Abstract
Macrophage and dendritic cell (DC) are hematopoietic cells found in all tissues in the steady state that share the ability to sample the environment but have distinct function in tissue immunity. Controversies remain on the best way to distinguish macrophages from DCs in vivo. In this Perspective, we discuss how recent discoveries in the origin of the DC and macrophage lineage help establish key functional differences between tissue DC and macrophage subsets. We also emphasize the need to further understand the functional heterogeneity of the tissue DC and macrophage lineages to better comprehend the complex role of these cells in tissue homeostasis and immunity.
History of the Discovery of the Mononuclear Phagocyte System
In 1908, Elie Metchnikoff (1845–1916), together with Paul Ehrlich, was awarded the Nobel Prize for his work on phagocytosis (Kaufmann, 2008). Metchnikoff classified phagocytes (named after the Greek “phago” for devour and “cytes” for cells) into “macrophages” (large eaters) and “microphages” (a smaller type of phagocytic cell, the polymorphonuclear leukocyte now known as granulocytes) and argued that both types of phagocytes played an important role in host resistance against infections. Metchnikoff also recognized the close relationship between mononuclear phagocytic cells in the spleen, lymph nodes, bone marrow, and connective tissues, leading him to introduce for the first time the term “macrophage system” (Gordon, 2008). Karl Albert Ludwig Aschoff, a German physician and pathologist, developed this concept further and grouped several cell types into what he called the reticulo-endothelial system and subsequently the reticulo-histiocyte system. The reticuloendothelial system included reticular cells (or fixed macrophages) of the spleen and lymph nodes, endothelial cells of the lymph and blood sinuses, monocytes, and histiocytes, a term referred to as “tissue wandering as opposed to fixed” macrophages. These cells were grouped on the basis of their capacity to uptake vital dye in vivo, an assay thought to measure phagocytic activity. However, poorly phagocytic cells, such as endothelial cells can also become labeled as a result of pinocytosis, especially when large amounts of dye are applied. Such labeling is therefore unreliable as a criterion for the identification of mononuclear phagocytes and a new classification was devised in 1969 to group only highly phagocytic cells and their precursors in one system called the mononuclear phagocyte system (MPS), a term that is still in use today (van Furth et al., 1972).
In the early 1970s, Ralph Steinman and Zanvil Cohn identified a population of hematopoietic cells in the mouse spleen that excel at antigen presentation and T cell stimulation, named dendritic cells (DCs) (Steinman and Cohn, 1973, 1974). Similar to macrophages, DCs are derived from hematopoietic progenitors and populate most lymphoid and nonlymphoid tissues. Initially thought to be poorly phagocytic, there is now evidence that DCs have a potent phagocytic activity that is mostly dedicated to inform the adaptive immune system about peripheral tissue cues through their unique ability to sample tissue antigens, migrate to the draining lymph nodes, present extracellular antigens, and initiate tissue-specific T cell immunity (Mellman and Steinman, 2001).
Accumulating evidence now suggests that macrophages and DCs are tissue phagocytic cells that specialize in the sensing and sampling of the tissue environment. Although the term MPS has created a framework that helped our understanding of tissue phagocyte biology, it also led to the assumption that all tissue phagocytes are identical in origin and function. Here, we describe the limitation of the MPS definition and its failure to account for the heterogeneous origin of tissue phagocytes. We also argue that defining DC and macrophage population on the basis of their origin should help understand the discrete role of these cells in tissue immunity and homeostasis.
Heterogeneity of Tissue-Resident DCs
DCs are classified into two main subsets that include classical DCs and plasmacytoid DCs (Heath and Carbone, 2009). Because the main theme of this Perspective is to discuss tissue phagocyte heterogeneity, plasmacytoid DCs will not be discussed here, considering that the ability of these cells to populate nonlymphoid tissues in the steady state as well as to phagocytose, process, and present extracellular tissue antigens is limited (Villadangos and Young, 2008).
Classical DCs (referred to as DCs here) form a heterogeneous population of hematopoietic cells that reside in all lymphoid, interface, and connective tissues (Figure 1). One of the biggest challenges to understand the molecular mechanisms that control DC function has been the lack of specific markers to define the DC lineage. DCs continue to be defined to date as hematopoietic cells lacking hematopoietic lineage markers while expressing high amounts of MHC class II (shared by B cells and activated macrophages) and the integrin CD11c (also expressed by some macrophage population, activated T cells, B cells and NK cells).
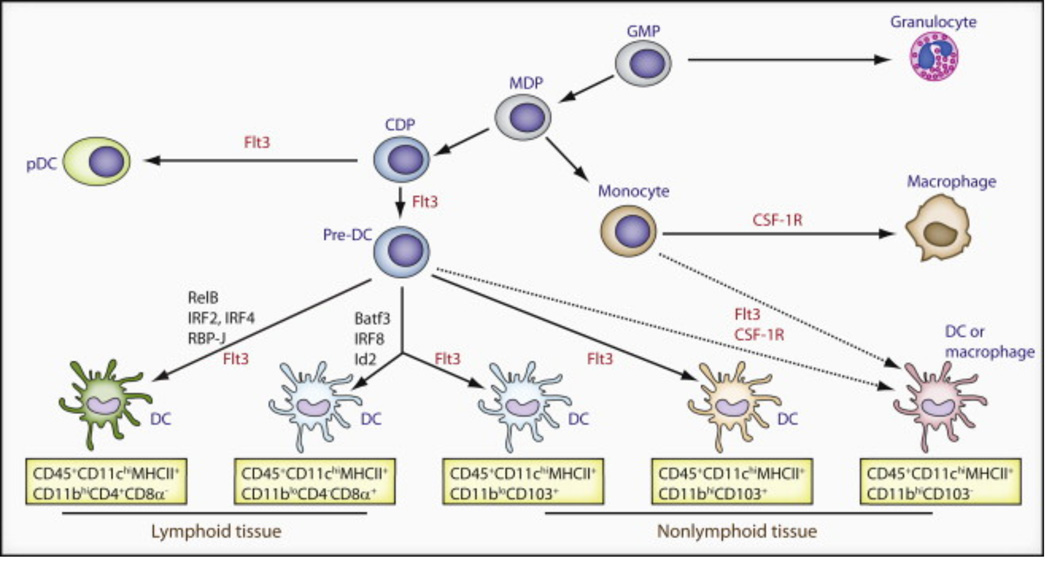
Figure 1. Origin of Batf3-IRF8-Id2-Dependent and -Independent Tissue-Resident DCs in Mice.
Lymphoid and peripheral tissues have at least four distinct resident DC subsets: CD4− CD8α+, CD4+ CD8α−, CD103+ CD11b−, and CD103− CD11b+ DCs. A CD103+ CD11b+ DC subset has been best characterized within the small intestine. This figure illustrates the precursors, transcription factors (black), and cytokine receptors (red) required for the development of each population. Commitment to the mononuclear phagocyte lineage is determined at the stage of the macrophage-dendritic cell progenitor (MDP). Granulocyte/macrophage progenitor (GMP) give rise to MDP, which has lost granulocyte potential and gives rise only to monocytes and DCs restricted precursors (common DC precursors [CDPs]). CDPs have lost monocyte-macrophage differentiation potential and give rise exclusively to plasmacytoid DCs (pDCs) and pre-DCs circulating precursors that migrate to lymphoid tissues where they differentiate into lymphoid tissue CD4−CD8α+ and CD4+CD8α DCs and to nonlymphoid tissue to give rise to CD103+CD11b− DCs and CD103+CD11b+ DCs. Flt3 controls myeloid precursors commitment to the DC lineage as well as the differentiation of mature DCs in tissue. The transcription factors Batf3 Id2 and IRF8 control the differentiation of lymphoid tissue CD8+ DC and nonlymphoid tissues CD103+CD11b− DC but do not control CD11b+ DC differentiation in lymphoid and nonlymphoid tissues.
Progress has been made in recent years in understanding the origin and the transcriptional program that control DC differentiation in vivo. These results allowed the study of homogenous cell populations that derive from committed precursors along a distinct differentiation program shifting the focus from studies on phenotypically defined subsets to developmentally regulated subsets. Below we discuss the importance of revisiting the role of DCs by separating the contribution of these developmentally distinct DC subsets in tissue immunity.
The Batf3-IRF8-Id2 DC Lineage
The Batf3-IRF8-Id2 DC lineage refers to a group of DCs located in lymphoid and non-lymphoid tissues that share a common origin, phenotype and function.
Lymphoid Tissue CD8+ DCs and Nonlymphoid Tissue CD103+CD11b− DCs Have a Common Origin
As shown in Figure 1, the presence in lymphoid tissues of a discrete population of DCs that expressed the CD8αα homodimer but lacked the CD8β chain and CD11b marker, most commonly referred to as CD8+ DCs, has been known for more than a decade (Shortman and Heath, 2010). Several studies have now established that CD8+ DCs arise from a distinct differentiation program that is dependent on key transcription factors that include Batf3 (Hildner et al., 2008) and IRF8 (Aliberti et al., 2003; Schiavoni et al., 2002; Tailor et al., 2008) and the inhibitor of DNA protein Id2 (Hacker et al., 2003). Absence of any of these factors prevents the differentiation of CD8+ DCs in lymphoid organs, but does not affect the differentiation of CD8− DCs (Hacker et al., 2003; Hildner et al., 2008; Tailor et al., 2008). Data have also shown that the mammalian target of rapamycin (mTOR), promotes the differentiation of CD8+ DCs and that the loss of the tumor suppressor gene PTEN, a negative regulator of mTOR, expands the number of CD8+ DCs in murine lymphoid tissues (Sathaliyawala et al., 2010).
A DC subset equivalent to lymphoid tissue CD8+ DCs has been identified in nonlymphoid tissues. These cells lack the CD8 marker and express the integrin CD103 (Bedoui et al., 2009; del Rio et al., 2007; Sung et al., 2006). Similar to CD8+ DCs, they express low amounts CD11b (del Rio et al., 2007; GeurtsvanKessel et al., 2008; Sung et al., 2006) and lack macrophage markers such as CD172a, F4/80, and CX3CR1 (Ginhoux et al., 2009). Nonlymphoid tissue CD103+CD11b− DCs share the same differentiation requirements as CD8+ DCs. They are absent in Batf3-deficient (Edelson et al., 2010), Id2-deficient (Ginhoux et al., 2009; Edelson et al., 2010) and IRF8-deficient mice (Ginhoux et al., 2009) and expand in PTEN-deficient mice (Sathaliyawala et al., 2010). Of note, CD103 is not a specific marker of Batf3-IRF8-Id2-dependent DCs as CD103 is also expressed on a subset of CD11b+ DCs in the lamina propria (Coombes et al., 2007; Jaensson et al., 2008; Johansson-Lindbom et al., 2005; Sun et al., 2007). Lamina propria CD103+CD11b+ DCs develop normally in Batf3-deficient and IRF8 deficient mice (Edelson et al., 2010) and in Id2-deficient mice (M. Bogunovic, F. Ginhoux and M.M., unpublished data) as discussed below.
Importantly, lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+CD11b− DCs, which will be referred to as Batf3-IRF8-Id2 DCs in this Perspective, are critically dependent on the cytokine fms-like thyrosine kinase 3 ligand (Flt3L) and its receptor (Flt3) for their development and homeostasis (Figure 1, Table 1). Flt3 and Flt3L are key regulators of DC commitment in hematopoiesis. Flt3 is expressed on short-term repopulating hematopoietic stem cells and is progressively extinguished on most hematopoietic cell lineages with the exception of the DC precursors (Merad and Manz, 2009). Loss of Flt3 expression in hematopoietic progenitors correlates with the loss of DC differentiation potential, whereas enforcement of Flt3 expression on Flt3-negative progenitors rescues their ability to differentiate into DC (Onai et al., 2006). Flt3 expression is maintained in lymphoid tissue DCs and Flt3L has been shown to control the proliferation and homeostasis of DCs in lymphoid organs (Liu et al., 2007; Waskow et al., 2008). Importantly, nonlymphoid tissue CD103+CD11b− DCs also express high amounts of Flt3 receptor on the cell surface and are more severely reduced in mice reconstituted with Flt3-deficient hematopoietic progenitors compared to CD103− DCs or tissue macrophages (Ginhoux et al., 2009). It is interesting to note that in bone marrow chimeric mice reconstituted with Flt3-deficient hematopoietic progenitors, CD103+CD11b− DCs are completely absent in nonlymphoid tissue whereas CD8+ DCs are reduced by one-third in lymphoid tissues (Ginhoux et al., 2009). These results are likely to reflect the more stringent role of Flt3 in maintaining DC homeostasis in nonlymphoid tissues compared to lymphoid organs.
Table 1.
Dendritic Cell Heterogeneity
| Pattern recognition receptors | |||||||
|---|---|---|---|---|---|---|---|
| TLR3 | TLR7 | TLR11–12 | RIG-I | MDA5 | NOD1 | NLRC4 | |
| Batf3-dependent DC | + | − | + | − | − | − | − |
| Batf3-independent DC | − | −/+ | −/+ | + | + | −/+ | −/+ |
| References | Edwards 2003 | Edwards 2003 | Yarovinsky 2005 | Luber 2010 | Luber 2010 | Luber 2010 | Luber 2010 |
| Lectin-like receptors | ||||||||
|---|---|---|---|---|---|---|---|---|
| CD205 | CD207 | CD209 | 33D1 | Clec7a | Clec9a | Clec12a | CD24 | |
| Batf3-dependent DC | ++ | int/− | − | − | − | + | ++ | ++ |
| Batf3-independent DC | + | − | + | −/+ | + | − | + | + |
| References | Shortman 2010 | Shortman 2010 | Our data | Dudziak 2007 | Shortman 2010 | Shortman 2010 | Shortman 2010 | Shortman 2010 |
| Other differential cell markers | |||||||
|---|---|---|---|---|---|---|---|
| CD36 | Sirpa | CX3CR1 | F4/80 | XCR1 | Cystatin C | Necl2 | |
| Batf3-dependent DC | + | − | − | − | + | + | − |
| Batf3-independent DC | − | + | −/+ | + | − | − | + |
| References | Shortman 2010 | Lahoud 2006 | Ginhoux 2009 | Ginhoux 2009 | Domer 2009 | Shortman 2010 | Galibert 2005 |
This table summarizes published data on differential expression of Toll-like receptors (Edwards et al., 2003; Luber et al., 2010; Yarovinsky et al., 2005), lectin-like receptors (Dudziak et al., 2007; Shortman and Heath, 2010), and other cell markers (Dorner et al., 2009; Galibert et al., 2005; Ginhoux et al., 2009; Lahoud et al., 2006; Shortman and Heath, 2010) between Batf3-IRF8-Id2 dependent DCs (including CD8+and/or CD103+DCs) and Batf3-IRF8-Id2- independent DCs.
In contrast, lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+CD11b− DCs lack the receptor for colony stimulating factor 1 (Csf-1), a cytokine that controls the differentiation of many tissue macrophages in vivo (Chitu and Stanley, 2006) and remain unaffected in mice that lack Csf-1 receptor (Csf-1R) and in mice that are reconstituted with Csf-1 receptor (Csf-1R)-deficient hematopoietic progenitors (Ginhoux et al., 2009).
Lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+CD11b− DCs are derived from DC-restricted precursors that have been identified in mice blood and bone marrow. Commitment to the mononuclear phagocyte lineage is determined at the stage of the macrophage-dendritic cell progenitor (MDP) (Auffray et al., 2009; Fogg et al., 2006), at which point, erythroid, megakaryocyte, lymphoid, and granulocyte fates have been precluded. MDPs give rise to monocytes and DCs restricted precursors (also called common DC precursors [CDPs]), which have lost monocyte-macrophage differentiation potential. CDPs give rise exclusively to plasmacytoid DCs and pre-DCs, circulating precursors that migrate to lymphoid tissues where they differentiate into lymphoid tissue CD8+ and CD8− DCs (Naik et al., 2007; Onai et al., 2007). Subsequent studies revealed that pre-DCs also migrate to nonlymphoid tissues and give rise to nonlymphoid tissue CD103+CD11b− DCs but also to CD103+CD11b+ DCs (Bogunovic et al., 2009; Ginhoux et al., 2009; Varol et al., 2009) (Figure 1).
Interestingly, Batf3 Id2 and IRF8 are differentially expressed along the DC lineage (Edelson et al., 2010; Jackson et al., 2011). IRF8 gene is expressed in DC-restricted precursors in the bone marrow and remained expressed in circulating precursors and in terminally differentiated CD8+ and CD103+ DC subsets, whereas its expression is extinguished in lymphoid tissue-resident CD8− DCs, which also derive from DC-restricted precursors (Edelson et al., 2010; Jackson et al., 2011). Reporter mice and expression array data revealed that Id2 is absent in DC-restricted precursors in the bone marrow, starts to be expressed in pre-DCs that reach the spleen, increases in all DC subsets but reach higher amounts in CD8+ and CD103+ DCs (Edelson et al., 2010; Jackson et al., 2011). In contrast, Batf3 gene becomes expressed only in the terminal stages of DC development and is highly expressed by all DCs (Edelson et al., 2010; Jackson et al., 2011). The overlap of high IRF8, Id2, and Batf3 expression was only found within the CD8+ and CD103+ DC subsets, strongly suggesting that it is the combination of these three factors that controls the differentiation and/or function of lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+CD11b− DCs.
CD8+ and CD103+CD11b− DCs Share a Similar Gene Expression Pattern and a Similar Sensing Repertoire
Transcriptional analysis of purified lymphoid tissue CD8+ and nonlymphoid tissue CD103+CD11b− DCs isolated from different tissue sites share a remarkably similar gene expression pattern (Edwards et al., 2003; Edelson et al., 2010) (see also www.immgen.org). Lymphoid tissue-resident CD8+ DCs share several sensing receptors with nonlymphoid tissue CD103+CD11b− DCs. For example, lymphoid tissue CD8+ DCs are the only lymphoid tissue DCs expressing toll-like receptor 3 (TLR3) and TLR11– TLR12 but they lack TLR7 (Edwards et al., 2003; Shortman and Heath, 2010; Yarovinsky et al., 2005). Similarly, skin migratory CD103+CD11b− DCs are the only migratory DCs expressing TLR3 (Jelinek et al., 2011); they also express high amounts of TLR11 transcripts and lack TLR7. Lymphoid tissue CD8+ DCs also lack the retinoic acid inducible gene (RIG-I), a sensor required to sense certain viruses in the cytoplasm (Luber et al., 2010), and similarly, RIG-I transcripts are expressed at much lower levels in nonlymphoid tissue CD103+CD11b− DCs compared to other nonlymphoid tissue DC (www.immgen.org) (Table 1).
Lymphoid tissue CD8+ DCs express the lectin-like receptors CD207 and CD205, and the recently described Clec9A specialized in the sensing of necrotic bodies, whereas these lectins are absent or expressed at lower amounts in CD8− DCs (Dudziak et al., 2007; Sancho et al., 2009; Shortman and Heath, 2010). Similarly, nonlymphoid tissue CD103+CD11b− DCs express CD205 and can express CD207 at least in some tissues including skin and lung, whereas CD207 is always absent from lung and skin CD103−CD11b+ DCs (Helft et al., 2010). Our unpublished data also suggest that Clec9A is expressed specifically by nonlymphoid tissue CD103+CD11b− DC and is absent from CD103−CD11b+ DC isolated from the same tissues.
Lymphoid tissue CD8+ DCs (Dorner et al., 2009) and nonlymphoid tissue CD103+CD11b− DCs (J. Helft and M.M., unpublished data) also uniquely express the chemokine receptor XCR1.
CD8+ and CD103+ CD11b− DCs Share a Common Function
Lymphoid tissue CD8+ DCs and nonlymphoid tissue CD103+ CD11b− DCs are very potent at producing interleukin-12 (IL-12), cross-presenting antigens to CD8+ T cells, and inducing the differentiation of effector CD8+ T cells (Sung et al., 2006; del Rio et al., 2007; GeurtsvanKessel et al., 2008; Henri et al., 2010; Kim and Braciale, 2009; Bedoui et al., 2009; Heath and Carbone, 2009; Shortman and Heath, 2010). The GTPase Rac2 specifically expressed by CD8+ DCs and absent from CD8− DCs is thought to control antigen processing and cross-presentation through the regulation of phagosomal pH and oxidation (Savina et al., 2006; Savina et al., 2009). Although likely, it remains to be determined whether a similar machinery is also in place in nonlymphoid tissue CD103+CD11b− DCs. The increased ability to interact with CD8+ T cells was also recently shown to be dependent on the expression of XCR1, a chemokine receptor which ligand XCL1 is produced by naive CD8+ T cells (Dorner et al., 2009). XCR1 is restricted to lymphoid tissue CD8+ DCs (Dorner et al., 2009) and nonlymphoid tissue CD103+CD11b− DCs (unpublished data) and is absent from Batf3-IRF8-Id2 independent DCs.
In agreement with their unique TLR3 expression, CD8+ DCs and CD103+CD11b− DCs share a superior ability to respond to TLR3 ligand adjuvant (Hochrein et al., 2001; Jelinek et al., 2011; Longhi et al., 2009; Sung et al., 2006). In addition, recent data revealed that CD8+ DCs are also the only lymphoid tissue DC subset that produces interferon lambda in response to the TLR3 ligand Poly: I:C (Lauterbach et al., 2010), and similar results were found for lung CD103+CD11b− DC (unpublished data).
Overall, these results show that Batf3-IRF8-Id2-dependent DCs regardless of their tissue localization share a similar origin, differentiation program, sensing repertoire, and T cell stimulatory function, suggesting that at least some diversity of the DC lineage is determined at the precursor stage.
Batf3-IRF8-Id2 DC Equivalent in Humans and Other Species
In comparison to mice DCs, considerably less is known about the origin of human DCs, their differentiation program and their functional differentiation in situ due to their rarity in blood and poor accessibility of human tissues, with the exception of the skin. Circulating DC subsets have been distinguished based on three cell surface markers including CD1c (or BDCA1) expressed on the majority of circulating DCs in humans, CD141 (also called BDCA3) expressed on a minute population, and Fcγ receptor III (CD16) (Dzionek et al., 2000; Huysamen et al., 2008; Schäkel et al., 2002; Ueno et al., 2010).
Similar to Batf3-IRF8-Id2-dependent DCs in mice, CD141+ CD1c− DCs were found to uniquely express the lectin Clec9A and the chemokine receptor XCR1 and excel in the production of IL-12 and the cross-priming of CD8+ T effector cells (Bachem et al., 2010; Crozat et al., 2010; Jongbloed et al., 2010; Lauterbach et al., 2010; Poulin et al., 2010). CD141+CD1c− DCs were also found to uniquely express TLR3 and to be the main producers of interferon lambda in response to poly:I:C activation (Lauterbach et al., 2010). However, the human CD1c+ DC subset also shared some functional properties with mice Batf3-IRF8-Id2 DCs given that they were found to produce high amounts of IL-12 and to cross-prime antigens to CD8+ T cells (Mittag et al., 2011). Interestingly, the genetic analysis of patients with disseminated mycobacterial infection identified two distinct mutations affecting IRF8 transcriptional activity. One mutation led to the complete depletion of DCs and circulating monocytes (Hambleton et al., 2011). A distinct mutation led to the specific depletion of circulating CD1c+ DCs, whereas CD141+ DCs remained intact in these patients (Hambleton et al., 2011). These results suggest that IRF8 transcription factors also controls DC differentiation in humans but, in contrast to mice IRF8, is also involved in the development of monocytes. Further studies are required to fully understand the transcriptional program that controls DC development in humans, which could potentially differ from mice. More studies are also required to further understand the functional heterogeneity of human DC subsets and to assess whether the phenotypic markers currently used to define these subsets in the blood help define functionally distinct populations.
Interestingly, analysis of skin migratory DC isolated from the sheep skin draining lymphatic vessels revealed that the presence of a DC subset, which, similar to mouse Batf3-IRF8-Id2 DCs, lacks the macrophage marker CD172a, expresses high amounts of CD205, Clec9A, and XCR1, and excels in the cross-presentation of soluble antigens and in the differentiation of CD8+ effector T cells (Contreras et al., 2010). The molecular program that controls the development of this functional and phenotypically distinct DC subset remains to be analyzed but if identical to mice, these results will support the presence of an evolutionary conserved DC subset further emphasizing the functional relevance of this DC population to tissue immunity.
The Batf3-IRF8-Id2-Independent DC Lineage
The Batf3-IRF8-Id2 independent DC lineage refers to DCs that develop independently of Batf3, Id2, and IRF8. Phenotypically, Batf3-IRF8-Id2 independent DCs are more heterogeneous than Batf3-IRF8-Id2 -dependent DCs. Although they always express CD11b and lack CD8, they are heterogenous for CD103, CD172a, F4/80, and CX3CR1 expression (Ginhoux et al., 2009; Helft et al., 2010). The transcription factors IRF4, IRF2 (Ichikawa et al., 2004; Suzuki et al., 2004), and RBPJ (Caton et al., 2007) partly control the differentiation of CD8− DCs in lymphoid organs, but the molecular program that controls the differentiation of Batf3-IRF8-Id2 -independent DCs in nonlymphoid tissues needs to be clarified and the exact relationship between Batf3-IRF8-Id2 -independent DCs in lymphoid versus nonlymphoid tissues remains unclear.
The heterogeneity of Batf3-IRF8-Id2 independent DCs has been most clearly identified in the lamina propria, where they consist of two distinct DC populations best distinguished on the basis of CD103 expression. Lamina propria Batf3-IRF8-Id2 independent DCs include CD103+CD11b+ DCs and CD103−CD11b+ DC subsets. CD103+CD11b+ lamina propria DCs express low amounts of the macrophage markers CD172a, F4/80 and CX3CR1 and low Csf-1R. They express high amounts of Flt3 receptor and are dependent on Flt3L and GM-CSF (also called Csf-2) for their development, whereas they are unaffected in Csf-1R deficient mice (Bogunovic et al., 2009; Schulz et al., 2009; Varol et al., 2009). Adoptive transfer studies of purified precursors revealed that lamina propria CD103+CD11b+ DCs derive from DC-restricted precursors (Bogunovic et al., 2009; Varol et al., 2009) and potently migrate to the draining lymph nodes upon oral microbial stimuli (Bogunovic et al., 2009; Schulz et al., 2009).
In contrast, lamina propria CD103− CD11b+ cells express the macrophage markers CD172a, F4/80, CX3CR1, and the Csf-1R. They are derived from circulating monocytes and not from DC restricted precursors and require Csf-1R but not Flt3, Flt3L, Csf-2, or Csf-2R for their development (Bogunovic et al., 2009; Schulz et al., 2009; Varol et al., 2009). They are much less potent than CD103+CD11b+ DCs at stimulating T cell proliferation (Schulz et al., 2009) and migrate poorly to the draining lymph nodes during oral infection (Bogunovic et al., 2009; Ginhoux et al., 2009; Varol et al., 2009), suggesting that this population despite expression of high MHC II and CD11c, probably corresponds to tissue macrophages. Altogether, CD103 expression helps decipher the heterogeneity of Batf3-IRF8-Id2-independent CD11b+ DCs in the lamina propria, which consists of CD103+CD11b+ DC and CD103− CD11b+ macrophages.
However, such markers are currently lacking in other tissues. For example in the lung, CD11b+ DCs lack CD103 but are heterogeneous for macrophage markers and for the DC marker CD24. They are partly reduced in mice reconstituted with Flt3−/− or Csf1r−/− hematopoietic progenitors (Ginhoux et al., 2009), which probably reflect the heterogeneity of this DC population. The use of Flt3 or Csf-1R as markers of heterogeneity as well as other profiling tools together with lineage tracing and functional genomic studies as discussed below should help clarify the regulatory and functional attributes of this DC subset.
The Langerhans Cell Exception
Langerhans cells (LCs) refer to the DCs that reside in the epidermal layer of the skin (Merad et al., 2008). These cells stand apart from other tissue DCs because they have unique ontogeny and homeostatic properties (Ginhoux and Merad, 2010). Phenotypically, LCs express lower MHC II, intermediate CD11c, and very high amounts of the lectin receptor CD207, which is also expressed, although at lower amounts, by Batf3-IRF8-Id2-dependent DCs (Merad et al., 2008). They express CD11b, F4/80 and lack CX3CR1 (Merad et al., 2008). In contrast to most tissue DCs, they arise from embryonic precursors that are recruited to the skin prior to birth and are maintained in the skin throughout life in steady-state conditions (Chang-Rodriguez et al., 2005; Chorro et al., 2009; Merad et al., 2002). LCs are absent in Id2-deficient mice (Hacker et al., 2003) and have been reported to be reduced in IRF8-deficient mice (Schiavoni et al., 2004). However, epidermal LCs remained unaffected in patients with mutations impairing IRF8 gene function (Hambleton et al., 2011), which is consistent with our unpublished data showing that epidermal LC are not reduced in numbers in IRF8-deficient mice but express reduced MHC II levels.
In agreement with their distinct origin and homeostasis, LCs express a very distinct gene expression pattern compared to other DCs (www.immgen.org). In contrast to most DCs, LCs lack Flt3 and develop independently of the DC cytokine Flt3 ligand but require Csf-1R but not Csf-1 for their development (Ginhoux et al., 2006). These results came as a surprise given that Csf-1R was thought to control mainly macrophage development (Chitu and Stanley, 2006). In the epidermis, LCs are highly phagocytic and have low MHC class II expression on the cell surface, but migratory LC that reach the lymph nodes become indistinguishable phenotypically from other migratory DC (Merad et al., 2008). LC are also thought to modulate skin contact hypersensitivity response (Kaplan et al., 2005) and are required to induce antigen specific T helper 17 cell differentiation in response to skin candida albicans infection (Igyártó et al., 2011). In vitro studies using primary LCs isolated from mice and humans also suggest that LCs are very potent at initiating T cell response and at cross-presenting cell-associated antigens (Klechevsky et al., 2008; Stoitzner et al., 2008).
Monocyte-Derived DC and Tip DC
Monocytes can easily be differentiated into DCs in vitro (Sallusto and Lanzavecchia, 1994,) but the exact contribution of monocytes to the DC pool in the steady state remains unclear. In contrast to the steady state, monocyte differentiation into DC dramatically increases in inflamed tissues. The term TNF-iNOS DC (Tip DC) was first used to describe monocytes that differentiate into DCs in the spleens of listeria-infected animals (Serbina et al., 2003). Tip DCs were described as cells expressing Gr-1 (Ly6C), CD163, intermediate MHC II, and the integrin CD11c and producing TNF and iNOS (Serbina et al., 2003). This term was subsequently extended to all inflammatory monocyte-derived DCs without always providing supportive data showing TNF and iNOS secretion. Recent studies identified new means to distinguish monocyte-derived DCs in inflamed lymphoid organs, which should help the purification and the study of these cells (Cheong et al., 2010). The cytokines and transcription factors that control monocyte-derived DC differentiation remain unclear. Csf-2, a cytokine essential for the generation of in vitro-derived DCs, is thought to be critical for the differentiation of inflammatory DCs, although definite proof of the role of this cytokine in DC differentiation in vivo remains to be established (Shortman and Naik, 2007).
In Vitro-Derived DC
The identification of in vitro culture conditions that promote the differentiation of DCs contributed substantially to the expansion of the DC field (Caux et al., 1992; Inaba et al., 1992; Sallusto and Lanzavecchia, 1994). In mice, bone marrow progenitors represent the main source of in vitro-derived DCs, whereas in humans, DCs are generated from circulating monocytes or CD34+ hematopoietic progenitors. In DC cultures, Csf-2 is a key cytokine to drive DC differentiation (Caux et al., 1992; Inaba et al., 1992; Sallusto and Lanzavecchia, 1994). However, DC populations obtained in Csf-2-driven cultures do not faithfully resemble DC subsets identified in vivo. These results together with the realization that Flt3L rather than Csf-2 drives DC differentiation in vivo (Vremec et al., 1997) have led to substitution of Flt3L for Csf-2 in DC differentiation cultures (Naik et al., 2007). Bone marrow-derived DCs cultured in the presence of Flt3L leads to the differentiation of DC subsets that phenotypically and functionally resemble CD8+ and CD8− DCs (del Rio et al., 2008; Naik et al., 2007). Further analysis of these DC populations both at the transcriptional and functional levels should help establish the physiological relevance of this in vitro differentiation DC system.
Heterogeneity of Tissue Macrophages
Macrophages are hematopoietic cells that populate every tissue including the brain. Whereas DCs are focused on initiating tissue immune responses, macrophages main role is to ensure tissue integrity. Functions shared by most tissue macrophages include a high phagocytic function and degradative potential, allowing them to clear foreign and damaged cells (Gordon and Taylor, 2005). Macrophages also participate in the induction of innate immunity in response to tissue infection, which plays a critical role in the killing of micro-organisms and their pathogenic factors (Gordon and Taylor, 2005). They can also load extracellular antigen in MHC class II compartments but they are less efficient than DCs at priming naive T cells and mostly interact with effector CD4+ T cells. Different names have been used to identify macrophage populations in vivo, which are mostly defined on the basis of a set of defined cell surface markers (Table 2). These names can reflect those of the scientists who discovered these cells (i.e., Kupffer cells in the liver) or the location in which they reside (i.e., marginal zone macrophages and the red pulp macrophages of the spleen, subcapsular sinus, and medullary macrophages in lymph nodes), but are rarely informative of the cells’ origin and function. Similar to DCs, the lack of genetic tools targeting specifically the macrophage lineage compromises our understanding of the regulatory mechanisms that control macrophage development and function in vivo. The recent development of engineered mouse models to track, deplete, modify, and alter the macrophage compartment will undoubtedly transform our view of the macrophage lineage.
Table 2.
Heterogeneity of Tissue Macrophages in Mice
| Lymphoid tissue macrophages | ||||
|---|---|---|---|---|
| Spleen | Lymph node | Bone marrow | Thymus | |
| Phenotype and Localization | Red pulp MØ -Entire red pulp -F4/80hiCD11bloCD169loMHCIIlo CD163+CD68+CD115+CD172a+ |
Subcapsular sinus MØ -Lining subcapsular sinus -F4/80loCD11b+CD169+CD11clo MARCO+ |
Bone marrow CD169+ MØ -Distributed throughout the marrow but enriched around the hematopoietic stem cell niche F4/80+CD11bloCD169+CD11clo CD68+CX3CR1−CD115+ |
Subcapsular MØ -MHCII+F4/80+Mac-2−FcγRII+FcγRIII+ |
| Marginal zone MØ -Outer layer of marginal zone sinus -F4/80−SIGN-R1+MARCO+ |
Medullar MØ -Lining medullar sinus -F4/80hiCD11b+CD169+CD11clo |
Cortex MØ -MHCII−F4/80+Mac-2+FcγRII+FcγRIII+ |
||
| Marginal zone metallophilic MØ -Inner layer of marginal zone sinus -F4/80−CD169+ |
CD11chiCD169+ MØ -Boundary between the sinus and the T cell zone or B cell follicle -MHCII+F4/80+CD169+CD11chiCD8+ |
Cortico-medullary MØ -MHCII+F4/80+Mac-2+ FcγRIIhiFcγRIIIhi |
||
| Tingible body MØ -Germinal center -F4/80−CD11b−CD68+MFG-E8+ |
Tingible body MØ -Germinal center -F4/80−CD11b−CD68+MFG-E8+ |
Medulla MØ -Germinal center -MHCIIhiF4/80+Mac-2+ FcγRII+FcγRIII+ |
||
| References |
Lloyd et al., 2008 Kohyama et al., 2009 Miyake et al., 2007 You et al., 2011 Hanayama et al., 2004 Rabinowitz et al., 1991 |
Phan et al., 2009 Asano et al., 2011 |
Chow et al., 2011 Crocker and Gordon, 1986 |
Soga et al., 1997 |
| Non-lymphoid tissue macrophages | |||||
|---|---|---|---|---|---|
| Liver | Lung | Gut | Skin | Kidney | |
| Phenotype and Localization | Kupffer cell -In the liver sinusoid -F4/80hiCD11bloCD169+ CD68+Mac-2+ |
Alveolar MØ -Alveolar airspace -F4/80+CD11bloCD169+ CD11chiCD68+SiglecF+ MARCO+Mac-2+ |
Lamina propria MØ -Intestinal lamina propria -MHCII+F4/80+CD11b+ CD11c+CD103−CD115+ CX3CR1+CD172a+ |
Dermal MØ -F4/80+CD11b+CD11clo mMGL+CD206+MHCIIlo In deep dermis, MØ can express CD169+ |
Kidney MØ -F4/80+CD11bloCD103− CX3CR1+CD172a+ |
| Interstitial MØ -Alveolar interstitium -CD11c−F4/80+CD68+ MHCII+ |
Serosal DC or MØ -Intestinal muscular layer and serosa -MHCIIhiF4/80+CD11b+ CD169+CD11cloCX3CR1+ CD103−CD115+ |
||||
| References |
Flotte et al., 1983 Kinoshita et al., 2010 Crocker and Gordon, 1989 |
Flotte et al., 1983 Tateno et al., 2007 Ducreux et al., 2009 Palecanda et al., 1999 Bedoret et al., 2009 Lagranderie et al., 2003 |
Bogunovic et al., 2009 Flores-Langarica et al., 2005 |
Dupasquier et al., 2004 Dupasquier et al., 2006 |
Our unpublished data |
This table describes the name, localization, and phenotype of macrophage populations that reside in the spleen (Hanayama et al., 2004; Kohyama et al., 2009; Lloyd et al., 2008; Miyake et al., 2007; Rabinowitz and Gordon, 1991; You et al., 2011), lymph nodes (Phan et al., 2009); (Asano et al., 2011), bone marrow (Chow et al., 2011; Crocker and Gordon, 1986), thymus (Soga et al., 1997), liver (Crocker and Gordon, 1989; Flotte et al., 1983; Kohyama et al., 2009), lungs (Bedoret et al., 2009; Ducreux et al., 2009; Flotte et al., 1983; Lagranderie et al., 2003; Palecanda et al., 1999; Tateno et al., 2007), gut (Bogunovic et al., 2009; Flores-Langarica et al., 2005), and skin (Dupasquier et al., 2004; Dupasquier et al., 2006).
Regulation of Macrophage Development
Although the current dogma suggests that most tissue macrophages derive from circulating monocytes that originate in the bone marrow, direct evidence that circulating monocytes contribute to all tissue macrophages is lacking. Adoptive transfer studies revealed that monocytes contribute to the red pulp macrophages (Fogg et al., 2006) and the mucosal macrophage (Varol et al., 2007; Bogunovic et al., 2009; Varol et al., 2009) pool, but further studies are needed to confirm the contribution of circulating monocytes to other tissue macrophage subsets. There is now clear evidence that macrophages can proliferate locally in the steady state and in response to specific tissue injury, and the contribution of circulating precursors versus self-renewal to tissue macrophage homeostasis remains to be established (Gordon and Taylor, 2005; Jenkins et al., 2011; Randolph, 2011). The capacity of macrophage to self-renew and contribute to their homeostasis highlights one of the limitations of the MPS definition that suggests that all tissue phagocytes are derived from a mobile pool of circulating monocytes.
The dramatic reduction of tissue macrophages observed in Csf1r−/− mice, that lack the Csf-1R (Dai et al., 2002) and Csf1op/op mice (Yoshida et al., 1990), that have a natural null mutation in the Csf-1 cytokine gene, established the key role of Csf-1 and its receptor in macrophage homeostasis in vivo (Pixley and Stanley, 2004). However, the precise role of Csf-1 and its receptor in monocyte commitment remains controversial. One hypothesis is that Csf-1 drives the differentiation of monocytes into tissue macrophages (Metcalf, 1985), whereas a different hypothesis suggests that Csf-1 provides a survival signal to the differentiating monocytes and that surviving cells utilize an intrinsic developmental program to become mature macrophages (Korn et al., 1973; Lagasse and Weissman, 1997; Nakahata et al., 1982). Importantly, some tissue macrophages are more profoundly affected in the absence of Csf-1R than in the absence of its ligand Csf-1. For example, the brain resident macrophages, also called microglia and the epidermal LCs, develop normally in Csf1op/op mice but are absent in Csf1r−/− mice (Ginhoux et al., 2010; Ginhoux et al., 2006). Interestingly, a second CSF-1R ligand called interleukin 34 (IL-34) has been identified (Lin et al., 2008). Evolutionarily conserved IL-34 has been identified in humans, mice, and birds (Garceau et al., 2010). IL-34 is highly expressed in the brain of postnatal mice (Wei et al., 2010) and the exact role of IL-34 in the development and homeostasis of microglia and other tissue macrophages remains to be determined.
The transcriptional program that control monocyte-macrophage differentiation has been reviewed elsewhere (Auffray et al., 2009; Friedman, 2007; Geissmann et al., 2010). However, most of the studies on the transcriptional regulation of macrophage differentiation were done in vitro with progenitor-enriched populations, and the exact role of these factors in driving the differentiation of monocytes into distinct macrophage subsets in vivo remains to be examined. This is particularly important given that functional macrophage specialization is likely to be regulated at the tissue levels and not at the progenitor level.
Functional Specialization of Tissue Macrophages
Although the developmental pathways that give rise to distinct macrophage subsets are still unclear, there is evidence that different macrophage populations play distinct roles in vivo. In most tissues, macrophages promote tumor surveillance through their ability to capture and kill malignant hematopoietic cells (Jaiswal et al., 2010), but can also promote tumor growth through their ability to increase angiogenesis and immunosupression (Qian and Pollard, 2010). Osteoclasts represent a population of bone macrophages that specialize in bone resorption and are critical to maintain bone mass homeostasis (Edwards and Mundy, 2011). Bone marrow macrophages promote the retention of hematopoietic stem cells and the depletion of bone marrow macrophages mobilizes hematopoietic progenitors to the blood, enhancing yields of clinical mobilization regimen for hematopoietic cell transplants (Chow et al., 2011). In allogeneic hematopoietic cell transplant recipient, host lymphoid tissue macrophages reduce the allogeneic T cell pool and limit graft versus host reactions through their ability to capture live allogeneic T cells and inhibit T cell proliferation (Hashimoto et al., 2011). In lymph nodes, subcapsular macrophages provide a key barrier that protects from the systemic dissemination of viruses (Iannacone et al., 2010) and promote the presentation of viral antigens to B cells (Phan et al., 2009; Phan et al., 2007). These results emphasize the functional diversity of the macrophage network, which may be partly dictated by the environment in which the macrophage resides. In support of this hypothesis are the findings showing that whereas Csf-1R and the transcription factor PU.1 (Friedman, 2007) control the commitment to a common macrophage program, the transcription factors c-fos and MITF control specifically osteoclast differentiation (Edwards and Mundy, 2011), and the transcription factor Spi-C selectively controls the development of red pulp macrophages but no other macrophage population (Kohyama et al., 2009). Therefore, one of the biggest challenges in the macrophage field is to continue to explore the origin of the precursor cell population and the molecular program that control global macrophage commitment in bone marrow progenitors, but in addition, to decipher the regulatory program that controls macrophage specialization and function in the periphery.
The Microglia Exception
Microglia cells (from the Greek “glia” for glue) refer to cells in the brain that are nonneuronal and nonastrocytic playing a supportive role in brain homeostasis. Although microglia share many key features with other tissue macrophages, they have a distinct origin in that they derive from embryonic precursors that have been recruited to the brain prior to birth (Ransohoff and Perry, 2009). Using in vivo cell fate mapping studies, recent data suggest that primitive macrophages are the main precursors of adult microglia that populate the adult brain (Ginhoux and Merad, 2010). In addition and in contrast to most adult tissue macrophages, some studies have reported that microglia are maintained throughout life independently of any blood input and can resist high dose γ-ray irradiation (Ajami et al., 2007; Mildner et al., 2007), although arguments against this hypothesis have also been presented (Soulet and Rivest, 2008). Similar results were found for epidermal LCs (Merad et al., 2002). Importantly, microglia and LCs can be repopulated by circulating monocytes upon brain or skin injuries (Ginhoux et al., 2006; Mildner et al., 2007). Whether embryonic-derived cells that populate the normal skin and brain have a similar function to that of monocyte-derived cells that arise in injured tissues remains to be examined.
Altogether, these results emphasize yet another limitation of the MPS definition that suggests that two prominent populations of phagocytes that reside in the brain and skin are in fact derived from embryonic myeloid precursors and not from bone marrow-derived monocytes.
Distinguishing DCs from Macrophages In Vivo
Most cells types in vivo are identified on the basis of their ontogeny, phenotype, and function. In contrast, the definition of tissue DCs and macrophages remains mostly phenotypic and the use of the cell surface markers CD11c and F4/80 as surrogate for origin and function have lead to confusion on the exact contribution of DCs versus macrophages to tissue immunity. Indeed, the CD11c integrin can be expressed on macrophages and monocytes, and therefore the role of CD11c+ cells does not always reflect the role of DCs in vivo. Similarly, F4/80 is also expressed by eosinophils in the gut and neutrophils in the bone marrow and is not a specific marker of macrophages. Whereas differentiating macrophages from Batf3-IRF8-Id2-independent MHCII+ CD11c+ cells continues to be difficult probably because these cells as currently defined are heterogeneous and include tissue macrophages as discussed above, Batf3-IRF8-Id2 DCs are easily distinguished from macrophages as described above and summarized below.
Origin: Batf3-IRF8-Id2 DCs arise from CDP in an Flt3L-dependent and Csf-1R ligand-independent manner, whereas macrophages arise independently of CDP and require Csf-1R ligand but not Flt3L for their development.
Phenotype: Batf3-IRF8-Id2 DCs lack Csf-1R, CD11b, CX3CR1, F4/80, and CD172a, whereas macrophages always express some or all of these markers.
Function: Batf3-IRF8-Id2 DCs are much more potent at initiating T cell differentiation compared to macrophages. Batf3-IRF8-Id2 DCs have a much lower degradative potential compared to macrophages and excel in antigen cross-presentation to CD8+ T cells, whereas macrophages can only cross-present very large amount of antigens.
Migratory Potential : Batf3-IRF8-Id2 DCs are much more potent in migrating to the draining LN compared to macrophages.
In sum, prior confusion on DCs and macrophages contribution to tissue immunity is partly due to the fact that lineage marker − MHCII+ CD11c+ cells are not synonymous of DCs and an effort to better define the heterogeneity of Batf3-IRF8-Id2-independent MHCII+ CD11c+ cells is critically needed to help clarify the role of these cells in the modulation of tissue immune responses.
Focusing on DCs and Macrophage Heterogeneity Rather than Similarities
The term mononuclear phagocyte system was critical to alert the scientific community of the importance of phagocytes in innate immune defense. Stressing the commonality of a cellular system widely distributed throughout the body to promote host defense against foreign invasion facilitated the acceptance of a new cellular lineage by the scientific community. More than a century after their discovery, the identification of human diseases due to compromised phagocyte function together with the development of mouse models with engineered phagocyte-specific genetic defects established the key role of phagocytes in tissue immunity and homeostasis. However, these studies have also revealed the complexity and heterogeneity of this cellular system. In fact, while Aschoff stressed the importance of a certain fundamental resemblance between the cells with respect to phagocytosis and storage, he insisted that similarities do not amount to identity and expected that future research would make it possible to distinguish more accurately between different populations of tissue phagocytes.
There have been controversies on whether cross-presentation function is unique to DCs, given that macrophages can also cross-present extracellular antigens to CD8+ T lymphocytes if incubated with large amount of soluble antigens. Subsequent studies using genetically engineered mouse models revealed that absence of Batf3-IRF8-Id2-dependent DCs abolishes the cross-presentation of cell-associated antigens despite the presence of normal tissue macrophages (Hildner et al., 2008). These results are consistent with in vitro studies showing that macrophages are unable to cross-present limiting amounts of soluble antigens. These data also suggest that in considering differences among tissue phagocytes, it is critical not only to assess the capacity of the cells to perform a function but also the efficiency with which this function is performed. Focusing on differences in cross-presentation among DCs and macrophages has helped investigators identify pathways that promote DC superior ability to cross-present tissue antigens (Savina et al., 2006; Savina et al., 2009). These studies have also helped the development of targeting strategies to promote T cell immunity in vivo (Bonifaz et al., 2002; Bonifaz et al., 2004; Ueno et al., 2011). There are also controversies on whether tissue cell migration to the draining lymph node is unique to DCs because macrophages also can migrate to tissue draining lymph nodes in response to tissue injury, although this occurs at low frequency, and thus its functional relevance is not known. DCs, in contrast to macrophages, can migrate to the draining lymph nodes in the absence of tissue injury, and steady state migration to the lymph nodes and presentation of self-antigens provides a mechanism to promote peripheral tolerance (Hemmi et al., 2001; Huang et al., 2000; Mellman and Clausen, 2010; Ohl et al., 2004; Pugh et al., 1983) that could be exploited for the treatment of autoimmune diseases (Steinman et al., 2003). DCs’ ability to migrate to the draining LN in response to tissue injury augments many folds, is controlled in part by CCR7 ligands, and is required to induce adaptive immune response to peripheral tissue antigens (Ohl et al., 2004; Randolph et al., 2008).
What Next in MPS Research?
Although we have highlighted the importance of acknowledging phagocyte heterogeneity in vivo, it is critical to distinguish between phenotypic plasticity and discrete mononuclear phagocyte subsets. The definition of a mononuclear phagocyte subset should only be based on developmental and functional evidence and not on a set of phenotypical differences. System-wide investigation of gene expression at the mRNA transcript level should help revisit phagocyte subset classification and identify gene regulatory networks that control MPS development, differentiation, and function. Quantitative proteomics also holds great promise to enhance or complement the picture of gene expression in cells, and thus to contribute to the understanding of tissue phagocytes’ molecular program. The paucity of available tissue phagocytes especially in nonlymphoid tissues remains, however, a great limitation.
System biology studies should also help the development of genetic tools allowing inducible gene regulation in vivo and lineage tracing of genetically marked, defined myeloid precursor population to further comprehend the developmental complexity of the phagocyte system. RNA interference (RNAi) is the most rapid means to identify gene function in human and mouse cells. At present, high-throughput screening is only feasible in cell culture, which limits the approach to in vitro derived macrophage and DC with the limitation discussed above as primary tissue phagocytes cannot be easily manipulated in culture. However, as we learn more about the developmental requirements of macrophages and DCs, its hould be possible to generate better in vitro and in vivo models for performing gain- and loss-of-function studies. These studies will then need to be validated in the human setting using similar genetics approaches. Unraveling the transcriptional program that controls macrophage and DC functional heterogeneity will undoubtedly opens new strategies for taking MPS biology into medicine (Steinman and Banchereau, 2007).
ACKNOWLEDGMENTS
M.M. is supported by the National Institutes of Health grants HL086899, AI095611, and CA154947.
REFERENCES
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, Sher A. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- Asano K, Nabeyama A, Miyake Y, Qiu CH, Kurita A, Tomura M, Kanagawa O, Fujii S, Tanaka M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity. 2011;34:85–95. doi: 10.1016/j.immuni.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Auffray C, Sieweke MH, Geissmann F. Blood monocytes: Development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- Bachem A, Güttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, Salama A, Movassaghi K, Opitz C, Mages HW, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoret D, Wallemacq H, Marichal T, Desmet C, Quesada Calvo F, Henry E, Closset R, Dewals B, Thielen C, Gustin P, et al. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Invest. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S, Whitney PG, Waithman J, Eidsmo L, Wakim L, Caminschi I, Allan RS, Wojtasiak M, Shortman K, Carbone FR, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 2009;10:488–495. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 2002;196:1627–1638. doi: 10.1084/jem.20021598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, Brimnes MK, Moltedo B, Moran TM, Steinman RM. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J. Exp. Med. 2004;199:815–824. doi: 10.1084/jem.20032220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Chang-Rodriguez S, Hoetzenecker W, Schwärzler C, Biedermann T, Saeland S, Elbe-Bürger A. Fetal and neonatal murine skin harbors Langerhans cell precursors. J. Leukoc. Biol. 2005;77:352–360. doi: 10.1189/jlb.1004584. [DOI] [PubMed] [Google Scholar]
- Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr. Opin. Immunol. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, Malissen B, Kissenpfennig A, Barbaroux JB, Groves R, Geissmann F. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med. 2009;206:3089–3100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Lucas D, Hidalgo A, Mendez-Ferrer S, Hashimoto D, Scheiermann C, Battista M, Leboeuf M, Prophete C, van Rooijen N, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J. Exp. Med. 2011;208:261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras V, Urien C, Guiton R, Alexandre Y, Vu Manh TP, Andrieu T, Crozat K, Jouneau L, Bertho N, Epardaud M, et al. Existence of CD8a-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J. Immunol. 2010;185:3313–3325. doi: 10.4049/jimmunol.1000824. [DOI] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Gordon S. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J. Exp. Med. 1986;164:1862–1875. doi: 10.1084/jem.164.6.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Gordon S. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J. Exp. Med. 1989;169:1333–1346. doi: 10.1084/jem.169.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, Vu Manh TP, Baranek T, Storset AK, Marvel J, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- del Rio ML, Rodriguez-Barbosa JI, Kremmer E, Förster R. CD103− and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 2007;178:6861–6866. doi: 10.4049/jimmunol.178.11.6861. [DOI] [PubMed] [Google Scholar]
- del Rio ML, Rodriguez-Barbosa JI, Bölter J, Ballmaier M, Dittrich-Breiholz O, Kracht M, Jung S, Förster R. CX3CR1+ c-kit+ bone marrow cells give rise to CD103+ and CD103− dendritic cells with distinct functional properties. J. Immunol. 2008;181:6178–6188. doi: 10.4049/jimmunol.181.9.6178. [DOI] [PubMed] [Google Scholar]
- Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–833. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Ducreux J, Crocker PR, Vanbever R. Analysis of sialoadhesin expression on mouse alveolar macrophages. Immunol. Lett. 2009;124:77–80. doi: 10.1016/j.imlet.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Dupasquier M, Stoitzner P, van Oudenaren A, Romani N, Leenen PJ. Macrophages and dendritic cells constitute a major subpopulation of cells in the mouse dermis. J. Invest. Dermatol. 2004;123:876–879. doi: 10.1111/j.0022-202X.2004.23427.x. [DOI] [PubMed] [Google Scholar]
- Dupasquier M, Stoitzner P, Wan H, Cerqueira D, van Oudenaren A, Voerman JS, Denda-Nagai K, Irimura T, Raes G, Romani N, et al. The dermal microenvironment induces the expression of the alternative activation marker CD301/mMGL in mononuclear phagocytes, independent of IL-4/IL-13 signaling. J. Leukoc. Biol. 2006;80:838–849. doi: 10.1189/jlb.1005564. [DOI] [PubMed] [Google Scholar]
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: Three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, Mundy GR. Advances in osteoclast biology: Old findings and new insights from mouse models. Nat. Rev. Rheumatol. 2011;7:235–243. doi: 10.1038/nrrheum.2011.23. [DOI] [PubMed] [Google Scholar]
- Edwards AD, Diebold SS, Slack EM, Tomizawa H, Hemmi H, Kaisho T, Akira S, Reis e Sousa C. Toll-like receptor expression in murine DC subsets: Lack of TLR7 expression by CD8 alpha+ DC correlates with unresponsiveness to imidazoquinolines. Eur. J. Immunol. 2003;33:827–833. doi: 10.1002/eji.200323797. [DOI] [PubMed] [Google Scholar]
- Flores-Langarica A, Meza-Perez S, Calderon-Amador J, Estrada-Garcia T, Macpherson G, Lebecque S, Saeland S, Steinman RM, Flores-Romo L. Network of dendritic cells within the muscular layer of the mouse intestine. Proc. Natl. Acad. Sci. USA. 2005;102:19039–19044. doi: 10.1073/pnas.0504253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotte TJ, Springer TA, Thorbecke GJ. Dendritic cell and macrophage staining by monoclonal antibodies in tissue sections and epidermal sheets. Am. J. Pathol. 1983;111:112–124. [PMC free article] [PubMed] [Google Scholar]
- Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, Cumano A, Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- Galibert L, Diemer GS, Liu Z, Johnson RS, Smith JL, Walzer T, Comeau MR, Rauch CT, Wolfson MF, Sorensen RA, et al. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 2005;280:21955–21964. doi: 10.1074/jbc.M502095200. [DOI] [PubMed] [Google Scholar]
- Garceau V, Smith J, Paton IR, Davey M, Fares MA, Sester DP, Burt DW, Hume DA. Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J. Leukoc. Biol. 2010;87:753–764. doi: 10.1189/jlb.0909624. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel CH, Willart MA, van Rijt LS, Muskens F, Kool M, Baas C, Thielemans K, Bennett C, Clausen BE, Hoogsteden HC, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J. Exp. Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Merad M. Ontogeny and homeostasis of Langerhans cells. Immunol. Cell Biol. 2010;88:387–392. doi: 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Tacke F, Angeli V, Bogunovic M, Loubeau M, Dai XM, Stanley ER, Randolph GJ, Merad M. Langerhans cells arise from monocytes in vivo. Nat. Immunol. 2006;7:265–273. doi: 10.1038/ni1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Elie Metchnikoff: Father of natural immunity. Eur. J. Immunol. 2008;38:3257–3264. doi: 10.1002/eji.200838855. [DOI] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, Kuhl C, Jorgas T, Kurz SM, Rose-John S, Yokota Y, Zenke M. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, Nagata S. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Greter M, Saenger Y, Kwan WH, Leboeuf M, Ginhoux F, Ochando JC, Kunisaki Y, van Rooijen N, et al. Pre-transplant CSF-1 therapy expands recipient macrophages and ameliorates GVHD after allogeneic hematopoietic cell transplantation. J. Exp. Med. 2011 doi: 10.1084/jem.20101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 2009;10:1237–1244. doi: 10.1038/ni.1822. [DOI] [PubMed] [Google Scholar]
- Helft J, Ginhoux F, Bogunovic M, Merad M. Origin and functional heterogeneity of non-lymphoid tissue dendritic cells in mice. Immunol. Rev. 2010;234:55–75. doi: 10.1111/j.0105-2896.2009.00885.x. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Yoshino M, Yamazaki H, Naito M, Iyoda T, Omatsu Y, Shimoyama S, Letterio JJ, Nakabayashi T, Tagaya H, et al. Skin antigens in the steady state are trafficked to regional lymph nodes by transforming growth factor-beta1-dependent cells. Int. Immunol. 2001;13:695–704. doi: 10.1093/intimm/13.5.695. [DOI] [PubMed] [Google Scholar]
- Henri S, Poulin LF, Tamoutounour S, Ardouin L, Guilliams M, de Bovis B, Devilard E, Viret C, Azukizawa H, Kissenpfennig A, Malissen B. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein H, Shortman K, Vremec D, Scott B, Hertzog P, O’Keeffe M. Differential production of IL-12, IFN-alpha, and IFN-gamma by mouse dendritic cell subsets. J. Immunol. 2001;166:5448–5455. doi: 10.4049/jimmunol.166.9.5448. [DOI] [PubMed] [Google Scholar]
- Huang FP, Platt N, Wykes M, Major JR, Powell TJ, Jenkins CD, MacPherson GG. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J. Exp. Med. 2000;191:435–444. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J. Biol. Chem. 2008;283:16693–16701. doi: 10.1074/jbc.M709923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannacone M, Moseman EA, Tonti E, Bosurgi L, Junt T, Henrickson SE, Whelan SP, Guidotti LG, von Andrian UH. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465:1079–1083. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa E, Hida S, Omatsu Y, Shimoyama S, Takahara K, Miyagawa S, Inaba K, Taki S. Defective development of splenic and epidermal CD4+ dendritic cells in mice deficient for IFN regulatory factor-2. Proc. Natl. Acad. Sci. USA. 2004;101:3909–3914. doi: 10.1073/pnas.0400610101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igyártó BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, Kaplan DH. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JT, Hu Y, Liu R, Masson F, D’Amico A, Carotta S, Xin A, Camilleri MJ, Mount AM, Kallies A, et al. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 2011;30:2690–2704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg PL, Davidsson T, Powrie F, Johansson-Lindbom B, Agace WW. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Chao MP, Majeti R, Weissman IL. Macrophages as mediators of tumor immunosurveillance. Trends Immunol. 2010;31:212–219. doi: 10.1016/j.it.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek I, Leonard JN, Price GE, Brown KN, Meyer-Manlapat A, Goldsmith PK, Wang Y, Venzon D, Epstein SL, Segal DM. TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J. Immunol. 2011;186:2422–2429. doi: 10.4049/jimmunol.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Förster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, Chen CJ, Dunbar PR, Wadley RB, Jeet V, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23:611–620. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH. Immunology’s foundation: The 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nat. Immunol. 2008;9:705–712. doi: 10.1038/ni0708-705. [DOI] [PubMed] [Google Scholar]
- Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE. 2009;4:e4204. doi: 10.1371/journal.pone.0004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, Briere F, Chaussabel D, Zurawski G, Palucka AK, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 2008;29:497–510. doi: 10.1016/j.immuni.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, Frazier WA, Murphy TL, Murphy KM. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn AP, Henkelman RM, Ottensmeyer FP, Till JE. Investigations of a stochastic model of haemopoiesis. Exp. Hematol. 1973;1:362–375. [PubMed] [Google Scholar]
- Lagasse E, Weissman IL. Enforced expression of Bcl-2 in monocytes rescues macrophages and partially reverses osteopetrosis in op/op mice. Cell. 1997;89:1021–1031. doi: 10.1016/s0092-8674(00)80290-1. [DOI] [PubMed] [Google Scholar]
- Lagranderie M, Nahori MA, Balazuc AM, Kiefer-Biasizzo H, Lapa e Silva JR, Milon G, Marchal G, Vargaftig BB. Dendritic cells recruited to the lung shortly after intranasal delivery of Mycobacterium bovis BCG drive the primary immune response towards a type 1 cytokine production. Immunology. 2003;108:352–364. doi: 10.1046/j.1365-2567.2003.01609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoud MH, Proietto AI, Gartlan KH, Kitsoulis S, Curtis J, Wettenhall J, Sofi M, Daunt C, O’Keeffe M, Caminschi I, et al. Signal regulatory protein molecules are differentially expressed by CD8- dendritic cells. J. Immunol. 2006;177:372–382. doi: 10.4049/jimmunol.177.1.372. [DOI] [PubMed] [Google Scholar]
- Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, Freudenberg MA, Davey GM, Vremec D, Kallies A, et al. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J. Exp. Med. 2010;207:2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- Lloyd CM, Phillips AR, Cooper GJ, Dunbar PR. Three-colour fluorescence immunohistochemistry reveals the diversity of cells staining for macrophage markers in murine spleen and liver. J. Immunol. Methods. 2008;334:70–81. doi: 10.1016/j.jim.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M, Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Mellman I, Clausen BE. Immunology. Beta-catenin balances immunity. Science. 2010;329:767–769. doi: 10.1126/science.1194185. [DOI] [PubMed] [Google Scholar]
- Mellman I, Steinman RM. Dendritic cells: Specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- Merad M, Manz MG. Dendritic cell homeostasis. Blood. 2009;113:3418–3427. doi: 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat. Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- Metcalf D. The granulocyte-macrophage colony-stimulating factors. Science. 1985;229:16–22. doi: 10.1126/science.2990035. [DOI] [PubMed] [Google Scholar]
- Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Brück W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K, Wu L, Harrison LC. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 2011;186:6207–6217. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, Tanaka M. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J. Clin. Invest. 2007;117:2268–2278. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O’Keeffe M, Bahlo M, Papenfuss A, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Nakahata T, Gross AJ, Ogawa M. A stochastic model of self-renewal and commitment to differentiation of the primitive hemopoietic stem cells in culture. J. Cell. Physiol. 1982;113:455–458. doi: 10.1002/jcp.1041130314. [DOI] [PubMed] [Google Scholar]
- Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Förster R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J. Exp. Med. 2006;203:227–238. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacy-toid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Palecanda A, Paulauskis J, Al-Mutairi E, Imrich A, Qin G, Suzuki H, Kodama T, Tryggvason K, Koziel H, Kobzik L. Role of the scavenger receptor MARCO in alveolar macrophage binding of unopsonized environmental particles. J. Exp. Med. 1999;189:1497–1506. doi: 10.1084/jem.189.9.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 2009;10:786–793. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: Complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, Keller AM, Joffre O, Zelenay S, Nye E, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh CW, MacPherson GG, Steer HW. Characterization of nonlymphoid cells derived from rat peripheral lymph. J. Exp. Med. 1983;157:1758–1779. doi: 10.1084/jem.157.6.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph GJ. Immunology. No need to coax monocytes. Science. 2011;332:1268–1269. doi: 10.1126/science.1208480. [DOI] [PubMed] [Google Scholar]
- Randolph GJ, Ochando J, Partida-Sánchez S. Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 2008;26:293–316. doi: 10.1146/annurev.immunol.26.021607.090254. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH. Microglial physiology: Unique stimuli, specialized responses. Annu. Rev. Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D, Joffre OP, Keller AM, Rogers NC, Martínez D, Hernanz-Falcón P, Rosewell I, Reis e Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, O’Gorman WE, Greter M, Bogunovic M, Konjufca V, Hou ZE, Nolan GP, Miller MJ, Merad M, Reizis B. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 2010;33:597–606. doi: 10.1016/j.immuni.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Duménil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Savina A, Peres A, Cebrian I, Carmo N, Moita C, Hacohen N, Moita LF, Amigorena S. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8(+) dendritic cells. Immunity. 2009;30:544–555. doi: 10.1016/j.immuni.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Schäkel K, Kannagi R, Kniep B, Goto Y, Mitsuoka C, Zwirner J, Soruri A, von Kietzell M, Rieber E. 6-Sulfo LacNAc, a novel carbohydrate modification of PSGL-1, defines an inflammatory type of human dendritic cells. Immunity. 2002;17:289–301. doi: 10.1016/s1074-7613(02)00393-x. [DOI] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC, 3rd, Belardelli F, Gabriele L. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J. Exp. Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Borghi P, Sestili P, Venditti M, Morse HC, 3rd, Belardelli F, Gabriele L. ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood. 2004;103:2221–2228. doi: 10.1182/blood-2003-09-3007. [DOI] [PubMed] [Google Scholar]
- Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Shortman K, Heath WR. The CD8+ dendritic cell subset. Immunol. Rev. 2010;234:18–31. doi: 10.1111/j.0105-2896.2009.00870.x. [DOI] [PubMed] [Google Scholar]
- Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 2007;7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- Soga H, Nakamura M, Yagi H, Kayaba S, Ishii T, Gotoh T, Itoh T. Heterogeneity of mouse thymic macrophages: I. Immunohistochemical analysis. Arch. Histol. Cytol. 1997;60:53–63. doi: 10.1679/aohc.60.53. [DOI] [PubMed] [Google Scholar]
- Soulet D, Rivest S. Bone-marrow-derived microglia: Myth or reality? Curr. Opin. Pharmacol. 2008;8:508–518. doi: 10.1016/j.coph.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. II. Functional properties in vitro. J. Exp. Med. 1974;139:380–397. doi: 10.1084/jem.139.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu. Rev. Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- Stoitzner P, Green LK, Jung JY, Price KM, Tripp CH, Malissen B, Kissenpfennig A, Hermans IF, Ronchese F. Tumor immunotherapy by epicutaneous immunization requires langerhans cells. J. Immunol. 2008;180:1991–1998. doi: 10.4049/jimmunol.180.3.1991. [DOI] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung SS, Fu SM, Rose CE, Jr, Gaskin F, Ju ST, Beaty SR. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 2006;176:2161–2172. doi: 10.4049/jimmunol.176.4.2161. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alphadendritic cell development. Proc. Natl. Acad. Sci. USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P, Tamura T, Morse HC, 3rd, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, Paulson JC. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol. Cell Biol. 2007;27:5699–5710. doi: 10.1128/MCB.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Schmitt N, Klechevsky E, Pedroza-Gonzalez A, Matsui T, Zurawski G, Oh S, Fay J, Pascual V, Banchereau J, Palucka K. Harnessing human dendritic cell subsets for medicine. Immunol. Rev. 2010;234:199–212. doi: 10.1111/j.0105-2896.2009.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Klechevsky E, Schmitt N, Ni L, Flamar AL, Zurawski S, Zurawski G, Palucka K, Banchereau J, Oh S. Targeting human dendritic cell subsets for improved vaccines. Semin. Immunol. 2011;23:21–27. doi: 10.1016/j.smim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. Mononuclear phagocytic system: New classification of macrophages, monocytes and of their cell line. Bull. World Health Organ. 1972;47:651–658. [PMC free article] [PubMed] [Google Scholar]
- Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 2007;204:171–180. doi: 10.1084/jem.20061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Villadangos JA, Young L. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity. 2008;29:352–361. doi: 10.1016/j.immuni.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur. J. Immunol. 1997;27:40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- Waskow C, Liu K, Darrasse-Jèze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, Williams LT, Lin H, Stanley ER. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J. Leukoc. Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–1629. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- You Y, Myers RC, Freeberg L, Foote J, Kearney JF, Justement LB, Carter RH. Marginal zone B cells regulate antigen capture by marginal zone macrophages. J. Immunol. 2011;186:2172–2181. doi: 10.4049/jimmunol.1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]