Summary
Advances in pluripotent stem cell and reprogramming technologies have given hope of generating hematopoietic stem cells (HSC) in culture. To succeed, greater understanding of the self-renewing HSC during human development is required. We discovered that glycophosphatidylinositol-anchored surface protein GPI-80 defines a subpopulation of human fetal liver hematopoietic stem/progenitor cells (HSPC) with self-renewal ability. CD34+CD38lo/−CD90+GPI-80+ HSPC were the sole population that maintained proliferative potential and undifferentiated state in stroma co-culture and engrafted in immunodeficient mice. GPI-80 expression also enabled tracking of HSPC once they have emerged from endothelium and migrate between human fetal hematopoietic niches. GPI-80 co-localized on the surface of HSPC with Integrin alpha-M (ITGAM), which in leukocytes cooperates with GPI-80 to support migration. Knockdown of GPI-80 or ITGAM was sufficient to compromise HSPC expansion in culture and engraftment in vivo. These findings indicate that human fetal HSC employ mechanisms used in leukocyte adhesion and migration to mediate HSC self-renewal.
Introduction
The ability to replenish blood and immune cells relies on rare hematopoietic stem cells (HSC) that can differentiate into all blood cell types, self-renew and engraft upon transplantation (Morrison et al., 1995a; Weissman, 2000). HSC hold immense therapeutic value for treating hematological disorders (Bordignon, 2006; Shenoy, 2013); however, there is a shortage of immunocompatible HSC donors, particularly for patients of minority descent or mixed ethnic background (Dehn et al., 2008). Use of induced pluripotent stem (iPS) cells or lineage reprogramming strategies provide a promising avenue for the generation of patient specific HSC (Dravid and Crooks, 2011; Risueño et al., 2012). However, better understanding of HSC emergence and expansion during human development is critical for identifying programs necessary for the in vitro generation and maintenance of HSC that fulfill the functional and safety criteria for transplantation to patients.
The first hematopoietic cells in the embryo emerge in the yolk sac and first generate a cohort of primitive erythroblasts, followed by erythro-myeloid progenitors that initiate fetal liver erythropoiesis (Mikkola and Orkin, 2006). Subsequently, HSC emerge in the dorsal aorta, vitelline and umbilical arteries, the yolk sac and the placenta from specialized, “hemogenic” endothelium (Chen et al., 2009; Rhodes et al., 2008; Zovein et al., 2008). HSC then seed the fetal liver (FL) for expansion, after which they migrate to their lifelong niche in the bone marrow (BM) where they become predominantly quiescent (Ciriza et al., 2013). Although the sites for HSC emergence and expansion were first established in mice, the same anatomical sites have been shown to support hematopoiesis during human development (Van Handel et al., 2010; Ivanovs et al., 2011; Robin et al., 2009; Tavian et al., 2010).
Insufficient knowledge of the surface proteins specific to human HSC has hampered studies of their regulation. Many well-established murine HSC markers such as Sca1 and the SLAM markers are not conserved in human ((Larochelle et al., 2011)). Moreover, HSC surface markers change during development as shown in mouse embryos (McKinney-Freeman et al., 2012; Mikkola and Orkin, 2006). Previous studies suggest that human long-term repopulating HSC reside in the CD34+CD38lo/−CD90+ population in both cord blood and fetal hematopoietic tissues (Hogan et al., 2002; Magnusson et al., 2013; Majeti et al., 2007; McKenzie et al., 2007). However, as both endothelium and HSC co-express CD34 and CD90, these markers do not distinguish HSC from endothelium. Thus, markers that demarcate human HSC from short-lived progenitors or endothelium remain elusive.
Here we define GPI-80 as a novel surface marker for HSC during human development by functionally validating surface proteins highly enriched in undifferentiated fetal liver HSPC. We discovered that GPI-80 distinguishes a population of self-renewing HSC, and allows tracking HSC in multiple developmental niches. Molecular analysis of GPI-80 HSPC provides new insight into the regulatory machinery in the highly self-renewing fetal HSC, and reveals that fetal HSC preserve self-renewal ability using mechanisms utilized in leukocyte adhesion and migration.
Results
GPI-80 expression defines a subpopulation of human fetal liver HSPC
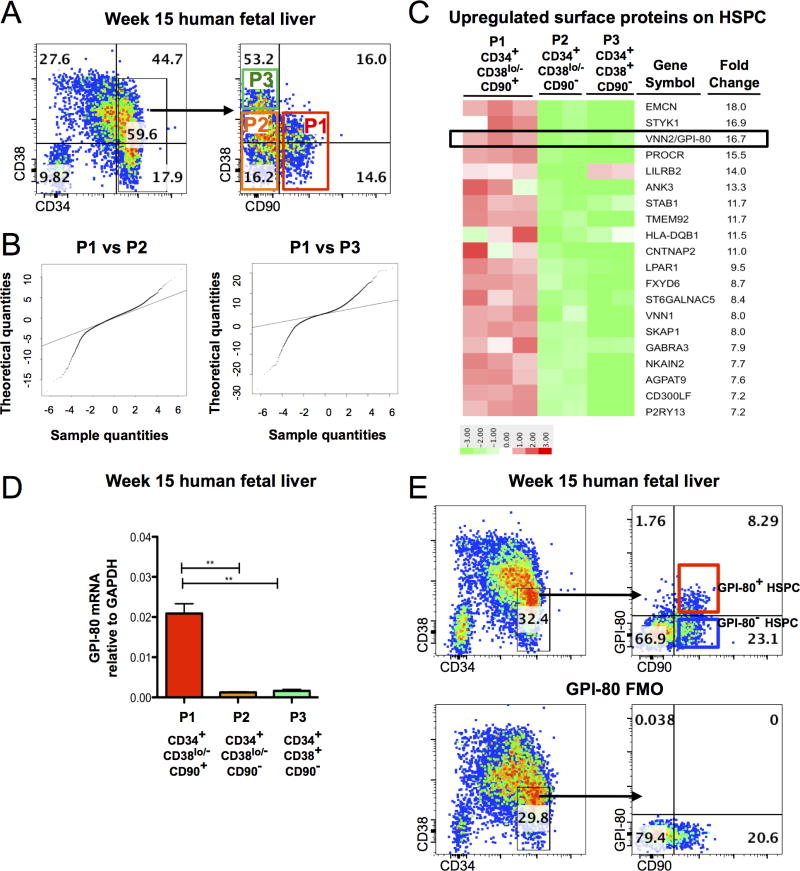
Our goal was to define the identity and molecular properties of fetal liver HSC, which are highly self-renewing, and the most potent human HSC when transplanted to immunodeficient mice (Holyoake et al., 1999). Fetal liver (FL) HSC are ontologically closer to PSC-derived hematopoietic cells than adult bone marrow (BM) or cord blood (CB), thereby representing an ideal model to study human HSC self-renewal and the molecular blocks hampering in vitro HSC generation. To identify novel candidate surface markers for the highly self-renewing fetal HSC, we performed gene expression analysis on HSPC subsets isolated from second trimester (week 15–18 of developmental age) human fetal liver. An HSC enriched population and their downstream progeny were isolated from Ficoll-purified, CD34 enriched FL cells using previously established HSPC surface markers: CD34+CD38lo/−CD90+ (P1, most undifferentiated), CD34+CD38lo/−CD90− (P2), and CD34+CD38+CD90− (P3) (Figure 1A). As expected, 16 weeks after transplantation into NOD-scid IL2Rγ-null (NSG) mice, robust BM engraftment was detected only in mice transplanted with P1 cells (Figure 1B). These data verified that fetal liver HSC engraftment ability is restricted to CD34+CD38lo/−CD90+ (P1) population, henceforth referred to as HSPC, while P2 and P3 consist of hematopoietic progenitor cells (HPC).
Figure 1. A subpopulation of human fetal liver HSPC express GPI-80/VNN2.
A. Flow cytometry plot of hematopoietic populations from Ficoll-purified week 15 human FL with CD34, CD38 and CD90 documenting subfractionation of CD34+ cells to P1 (CD34+CD38lo/−CD90+), P2 (CD34+CD38lo/−CD90−) and P3 (CD34+CD38+CD90−). Quadrants were drawn according to FMO (fluorescence minus one) control. B. Quantification of engrafted cells in murine bone marrow at 16 weeks post-transplantation with representative flow cytometry plots (n=3 mice/group). C. Quantile-Quantile plots comparing differentially expressed genes between P1 (CD34+CD38lo/−CD90+) vs. P2 (CD34+CD38lo/−CD90−) or P3 (CD34+CD38+CD90−). D. 20 cell surface proteins upregulated in CD34+CD38lo/−CD90+ HSPC vs. CD34+CD38lo/−CD90− HPC. E. qRT-PCR of GPI-80 mRNA in CD34+CD38lo/−CD90+ population as compared to CD34+CD38lo/− CD90− and CD34+CD38+CD90− cells (p≤0.01) (n=3 different donor tissues). Error bars represent mean ± SEM. F. Representative flow cytometry plot of Ficoll-purified, CD34+ enriched cells from 15 week developmental age FL stained for CD34, CD38, CD90 and GPI-80. FMO control for GPI-80 is shown. See also Figure S1 and Table S1.
To generate candidates for further purification of HSC, we used Affymetrix microarray analysis to identify surface proteins enriched in CD34+CD38lo/−CD90+ HSPC as compared to downstream HPC. A total of 1274 genes were upregulated > 2-fold and 1369 genes were > 2 fold down-regulated in P1 HSPC as compared to P2 HPC (p<0.05). Quantile-Quantile (Q-Q) analysis of differentially expressed genes confirmed that the P1 population was more similar to P2 than P3 (Figure 1C). Candidate HSC surface proteins were selected from the most highly up-regulated membrane proteins on P1 HSPC (Figure 1D, Table S1). Of these, EMCN (Endomucin) and PROCR (Endothelial Protein C Receptor, EPCR) have been previously identified as markers that enrich for HSC (Balazs et al., 2006; Matsubara et al., 2005).
Vanin-2 (VNN2, glycosylphosphatidylinositol-anchored surface protein, GPI-80; Figure 1D) a molecule that governs neutrophil adhesion and transendothelial migration(Suzuki et al., 1999), was 16.7-fold upregulated in P1 HSPC. Upregulation of GPI-80 mRNA in P1 HSPC as compared to downstream HPC (P2 and P3) was verified by qRT-PCR (Figure 1E). Flow cytometry analysis indicated that GPI-80 surface expression subfractionated FL HSPC into a distinct GPI-80+ subpopulation (32%±7.0% of P1) (Figure 1F). Minimal GPI-80 surface expression was found in CD34+CD38lo/−CD90− (P2) or CD34+CD38+CD90− (P3) HPC, confirming its expression within Ficoll-purified FL CD34+ compartment was restricted to most undifferentiated HSPC. However, some expression was detected in more differentiated CD34− cells (Figure S1A). The expected expression of GPI-80 in myeloid cells in fetal liver and adult peripheral blood was confirmed by flow cytometry (Figure S1B). Markers associated with HSPC differentiation such as CD45RA (Figure S1C), or myelo-erythoid lineage markers CD14, CD66, CD13 and CD235, were not co-expressed with GPI-80 HSPC, and therefore would not further purify this subset, in Ficoll-purified, CD34 enriched fetal liver; however, if total fetal liver was analyzed, cells co-expressing myeloid markers were found within the GPI-80+ HSPC population (Figure S1D). The percentage of CD45 cells that have the GPI-80+CD34+CD38lo/−CD90+ phenotype in total FL ranged from 0.03%±0.01%; however, over 60% of these cells co-express at least one myeloid marker. In the Ficoll-purified, CD34 enriched fraction, the frequency of GPI-80+ HSPC was 1.4%±1.2%. To ensure maximum purity of HSPC, Ficoll-purified FL was used for all the assays unless otherwise noted. These data implied that, in addition to having a function in the myeloid compartment, GPI-80 is also expressed in undifferentiated human fetal HSPC.
GPI80+ HSPC exhibit robust multi-lineage differentiation potential
GPI-80+ and GPI-80− fractions of FL HSPC were FACS sorted and evaluated for key HSC properties: multipotency, self-renewal and engraftment ability. Both HSPC fractions generated myeloid and erythroid colonies, although GPI-80+ HSPC formed fewer colonies than GPI-80− cells (Figure S2A). When B- and T-lymphoid differentiation potential was assayed on OP9M2 (Magnusson et al., 2013; Nakano et al., 1994) and OP9DL1 stroma (Schmitt and Zúñiga-Pflücker, 2002), respectively, as bulk cultures, both GPI-80+ and GPI-80− cells formed CD19+ B-cells (Figure S2B) and CD4/CD8+ T-cells (Figure S2C).
To investigate multilineage differentiation ability at clonal level, GPI-80+ and GPI-80− HSPC were sorted as single cells and cultured on OP9M2 in conditions that support both myeloid and B-cell differentiation. After two weeks, expanded clones were enumerated and scored as myeloid (CD13+, CD14+ or CD66+ cells), lymphoid (CD19+ cells), myelo-lymphoid or undifferentiated (>30% CD34+ cells with minimal expression of differentiation markers). Clones were scored as proliferative if >200 daughter cells after 14 days was observed. GPI-80+ HSPC averaged 33% proliferating clones whereas GPI-80− HSPC averaged 25% (Figure S2D). Furthermore, GPI-80+ HSPC demonstrated enrichment in clones that remained largely undifferentiated or displayed myelo-lymphoid differentiation, whereas GPI-80− HSPC clones gave predominantly rise to myeloid progeny. These data imply that, although both GPI-80+ and GPI-80− HSPC possess multilineage differentiation ability in bulk culture, only GPI-80+ HSPC display robust multipotency at clonal level.
In vitro expansion ability is restricted to GPI-80+ HSPC
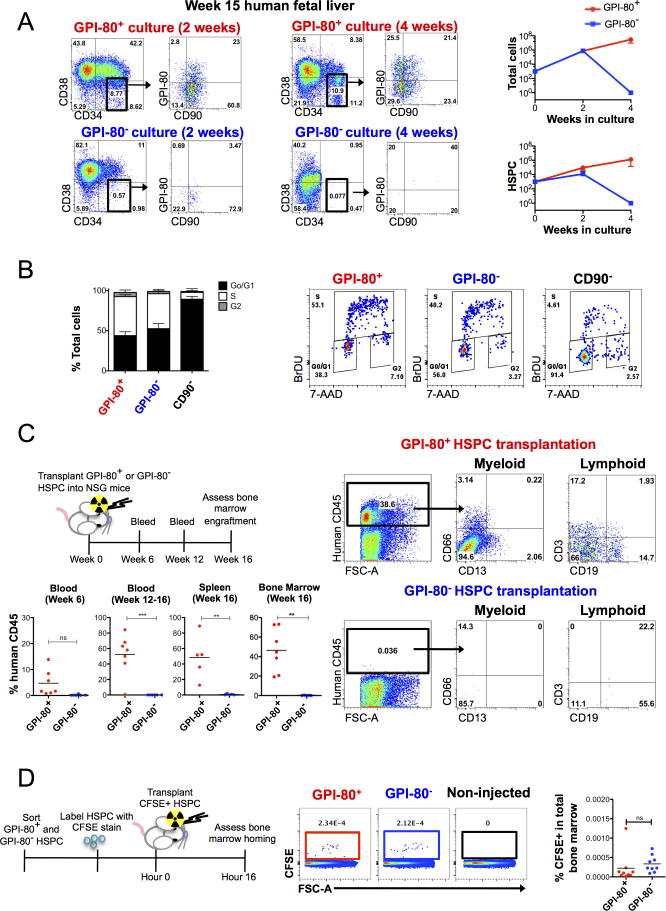
We next evaluated in vitro self-renewal ability of GPI-80+ and GPI-80− HSPC on OP9M2 stroma; we have previously shown that this mesenchymal stem cell (MSC)-like stroma line supports robust expansion of human FL HSPC over several weeks and maintains transplantable HSC for over 2 weeks in culture (Magnusson et al., 2013). After 2 and 4 weeks, the differentiation status of GPI80+ and GPI-80− HSPC was assessed by cell surface phenotype. Strikingly, only GPI-80+ HSPC showed robust expansion of HSPC (>1000 fold in 4 weeks), a subpopulation of which continued to express GPI-80 (Figure 2A). In contrast, GPI-80− cells were unable to maintain HSPC in culture, and exhibited limited proliferative potential. These data indicate that the GPI-80+ subpopulation is fully responsible for the in vitro expansion of undifferentiated CD34+CD38lo/−CD90+ HSPC.
Figure 2. GPI-80 expression defines HSPC with self-renewal and engraftment ability.
A. In vitro self-renewal/expansion assay of GPI-80+ and GPI-80− HSPC co-cultured on OP9M2 stroma and analyzed at 2 and 4 weeks for the presence of undifferentiated HSPC is shown. Growth of total cells and expansion of CD34+CD38lo/−CD90+ HSPC on OP9M2 are shown (n=3 different donor tissues, which are representative examples of 10 independent experiments that showed the same result). Flow cytometry plots of hematopoietic populations after 2 and 4 weeks of co-culture on OP9M2 stroma are shown. Error bars represent mean ± SD. B. Cell cycle analysis of FACS sorted FL HSPC cultured on OP9M2 overnight and pulsed with BRDU is shown (n=3 different donor tissues analyzed in 2 independent experiments). Representative flow cytometry plots assessing BRDU incorporation in GPI-80+ and GPI-80− cells are shown. Error bars represent mean ± SEM. C. Scheme of transplant assay for analysis of engraftment ability of GPI-80+ and GPI-80− HSPC. Each mouse was transplanted with sorted HSPC subset originating from 50,000 CD34+ cells. Flow cytometry analysis of human hematopoietic engraftment in peripheral blood, spleen and BM of mice transplanted with GPI-80+ and GPI-80−HSPC from human FL is shown (p≤0.01)(n=3 donor tissues assessed in 3 independent experiments). D. Scheme of transplant assay for CFSE labeled HSPC. Flow cytometry analysis of murine bone marrow 16 hours post transplant is shown (n=3 donor tissues assessed in 2 independent experiments). See also Figure S2 and Table S2.
Cell cycle analysis using BrdU incorporation in HSPC in OP9M2 co-culture (Figure 2B) revealed that both GPI-80+ and GPI-80− HSPC show similar distribution between cell cycle phases, while CD34+CD38lo/−CD90− cells exhibit a reduction of S-phase cells. These data suggest that the poor expansion of GPI-80− HSPC in culture is not due to lower proliferation rate, but rather due to inability of GPI-80− HSPC to undergo self-renewal divisions to maintain/expand undifferentiated HSPC.
GPI-80 identifies in vivo engraftable fetal HSC
To assess the in vivo engraftment and reconstitution ability of GPI-80+ HSPC, xenotransplantation assays were performed. NSG mice were sublethally irradiated and transplanted with GPI-80+ or GPI-80− HSPC (respective populations from 50,000 CD34+ cells) (Figure 2C). Mice were bled retro-orbitally at 6 weeks and 12–16 weeks to track peripheral blood reconstitution, and hematopoietic organs were assessed for human engraftment at 16 weeks. Robust populations of human CD45+ cells were detected in peripheral blood of all mice transplanted with GPI-80+ HSPC. The average human CD45+ bone marrow chimerism of GPI-80+ transplants after 16 weeks was 46.4±22.1% of total cells, CD13+or CD66+ myeloid cells constituted 11.2±9.4%, CD19+ B cells 17.7±16.4% and CD3+ T-cells 5.6 ±12.4%(Table S2). In contrast, GPI-80− HSPC transplants did not yield detectable engrafted cells (0.2±0.2%). Transplantation of CFSE-labeled HSPC showed that both GPI-80+ and GPI-80− HSPC could be found in the BM shortly after transplantation, implying defective engraftment/self-renewal rather than lack of BM homing as the key difference between GPI-80+ and GPI-80− HSPC (Figure 2D). These results indicate that only the GPI-80+ subset of human FL HSPC displays capacity for in vivo engraftment and multilineage hematopoietic reconstitution.
GPI-80 can be used to track immunophenotypic HSPC with high proliferative potential through early human development
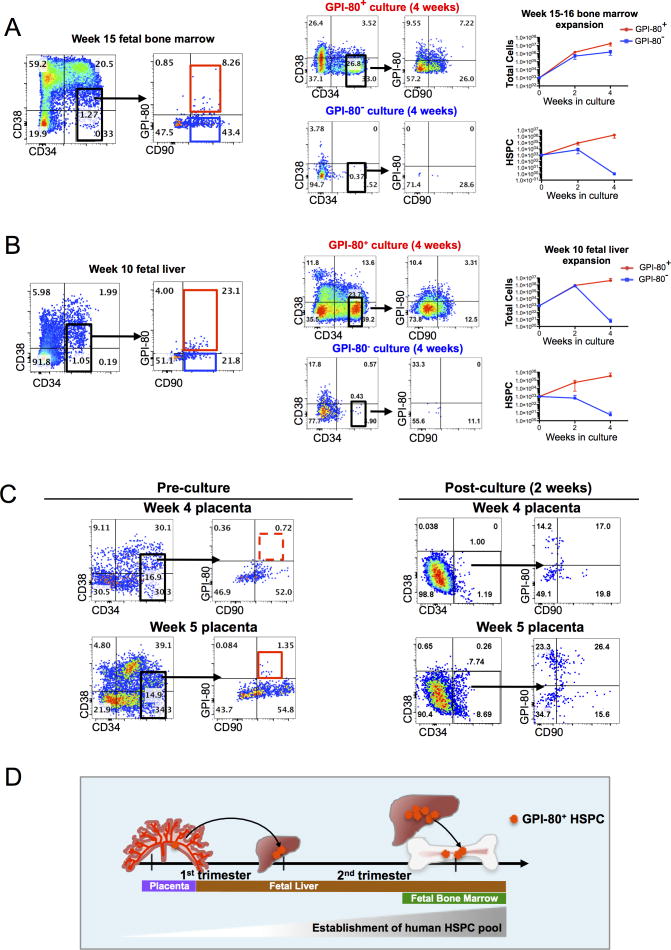
We next assessed whether GPI-80 expression marks undifferentiated HSPC also in fetal bone marrow (FBM), which is colonized after the fetal liver. FACS analysis demonstrated the presence of GPI-80+ HSPC in FBM by 15 weeks of development. In vitro expansion assays revealed that only GPI-80 HSPC harbored in vitro expansion ability in FBM, whereas GPI-80− HPSC were not sustained in culture (Figure 3A). Flow cytometry analysis showed that some of GPI-80+ HSPC obtained from total BM co-express myeloid differentiation markers; however, Ficoll-purification of FBM largely depleted hematopoietic cells that co-expressed myeloid and HSPC markers, and enriched for the undifferentiated GPI-80+ HSPC (Figure S3A).
Figure 3. GPI-80 tracks self-renewing HSPC during human development.
A. Flow cytometry plot of 15 week fetal BM with CD34, CD38, CD90 and GPI-80 is shown. Representative FACS plots and growth curves of cultured GPI-80+ and GPI-80− HSPC from fetal BM are shown (n=3 independent experiments with different donor tissues). Error bars represent mean ± SD. B. Representative flow cytometry plots of 10 week human FL with CD34, CD38, CD90 and GPI-80 is shown. Representative FACS plots and growth curves of cultured GPI-80+ and GPI-80− HSPC from 10 week FL are shown (n=3 independent experiments with different donor tissues). Error bars represent mean ± SD. C. Representative flow cytometry plots of 4 and 5 week placenta stained with CD34, CD38, CD90 and GPI-80 are shown. Representative flow cytometry plots of CD34+ placental cells after 2 week in culture on OP9M2 are shown. D. Model of timeline of migration of GPI-80+ HSPC in first and second trimester human hematopoietic tissues. See also Figure S3.
We then assessed GPI-80 expression in 1st trimester hematopoietic tissues. FACS analysis of 10 week FL showed the presence of GPI-80+ immunophenotypic HSPC (Figure 3B). Analysis of in vitro expansion ability of immunophenotypic HSPC sorted based on GPI-80 confirmed that, similar to the 2nd trimester FL and BM, in vitro self-renewal capacity in 10 week FL was restricted to GPI-80+ cells (Figure S3A). In contrast, 8–10 week FBM showed the absence of a robust population of undifferentiated GPI-80+ HSPC, although some GPI-80 expression was observed in more differentiated cells that co-expressed lineage markers (data not shown). Moreover, there was no expansion of CD34+ cells from first trimester FBM in culture (data not shown), consistent with the finding that in vitro expansion potential is restricted to GPI-80+ HSPC.
To determine whether GPI-80 is expressed at a site of HSPC emergence, 1st trimester human placentas were analyzed. The first GPI-80+ immunophenotypic HSPC appeared in the placenta between 4 and 5 weeks of development; by 5 weeks, a distinct population of GPI-80+ HSPC was reproducibly observed (Figure 3C). Coinciding with the presence of a GPI-80+ immunophenotypic HSPC, cultured CD34+ cells from 5 week placenta maintained undifferentiated HSPC, many of which co-expressed GPI-80 (Figure 3C).
Co-staining for GPI-80 and endothelial and hematopoietic markers indicated that in the placenta, similar to FL and FBM, GPI-80+ cells were largely confined to the hematopoietic population that expressed CD45, and lacked high VE-Cadherin and CD31/PECAM expression (Figure S3C). Thus, unlike many HSC markers, GPI-80 expression can distinguish HSPC from endothelium.
In summary, analysis of fetal hematopoietic tissues during early human development indicates that in all niches studied, GPI-80 expression correlates with the acquisition of HSPC high proliferative potential, and can be used to track the migration of candidate HSC from the placenta to fetal liver during first trimester, and to fetal bone marrow by the beginning of the second trimester (Figure 3D).
GPI-80 and ITGAM are critical for human fetal HSPC expansion
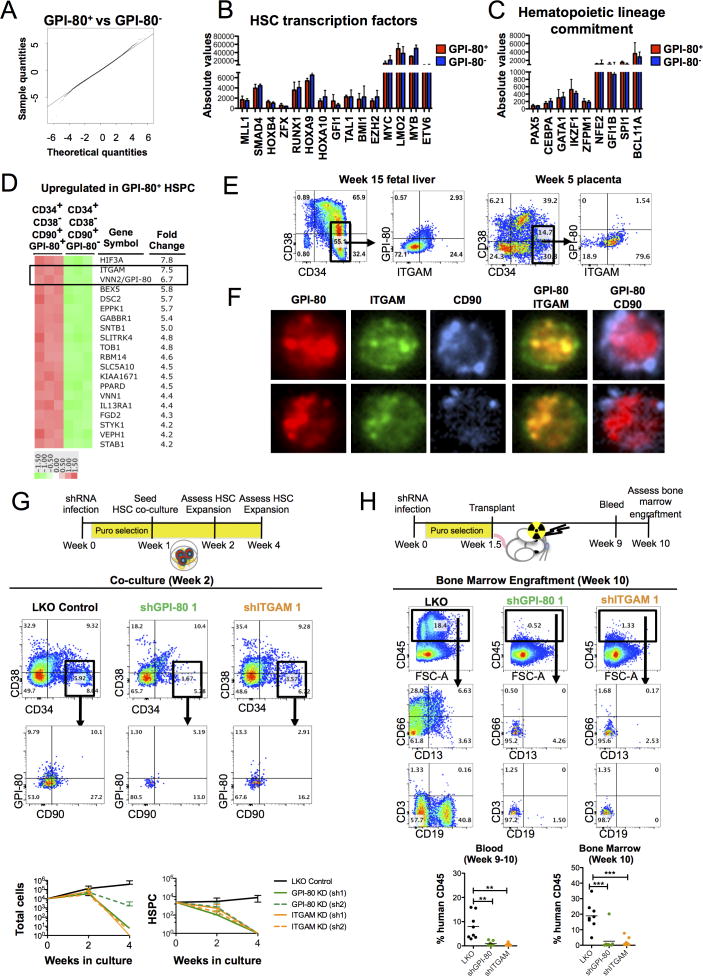
To provide insights into the mechanisms of how human fetal HSC sustain self-renewal, Affymetrix microarray analysis was performed to identify genetic networks enriched in GPI-80+ HSPC as compared to their immediate progeny. Q-Q probability plot revealed remarkable similarity between GPI-80+ and GPI-80− HSPC (Figure 4A), in contrast to the more obvious differences between CD90+ HSPC and downstream HPC (Figure 1B). Similar expression levels of transcription factors governing HSC specification (TAL1/SCL, LMO2, RUNX1/AML, MYB) and maintenance (MLL1, MYC, SMAD4, HOXB4, HOXA9, BMI1, GFI1, ETV6/TEL Figure 4B) or lineage commitment (PU.1, BCL11A, CEPBA, GATA1, IKAROS, Figure 4C) was observed, indicating that GPI-80− HSPC have not yet committed to specific downstream fates. Only 574 genes were upregulated on GPI-80+ HSPC while 246 genes were downregulated (>2 fold, p<0.05; Table S3). Of the 20 most differentially expressed annotated genes, three were transcription factors (HIF3α, TOB1, KLF4) with no known function in HSC.
Figure 4. GPI-80 and ITGAM are required for maintaining undifferentiated HSPC.
A. Quantile-quantile plot comparing differentially expressed genes between GPI-80+ and GPI-80− HSPC is shown. B. Bar graphs showing the relative expression of known transcription factors required for HSC development or maintenance. C. Bar graphs showing the relative expression of transcription factors responsible for lineage differentiation. D. Heat map illustrating top 20 annotated genes significantly and consistently up-regulated among replicates (n=3 fetal livers, p<0.05) in GPI-80+ HSPC. E. Representative 15 week FL and 5 week placental tissue stained for CD34, CD38, CD90, GPI-80 and ITGAM are shown. F. Representative ImageStream images of 15 week CD34+ FL cells stained for CD90, GPI-80 and ITGAM are shown. G. Representative flow cytometry plots of cultured CD34+ FL cells after lentiviral knockdown of GPI-80 or ITGAM are shown. Quantification of expansion of total cells and HSPC population after knockdown of GPI-80 or ITGAM with two different shRNA vectors are shown (n=3 donor tissues analyzed in two independent experiments). Error bars represent mean ± SD. H. Representative flow cytometry plots of mouse BM 10 weeks after transplantation with human FL CD34+ cells transduced with control LKO vector, shGPI-80 vector or shITGAM vector. Quantification of engrafted cells in blood and BM is shown (n=3 donor tissues analyzed in 2 independent experiments). See also Figure S4, Table S3, and Table S4.
The second most highly upregulated transcript was ITGAM (Integrin α-M, CD11b) (Figure 4D), an α integrin component of the MAC-1 (CR3) complex that consists of ITGAM and ITGB2 (CD18). ITGAM co-localizes with GPI-80 on neutrophils, enabling their adherence and extravasation (Huang et al., 2004). Flow cytometry of fetal liver and placental HSPC demonstrated robust co-expression of ITGAM and ITGB2 on GPI-80+ HSPC (Figure 4E, Figure S4A). ImageStream analysis was used localize ITGAM relative to GPI-80 on HSPC surface. Distinct regions of co-localization between GPI-80 and ITGAM were visualized, as described on neutrophils (Huang et al., 2004), while there was limited co-localization between GPI-80 and CD90, another GPI-anchored surface protein (Figure 4F). These data suggested that GPI-80 and ITGAM function together in HSC.
To assess whether GPI-80 and/or ITGAM are required for maintaining HSC properties, lentiviral-mediated shRNA knockdown of GPI-80 and ITGAM was performed on FL CD34+ cells. FACS analysis confirmed a reduction of GPI-80 and ITGAM in HSPC by 1 week in culture with two independent shRNAs (Figure S4B). Knockdown of either protein led to progressive depletion of undifferentiated HSPC on OP9M2 stroma (Figure 4G), but did not impair their ability to differentiate into myeloid, erythroid or lymphoid cells (Figure S4C). Moreover, transplantation of shRNA transduced HSPC into NSG mice showed that loss of GPI-80 or ITGAM severely compromised their engraftment/reconstitution ability in vivo (Figure 4H, Table S4). Collectively, these data suggest that GPI80 and ITGAM are not only markers for the highly self-renewing fetal HSC, but also required for their proper function.
Discussion
Uncovering the identity and regulation of the self-renewing HSC as they emerge and expand during embryogenesis is of paramount significance, both for understanding the fundamental mechanisms governing “stemness”, and for bringing pluripotent stem cell derivatives to clinical applications. We identified GPI-80 as a novel surface protein that demarcates HSC from non-self-renewing progenitors during human development. Although the GPI-80− subset of FL HSPC also displays a surface immuophenotype associated with human HSC (e.g. CD34+CD38lo/−CD90+) and exhibits striking similarity with GPI-80+ HSPC in global gene expression, they are profoundly different functionally by lacking the ability to self-renew and engraft.
Analysis of GPI-80 expression at different stages and anatomical sites during human development revealed a correlation of GPI-80 expression with the capacity to expand undifferentiated HSPC in MSC stroma and engraft in vivo. Thus, GPI-80 expression provides a unique tool for temporal and spatial tracking of candidate HSPC with high proliferative potential as they migrate between fetal hematopoietic organs. GPI-80+ HSPC appear in the human placenta by 5 weeks of development, which parallels the time when transplantable HSC populate the mouse placenta (E11.5) (Gekas et al., 2005). In human placentas, transplantable HSC have been detected as early as 6 weeks of human development (Robin et al., 2009), although another study did not detect robust reconstitution of placental cells until the 2nd trimester, while AGM cells from 1st trimester tissues engrafted readily (Ivanovs et al., 2011). Due to the techniques used for the elective terminations to obtain tissues, it was not possible to obtain intact embryos to analyze GPI-80 expression in human AGM. Future studies will be needed to investigate whether GPI-80 is already expressed by the AGM HSPC as they emerge from hemogenic endothelium, or only after HSPC have migrated to the placenta and FL. In contrast to CD34 and CD90 which, similar to many HSC markers, are also expressed in endothelium, GPI-80 expression was not observed in VE-cadherinhigh endothelial cells, but only after HSPC co-express the pan-hematopoietic marker CD45. Our studies also show that GPI-80 is not expressed in hemogenic endothelial precursors isolated from embryoid bodies during human ES cell differentiation; however, GPI-80 can be induced in a subset of CD34+CD38−CD90+CD45+ cells during their developmental maturation on OP9M2 stroma, further suggesting that GPI-80 expression is turned on after departure from hemogenic endothelium (Dou, Mikkola, manuscript in preparation). The finding that GPI-80 expression can be detected in PSC derived HPC that are defective in self-renewal implies that GPI-80 expression is necessary, but not sufficient, for self-renewal ability. Nevertheless, GPI-80 now represents an attractive candidate to track the appearance and developmental maturation of PSC-derived HSC precursors.
An intriguing property of GPI-80 compared to many known HSC surface markers is that it is functionally required for HSC self-renewal; this observation opens new avenues to understand HSC self-renewal mechanisms during human development. Although GPI-80 does not possess an intracellular domain to convey a signal from the niche, it associates with the MAC1 (CR3) integrin complex that consists of ITGAM/CD11b and ITGB2/CD18, which in leukocytes co-operate with GPI-80 to mediate their migration and extravasation (Huang et al., 2004). Mac1 is also expressed on mouse fetal hematopoietic stem cells (Morrison et al., 1995b) and down-regulated in adult BM HSC (Morrison and Weissman, 1994). Our finding that GPI-80 and ITGAM co-localize on highly self-renewing human fetal HSPC, and that disruption of the expression of either leads to loss of undifferentiated HSPC in culture and prevents engraftment in vivo, suggests that they function together to protect HSC. These findings introduce the unanticipated concept that HSC, the most undifferentiated cells in hematopoietic hierarchy, utilize mechanisms employed by terminally differentiated leukocytes during immune response, for maintaining “stemness”.
In stark contrast to the profound functional differences between GPI-80+ versus GPI-80− HSPC, gene expression profiling showed striking molecular similarity, including fundamental hematopoietic programs known to govern HSC specification and function. Nevertheless, our analysis uncovered unique differentially expressed genes that are novel candidates for governing HSC self-renewal. HIF3-α, a transcription factor whose expression declines during culture (Magnusson et al., 2013) when HSPC begin to lose their functional properties, was the most differentially expressed gene between GPI-80+ and GPI-80− HSPC. We believe that deciphering the function of genes upregulated in highly self-renewing GPI-80+ fetal HSC will provide an invaluable resource for dissecting the genetic programs that govern “stemness” in HSC. Inducing these programs in human ES and iPSC-derived HPC may bring us closer to creating self-renewing HSC in vitro for clinical applications.
EXPERIMENTAL PROCEDURES
Ethical statement
Experimental protocols involving mice were reviewed and approved by the UCLA Animal Research Committee (Protocol number 2005–109). Fetal hematopoietic tissues were discarded material devoid of personal identifiers obtained from elective terminations performed by UCLA Medical Center, or purchased from Novogenix. All material was used with written informed consent.
Tissues, cell culture and hematopoietic assays
In vitro HSPC expansion was assayed on OP9M2 stroma and. B and T cell assays were performed on OP9M2 and OP9DL1, respectively, as described in (Magnusson et al., 2013) and supplemental materials. Processing and transplantation of hematopoietic tissues is described in supplemental material.
Lentiviral shRNA knockdown
pLKO lentiviral vectors from the TRC library (Sigma) were produced and transduced in fetal liver CD34+ cells as described in supplemental material. Two different shRNAs with confirmed knockdown of GPI-80 or ITGAM were used: TRCN0000158666 (GPI-80 1), TRCN0000161721 (GPI-80 2), TRCN0000029051 (ITGAM 1), and TRCN0000029050 (ITGAM 2).
Graphical and statistical analysis
Graphs were generated and statistics were analyzed using GraphPad Prism software. Student’s two-tailed t tests were used to calculate p values.
Gene expression, flow cytometry analysis and Image Stream: see supplemental material.
Supplementary Material
Figure S1, related to Figure 1. GPI-80 expression in HSPC and myeloid cells. A. Expression of GPI-80 in hematopoietic populations in Ficoll-purified, CD34+ enriched fetal liver cells. B. Flow cytometry of total fetal liver and adult peripheral blood with CD66, CD14 and GPI-80 verifies expression of GPI-80 in myeloid cells. Fetal liver cells are gated from CD45+CD34− population. C. CD45RA expression in fetal liver CD34+ cells is restricted CD90− GPI-80− cells. D. Flow cytometry analysis of 18 week fetal liver with lineage markers CD13, CD66, CD235, and CD14 with and without Ficoll purification shows depletion of Lin+ cells with Ficoll purification.
Figure S2, related to Figure 2. Multipotency of GPI-80+ HSPC. A. Myelo-erythroid differentiation potential of GPI-80+ and GPI-80− HSPC on methylcellulose assay is shown (n=6 donors). Error bars represent mean ± SEM. B. Flow cytometry analysis for B-cell marker CD19 on GPI-80+ and GPI-80− HSPC after 2 weeks culture on OP9M2 is shown (n=3 donors). Error bars represent mean ± SEM. C. Flow cytometry analysis of T cell markers CD4 and CD8 after 2 weeks of culture on OP9-DII1 (n=3 donors). Error bars represent mean ± SEM. D. Myeloid and lymphoid potential of GPI-80+ and GPI-80− HSPC at the single cell level is shown. Quantification of proliferating clones (defined as >200 cells, n=2 donors), distribution of clone types (40 clones analyzed), and representative clones from GPI-80+ and GPI-80− HSPC after two weeks of culture on OP9M2 are shown. Though bulk cultures demonstrate multilineage potential of GPI-80− HSPC, single cell analysis reveals enrichment of multipotent cells in GPI-80+ population. Error bars represent mean ± SEM.
Figure S3, related to Figure 3. GPI-80 expression in multiple fetal hematopoietic sites. A. Lineage analysis of total vs ficoll purified, CD34+ enriched second trimester bone marrow with lineage markers CD13, CD66, CD235a, and CD14 shows depletion of Lin+ cells with Ficoll purification. B. Lineage analysis of 5 week total placenta with differentiation markers CD13, CD66, CD235a, and CD14 shows the presence of a subpopulation of GPI-80 HSPC that are devoid of lineage marker expression. C. Representative flow cytometry plots of endothelial cells show that GPI-80 expression in the placenta, fetal liver and fetal bone marrow is confined to hematopoietic cells.
Figure S4, related to Figure 4. Lentiviral shRNA knockdown of GPI-80 and ITGAM. A. Representative flow cytometry plot of fetal liver showing expression of CD11b(ITGAM) and CD18(ITGB2) on GPI-80+ HSPC. B. Representative flow cytometry plots of GPI-80 and ITGAM expression one week after lentiviral transduction, documenting reduction of GPI-80 and ITGAM protein on HSPC with two different shRNA vectors. C. Differentiation ability of HSPC after knockdown of GPI-80 or ITGAM (n=4 donors). Error bars represent mean ± SEM.
Table S1. Gene expression analysis of fetal liver hematopoietic subsets, Related to Figure 1. Gene expression analysis shows the comparison between CD34+CD38lo/−CD90+ HSPC and CD34+CD38lo/−CD90− HPC in human fetal liver. Significantly upregulated and downregulated genes (≥2 fold, p<0.05) are shown.
Table S2. Human engraftment in the bone marrow of NSG mice transplanted with GPI-80+ and GPI-80− HSPC, Related to Figure 2. Human engraftment at 16 weeks post-transplantation is shown.
Table S3. Gene expression analysis of fetal liver GPI-80+ and GPI-80− HSPC, Related to Figure 4. Gene expression analysis shows comparison between CD34+CD38lo/−CD90+GPI-80+ and CD34+CD38lo/−CD90+GPI-80− HSPC. Significantly upregulated and downregulated genes (≥2 fold, p<0.05) are shown.
Table S4. Human engraftment in the bone marrow of NSG mice transplanted with human fetal liver hematopoietic cells transduced with LKO, shGPI-80, or shITGAM lentiviral vectors, Related to Figure 4. Human engraftment at 10 weeks post-transplantation is shown.
Acknowledgments
The authors thank the Felicia Codrea, Jessica Scholes, Min Zhou and Jesse Oakes at FACS Core facilities at BSCRC and JCCC at UCLA for assistance with cell sorting, and Ann George at Children’s Hospital Los Angeles, for Imagestream analysis; the UCLA Clinical Microarray Core for the microarray analysis; Owen Witte, Batoul Amir-Ahmady, Donny Johnson and Melissa McCracken for help with homing assays in mice, Alvin Welch at UCLA Warren Hall staff for animal care; and UCLA Tissue and Pathology Core, and Heather Martin at Novogenix LLC for provision of human hematopoietic tissue. This work was supported by NIH RO1s HL097766 and DK100959, CIRM New Faculty Award RN1-00557-1, Broad Stem Cell Center at University of California Research Award, Jonsson Cancer Center Foundation, and Leukemia & Lymphoma Society (LLS) Scholar award for H.K.A.M and NIH P01 HL073104 for G.C. S.L.P. was supported by the Howard Hughes Medical Institute Gilliam fellowship, C.Y. by the Howard Hughes Undergraduate Research scholarship, V.C by Leukemia & Lymphoma Society Special Fellow Award and M.M. by the Swedish Research Council and Tegger Foundation.
Footnotes
Authorship:
Contributions: S.L.P., C.Y., V.C., M.M. G.C. and H.K.A.M. designed experiments and interpreted data. S.L. P., C.Y., V.C., J.K., R.S. and M.M. performed experiments. S.L.P., C.Y., V.C and H.K.A.M. wrote the manuscript, which all authors edited and approved.
Conflict-of-interest disclosure: M.M. is a cofounder of and own significant financial stakes in Novogenix Laboratories, LLC. The remaining authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordignon C. Stem-cell therapies for blood diseases. Nature. 2006;441:1100–1102. doi: 10.1038/nature04962. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriza J, Thompson H, Petrosian R, Manilay JO, García-Ojeda ME. The migration of hematopoietic progenitors from the fetal liver to the fetal bone marrow: lessons learned and possible clinical applications. Experimental Hematology. 2013;41:411–423. doi: 10.1016/j.exphem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Dehn J, Arora M, Spellman S, Setterholm M, Horowitz M, Confer D, Weisdorf D. Unrelated donor hematopoietic cell transplantation: factors associated with a better HLA match. Biology of Blood and Marrow Transplantation: Journal of the American Society for Blood and Marrow Transplantation. 2008;14:1334–1340. doi: 10.1016/j.bbmt.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid GG, Crooks GM. The challenges and promises of blood engineered from human pluripotent stem cells. Advanced Drug Delivery Reviews. 2011;63:331–341. doi: 10.1016/j.addr.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lièvre F, Orkin SH, Mikkola HKA. The placenta is a niche for hematopoietic stem cells. Developmental Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Van Handel B, Prashad SL, Hassanzadeh-Kiabi N, Huang A, Magnusson M, Atanassova B, Chen A, Hamalainen EI, Mikkola HKA. The first trimester human placenta is a site for terminal maturation of primitive erythroid cells. Blood. 2010;116:3321–3330. doi: 10.1182/blood-2010-04-279489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan CJ, Shpall EJ, Keller G. Differential long-term and multilineage engraftment potential from subfractions of human CD34+ cord blood cells transplanted into NOD/SCID mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:413–418. doi: 10.1073/pnas.012336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoake TL, Nicolini FE, Eaves CJ. Functional differences between transplantable human hematopoietic stem cells from fetal liver, cord blood, and adult marrow. Experimental Hematology. 1999;27:1418–1427. doi: 10.1016/s0301-472x(99)00078-8. [DOI] [PubMed] [Google Scholar]
- Huang JB, Takeda Y, Araki Y, Sendo F, Petty HR. Molecular proximity of complement receptor type 3 (CR3) and the glycosylphosphatidylinositol-linked protein GPI-80 on neutrophils: effects of cell adherence, exogenous saccharides, and lipid raft disrupting agents. Molecular Immunology. 2004;40:1249–1256. doi: 10.1016/j.molimm.2003.11.033. [DOI] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Welch L, Anderson RA, Turner ML, Medvinsky A. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. The Journal of Experimental Medicine. 2011;208:2417–2427. doi: 10.1084/jem.20111688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle A, Savona M, Wiggins M, Anderson S, Ichwan B, Keyvanfar K, Morrison SJ, Dunbar CE. Human and rhesus macaque hematopoietic stem cells cannot be purified based only on SLAM family markers. Blood. 2011;117:1550–1554. doi: 10.1182/blood-2009-03-212803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M, Sierra MI, Sasidharan R, Prashad SL, Romero M, Saarikoski P, Van Handel B, Huang A, Li X, Mikkola HKA. Expansion on stromal cells preserves the undifferentiated state of human hematopoietic stem cells despite compromised reconstitution ability. PloS One. 2013;8:e53912. doi: 10.1371/journal.pone.0053912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara A, Iwama A, Yamazaki S, Furuta C, Hirasawa R, Morita Y, Osawa M, Motohashi T, Eto K, Ema H, et al. Endomucin, a CD34-like sialomucin, marks hematopoietic stem cells throughout development. The Journal of Experimental Medicine. 2005;202:1483–1492. doi: 10.1084/jem.20051325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Dick JE. Reversible cell surface expression of CD38 on CD34-positive human hematopoietic repopulating cells. Experimental Hematology. 2007;35:1429–1436. doi: 10.1016/j.exphem.2007.05.017. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, Loewer S, Naveiras O, Kathrein KL, Konantz M, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HKA, Orkin SH. The journey of developing hematopoietic stem cells. Development (Cambridge, England) 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annual Review of Cell and Developmental Biology. 1995a;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1995b;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HKA. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risueño RM, Sachlos E, Lee JH, Lee JB, Hong SH, Szabo E, Bhatia M. Inability of human induced pluripotent stem cell-hematopoietic derivatives to downregulate microRNAs in vivo reveals a block in xenograft hematopoietic regeneration. Stem Cells (Dayton, Ohio) 2012;30:131–139. doi: 10.1002/stem.1684. [DOI] [PubMed] [Google Scholar]
- Robin C, Bollerot K, Mendes S, Haak E, Crisan M, Cerisoli F, Lauw I, Kaimakis P, Jorna R, Vermeulen M, et al. Human placenta is a potent hematopoietic niche containing hematopoietic stem and progenitor cells throughout development. Cell Stem Cell. 2009;5:385–395. doi: 10.1016/j.stem.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Shenoy S. Umbilical cord blood: an evolving stem cell source for sickle cell disease transplants. Stem Cells Translational Medicine. 2013;2:337–340. doi: 10.5966/sctm.2012-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Watanabe T, Sakurai S, Ohtake K, Kinoshita T, Araki A, Fujita T, Takei H, Takeda Y, Sato Y, et al. A novel glycosylphosphatidyl inositol-anchored protein on human leukocytes: a possible role for regulation of neutrophil adherence and migration. Journal of Immunology (Baltimore, Md : 1950) 1999;162:4277–4284. [PubMed] [Google Scholar]
- Tavian M, Biasch K, Sinka L, Vallet J, Péault B. Embryonic origin of human hematopoiesis. The International Journal of Developmental Biology. 2010;54:1061–1065. doi: 10.1387/ijdb.103097mt. [DOI] [PubMed] [Google Scholar]
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. GPI-80 expression in HSPC and myeloid cells. A. Expression of GPI-80 in hematopoietic populations in Ficoll-purified, CD34+ enriched fetal liver cells. B. Flow cytometry of total fetal liver and adult peripheral blood with CD66, CD14 and GPI-80 verifies expression of GPI-80 in myeloid cells. Fetal liver cells are gated from CD45+CD34− population. C. CD45RA expression in fetal liver CD34+ cells is restricted CD90− GPI-80− cells. D. Flow cytometry analysis of 18 week fetal liver with lineage markers CD13, CD66, CD235, and CD14 with and without Ficoll purification shows depletion of Lin+ cells with Ficoll purification.
Figure S2, related to Figure 2. Multipotency of GPI-80+ HSPC. A. Myelo-erythroid differentiation potential of GPI-80+ and GPI-80− HSPC on methylcellulose assay is shown (n=6 donors). Error bars represent mean ± SEM. B. Flow cytometry analysis for B-cell marker CD19 on GPI-80+ and GPI-80− HSPC after 2 weeks culture on OP9M2 is shown (n=3 donors). Error bars represent mean ± SEM. C. Flow cytometry analysis of T cell markers CD4 and CD8 after 2 weeks of culture on OP9-DII1 (n=3 donors). Error bars represent mean ± SEM. D. Myeloid and lymphoid potential of GPI-80+ and GPI-80− HSPC at the single cell level is shown. Quantification of proliferating clones (defined as >200 cells, n=2 donors), distribution of clone types (40 clones analyzed), and representative clones from GPI-80+ and GPI-80− HSPC after two weeks of culture on OP9M2 are shown. Though bulk cultures demonstrate multilineage potential of GPI-80− HSPC, single cell analysis reveals enrichment of multipotent cells in GPI-80+ population. Error bars represent mean ± SEM.
Figure S3, related to Figure 3. GPI-80 expression in multiple fetal hematopoietic sites. A. Lineage analysis of total vs ficoll purified, CD34+ enriched second trimester bone marrow with lineage markers CD13, CD66, CD235a, and CD14 shows depletion of Lin+ cells with Ficoll purification. B. Lineage analysis of 5 week total placenta with differentiation markers CD13, CD66, CD235a, and CD14 shows the presence of a subpopulation of GPI-80 HSPC that are devoid of lineage marker expression. C. Representative flow cytometry plots of endothelial cells show that GPI-80 expression in the placenta, fetal liver and fetal bone marrow is confined to hematopoietic cells.
Figure S4, related to Figure 4. Lentiviral shRNA knockdown of GPI-80 and ITGAM. A. Representative flow cytometry plot of fetal liver showing expression of CD11b(ITGAM) and CD18(ITGB2) on GPI-80+ HSPC. B. Representative flow cytometry plots of GPI-80 and ITGAM expression one week after lentiviral transduction, documenting reduction of GPI-80 and ITGAM protein on HSPC with two different shRNA vectors. C. Differentiation ability of HSPC after knockdown of GPI-80 or ITGAM (n=4 donors). Error bars represent mean ± SEM.
Table S1. Gene expression analysis of fetal liver hematopoietic subsets, Related to Figure 1. Gene expression analysis shows the comparison between CD34+CD38lo/−CD90+ HSPC and CD34+CD38lo/−CD90− HPC in human fetal liver. Significantly upregulated and downregulated genes (≥2 fold, p<0.05) are shown.
Table S2. Human engraftment in the bone marrow of NSG mice transplanted with GPI-80+ and GPI-80− HSPC, Related to Figure 2. Human engraftment at 16 weeks post-transplantation is shown.
Table S3. Gene expression analysis of fetal liver GPI-80+ and GPI-80− HSPC, Related to Figure 4. Gene expression analysis shows comparison between CD34+CD38lo/−CD90+GPI-80+ and CD34+CD38lo/−CD90+GPI-80− HSPC. Significantly upregulated and downregulated genes (≥2 fold, p<0.05) are shown.
Table S4. Human engraftment in the bone marrow of NSG mice transplanted with human fetal liver hematopoietic cells transduced with LKO, shGPI-80, or shITGAM lentiviral vectors, Related to Figure 4. Human engraftment at 10 weeks post-transplantation is shown.