice and control littermates were processed for immunofluorescence and labeled with antibodies against PDI, a marker for the endoplasmic reticulum (ER). Previous studies have shown that the ER is rapidly degraded at the edge of the OFZ.26 The disappearance of PDI staining at the border of the wild-type OFZ was confirmed in the current study. The OFZ also formed normally in MLR10tg/−;Birc7flox/flox mice (Fig. 11), indicating that Livin is not necessary for the removal of the endoplasmic reticulum (nor the fiber cell nuclei; see Supplementary Fig. S4).
Figure 11.
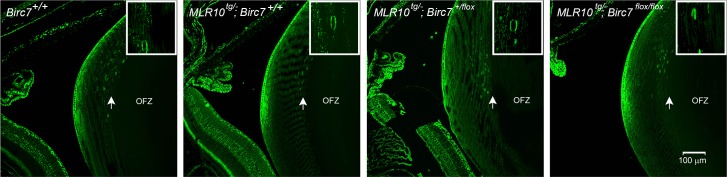
Degradation of the endoplasmic reticulum occurs normally in the absence of Livin. The distribution and fate of the endoplasmic reticulum was visualized with anti-PDI. In wild-type lenses, PDI immunofluorescence is strong in the lens epithelium and outer cortical fibers. The abrupt disappearance of PDI staining marks the boundary of the OFZ. The loss of PDI immunoreactivity in cells bordering the OFZ was indistinguishable in wild-type or conditional knockout lenses.
Previous studies have shown that the lens can be surprisingly resilient to the inactivation of even relatively abundant genes. Although deletion of membrane30 or cytoskeletal proteins31 does not necessarily result in loss of transparency it often disrupts the organization of the fiber cell stacks. To examine whether the absence of Livin affected the cytoarchitecture of the lens, we used a novel multiphoton imaging technique. For this application, we utilized Birc7-null mice,18 which were crossed with the mT/mG reporter strain. In mT/mG mice, membrane-targeted TdTomato is expressed in cells throughout the body, including the lens. Endogenous expression of TdTomato in the plasma membranes of all fiber cells allowed fiber morphology to be imaged in three dimensions in intact, living lenses. Using a long working distance water immersion lens, lens fiber cells were visualized at depths of up to 500 μm below the equatorial lens surface. This allowed cells to be inspected in the EP, OC, IC, and OFZ regions (i.e., before, during, and after the period of Livin expression in wild-type lenses). Image stacks collected from 1-month-old wild-type or Birc7-knockout lenses were indistinguishable (Fig. 12). In each case, fiber cells were organized into radial cell columns and the cells had characteristic shapes that varied with depth, as described previously.6 Thus, the absence of Livin was not associated with a gross disturbance in the cellular organization of cortical lens fiber cells.
Figure 12.
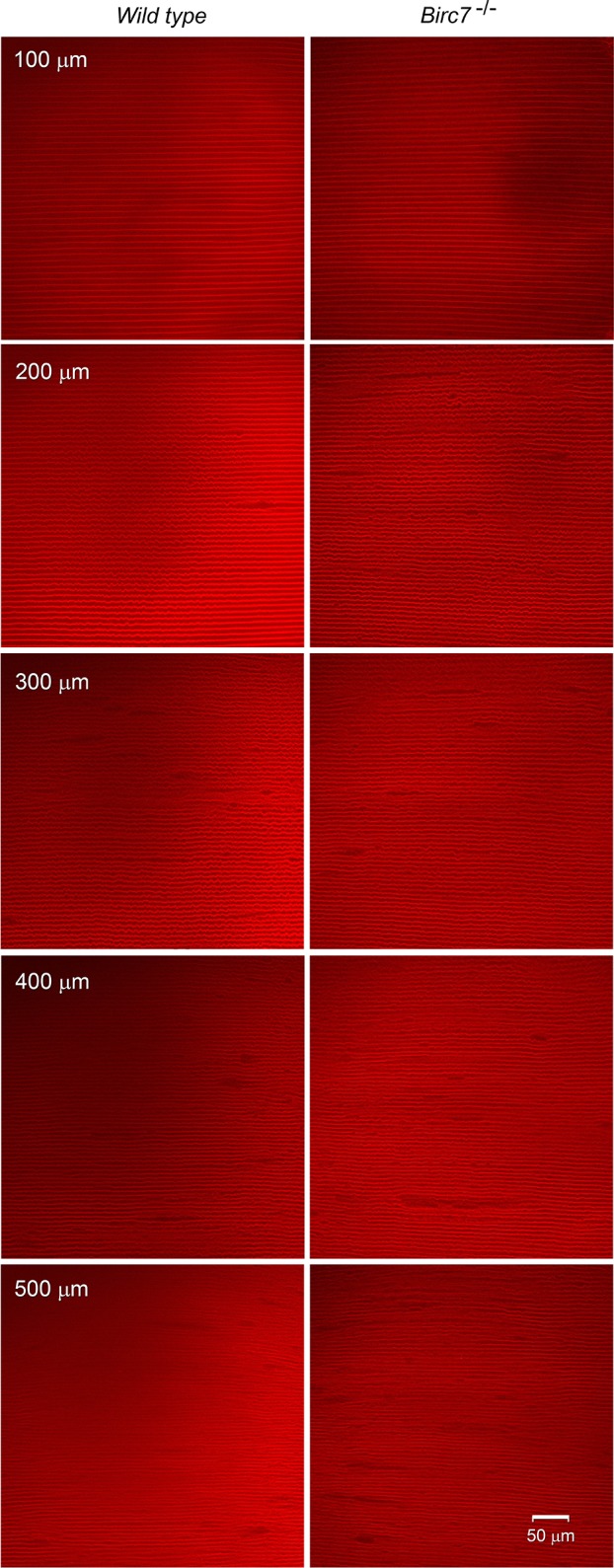
Multiphoton microscopic analysis of cellular organization in lenses from wild-type or Birc7-null animals. Crosses were established with mT/mG mice (a line in which the fiber cell membranes are endogenously labeled with TdTomato) to allow the morphology of individual cells located at various depths below the lens surface to be visualized. Individual optical sections are shown. Note that the border of the OFZ is located approximately 200 μm below the lens equatorial surface. There was no obvious difference in cell morphology or packing arrangement on either side of the OFZ border.
Discussion
Microarray analysis of laser microdissected lens tissue was used to screen for genes expressed late in the fiber cell differentiation program. A similar strategy was employed previously by Ivanov et al.,20 who identified several genes with modulated expression in mature fibers. In the current work, we confirmed the expression of previously identified late fiber genes, including lgsn and Dnase2B. However, in our study, the most upregulated transcript in inner cortical fibers was Livin, encoded by the Birc7 gene. Livin expression has been noted previously in human lens samples, as part of the expressed sequence tag NEI bank project,32 and in a recent RNA-Seq study,33 but a detailed analysis of its expression and function in the lens has not been reported.
Livin is a member of the inhibitor of apoptosis family of proteins. These proteins are characterized by the presence of one or more BIR motifs (baculovirus inhibitor of apoptosis protein repeat) that interact directly with caspases to suppress apoptotic cell death. Livin contains a single BIR domain and a RING-domain with putative E3 ubiquitin ligase activity. Livin was first isolated from malignant melanoma cells and its expression level in most nontransformed cell types is believed to be very low.34 Its antiapoptotic action is believed to be mediated via inhibition of caspase-3, -7, and -934 and by its ability to promote the degradation of Smac/Diablo, an endogenous regulator of the IAP family.35
Two splice variants (α and β) of Livin have been described.21 In the lens, the α variant dominates. In apoptotic cells, Livin is cleaved by caspases, generating a truncated version (tLivin) that redistributes to the Golgi apparatus.36 Paradoxically, under some circumstances tLivin may promote cell death.37 In the lens, Livin was evenly distributed throughout the cytoplasm. Several Livin species were observed by immunoblotting. Presumably the multiple bands represented full length and truncated α and β variants. Immunopositive bands were not detected in lens samples from MLR10tg/−;Birc7flox/flox mice, indicating that the wild-type labeling pattern represents authentic Livin immunoreactivity rather than nonspecific antibody binding. The identity of the various species and their functional significance, if any, has yet to be determined. Lens cells are retained throughout life and by isolating successive layers of tissue, it was possible to follow the fate of Livin protein during the fiber differentiation process. Such analysis indicated that, over time, full-length Livin disappeared and that the inner cortical fiber cells contained only the 30-kDa isoform. The progressive cleavage of lens proteins is a well-established phenomenon. For example, Aqp0, the most abundant integral membrane protein of the lens is extensively cleaved in the lens core38 and the cytoskeletal protein alpha-spectrin, is cleaved to a 150-kDa fragment in the inner cortical region.39 The protease(s) responsible for degrading full-length Livin in differentiating lens fiber cells is unknown but several proteases are activated in cells bordering the OFZ, including the calcium-activated protease, calpain 3.9
Livin expression began in primary lens fiber cells sometime between E12.5 and E16.5. It is noteworthy that elimination of cytoplasmic organelles in the innermost fiber cells begins in the period E15 to E18.29 The strict spatial/temporal relationship between organelle loss and Livin expression was maintained in the secondary fiber cells of postnatal lenses, where Livin expression in the inner cortex immediately preceded organelle degradation. Although Livin expression is not essential for formation of the OFZ, it is possible that Livin has a role in organelle degradation. The ubiquitin proteasome pathway has been implicated in organelle degradation during formation of the OFZ.40,41 In the ubiquitin proteasome pathway, protein substrates are recognized by E3 ubiquitin ligases which mediate the transfer of one or more ubiquitin molecules to the substrate, thereby targeting it for degradation via the proteasome. Although the human genome encodes >600 RING domain-containing E3 ligases,42 the identities of the E3 ligases operating during organelle degradation have yet to be determined. The specific expression of Livin (a putative ubiquitin E3 ligase) in cells bordering the OFZ suggests that it may function in the process.
Livin was expressed specifically by the innermost nucleated fiber cells. In human uveal melanoma samples, the expression of Livin followed a broadly similar pattern, with low-level expression in the outer tissue layers and enhanced expression in the inner layers. Recent studies suggest that Livin is induced in the hypoxic interior of solid tumors through binding of hypoxia-inducible factor 1α (Hif-1α) to the Livin promoter.43 A similar mechanism may operate in the avascular lens. The concentration of oxygen in the central region of the lens is extremely low, due to consumption by the overlying fiber cells.3 Thus, during terminal differentiation, fiber cells necessarily traverse a steep standing gradient of oxygen. The inner cortical cells are the most hypoxic of all the nucleated cells in the lens. The concentration of oxygen in the interior of the mouse lens is unknown; but in bovine lens the partial pressure of oxygen (pO2) in the central region is 1 to 2 mm Hg3 and in rabbit lenses ≈10 mm Hg.44 For comparison, mean pO2 values in human melanoma xenografts typically range from 1 to 10 mm Hg.45 The role of Hif-1α in the lens is not well defined, but conditional deletion of Hif-1α is associated with widespread lens cell death.46
To test the physiological role of Livin, we conditionally inactivated the Birc7 locus by crossing mice carrying a floxed Birc7 allele with either LeCre or MLR10 mice, two strains that express Cre recombinase in the lens. Unexpectedly, the resulting lens phenotypes depended on the Cre line that was used. LeCretg/−;Birc7flox/flox mice showed a highly penetrant lens phenotype, with vacuolated fiber cells and widespread apoptosis in the lens. However, a similar lens phenotype was also observed in many (37%) control (LeCretg/−;Birc7+/+) animals. Thus, on the mixed background used here, mice hemizygous for the LeCre transgene exhibited ocular phenotypes that were independent of the floxed Birc7 allele. This finding is consistent with a recent report in which, depending on the genetic background, hemizygous LeCre mice developed severe eye abnormalities even in the absence of LoxP sites.47 The precise mechanism remains to be determined but, presumably, reflects an interaction between the LeCre transgene and modifier genes present in certain backgrounds. The LeCre mouse has been widely used in developmental studies of the lens. The current findings, together with those of Dora et al.,47 indicate that conclusions of earlier studies utilizing the LeCre strain should be interpreted with caution.
To control for the confounding effect of the LeCre transgene, we generated conditional knockouts using MLR10 mice and we also examined the lens phenotype of germline Birc7 knockout mice. There was no discernible lens phenotype in either case. The absence of a phenotype indicated that despite its strong expression in lens fiber cells, Livin is not required for normal lens growth, clarity, or cellular organization. It is possible that expression of other Birc family members could compensate for the loss of Livin in the Birc7 knockouts. However, our microarray data showed that the expression of other family members is usually low in the lens relative to Birc7 (data not shown) and, in all cases, expression was found to diminish during fiber cell differentiation in wild type lenses.
In conclusion, we have identified a new member of the late fiber cell gene grouping. Livin, known previously for its expression in melanoma and other solid tumors, is strongly expressed by lens fiber cells as they approach the OFZ. To the best of our knowledge, this is the only site in the body where constitutively high level expression of Livin has been reported. The role of Livin in the lens is enigmatic, since conditional inactivation of the Birc7 locus did not produce an overt lens phenotype. It is possible that Livin serves to block apoptosis under conditions of cellular stress or, alternatively, contributes to organelle breakdown by targeting organelle components for proteolysis. Future experiments will test these possibilities explicitly.
Supplementary Material
Acknowledgments
The authors thank Seta Dikranian for technical assistance; Mike Casey and Sue Penrose of the Vision Core; Bill Harbour for providing uveal melanoma histopathology samples; Seth Crosby and members of the Genome Technology Access Center (GTAC) at Washington University for their help with the microarray analysis; and Domagoj Vucic (Genentech, Inc., San Francisco, CA, USA) for providing ML-IAP antibody and germline knockout mice.
Supported by National Institutes of Health Grant R01EY09852 (SB); Core Grant for Vision Research P30 EY02687; and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness.
Disclosure: A. De Maria, None; S. Bassnett, None
References
- 1. Kuszak JR. The ultrastructure of epithelial and fiber cells in the crystalline lens. Int Rev Cytol. 1995; 163: 305–350. [DOI] [PubMed] [Google Scholar]
- 2. Bassnett S,, Beebe DC. Lens fiber differentiation. : Lovicu FJ,, Robinson ML, Development of the Ocular Lens. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- 3. McNulty R,, Wang H,, Mathias RT,, Ortwerth BJ,, Truscott RJ,, Bassnett S. Regulation of tissue oxygen levels in the mammalian lens. J Physiol. 2004; 559: 883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winkler BS,, Riley MV. Relative contributions of epithelial cells and fibers to rabbit lens ATP content and glycolysis. Invest Ophthalmol Vis Sci. 1991; 32: 2593–2598. [PubMed] [Google Scholar]
- 5. Bassnett S,, Beebe DC. Coincident loss of mitochondria and nuclei during lens fiber cell differentiation. Dev Dyn. 1992; 194: 85–93. [DOI] [PubMed] [Google Scholar]
- 6. Bassnett S,, Shi Y,, Vrensen GF. Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci. 2011; 366: 1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faulkner-Jones B,, Zandy AJ,, Bassnett S. RNA stability in terminally differentiating fibre cells of the ocular lens. Exp Eye Res. 2003; 77: 463–476. [DOI] [PubMed] [Google Scholar]
- 8. Vasiliev O,, Rhodes SJ,, Beebe DC. Identification and expression of Hop an atypical homeobox gene expressed late in lens fiber cell terminal differentiation. Mol Vis. 2007; 13: 114–124. [PMC free article] [PubMed] [Google Scholar]
- 9. De Maria A,, Shi Y,, Kumar NM,, Bassnett S. Calpain expression and activity during lens fiber cell differentiation. J Biol Chem. 2009; 284: 13542–13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishimoto S,, Kawane K,, Watanabe-Fukunaga R,, et al. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003; 424: 1071–1074. [DOI] [PubMed] [Google Scholar]
- 11. Nakahara M,, Nagasaka A,, Koike M,, et al. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007; 274: 3055–3064. [DOI] [PubMed] [Google Scholar]
- 12. Wyatt K,, Gao C,, Tsai JY,, Fariss RN,, Ray S,, Wistow G. A role for lengsin, a recruited enzyme, in terminal differentiation in the vertebrate lens. J Biol Chem. 2008; 283: 6607–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang JM,, Sim SM,, Kim HY,, Park GT. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol. 2010; 89: 537–546. [DOI] [PubMed] [Google Scholar]
- 14. Lazar I,, Perlman R,, Lotem M,, Peretz T,, Ben-Yehuda D,, Kadouri L. The clinical effect of the inhibitor of apoptosis protein Livin in melanoma. Oncology. 2012; 82: 197–204. [DOI] [PubMed] [Google Scholar]
- 15. Yan B. Research progress on Livin protein: an inhibitor of apoptosis. Mol Cell Biochem. 2011; 357: 39–45. [DOI] [PubMed] [Google Scholar]
- 16. Ashery-Padan R,, Marquardt T,, Zhou X,, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000; 14: 2701–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao H,, Yang Y,, Rizo CM,, Overbeek PA,, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004; 45: 1930–1939. [DOI] [PubMed] [Google Scholar]
- 18. Varfolomeev E,, Moradi E,, Dynek JN,, et al. Characterization of ML-IAP protein stability and physiological role in vivo. Biochem J. 2012; 447: 427–436. [DOI] [PubMed] [Google Scholar]
- 19. Muzumdar MD,, Tasic B,, Miyamichi K,, Li L,, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007; 45: 593–605. [DOI] [PubMed] [Google Scholar]
- 20. Ivanov D,, Dvoriantchikova G,, Pestova A,, Nathanson L,, Shestopalov VI. Microarray analysis of fiber cell maturation in the lens. FEBS Lett. 2005; 579: 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashhab Y,, Alian A,, Polliack A,, Panet A,, Ben Yehuda D. Two splicing variants of a new inhibitor of apoptosis gene with different biological properties and tissue distribution pattern. FEBS Lett. 2001; 495: 56–60. [DOI] [PubMed] [Google Scholar]
- 22. Bustin SA,, Benes V,, Garson JA,, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009; 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 23. Andersen CL,, Jensen JL,, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004; 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 24. Shi Y,, Tu Y,, De Maria A,, Mecham RP,, Bassnett S. Development composition, and structural arrangements of the ciliary zonule of the mouse. Invest Ophthalmol Vis Sci. 2013; 54: 2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi Y,, De Maria A,, Bennett T,, Shiels A,, Bassnett S. A role for epha2 in cell migration and refractive organization of the ocular lens. Invest Ophthalmol Vis Sci. 2012; 53: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassnett S. The fate of the Golgi apparatus and the endoplasmic reticulum during lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 1995; 36: 1793–1803. [PubMed] [Google Scholar]
- 27. Berthelet J,, Dubrez L. Regulation of apoptosis by inhibitors of apoptosis (IAPs). Cells. 2013; 2: 163–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vucic D,, Stennicke HR,, Pisabarro MT,, Salvesen GS,, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr Biol. 2000; 10: 1359–1366. [DOI] [PubMed] [Google Scholar]
- 29. Vrensen GF,, Graw J,, De Wolf A. Nuclear breakdown during terminal differentiation of primary lens fibres in mice: a transmission electron microscopic study. Exp Eye Res. 1991; 52: 647–659. [DOI] [PubMed] [Google Scholar]
- 30. De Maria A,, Shi Y,, Luo X,, Van Der Weyden L,, Bassnett S. Cadm1 expression and function in the mouse lens. Invest Ophthalmol Vis Sci. 2011; 52: 2293–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alizadeh A,, Clark JI,, Seeberger T,, et al. Targeted genomic deletion of the lens-specific intermediate filament protein CP49. Invest Ophthalmol Vis Sci. 2002; 43: 3722–3727. [PubMed] [Google Scholar]
- 32. Wistow G,, Bernstein SL,, Wyatt MK,, et al. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002; 8: 171–184. [PubMed] [Google Scholar]
- 33. Hoang TV,, Kumar PK,, Sutharzan S,, Tsonis PA,, Liang C,, Robinson ML. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol Vis. 2014; 20: 1491–1517. [PMC free article] [PubMed] [Google Scholar]
- 34. Kasof GM,, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001; 276: 3238–3246. [DOI] [PubMed] [Google Scholar]
- 35. Ma L,, Huang Y,, Song Z,, et al. Livin promotes Smac/DIABLO degradation by ubiquitin-proteasome pathway. Cell Death Differ. 2006; 13: 2079–2088. [DOI] [PubMed] [Google Scholar]
- 36. Nachmias B,, Lazar I,, Elmalech M,, et al. Subcellular localization determines the delicate balance between the anti- and pro-apoptotic activity of Livin. Apoptosis. 2007; 12: 1129–1142. [DOI] [PubMed] [Google Scholar]
- 37. Nachmias B,, Ashhab Y,, Bucholtz V,, et al. Caspase-mediated cleavage converts Livin from an antiapoptotic to a proapoptotic factor: implications for drug-resistant melanoma. Cancer Res. 2003; 63: 6340–6349. [PubMed] [Google Scholar]
- 38. Ball LE,, Garland DL,, Crouch RK,, Schey KL. Post-translational modifications of aquaporin 0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004; 43: 9856–9865. [DOI] [PubMed] [Google Scholar]
- 39. Lee A,, Morrow JS,, Fowler VM. Caspase remodeling of the spectrin membrane skeleton during lens development and aging. J Biol Chem. 2001; 276: 20735–20742. [DOI] [PubMed] [Google Scholar]
- 40. Zandy AJ,, Bassnett S. Proteolytic mechanisms underlying mitochondrial degradation in the ocular lens. Invest Ophthalmol Vis Sci. 2007; 48: 293–302. [DOI] [PubMed] [Google Scholar]
- 41. Caceres A,, Shang F,, Wawrousek E,, et al. Perturbing the ubiquitin pathway reveals how mitosis is hijacked to denucleate and regulate cell proliferation and differentiation in vivo. PLoS One. 2010; 5: e13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deshaies RJ,, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009; 78: 399–434. [DOI] [PubMed] [Google Scholar]
- 43. Hsieh CH,, Lin YJ,, Wu CP,, Lee HT,, Shyu WC,, Wang CC. Livin contributes to tumor hypoxia-induced resistance to cytotoxic therapies in glioblastoma multiforme. Clin Cancer Res. 2014; 21: 460–470. [DOI] [PubMed] [Google Scholar]
- 44. Barbazetto IA,, Liang J,, Chang S,, Zheng L,, Spector A,, Dillon JP. Oxygen tension in the rabbit lens and vitreous before and after vitrectomy. Exp Eye Res. 2004; 78: 917–924. [DOI] [PubMed] [Google Scholar]
- 45. Brurberg KG,, Graff BA,, Olsen DR,, Rofstad EK. Tumor-line specific pO(2) fluctuations in human melanoma xenografts. Int J Rad Oncol Biol Phys. 2004; 58: 403–409. [DOI] [PubMed] [Google Scholar]
- 46. Shui YB,, Arbeit JM,, Johnson RS,, Beebe DC. HIF-1: an age-dependent regulator of lens cell proliferation. Invest Ophthalmol Vis Sci. 2008; 49: 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dora NJ,, Collinson JM,, Hill RE,, West JD. Hemizygous Le-Cre transgenic mice have severe eye abnormalities on some genetic backgrounds in the absence of LoxP sites. PLoS One. 2014; 9: e109193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.