Abstract
Background
Hepatocellular carcinoma (HCC) is the second most frequent cause of cancer death worldwide. Sulfatase 1 (SULF1) functions as a tumor suppressor in HCC cell lines in vitro but also has an oncogenic effect in some HCCs in vivo.
Aim
The purpose of this study was to examine the mechanisms regulating SULF1 and its function in HCC.
Methods
First, SULF1 mRNA and protein expression were examined. Second, we examined SULF1 gene copy numbers in HCC cells. Third, we assessed whether DNA methylation or methylation and/or acetylation of histone marks on the promoter regulate SULF1 expression. Finally, we examined the effect of 5-aza-2′-deoxycytidine (5-Aza-dC) on sulfatase activity and drug-induced apoptosis.
Results
SULF1 mRNA was downregulated in nine of eleven HCC cell lines, but only in six of ten primary tumors. SULF1 mRNA correlated with protein expression. Gene copy number assessment by fluorescence in situ hybridization showed intact SULF1 alleles in low-SULF1-expressing cell lines. CpG island methylation in the SULF1 promoter and two downstream CpG islands did not show an inverse correlation between DNA methylation and SULF1 expression. However, chromatin immunoprecipitation showed that the SULF1 promoter acquires a silenced chromatin state in low-SULF1-expressing cells through an increase in di/trimethyl-K9H3 and trimethyl-K27H3 and a concomitant loss of activating acetyl K9, K14H3 marks. 5-Aza-dC restored SULF1 mRNA expression in SULF1-negative cell lines, with an associated increase in sulfatase activity and sensitization of HCC cells to cisplatin-induced apoptosis.
Conclusion
SULF1 gene silencing in HCC occurs through histone modifications on the SULF1 promoter. Restoration of SULF1 mRNA expression by 5-Aza-dC sensitized HCC cells to drug-induced apoptosis.
Key Words: Sulfatase 1, Genetic regulation, Epigenetic regulation, Hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most frequent cancer and the second most common cause of cancer death worldwide [1]. Although HCC is less common in developed countries, its incidence has been rising in the USA, Japan, and Western Europe [2,3,4]. Major risk factors for HCC include chronic hepatitis and cirrhosis secondary to hepatitis B virus or hepatitis C virus infection, alcoholic cirrhosis, and nonalcoholic steatohepatitis, whereas less common risk factors include hereditary hemochromatosis and α1-antitrypsin deficiency [5,6,7,8]. The mechanisms of HCC development and progression are not completely elucidated. Therefore, an improved understanding of the pathogenesis of HCC will allow a more rational development of therapies for HCC.
The sulfatase 1 (SULF1) gene, which is located on chromosome 8, consists of 23 exons. Alternative splicing of the gene results in multiple transcript variants, including four variants (1-4) that possess long open reading frames. SULF1 is a heparan sulfate 6-O sulfatase which has been shown to act as a tumor suppressor in cell lines derived from several cancer types, including HCC cell lines. In contrast, there is also evidence that SULF1 is overexpressed in a proportion of primary tumors and may have oncogenic activity in primary tumors [9,10]. SULF1 desulfates cell surface heparan sulfate glycosaminoglycans (HSGAGs), which are complex linear polysaccharides composed of disaccharide repeats that are covalently attached to a polypeptide core to form heparan sulfate proteoglycans (HSPGs). HSGAGs regulate cellular signaling in at least two major and potentially opposing ways. First, HSGAGs act as coreceptors for heparin-binding growth factors such as fibroblast growth factor and vascular endothelial growth factor. The coreceptor function of HSGAGs is dependent on their sulfation state. Consequently, desulfation of HSGAGs by SULF1 inhibits ligand-receptor binding, decreases receptor phosphorylation, and downregulates heparin-binding growth factor signaling, thus rendering HCC cells more sensitive to chemotherapy-induced apoptosis [11,12,13,14]. Second, HSGAGs can act as sequestration or storage sites for growth factors and cytokines on the cell surface or in the extracellular matrix. In this model of HSGAG action, desulfation by SULF1 or the related protein SULF2 can release bound growth factors and cytokines and enhance cellular signaling [9,10,15,16].
The regulation of SULF1 in cancer appears to be complex. Complete loss or markedly diminished expression of SULF1 is relatively widespread among cell lines derived from epithelial cancers, including ovarian, breast, pancreas, kidney and hepatocellular cancers [14,17,18,19]. However, there is also evidence of overexpression of SULF1 in proportions of some cancer types or subtypes, including HCC, T-cell prolymphocytic leukemia, acute myeloid leukemia, renal carcinoma, lung adenocarcinoma, as well as brain, breast, head and neck, skin, testicular and gastric cancers [10,15,20,21].
In HCC, SULF1 has been shown to be downregulated in nine of eleven (82%) human HCC cell lines, but is upregulated in approximately 70% of primary HCC tissues examined [10,13]. Furthermore, approximately 40% of HCCs have loss of heterozygosity/allelic imbalance at the SULF1 locus [13,22]. The existence of a wide spectrum of SULF1 splice variants has also been demonstrated in canine HCC tumors [22]. Most recently, rescue of SULF1 expression has been proposed as a novel therapeutic option for the treatment of HCCs, since drug treatment combinations, such as the combination of the histone deacetylase inhibitor apicidin with doxorubicin, have been shown to be more effective in high-SULF1-expressing HCC cells and xenografts [23,24]. However, given the evidence for overexpression and a possible oncogenic function for SULF1 in a significant proportion of primary HCC tissues, it is not clear that rescue of SULF1 expression is appropriate for the treatment of all HCCs. It may be that the specific strategy for modulating SULF1 expression or activity will need to be tailored to the demonstrated functional effect of SULF1 in the particular tumor.
Examination of the SULF1 gene has not revealed any mutations in cancer; however, a functional polymorphism of the SULF1 gene promoter that regulates SULF1 expression has been shown to be associated with age at onset and progression-free survival of ovarian cancer patients. A polymorphism in the SULF1 gene region has also been associated with persistence of human papillomavirus and cervical precancer and cancer [19,25,26,27,28]. Transcriptional regulation of SULF1 expression has been shown to be enhanced by TGFβ1 and suppressed by hypoxia-inducible factor-1α and by variant hepatic nuclear factor 1 [19,27,28]. With respect to epigenetic regulation of SULF1 expression, the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-Aza-dC) restored SULF1 mRNA expression in SULF1-negative ovarian cancer cells [17]. A putative CpG island in intron 1 of the SULF1 gene was highly methylated in SULF1-negative ovarian cancer cells, confirming regulation of SULF1 expression by DNA hypermethylation. In addition, there were higher levels of histone H3 (K9) acetylation in a high-SULF1-expressing ovarian cancer cell line, and, in contrast, higher levels of histone H3 (K9) methylation in SULF1-negative ovarian cancer cells, suggesting a further level of epigenetic regulation of SULF1 in ovarian cancer through chromatin modification [17]. SULF1 has also been shown to be downregulated by CpG island hypermethylation in human breast and gastric cancer cell lines and resected tumors [29]. Consistent with the tumor suppressor effect of SULF1 in ovarian cancer, downregulation of SULF1 was significantly associated with chemotherapy resistance in ovarian cancer patients [17]. These findings suggest that the enhancement of heparin-binding growth and survival signaling produced by downregulation of SULF1 in cancer is clinically important. In contrast, recent evidence suggests that activation of SULF1 expression by promoter hypomethylation may be oncogenic in gastric cancer [21].
We have previously shown that treatment with 5-Aza-dC-restored SULF1 mRNA expression in three of five SULF1-negative HCC cell lines examined [13]. This suggests that the expression of SULF1 may be regulated by DNA hypermethylation in HCCs, similar to findings shown in ovarian, breast and gastric cancer. However, given the apparently opposing and contradictory evidence for SULF1 action in the recent literature, we examined the regulation of SULF1 expression in both HCC cell lines and tissues as well as the functional consequences in HCC cell lines more extensively. In the current study, we investigated the SULF1 mRNA and protein expression in HCC cells and tissues. Next, we assessed whether deletion of the SULF1 gene locus was the mechanism for the loss of SULF1 gene expression. We then examined whether SULF1 downregulation is the consequence of DNA hypermethylation in the SULF1 promoter region or whether it is rather due to epigenetic methylation and/or acetylation of histones interacting with the SULF1 promoter. Finally, we determined whether restoration of SULF1 gene expression by demethylation with 5-Aza-dC enhances the sensitivity of HCC cells to chemotherapy-induced apoptosis.
Materials and Methods
HCC Tissue Samples and Cell Lines
HCC tumors were resected from patients who were admitted to the Mayo Clinic between 1987 and 2003. Tissues were frozen in liquid nitrogen and stored at −80°C. Human HCC cell lines Hep3B, HepG2, PLC/PRF5, SKHep1, SNU182, SNU387, SNU398, SNU423, SNU499 and SNU475 were obtained from the American Type Culture Collection (ATCC). The Huh7 cell line was obtained from Dr. Gregory Gores.
DNA and RNA Extractions
DNA was extracted from cell lines and frozen tissues using the DNeasy kit (Qiagen Inc., Valencia, Calif., USA). RNA was extracted using Trizol reagent (Invitrogen, Life Technologies) followed by the RNeasy column clean-up procedure (Qiagen).
Drugs and Reagents
5-Aza-dC (Sigma, St. Louis, Mo., USA) was dissolved in dimethyl sulfoxide (DMSO) at 1 mM, stored at −20°C and diluted with 1 × PBS. In all experiments, the concentration of DMSO did not exceed 0.1%. Cisplatin (Sigma) was prepared immediately before use as a 1,000-fold concentrated solution in DMSO.
Real-Time PCR
Quantitative real-time PCR was performed on cDNA from ten pairs of benign and HCC tumor tissue and eleven HCC cell lines to examine SULF1 gene expression. Two mRNAs are transcribed from alternate promoters upstream of the SULF1 gene. The Applied Biosystems assay used in this study amplifies a sequence at the 3′ end of the SULF1 gene that is located on the boundary of exon 21-22 of the shorter mRNA or the boundary of exon 22-23 of the longer mRNA. This assay therefore recognizes both SULF1 mRNA transcripts. PCR reactions were performed in 96-well plates in a volume of 25 µl. ABI Assay-on-Demand gene expression kits (Applied Biosystems, Carlsbad, Calif., USA) for SULF1 (HS00290918-M1), 18S ribosomal RNA (4333760F), and 2 × TaqMan Universal Master Mix were used. The 18S ribosomal RNA level was used as an internal control. Reactions were run on an ABI 7300 Sequence Detection System, and results were analyzed using ABI 7300 software.
Immunohistochemistry of SULF1 in HCCs
Immunostaining was performed using purified rabbit polyclonal anti-human SULF1 antibody on 4-μm sections from paraffin-embedded HCC tissues on the EnVision+ system (Dako Corporation, Carpinteria, Calif., USA) [11]. A negative control was set up by replacing the primary antibody with 1% BSA-TBS. All negative control specimens showed no nonspecific staining.
Fluorescence in situ Hybridization
Cell Line Harvest. 25 μl of 0.001% colcemid and 25 μl of 0.1% ethidium bromide were added to cells growing in a 5-ml flask at approximately 70% confluency. After incubation at 37°C and 5% CO2 for 2 h, the cells were digested with trypsin. After brief centrifugation, the cells were mixed with 2.5 ml 0.8% sodium citrate and 2.5 ml 0.075 M KCl and incubated at room temperature for 15 min. Then, 5 ml of 3:1 methanol:acetic acid was added to the cells and centrifuged. The supernatant was discarded and cells were fixed with 70% ethanol. The cell suspension was dropped onto dry slides and placed in a Thermotron slide drying chamber set at 25°C and 45-55% relative humidity.
Bacterial Artificial Chromosome Probe Preparation. Bacterial artificial chromosome (BAC) clone RP11-131P18 containing the SULF1 gene was obtained from Research Genetics, Inc. (Huntsville, Ala., USA). DNA was isolated using a modified Qiagen Midi Kit protocol (Qiagen, Valencia, Calif., USA). The BAC DNA was labeled with Texas Red dUTP using a nick-translation kit (Boehringer Mannheim, Mannheim, Germany), precipitated with room-temperature isopropanol and then resuspended in TE.
Fluorescence in situ Hybridization. After precipitation, a labeled probe was dissolved at 20 ng/μl in a hybridization mixture containing 50% formamide, 10% dextran sulfate, 2 × SSC, 400-500 ng/μl Cot-1 DNA (Gibco BRL) and 0.5 μg/ml sheared herring sperm DNA. Before hybridization, slides were denatured at 72°C in 70% formamide in 0.6 × SSC for 3 min, followed by passage through an alcohol series. Hybridizations were performed as previously described [30], and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The chromosome enumeration probe 8 (CEP8; Vysis, Inc., part of Abbott Laboratories, Abbott Park, Ill., USA) and SULF1 signals in a dual-probe hybridization were counted in a total of 300 nuclei with a Diaplan microscope (Leitz, Wetzlar, Germany) equipped with a triple-pass filter.
Bisulfite Genomic Sequencing
Two CpG regions downstream of the transcription start site (TSS) were studied. We used SULF1 mRNA variant 3 as a reference sequence (NM_015170.2 from the National Center for Biotechnology Information). The most recent human genome assembly of the Genome Reference Consortium (GRCh38) was used to determine the locations of the bisulfite genomic sequencing (BGS) primers. The primers selected for SULF1 CpG region 1 are located at +69492682 (forward) and +69493024 (reverse) of chromosome 8. According to the numbering downstream of the SULF1 TSS, the primers for CpG region 1 amplify a 343-bp product in intron 1 (26,158-26,501). The primers for CpG region 2 are located at +69563760 (forward) and +69564131 (reverse) of chromosome 8. Based on the SULF1 TSS, primers for CpG region 2 amplify a 372-bp product in exon 5 (97,236-97,608). For the two CpG regions targeted in this study, the upstream CpG region 1 possesses more CpG dinucleotides than the downstream CpG region 2. Both HCC cell lines and HCC tissues were used for the BGS studies. HCC cell lines were cultured in media without demethylating agents. DNA isolated from HCC cell lines and HCC primary tissues was treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research, Orange, Calif., USA) and eluted in 20 µl of elution buffer. 1 µl of bisulfite-modified DNA was amplified in a 25-µl volume containing 12.5 µl of SyBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, Calif., USA), 0.1 µl (50 pmol/µl) of each SULF1 primer, and 11.3 µl of H2O. CpG region 1 is located in intron 1 and has previously been shown to have an increased frequency of DNA methylation in ovarian cell lines and ovarian primary tumors with suppressed SULF1 expression by Staub et al. [17]. CpG region 2 is located in exon 5 and has the next highest density of CpG dinucleotides in the region surrounding the human SULF1 promoter. The BGS primers for CpG region 1 were 5′-TATGGGGGTGTGTGATGATAATAATA-3′ (forward) and 5′-AAAACCACCTTCAAAATCTTCTCTAA-3′ (reverse), and for CpG region 2 they were 5′-TTGGAGTTTGGATTTTATTTTAGTT-3′ (forward) and 5′-CCTCAAAATATCTCCTAATAATCCT-3′ (reverse). The locations of these two primer sets are stated above. Amplification was done at 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 30 s, annealing at 58°C for 30 s and extension at 72°C for 45 s, followed by a final 10-min extension step. PCR products were ligated into pCR 2.1-TOPO cloning vector (Invitrogen, Carlsbad, Calif., USA). Blue and white selection was used, and white colonies were selected for standard PCR amplification using M13 primers [17-mer universal primers 5′-GTAAAACGACGGCCAGT-3′ (forward) and 5′-CAGGAAACAGCTATGAC-3′ (reverse)]. PCR products with SULF1 inserts were treated with ExoSAP-IT® solution (USB Corporation, Cleveland, Ohio, USA) to remove residual dNTPs, primers and any extraneous DNA. The samples were then sequenced using an ABI Prism 377 DNA Sequencer (Perkin Elmer, Boston, Mass., USA) to ascertain the methylation status of each CpG site.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was performed from nuclear extracts of SNU182, SNU449 and Hep3B cells using the Magna ChIP™ G Chromatin Immunoprecipitation Kit (Millipore, Billerica, Mass., USA). Antibodies against dimethyl K9 histone H3 (Millipore), trimethyl K9 histone H3 (Millipore), acetyl (K9, K14) histone H3 (Millipore) and trimethyl K27 histone H3 (Abcam, Cambridge, Mass., USA) and a control antibody were used. Specific primers for 170-bp [5′-GAGATGGCCCTTCCATAGACG-3′ (forward; −205 of TSS) and 5′-GAGAAGTGGGAGGTGCTACAG-3′ (reverse; −36 of TSS)] and 205-bp [5′-GCCACGATCTAATCAAAGAAAGG-3′ (forward; −348 of TSS) and 5′-CTCAGTGGGATGCACAGAAAC-3′ (reverse; −144 of TSS)] regions of the SULF1 promoter were utilized for PCR amplification. PCR products were separated on a 2% agarose gel.
Sulfatase Assay
Five days after treatment with 5-Aza-dC, SNU449 and Huh7 cells were washed in ice-cold PBS and lysed in SIE buffer (250 mM sucrose, 3 mM imidazole, pH 7.4, 1% ethanol) containing 1% (w/v) Nonidet P-40 and protease inhibitor cocktail (Roche Molecular Biochemicals). A total cellular protein of 100 μg was preincubated with 10 μM estrone-3-O-sulfamate (Sigma) at 37°C for 1 h to inhibit steroid sulfatases. 4-Methylumbelliferyl sulfate (4-MUS) was then added at 10 mM in 10 mM lead acetate in a 200-μl volume. After incubation for 24 h at 37°C, the reaction was terminated using 1 ml 0.5 M NaHCO3, pH 10.7. Fluorescence of the liberated 4-methylumbelliferone was measured using excitation and emission wavelengths of 365 and 450 nm, respectively.
Analysis of Apoptosis
Apoptosis was quantitated by assessing the number of cells with nuclear changes indicative of apoptosis (chromatin condensation and nuclear fragmentation) after staining with DAPI. SNU449 and Huh7 cells were treated with 5 μM 5-Aza-dC for 5 days; the medium was changed with a fresh drug every 3 days. The plates were washed and switched to serum-free medium. Cisplatin was then added at 5 μM. After 24 h of incubation at 37°C, DAPI was added at 5 μg/ml. After 20 min of incubation in the dark at 37°C, cells were examined by fluorescence microscopy (Nikon Eclipse TE200; Nikon Corp., Tokyo, Japan). An individual blinded to the experimental conditions counted at least 300 cells in six different high-power fields for each treatment. Each treatment was repeated at least three times in triplicate [13].
Results
SULF1 Is Downregulated in Most HCC Cell Lines but Variably Down- or Upregulated in Primary HCC Tumors
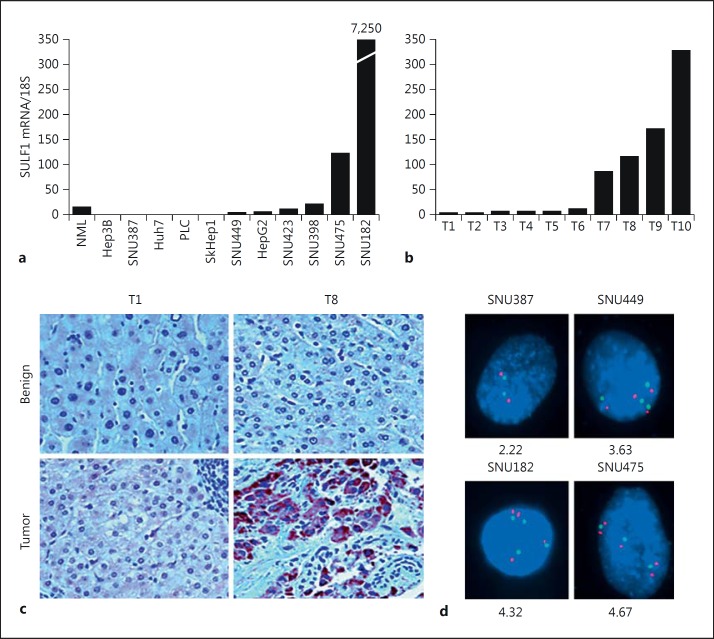
To determine the expression of SULF1 mRNA, we performed quantitative real-time PCR on cDNA from eleven HCC cell lines and ten pairs of benign and HCC tumor tissues. 18S ribosomal RNA was used as an internal control (fig. 1a). Since the target sequence for the SULF1 TaqMan gene expression assay is located at the boundaries of SULF1 exons 21-22 (for the shorter mRNA transcript) or exons 22-23 (for the longer mRNA transcript), the gene expression assay was able to detect both of the SULF1 transcripts in the HCC cells and HCC tumors. SULF1 expression was downregulated in 9 of the eleven HCC cell lines (82%) and upregulated in two cell lines, SNU475 and SNU182, which have high SULF1 expression. Of the primary tumors, SULF1 expression was downregulated in 6 of the 10 HCCs (60%), which were designated tumors T1-T6, and upregulated in 4 of the 10 (40%), which were designated T7-T10 (fig. 1b). To determine whether SULF1 protein levels correlated with the mRNA levels, SULF1 protein was assessed in HCC tumor and adjacent benign tissues by immunohistochemistry using a purified polyclonal SULF1 antibody (fig. 1c). The benign tissues adjacent to tumors T1 and T8 both showed low SULF1 protein expression, while their corresponding tumor tissues showed low and significantly higher levels of SULF1 expression, respectively. These results indicate that SULF1 staining at the protein level corresponds to the levels of SULF1 observed at the RNA level by qPCR.
Fig. 1.
a SULF1 mRNA expression in normal primary hepatocytes and eleven HCC cell lines. Total RNA was isolated from primary hepatocytes and eleven cell lines, and SULF1 mRNA was measured by real-time PCR. SULF1 mRNA levels were normalized by comparison with 18S ribosomal RNA levels measured by real-time PCR. Each measurement was performed in quadruplicate; standard curves prepared from dilutions of synthesized SULF1 and 18S standards were used. b Real-time PCR results from ten primary HCC tumors, measured as in a. c Immunohistochemistry showing SULF1 protein expression in HCC tumor and adjacent benign tissues. Immunostaining was performed on sections from paraffin-embedded HCC tissues using the purified SULF1 antibody. d FISH analysis of the SULF1 gene locus in four HCC cell lines. A BAC containing the SULF1 gene hybridized to metaphase preparations of the four HCC cell lines. Hybridization signals for the SULF1 BAC are shown in red; hybridization signals for the control centromere enumeration probe for chromosome 8 are shown in aqua. The average number of SULF1 copies per nucleus was calculated for each cell line.
The SULF1 Gene Locus Is Not Deleted in SULF1-Negative HCC Cell Lines and Overrepresented but Not Amplified in SULF1-Positive HCC Cell Lines
Since SULF1 mRNA was downregulated in the majority of cell lines and a proportion of primary tumors, but upregulated in the majority of primary HCCs, we examined whether the SULF1 gene locus was deleted in SULF1-negative HCC cells by hybridizing a BAC containing the SULF1 gene locus to metaphases from four HCC cell lines, two of which (SNU387 and SNU449) express very low levels of SULF1, while the other two (SNU182 and SNU475) express high levels of SULF1 (fig. 1d). The SULF1 gene locus was not deleted in any of the four HCC cell lines. The SNU387 cell line, which shows essentially no expression of SULF1, had 2.2 copies of the SULF1 gene locus per cell on average; SNU449, which expresses SULF1 but at a substantially lower level than the normal liver, had 3.6 copies per cell. The high-SULF1-expressing SNU182 and SNU475 cell lines had 4.3 and 4.6 copies of the SULF1 gene locus per cell, respectively. In additional experiments, we amplified the exons of the SULF1 gene in each of the cell lines. No exon deletions were identified (data not shown). Therefore, SULF1 is not downregulated or inactivated in HCC as a result of gene deletion and high-SULF1-expressing lines have increased copies of the SULF1 gene locus but do not show high-level amplification.
Expression of SULF1 Is Not Inversely Related to Methylation of SULF1 Gene CpG Regions 1 and 2 in HCC Cell Lines and Primary Tumors
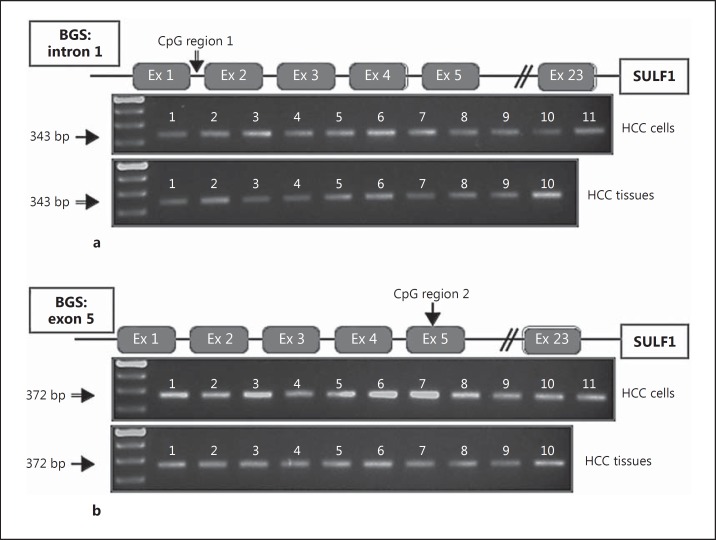
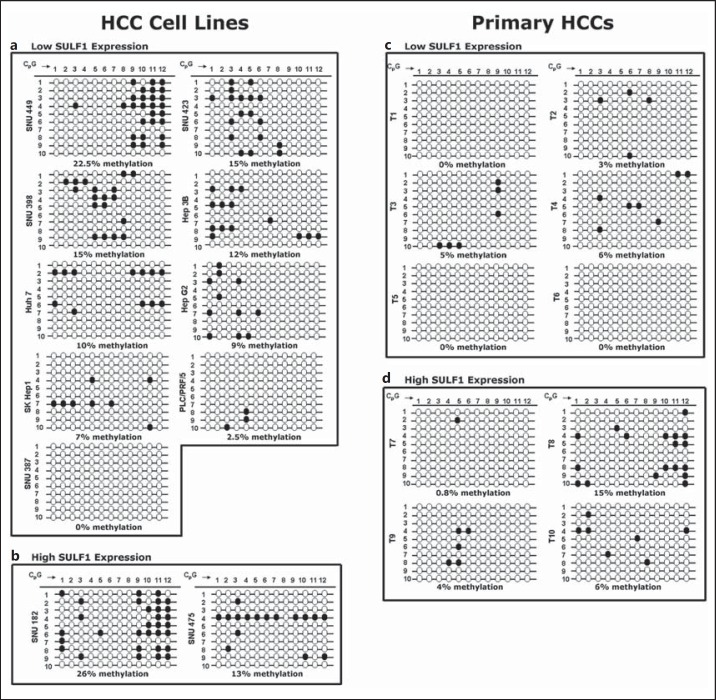
In silico analysis indicates the presence of two CpG-rich regions downstream of the SULF1 transcription start site. Methylation of the first of these sites has been shown to modulate chemoresistance in ovarian cancer [17]. To investigate the role of additional downstream gene methylation in the regulation of the SULF1 gene, we performed BGS of the two CpG-rich regions downstream of the transcription start site. The first downstream CpG region, designated CpG region 1, is located in intron 1 and has previously been identified as a DNA sequence that binds to CpG binding protein [31]. Region 1 has twelve CpG dinucleotides. After sodium bisulfite treatment of DNA samples from eleven HCC cell lines and ten primary HCCs, PCR was performed and fragments of the correct size were amplified (fig. 2a, upper and lower panels). This confirmed the amplification of region 1, and the amplified DNA was then subcloned and sequenced to reveal the spectrum of methylation changes in alleles from different cells. For each cell line or primary HCC, ten PCR products were cloned and sequenced using M13 primers to analyze the rates of CpG methylation (fig. 3). In contrast to the high levels of CpG methylation found in CpG region 1 in ovarian cancer cell lines and tumors, both HCC cell lines and primary HCCs had a relatively low methylation rate in CpG region 1. The methylation rates found in CpG region 1 for the low-SULF1-expressing cell line ranged from 0 to 22.5% (fig. 3a), while methylation rates for the two high-SULF1-expressing cell lines were 13-26% (fig. 3b). The average methylation rate was 10% in the low-SULF1-expressing cell lines and 19.5% in the high-SULF1-expressing cell lines. In tumors, the methylation rates found in CpG region 1 for the six low-SULF1-expressing tumors ranged from 0 to 6% (fig. 3c), while methylation rates for the four high-SULF1-expressing tumors ranged from 0.8 to 15% (fig. 3d). The average methylation rate was 2.3% in the low-SULF1-expressing tumors and 6.5% in the high-SULF1-expressing tumors. Therefore, the tumors and cell lines did not show the expected inverse correlation of CpG region 1 methylation rate with SULF1 mRNA expression, suggesting that this region is not epigenetically silenced via DNA methylation in HCC.
Fig. 2.
a PCR of CpG region 1 from eleven HCC cell lines after bisulfite treatment. Upper panel: lane 1, SNU387; 2, SNU398; 3, SNU423; 4, SNU449; 5, Huh7; 6, Hep3B; 7, SKHep1; 8, HepG2; 9, PLC/PRF/5; 10, SNU182; 11, SNU475. Lower panel: PCR of CpG region 1 for 10 HCC tumors. Lanes 1-10, primary HCC tumors. b PCR of CpG region 2 from eleven HCC cell lines after bisulfite treatment of DNA. This region contains the translation start site. Upper panel: lane 1, SNU387; 2, SNU398; 3, SNU423; 4, SNU449; 5, Huh7; 6, Hep3B; 7, SKHep1; 8, HepG2; 9, PLC/PRF/5; 10, SNU182; 11, SNU475. Lower panel: PCR of CpG region 2 for 10 HCC tumors. Lanes 1-10, primary HCC tumors.
Fig. 3.
Methylation status of CpG dinucleotides in CpG region 1 in eleven HCC cell lines and ten primary tumors determined using BGS. a Nine HCC cell lines with low SULF1 gene expression. b Two cell lines with high SULF1 expression. CpG numbers 1-12 (top) represent the CpG dinucleotides studied in this region. Numbers 1-10 (left) represent separate sequenced clones with twelve CpG dinucleotides. The percentages below each cell line represent the overall rate of methylation in the sample. c Six primary HCCs with low SULF1 gene expression. d Four primary HCCs with high SULF1 expression. Similarly, CpG numbers 1-12 (top) are the CpG dinucleotides studied in this region in each cell line, and numbers 1-10 (left) are separate sequenced clones. The percentages below each cell line represent the overall rate of methylation in the sample.
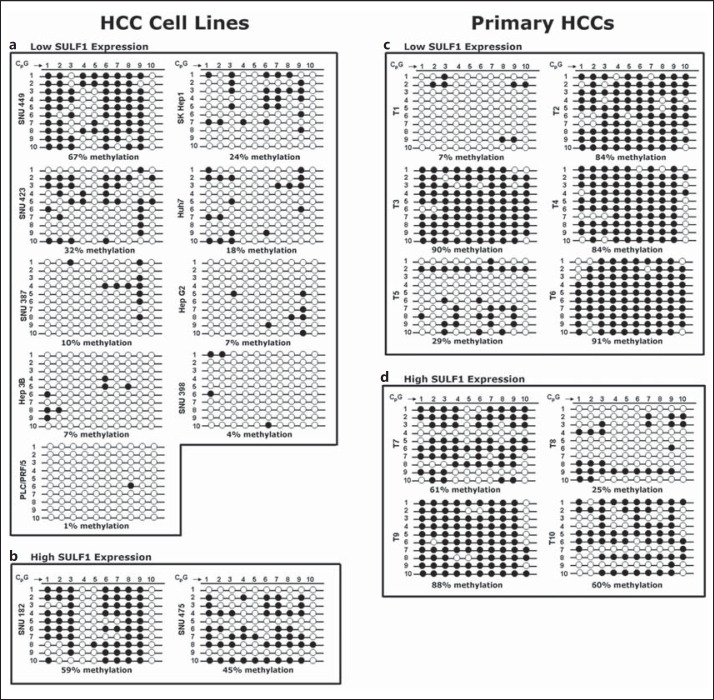
Next, we determined whether methylation at the second downstream region of high CpG content in exon 5 of the SULF1 gene adjacent to the translation start site, designated CpG region 2, was associated with expression of the SULF1 gene in HCC. Region 2 contains ten CpG dinucleotides. After sodium bisulfite treatment of DNA samples from eleven HCC cell lines and ten primary HCCs, the region was amplified and the correct fragment size was amplified in all samples. For each cell line or primary HCC, 10 PCR products were subcloned and sequenced using M13 primers to analyze the rates of CpG methylation (fig. 4). In contrast to the low levels of CpG methylation found in CpG region 1, this second region showed higher methylation rates. The methylation rates found in CpG region 2 for the low-SULF1-expressing cell lines ranged from 1 to 67% (fig. 4a), while methylation rates for the two high-SULF1-expressing cell lines ranged from 45 to 59%, respectively (fig. 4b). The average methylation rate was 19% in the low-SULF1-expressing cell lines and 52% in the high-SULF1-expressing cell lines. In tumors, the methylation rates found in CpG region 2 for the low-SULF1-expressing tumors ranged from 7 to 91% (fig. 4c), while methylation rates for the four high-SULF1-expressing tumors ranged from 25 to 88% (fig. 4d). The average methylation rate was 64% in the low-SULF1-expressing tumors and 58.5% in the high-SULF1-expressing tumors. Consequently, the expected inverse correlation between CpG methylation and mRNA expression was also not seen in region 2. For example, cell line SNU182, which expresses high levels of SULF1 mRNA, had essentially the same methylation rate as the low-expressing cell line SNU449. Thus, for neither downstream CpG region 1 nor CpG region 2 of the SULF1 gene was DNA methylation shown to account for the regulation of SULF1 expression in HCCs.
Fig. 4.
Methylation status of CpG dinucleotides in CpG region 2 in eleven HCC cell lines and ten primary tumors using BGS. a Nine HCC cell lines with low SULF1 gene expression. b Two cell lines with high SULF1 expression. CpG numbers 1-10 (top) represent the CpG dinucleotides studied in this region in each cell line. Numbers 1-10 (left) represent separate sequenced clones with ten CpG dinucleotides. The percentages below each cell line represent the overall rate of methylation. c Six primary HCCs with low SULF1 gene expression. d Four primary HCCs with high SULF1 expression. Similarly, CpG numbers 1-10 (top) are the CpG dinucleotides studied in this region in each cell line, and clone numbers 1-10 (left) are separate sequenced clones. The percentages below each cell line represent the overall rate of methylation.
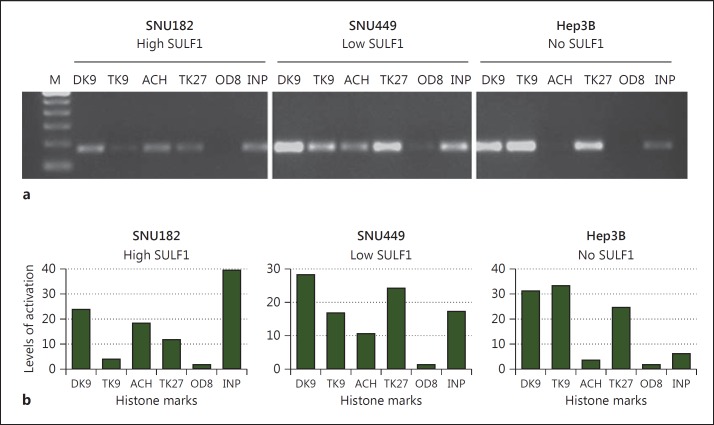
Low SULF1 Expression Corresponds to the Presence of Silencing Histone Marks on the SULF1 Promoter
Epigenetic modifications of not only DNA, but also histone proteins, which together with DNA comprise the core components of chromatin, serve as mechanisms for regulating gene expression. Previous studies have shown that treatment with 5-Aza-dC can reactivate gene expression without promoter demethylation [32,33]. Based on the Histone Code, covalent modifications of the N-terminal histone tails, which include acetylation, methylation, phosphorylation, ubiquitination and sumoylation, regulate key cellular processes such as gene transcription. Therefore, to investigate whether histone modifications present on the SULF1 promoter correlated with the level of SULF1 expression, we performed ChIP assays for various histone marks. For this purpose, we examined three HCC cell lines, one with high SULF1 expression (SNU182), one with low SULF1 expression (SNU449) and one with no SULF1 expression (Hep3B). For each of these cell lines, we performed a ChIP assay on the SULF1 promoter for histone modifications indicative of an ‘activated’ state (acetyl K9, K14 of histone H3) as well as marks for a ‘silenced’ chromatin state (dimethyl K9 histone H3, trimethyl K9 histone H3, and tri-methyl K27 histone H3). A nonspecific IgG antibody was used as a control. Interestingly, using two different sets of primers within the SULF1 promoter, the level of SULF1 expression corresponded with the histone modifications present on the promoter as shown by the ethidium bromide gels and semiquantitated levels of histone marks (fig. 5) from one primer set. The results of the other primer set revealed a similar pattern (data not shown). For example, in the high-expressing cell line, SNU182, the most prominent histone marks present on the promoter were acetyl K9, K14 H3 and dimethyl K9 histone H3 associated with a more permissive chromatin state. On the other hand, the low-expressing SNU449 cells acquired a more silenced chromatin state through an increase in di/trimethyl-K9H3 and trimethyl-K27H3. Finally, in Hep3B, the cell line with no SULF1 expression, a concomitant loss of the activating acetyl K9, K14H3 marks was observed in the presence of di/trimethyl-K9H3 and trimethyl-K27H3. Our negative control IgG antibody showed no amplification, demonstrating the specificity of our results.
Fig. 5.
SULF1 expression is regulated by histone modifications. Examination of histone marks demonstrates that the SULF1 promoter in low-expressing cells acquires a silenced chromatin state through an increase in trimethyl-K9H3 and trimethyl-K27H3 with a concomitant loss of activating acetyl K9, K14H3 marks. Positive amplification of PCR products is shown in the input DNA lanes (INP), demonstrating that this region of the SULF1 promoter is present in all samples before immunoprecipitation. M = Molecular weight marker; DK9 = dimethyl K9 histone H3; TK9 = trimethyl K9 histone H3; ACH = acetyl K9, K14 histone H3; TK27 = trimethyl K27 histone H3; OD8 = Omni-probe antibody (D-8), a mouse monoclonal IgG used as negative control.
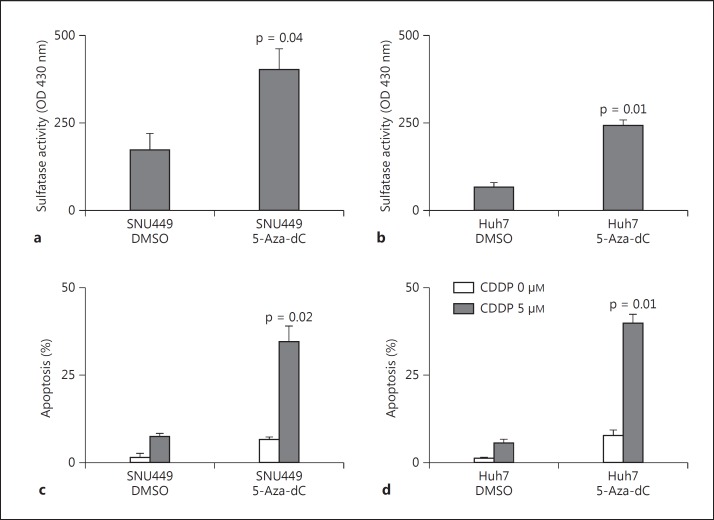
Restoration of Sulfatase Activity after 5-Aza-dC Treatment Is Associated with Increased Sensitivity of HCC Cells to Cisplatin-Induced Apoptosis
We have shown that treatment with 5-Aza-dC results in reactivation of SULF1 expression in the SNU449 and Huh7 cell lines, which have endogenous low levels of this transcript [13]. To determine whether restoration of SULF1 mRNA expression by DNA demethylation in cell lines with low expression of SULF1 results in synthesis of functional enzyme and increased sulfatase activity, we treated the SNU449 and Huh7 cell lines with 5-Aza-dC for 5 days. Nonsteroid sulfatase activity was then measured in whole cell lysates using 4-MUS as a substrate after pretreatment with EMATE to inhibit steroid sulfatase activity. Increased levels of sulfatase activity were observed in both SNU449 cells (fig. 6a) and Huh7 cells (fig. 6b) following treatment with 5-Aza-dC.
Fig. 6.
a, b Restoration of nonsteroid sulfatase activity of SULF1-negative HCC cells after 5-Aza-dC treatment. Sulfatase activity was measured in SNU449 Huh7 cells treated with DMSO only or with 5-Aza-dC after inhibition of steroid sulfatase activity using EMATE. Sulfatase activity was measured using 4-MUS as the substrate. The fluorescent reaction product was measured using excitation and emission wavelengths of 365 and 450 nm, respectively. c, d SNU449 and Huh7 cells were treated with either control DMSO or 5 μM 5-Aza-dC for 5 days. After day 5, the plates were washed and changed to serum-free medium, and cisplatin (CDDP; 0 or 5 μM) was added. After 24 h of incubation with CDDP at 37°C, DAPI (5 μg/ml) was added to each well. Apoptosis was then assessed by fluorescence microscopy using excitation and emission filters of 380 and 430 nm.
Since low levels of SULF1 expression have been associated with resistance to cisplatin-induced apoptosis [13], we examined whether reactivation of SULF1 gene expression had an effect on cisplatin sensitivity. For this purpose, the two 5-Aza-dC-responsive cell lines, SNU449 and Huh7, were treated with DMSO control or 5 μM of 5-Aza-dC. After 5 days, the plates were washed and switched to serum-free medium, and cisplatin (CDDP) was added at 0 and 5 μM, respectively. After 24 h, DAPI (5 μg/ml) was added to each well, and apoptosis was assessed by fluorescence microscopy. Treatment with 5 μM 5-Aza-dC resulted in increased sensitivity of both SNU449 and Huh7 cells to CDDP-induced apoptosis (fig. 6c, d). These results demonstrate that 5-Aza-dC treatment was able to restore synthesis of functional SULF1 enzyme and increase sulfatase activity, which also resulted in cisplatin sensitivity.
Discussion
The currently available evidence suggests that SULF1 functions as a heparan sulfate-editing sulfatase, with its primary effect as a 6-O sulfatase, removing sulfate moieties from HSGAG disaccharides. Because HSGAGs serve dual functions as either coreceptors or storage/sequestration sites for heparin-binding growth factors and cytokines, it is conceivable that the SULFs can have opposing actions on cellular functions such as growth, survival, motility, and invasiveness, depending on the relative dependence of the particular cell on different cell signaling pathways and the relative importance of the coreceptor or storage/sequestration functions of HSGAGs for cell function. Desulfation of HSGAGs by SULF1 profoundly inhibits HCC tumor growth and enhances sensitivity of HCC cells to chemotherapy induced-apoptosis by abrogating ligand-receptor binding, which is dependent on sulfation of cell surface HSGAG coreceptors. As a consequence of its effect on signaling by heparin-binding growth factors such as fibroblast growth factor-2, SULF1 reduces both anchorage-dependent and -independent cell growth, invasion and angiogenesis [14,18,34,35]. In addition, we have shown that SULF1 potentiates the anti-tumor effects of histone deacetylase inhibitors [11,23]. SULF1 therefore functions as a tumor suppressor in a proportion of HCC cell lines, and similar results have been shown in ovarian cancer, and head and neck squamous cell cancers [12,13,14].
As is usual for functional tumor suppressors, expression of SULF1 is downregulated in almost all HCC cell lines; SULF1 is also downregulated in approximately 30% of primary HCCs, but appeared to be paradoxically upregulated in 70% of HCCs [13]. To explore the mechanism for downregulation of SULF1 in HCC cell lines and primary tumors, we have previously shown an increased frequency of allelic imbalance (LOH) at the SULF1 locus in 94 primary HCC tumors [13]. Of the 31 HCCs in which SULF1 expression was previously examined by real-time PCR, 14 showed LOH at the SULF1 locus. Of the 14 tumors with LOH, 7 (50%) also showed downregulation of SULF1 mRNA expression. Similarly, SULF1 expression was restored by treatment of two SULF1-negative ovarian cancer cell lines with 5-Aza-dC [17].
Here, we studied the possible contributions of genetic (gene deletion) and epigenetic regulation (DNA hypermethylation and histone modifications) to suppression of SULF1 expression in HCC cell lines and primary tumors. First, we used real-time PCR to quantitate the downregulation of SULF1 expression in eleven HCC cell lines and ten primary HCC tumors, confirming the results of our previous real-time PCR analysis (fig. 1a, b) [13]. Second, we confirmed that tumor SULF1 protein levels reflect differences in mRNA expression, with data showing low SULF1 protein expression in a tumor with low SULF1 mRNA and significantly higher levels of SULF1 protein expression in a tumor with high SULF1 mRNA (fig. 1c). This result indicates that SULF1 expression in HCC may be regulated at the transcriptional level. Third, we confirmed that the downregulation of SULF1 in HCC cell lines is not due to decreased SULF1 gene copy numbers. Our fluorescence in situ hybridization (FISH) analysis showed that the SULF1 gene was not deleted in the four HCC cell lines tested, two with low SULF1 expression and the other two with high SULF1 expression. All four cell lines had at least diploid FISH signals, showing that there are at least a minimal number of copies of the SULF1 gene present (fig. 1d). We therefore hypothesized that the SULF1 gene is epigenetically regulated at the transcriptional level.
Analysis of the region immediately upstream of the SULF1 transcription start site showed that it was relatively CpG poor. Since CpG-poor sites can nevertheless be important for the regulation of DNA methylation, we examined the methylation status of this region in four HCC cell lines and four resected benign and tumor tissues using methylation-specific PCR. Due to the distances between the CpGs, it was technically only feasible to amplify a region containing six CpG sites for both methylation and hypomethylation (data inconclusive and not shown).
Previous analysis of bisulfite-modified genomic DNA from ovarian cancer cell lines and primary tumors has demonstrated an increased frequency of methylation of a CpG-rich region containing twelve CpG dinucleotides located in intron 1 of the SULF1 gene [17]. To determine whether SULF1 mRNA expression is regulated by epigenetic DNA methylation in HCCs, we analyzed two CpG-rich regions in the SULF1 gene locus downstream of the transcription start site. The first one, designated CpG region 1, contains twelve CpG dinucleotides and has previously been shown by Staub et al. [17] to be regulated by methylation in ovarian cancer. Bisulfite genomic sequencing of this region in eleven HCC cell lines and ten primary HCCs showed that the rates of methylation were much lower than those found in ovarian cancer (fig. 3) [17]. Therefore, although SULF1 is regulated by methylation of CpG region 1 in ovarian cancer, this does not appear to be the case for HCC. Subsequently, we explored the possibility that another CpG-rich region located in exon 5 is responsible for the epigenetic regulation of SULF1 in HCCs. This region, designated CpG region 2, containing ten CpG dinucleotides, is in close proximity to the translation start site (ATG). Region 2 was found to have much higher rates of CpG methylation than region 1; however, the expected inverse correlation of region 2 methylation with SULF1 mRNA expression was not shown (fig. 4). Although neither methylation of the CpG region 1 in intron 1 nor methylation of the CpG region 2 in exon 5 of the SULF1 gene showed a correlation with SULF1 mRNA expression. Nevertheless, non-promoter CpG island methylation has been shown to be susceptible to regulation by aberrant methylation [36]. In addition, previous studies have shown that treatment with 5-Aza-dC can reactivate gene expression without promoter demethylation [32,33]. For example, treatment with 5-Aza-dC can induce reexpression of the p21WAF1 tumor suppressor in human acute myeloid leukemia without evidence of p21WAF1 promoter methylation, but with increased acetylation of histone H3 at the unmethylated p21WAF1 promoter [32].
Similarly, in our current study, we demonstrated that the level of SULF1 expression corresponded with the histone modifications present on the promoter (fig. 5). Cells expressing no SULF1 mRNA, as compared to low- and high-SULF1-expressing cells, acquired a more silenced chromatin state through an increase in di/trimethyl-K9H3 and trimethyl-K27H3 with a concomitant loss of activating acetyl K9, K14H3 marks. Thus, our study demonstrates that, besides the previously identified mechanism for SULF1 downregulation by promoter DNA methylation in most other cancers, SULF1 can also be downregulated in a significant number of HCCs, in part as a result of the silenced state of chromatin on its promoter.
Finally, we determined that the restoration of SULF1 mRNA expression after 5-Aza-dC treatment in SNU449 and Huh7 cell lines is accompanied by increased sulfatase activity. In addition, treatment with 5 μM 5-Aza-dC resulted in increased sensitivity of both SNU449 and Huh7 cells to CDDP-induced apoptosis. Given that 5-Aza-dC treatment was associated with restoration of sulfatase activity and increased sensitivity to cisplatin-induced apoptosis, we suggest that 5-Aza-dC treatment can reactivate expression of SULF1 gene without promoter demethylation. In light of this data, we can propose a model to explain how 5-Aza-dC is able to mediate SULF1 reexpression independent of CpG island methylation. 5-Aza-dC may induce the reexpression of a transcription factor or chromatin-remodeling protein, which works according to the histone code hypothesis, to modify histones without the need of DNA methylation. The silencing histone marks observed on the SULF1 promoter, namely di/trimethyl-K9H3 and trimethyl-K27H3, are the products of two histone methyltransferase pathways, HP1-G9a-SUV39H1 and PRC2, respectively [37]. In fact, these two histone mark regulators and DNA methyltransferases form complexes inside the nucleus. For instance, EZH2, the histone methyltransferase of the Polycomb complex, interacts and induces the catalytic activity of DNMT1 [38]. A similar cooperation has been discovered between HP1 and DNMT1 [39]. Similar to our results, Buchi et al. [40] found that, although two distinct DNMT inhibitors could reactivate the IL3 gene, there was no significant change in DNA methylation status of IL3, but rather a loss of H3K27me3, and gain of acetylated histone H4 was responsible for its reactivation. Alternatively, 5-Aza-dC may be targeting an unknown regulatory element or enhancer region of SULF1, since it is known that many genes are regulated through the interaction of different regions of the genome in 3D. Thus, it is plausible that 5-Aza-dC targets a yet to be identified regulatory region outside of the ones studied here, which might work to regulate the SULF1 promoter in 3D [41]. Further experiments will explore the relative importance of this epigenetic regulation of SULF1 and its increased sensitivity to cisplatin-induced apoptosis as a target for therapy of the subset of HCCs with decreased SULF1 expression.
Conclusion
In this study, we have shown that HCC cell lines had intact SULF1 genes in their nucleus but low SULF1 mRNA expression due to the presence of histone modifications, indicative of silenced chromatin on the SULF1 promoter rather than DNA methylation. Finally, 5-Aza-dC treatment was associated with restoration of sulfatase activity and increased sensitivity to cisplatin-induced apoptosis.
Disclosure Statement
The authors have no competing interests to declare.
Acknowledgements
This work was supported by the Mayo Clinic and Mayo Clinic Cancer Center and by NIH Grants CA82862, CA100882, CA128633 and CA165076, an Industry Research Scholar Award and a Bridging Grant Award from the Foundation for Digestive Health and Nutrition and a Harold Amos Medical Faculty Development Award from the Robert Wood Johnson Foundation (to L.R.R.), as well as NIH grant P30DK084567 (Mayo Clinic Center for Cell Signaling in Gastroenterology). The authors thank Vicki Campion, Tammy Szewczynski and Jennifer Rud for secretarial assistance and Colin Boettcher for critical review of the manuscript.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Epidemiology of hepatocellular carcinoma in USA. Hepatol Res. 2007;37(suppl 2):S88–S94. doi: 10.1111/j.1872-034X.2007.00168.x. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MB, Abbruzzese JL. Opportunities for targeted therapies in hepatocellular carcinoma. J Clin Oncol. 2005;23:8093–8108. doi: 10.1200/JCO.2004.00.1537. [DOI] [PubMed] [Google Scholar]
- 5.Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–257. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 7.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene. 2011;30:1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309–2324. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhanasekaran R, Nakamura I, Hu C, Chen G, Oseini A, Seven E, Miamen A, Moser C, Zhou W, Mounajjed T, Fernandez-Zapico M, Roberts L. Activation of the transforming growth factor-β/SMAD transcriptional pathway underlies a novel tumor-promoting role of sulfatase1 in hepatocellular carcinoma. Hepatology 2014, Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 10.Yang JD, Sun Z, Hu C, Lai J, Dove R, Nakamura I, Lee JS, Thorgeirsson SS, Kang KJ, Chu IS, Roberts LR. Sulfatase 1 and sulfatase 2 in hepatocellular carcinoma: Associated signaling pathways, tumor phenotypes, and survival. Genes Chromosomes Cancer. 2011;50:122–135. doi: 10.1002/gcc.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai JP, Yu C, Moser CD, Aderca I, Han T, Garvey TD, Murphy LM, Garrity-Park MM, Shridhar V, Adjei AA, Roberts LR. Sulf1 inhibits tumor growth and potentiates the effects of histone deacetylase inhibitors in hepatocellular carcinoma. Gastroenterology. 2006;130:2130–2144. doi: 10.1053/j.gastro.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 12.Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, Smith DI, Roberts LR, Shridhar V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 13.Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, Montoya DP, Matthews TA, Nagorney DM, Cunningham JM, Smith DI, Greene EL, Shridhar V, Roberts LR. HSulf1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126:231–248. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 14.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 15.Bret C, Moreaux J, Schved JF, Hose D, Klein B. Sulfs in human neoplasia: implication as progression and prognosis factors. J Transl Med. 2011;9:72. doi: 10.1186/1479-5876-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, Scheerer M, Roberts LR, Molina J, Shridhar V. Epigenetic silencing of hsulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26:4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 18.Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, Shridhar V. Loss of hsulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 19.Khurana A, Liu P, Mellone P, Lorenzon L, Vincenzi B, Datta K, Yang B, Linhardt RJ, Lingle W, Chien J, Baldi A, Shridhar V. HSulf-1 modulates FGF2- and hypoxia-mediated migration and invasion of breast cancer cells. Cancer Res. 2011;71:2152–2161. doi: 10.1158/0008-5472.CAN-10-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junnila S, Kokkola A, Mizuguchi T, Hirata K, Karjalainen-Lindsberg ML, Puolakkainen P, Monni O. Gene expression analysis identifies over-expression of CXCL1, SPARC, SPP1, and SULF1 in gastric cancer. Genes Chromosomes Cancer. 2010;49:28–39. doi: 10.1002/gcc.20715. [DOI] [PubMed] [Google Scholar]
- 21.Hur K, Han TS, Jung EJ, Yu J, Lee HJ, Kim WH, Goel A, Yang HK. Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. J Pathol. 2012;228:88–98. doi: 10.1002/path.4055. [DOI] [PubMed] [Google Scholar]
- 22.Gill RB, Day A, Barstow A, Zaman G, Chenu C, Dhoot GK. Mammalian SULF1 RNA alternative splicing and its significance to tumour growth regulation. Tumour Biol 2012. [DOI] [PubMed]
- 23.Lai J, Sandhu D, Moser C, Cazanave S, Shire A, Oseini A, Shridhar V, Sanderson SR, Roberts LR. Additive effect of apicidin and doxorubicin in sulfatase 1 (SULF1)-expressing hepatocellular carcinoma in vitro and in vivo. Journal of Hepatology. 2009;50:1112–1121. doi: 10.1016/j.jhep.2008.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai JP, Thompson JR, Sandhu DS, Roberts LR. Heparin-degrading sulfatases in hepatocellular carcinoma: roles in pathogenesis and therapy targets. Future Oncol. 2008;4:803–814. doi: 10.2217/14796694.4.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han CH, Huang YJ, Lu KH, Liu Z, Mills GB, Wei Q, Wang LE. Polymorphisms in the SULF1 gene are associated with early age of onset and survival of ovarian cancer. J Exp Clin Cancer Res. 2011;30:5. doi: 10.1186/1756-9966-30-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M, Rodriguez AC, Sherman ME, Bratti C, Schiffman M, Wacholder S, Burk RD, Herrero R, Chanock SJ, Hildesheim A. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS One. 2010;5:e8667. doi: 10.1371/journal.pone.0008667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue X, Li X, Nguyen HT, Chin DR, Sullivan DE, Lasky JA. Transforming growth factor-β1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J Biol Chem. 2008;283:20397–20407. doi: 10.1074/jbc.M802850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P, Khurana A, Rattan R, He X, Kalloger S, Dowdy S, Gilks B, Shridhar V. Regulation of hsulf-1 expression by variant hepatic nuclear factor 1 in ovarian cancer. Cancer Res. 2009;69:4843–4850. doi: 10.1158/0008-5472.CAN-08-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Fan JQ, Li J, Li QS, Yan Z, Jia XK, Liu WD, Wei LJ, Zhang FZ, Gao H, Xu JP, Dong XM, Dai J, Zhou HM. Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer. 2009;124:739–744. doi: 10.1002/ijc.23960. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- 31.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 32.Scott SA, Dong WF, Ichinohasama R, Hirsch C, Sheridan D, Sanche SE, Geyer CR, Decoteau JF. 5-Aza-2′-deoxycytidine (decitabine) can relieve p21WAF1 repression in human acute myeloid leukemia by a mechanism involving release of histone deacetylase 1 (HDAC1) without requiring p21WAF1 promoter demethylation. Leuk Res. 2006;30:69–76. doi: 10.1016/j.leukres.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Schmelz K, Sattler N, Wagner M, Lubbert M, Dorken B, Tamm I. Induction of gene expression by 5-aza-2′-deoxycytidine in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) but not epithelial cells by DNA-methylation-dependent and -independent mechanisms. Leukemia. 2005;19:103–111. doi: 10.1038/sj.leu.2403552. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, Felix K, Giese T, Buchler MW, Friess H. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer. 2005;4:14. doi: 10.1186/1476-4598-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, Ramakrishnan S, Shridhar V. Hsulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66:6025–6032. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 36.Liang P, Song F, Ghosh S, Morien E, Qin M, Mahmood S, Fujiwara K, Igarashi J, Nagase H, Held WA. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011;12:231. doi: 10.1186/1471-2164-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nestorov P, Tardat M, Peters AH. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol. 2013;104:243–291. doi: 10.1016/B978-0-12-416027-9.00008-5. [DOI] [PubMed] [Google Scholar]
- 38.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 39.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchi F, Masala E, Rossi A, Valencia A, Spinelli E, Sanna A, Gozzini A, Santini V. Redistribution of H3K27me3 and acetylated histone H4 upon exposure to azacitidine and decitabine results in de-repression of the AML1/ETO target gene IL3. Epigenetics. 2014;9:387–395. doi: 10.4161/epi.27322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]