Abstract
Purpose
The blood–brain barrier (BBB) essentially restricts therapeutic drugs from entering into the brain. This study tests the hypothesis that brain endothelial cell derived exosomes can deliver anticancer drug across the BBB for the treatment of brain cancer in a zebrafish (Danio rerio) model.
Materials and Methods
Four types of exosomes were isolated from brain cell culture media and characterized by particle size, morphology, total protein, and transmembrane protein markers. Transport mechanism, cell uptake, and cytotoxicity of optimized exosome delivery system were tested. Brain distribution of exosome delivered anticancer drugs was evaluated using transgenic zebrafish TG (fli1: GFP) embryos and efficacies of optimized formations were examined in a xenotransplanted zebrafish model of brain cancer model.
Results
Four exosomes in 30–100 diameters showed different morphologies and exosomes derived from brain endothelial cells expressed more CD63 tetraspanins transmembrane proteins. Optimized exosomes increased the uptake of fluorescent marker via receptor mediated endocytosis and cytotoxicity of anticancer drugs in cancer cells. Images of the zebrafish showed exosome delivered anticancer drugs crossed the BBB and entered into the brain. In the brain cancer model, exosome delivered anticancer drugs significantly decreased fluorescent intensity of xenotransplanted cancer cells and tumor growth marker.
Conclusions
Brain endothelial cell derived exosomes could be potentially used as a carrier for brain delivery of anticancer drug for the treatment of brain cancer.
Keywords: blood–brain barrier, brain cancer, drug delivery, exosome, zebrafish
Introduction
Exosomes are a class of membrane secreted lipid vesicles (1,2). They are formed intracellularly by invaginations of the multi-vesicular body limiting membrane and then fused out of the cell plasma membrane. As exosomes are secreted by all cell types, they can be found in abundance in body fluids including blood, saliva, and urine (3). Theoretically, exosomes in the circulation should be more stable than those of other synthetic polymer based nanoparticle and liposome due to their endogenous origin and special surface composition (4,5). Although typically devoid of cellular organelles like the mitochondria or endoplasmic reticulum, exosomes have certain surface proteins on the phospholipid bilayer and interior contents surrounded by a lipid bilayer (3). These nanosize and bubble-like particles are capable of traveling from one to another cell releasing its contents such as proteins and microRNAs across the cell membrane and communicating with each other (4).
As intercellular communicators, exosomes have received much attention as not only a basic natural characteristic, transporting microRNAs and proteins among cells, but also a possible alternatives to traditional nanoparticle approaches as drug delivery vehicles with certain advantages: 1) biologicals and small drugs can be loaded into exosomes due to the natural presence of proteins and genetic materials; 2) exosome derived drug delivery vehicles have broader distribution in biological fluids, likely producing longer circulating time and possibly better efficacy; and 3) exosomes derived from tissue-specific cells can cross the physiological barrier and target the tissue via their natural surface proteins (1,4,6–9). Therefore, exosomes potentially have advantages over polymer and liposome based nano delivery systems, with a better safety profile and better selectivity (7,10). A variety of therapeutic agents have now been exploited using exosome-based delivery to particular tissues. The majority of studies are the transfer of interfering RNAs (siRNAs), while fewer studies investigate the potential of loading other types of therapeutic agents (5).
Since exosomes resemble liposomes in having a bilayer lipid membrane and an aqueous core, they could potentially be loaded with both hydrophilic and lipophilic drugs. However, natural exosomes as drug delivery vehicles will require different drug loading strategies in vitro. For hydrophilic molecules such as siRNA, loading could be achieved by a transient physical electroporation or chemical disruption of the exosome membrane for the uptake of the small molecules. Recently, Alvarez-Erviti et al. electroporated siRNA into dendritic cell-derived exosomes (7). A hydrophobic polyphenol curcumin was loaded into exosomes purified from an EL-4 mouse lymphoma cell line by simple incubation (11). To date, two clinical trials on exosomes loaded with curcumin are investigating for cancer treatment (5).
Chemotherapy is the gold standard for cancer treatment, but efficacy is limited because the drugs cannot effectively reach the brain across the blood–brain barrier (BBB) (12–14). The BBB restricts the penetration into the brain not only of large molecule drugs but also of more than 98% small molecule drugs, such as anticancer drugs paclitaxel, doxorubicin, methotrexate, and vincristine (14–16). So far, only two exosomes successful have been used for the brain delivery of therapeutic agents in rodent models of Alzheimer's disease and brain inflammatory diseases (7,10). Before exosomes can be considered for targeted drug delivery across biological membranes, additional molecular characterization of exosomes from different sources and understanding of their transport mechanisms across biological barriers are need (4). In this study, we tested exosome mediated drug delivery across the BBB in zebrafish (Danio rerio). Four types of exosomes from brain cells were isolated and characterized on particle size, morphology, and surface protein. The transport mechanism, cell uptake, and cytotoxic efficacy of anticancer drug delivered by exosomes were tested in brain cancer cells. In in vivo zebrafish study, exosomes released from brain endothelial cells delivered anticancer drug across the BBB, which subsequently exerted cytotoxic efficacy against brain cancer. These finding support the potential of exosome vesicles for targeted brain drug delivery in the treatment of brain cancer.
Methods and Materials
Materials
Cell lines, media, fetal bovine serum (FBS), penicillin plus streptomycin solution, and trypsin-EDTA were obtained from American Tissue Culture Collection (ATCC, Rockville, MD, USA). Exosome isolation solution, RNeasy Mini Kit, vascular endothelial growth factor (VEGF) forward and reverse primers, iTaq™ Universal SYBR® Green One-Step Kit, and biological agents were purchased from Life Technologies (Grand Island, NY, USA). CD9, CD63, and CD81 antibodies, ELISA kits, and exosome-depleted FBS were obtained from System Biosciences Inc. (Mountain View, CA, USA). Western Lightening Chemiluminescence reagents were purchased from Amersham Biosciences, Inc. (Piscataway, NJ, USA). Cell lysis buffer was purchased from Cell Signaling Technology Inc. (Boston, MA, USA). Rhodamine 123, paclitaxel, doxorubicin, tricaine, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, and other chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Cell Culture and Exosome Isolation
Brain neuronal glioblastoma-astrocytoma U-87 MG, endothelial bEND.3, neuroectodermal tumor PFSK-1, and glioblastoma A-172 cell lines were grown in recommended media supplemented with 10% FBS, 2 mM L-glutamine, 100 μg/ml penicillin plus 100 μg/ml streptomycin, in a humidified 37°C incubator with 5% CO2 according to ATCC protocols. When reaching a confluency of 60–80% in culture flasks, cells were cultured in exosome-depleted FBS media overnight. Exosomes were extracted from cell culture media using an Invitrogen® Total Exosome RNA and Protein Isolation Kit followed the recommended protocols. Briefly, after harvesting, cell media were collected and centrifuged at 2000×g for 30 min, 10 ml of culture media supernatant mixed with 5 ml of the total exosome isolation reagent was incubated overnight at 4°C. The exosomes were precipitated by centrifugation at 10,000×g for 60 min at 4°C using an Avanti JE centrifuge (Beckman Corp, Brea, CA, USA) and the supernatant was discarded. Finally, the pellet was resuspended in PBS and stored at −20°C until further characterization and study.
Exosome Characterization
Particle size, morphology, total protein, and surface proteins of exosome nanoparticles were characterized according to our previous studies (17,18). Particle sizes of exosome nanoparticles were measured using a Delsa™ Nano C nanosizing system (Beckman Coulter, Brea, CA, USA). A scanning electron microscope (AMRay, Bedford, MA, USA) was used to observe the morphology of exosomes.
Protein was extracted from the exosomes using a cell lysis buffer and total protein concentrations were measured with a Pierce BCA assay kit. Surface protein levels in exosomes were first analyzed by a western blotting method according to previously published procedures (18). After boiling, 50 μg of proteins were electrophoresed and transferred to a polyvinylidene difluoride membrane. After the membranes were treated with a primary CD9, CD63, or CD81 antibody and then a secondary antibody, signals for proteins were detected by Western Lightening Chemiluminescence reagents. The CD9, CD63, or CD81 protein level was quantified from the densiometric intensity of each band using a Bio-Rad Quantity software (Bio-Rad Laboratories, Hercules, CA, USA). The levels of surface proteins in exosomes were further characterized by ELISA according to the manufacturer's instructions. 50 μl of 400 μg/ml of each prepared exosome sample (quantified by total protein) was added into a micro-titer plate and incubated at 37°C overnight. After washing the plate three times, exosome specific primary antibody (CD9, CD63, or CD81) was added to each well and the plate was incubated at room temperature for 1 h with shaking. After adding substrate and stop buffer, the plate was quantitated by reading at 450 nm absorbance with a Synergy 4 microtiter plate reader (Biotek, Winooski, Vermont, USA).
Preparation of Drug Loaded Exosomes and Determination of Drug Loading
Drug loaded exosomes were prepared as previously described, with a slight modification for mixing drugs with exosome (8,11). The solution including 2 mg/ml of rhodamine 123, paclitaxel, or doxorubicin was added to exosomes (200 μg/ml of total proteins) in PBS and incubated at 37°C for 2 h. To determine drug loading efficiency, the mixture was centrifuged for 1 h at 100,000×g. Drug loaded exosomes were collected, washed, and resuspended in PBS. The concentration of drug in supernatant and in the exosomes was determined by high-performance liquid chromatography (Agilent 1220 DAD; Santa Clara, CA, USA) (19–21).
Transport Studies of Exosome Mediated Delivery
The fluorescent rhodamine 123 was used as a marker of exosome delivery and the transport mechanism for delivery from brain endothelial bEND.3 cell released exosomes was evaluated (22). The brain endothelial bEND.3 cells were seeded on a 100 mm petri dish with 10 ml of cell media at a density of 2×106 cells/ml counted by a Cellometer® Auto T4 Cell Counter (Nexcelom Bioscience LLC, Lawrence, MA, USA). After 2 h of incubation at 37 or 4°C, the buffer, 0.2 mg/ml rhodamine 123 alone or 0.2 mg/ml rhodamine 123 formulated with bEND.3 released exosomes (200 μg/ml total proteins) was added into the media. After incubation, the media with treatment solutions were removed and the cells were washed with 1× PBS three times. Cells were scraped, suspended, and diluted to 1×106 cells/ml and then analyzed with an Acuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA) using the FL3 detector position.
Cell Uptake with Exosome Mediated Delivery
Brain neuronal glioblastoma-astrocytoma U-87 MG cells were seeded on a 100 mm petri dish with 10 ml of cell media at a density of 2×106 cells/ml. After 24 h of incubation at 37°C and 5% CO2, the desired solutions including 0.2 mg/ml rhodamine 123 alone or 0.2 mg/ml rhodamine 123 formulated with bEND.3 exosomes (100 μg/ml or 200 μg/ml total proteins) were added into the media. After 18 h of incubation at 37°C, the media with treatment solution were removed and the cells were washed with 1× PBS three times. Cells were scraped and suspended in 1× PBS. Cell samples were diluted and then analyzed with an Acuri C6 flow cytometer using the FL3 detector position.
Cytotoxicity of Exosome Delivered Anticancer Drugs
Anticancer drugs doxorubicin and paclitaxel were formulated into exosomes derived from human brain neuronal glioblastoma-astrocytoma U-87 MG and brain endothelial bEND.3 cells with different protein concentrations (8). U-87 MG cells were grown in flat-bottom, 96-well microtiter tissue culture plates and cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described (17).
Brain Distribution of Exosome Mediated Delivery in Zebrafish Embryo
TG(fli1:GFP) zebrafish were incubated at 28°C in embryo medium according to standard zebrafish care and procedures (23). Rhodamine 123 (0.2 mg/ml), BODIPY® 564/570 conjugated paclitaxel (0.2 mg/ml), and doxorubicin (0.2 mg/ml) were formulated with bEND.3 exosomes (200 μg/ml total proteins). Microinjection and imaging protocols were modified from the previous publication (24). Briefly, 5 day post fertilization (dpf) zebrafish embryos were anesthetized with 0.2 mg/ml tricaine. Then 4.6 nl of solution was injected into the common cardinal vein of the anesthetized embryo using a Nanoject IITM Auto-Nanoliter Injector (Drummond Scientific Company, Broomall, PA, USA). Embryos were returned to embryo medium and incubator at 28°C. Embryos were anesthetized and imaged at 18 h post injection using an Olympus LSM 1000 confocal microscope with FluoView 10 software (Olympus Corp., Central Valley, PA, USA).
Brain Cancer Effects of Exosome Delivered Doxorubicin In Vivo
A model of primary brain cancer model was developed in zebrafish with a xenotransplantation method modified from previous studies (25). U-87 MG cells were labelled with Cell Brite® DiD (Biotium Inc., Hayward, CA, USA) according to standard procedures. Zebrafish embryos at 2 dpf were anesthetized with 0.2 mg/ml tricaine and 9.2 nl of labelled U-87 MG cells (2.5×107 cells/ml) were injected into the brain ventricle. Embryos were placed in individual wells in a 24-well plate at 28°C in embryo medium for 2 days. At 4 dpf, the embryos with brain cancer xenografts were anesthetized and treated with 2.3 nl of 0.2 mg/ml doxorubicin in PBS, 0.2 mg/ml doxorubicin formulated with 200 μg/l total protein of bEND.3 exosomes, or PBS alone. Injections were made into the common cardinal vein. Embryos were incubated in embryo medium individually in a 24-well plate and imaged at 7 dpf with an Olympus LSM 1000 confocal microscope for tumor size. After imaging, total RNA of fish was extracted using RNeasy Mini Kit and qualified by reading at 260/280 nm absorbance with a Take 3 micro-volume plate in Synergy 4 microtiter plate reader (Biotek, Winooski, Vermont, USA). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) were used to detect the RNA levels of vascular endothelial growth factor (VEGF), a marker of cancer cell growth using iTaq™ Universal SYBR® Green One-Step Kit according to the instruction in an iQ™5 Multicolor Real-Time PCR Detection System (Bio Rad, Philadelphia, PA, USA).
All animal studies were approved by the University of Maine at Orono Animal Care and Use Committee (IACUC Protocol #2013-11-02) and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Data Analysis
All experiments were repeated three times and each experiment had at least n=3 measurements in in vitro experiments, and n= 20 in in vivo experiments. One-way ANOVA was used to compare the data. When the differences in the means were significant, post-hoc pair wise comparisons were conducted using Newman-Keuls multiple comparison (GraphPad Prism, version 3.03, GraphPad Software, San Diego, CA). Differences in p-values less than 0.05 were considered statistically significant.
Results
Exosome Characterization
Mean diameters of the four exosomes ranged from 30 to 100 nm. Exosomes released from neuroectodermal tumor PFSK-1 cells were the biggest particles (74.5±1.5 nm) (Fig. 1a). Mean particle sizes of exosomes from brain endothelial bEND.3, glioblastoma A-172, and brain neuronal glioblastoma-astrocytoma U-87 MG cells were all less than 50 nm diameter, but bEND.3 exosomes were significantly bigger than the other two (p<0.05). The size scale provided by the scanning electron microscope (SEM) showed the exosomes were less than 150 nm in diameter, which was consistent with the results measured by the particle sizing system. All exosomes showed slightly different morphologies with a smooth or irregular outer membrane and a spherical or oval shaped appearance (Fig. 1b). Additionally, morphological studies conducted using SEM confirmed that the exosomes were solid nanospheres similar to liposomes or other polymeric nanocarriers and agglomerates were not detected (Fig. 1b). These results show that the isolated exosomes from different brain cells have different particle size and morphology.
Fig. 1.
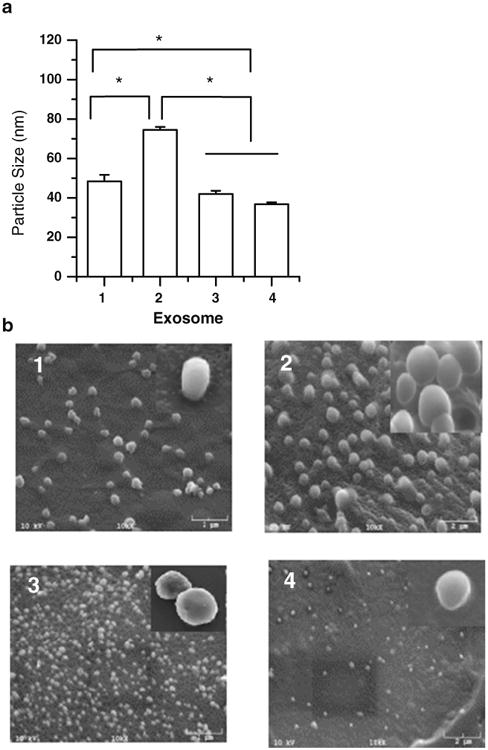
Particle size (a) and scanning electron micrograph (b) of exosomes isolated from (1) brain endothelial bEND.3, (2) neuroectodermal tumor PFSK-1, (3) glioblastoma A-172, and (4) glioblastoma -astrocytoma U-87 MG cells (* Results are significantly different, p<0.05).
Three tetraspanin surface proteins, CD9, CD63, and CD81 were characterized by ELISA and western blot methods. Western blotting imaged only detectable CD63 protein in bEND.3 cell derived exosomes (Fig. 2). ELISA showed the presence of CD63 in all four exosome types, and the amount in bEND.3 exosomes was significantly higher than in other types of exosomes (p<0.05, Fig. 2a). CD9 and CD81 were not detected by western blotting in any of exosomes. Levels of CD9 and CD81 were detected and quantified by ELISA in all four exosomes, and there were no significant differences among exosome types (p>0.05, Fig. 2b and c). Thus, only the high presence of CD63 in bEND.3 exosomes suggests that these exosome nanovesicles might be differently implicated in receptor-mediated transport across the BBB. The introduction of rhodamine 123, paclitaxel, doxorubicin into the exosomes was determined by HPLC. The procedure caused the loading of 7.9±1.4 ng rhodamine 123, 7.3±1.1 ng paclitaxel, and 132.2±2.9 ng doxorubicin into 1 μg of bEnd.3 exosomes. The loading efficiency of doxorubicin in the bEND.3 exosome was similar to previous reports where doxorubicin was encapsulated in exosomes released by human umbilical vein endothelial cells (8).
Fig. 2.
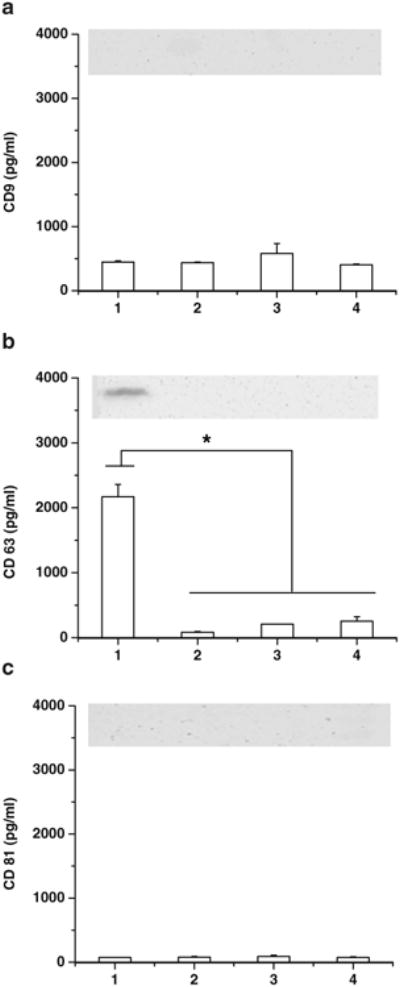
Western blots and ELISA results of (a) CD9, (b) CD63, and (c) CD81 in exosomes isolated from (1) bEND.3, (2) PFSK-1, (3) A-172, and (4) U-87 MG cells (* Results are significantly different, p<0.05).
Transport Mechanisms of Exosome Mediated Delivery
For quantitative analysis of cellular transport of exosomes, the cellular uptake fluorescence intensity from the PBS control, fluorescent rhodamine 123 alone, and rhodamine 123 loaded exosomes was evaluated in bEND.3 cells at 37 and 4°C. At 37°C, there was significantly greater cellular fluorescence with rhodamine 123 formulations compared to that of the control (p<0.05) (Fig. 3a). While some free rhodamine 123 may diffuse across the cell membrane into bENd.3 cells, the fluorescence with exosome delivered rhodamine 123 was significantly greater (p<0.05). There were no significant differences among groups at 4°C, where active uptake endocytosis of rhodamine 123 was inhibited while passive diffusion was not affected (Fig. 3b). In comparison to the uptake at 37°C, the accumulation of rhodamine 123 in cells was lower throughout the entire course of the study at 4°C (p<0.05) for both formulations of rhodamine 123 with and without exosomes. The differences in cell uptake of exosome delivered rhodamine 123 between 37 and at 4°C can be explained by the involvement of active receptor-mediated endocytosis process (energy-dependent internalization) in exosome delivered formulations.
Fig. 3.
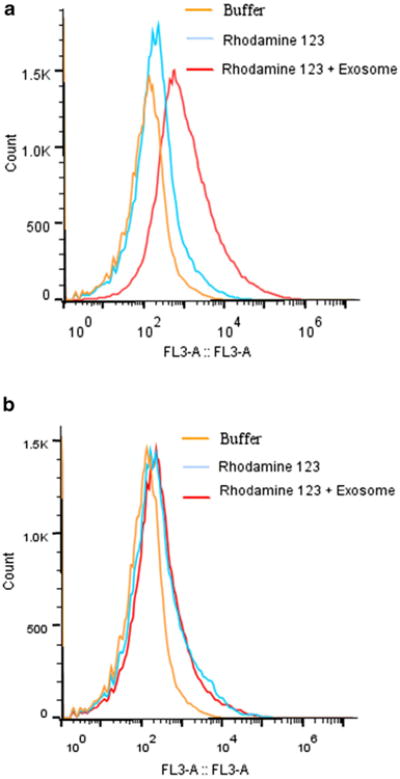
Flow cytometric analysis of rhodamine 123 in bEND.3 cell incubated at (a) 37°C and (b) 4°C.
Cell Uptake of Exosome Delivered Rhodamine 123
In order to evaluate the cellular delivery capacity and specificity of the bEND.3 exosomes, the cellular uptake of exosome delivered rhodamine 123 was also investigated in U-87 MG cells. Cell uptake studies using the C6 flow cytometer showed that exosomes enhanced the cell uptake of rhodamine 123 (Fig. 4a). Treated cells incubated with exosome delivered rhodamine 123 showed a higher fluorescence in comparison with cells treated with rhodamine 123 alone (without exosome) (Fig. 4a). Moreover, rhodamine 123 delivered by 200 μg/ml of bEND.3 exosomes showed higher uptake into the cell compared to that delivered by 100 μg/ml of bEND.3 exosomes. Greater intracellular fluorescence with rhodamine 123 delivered by the highest concentration of exosome was confirmed by confocal microscopy (Fig. 4b). The overlay results also supported that exosome delivery increased cellular uptake of rhodamine 123 as less fluorescence presented on the cell surface.
Fig. 4.
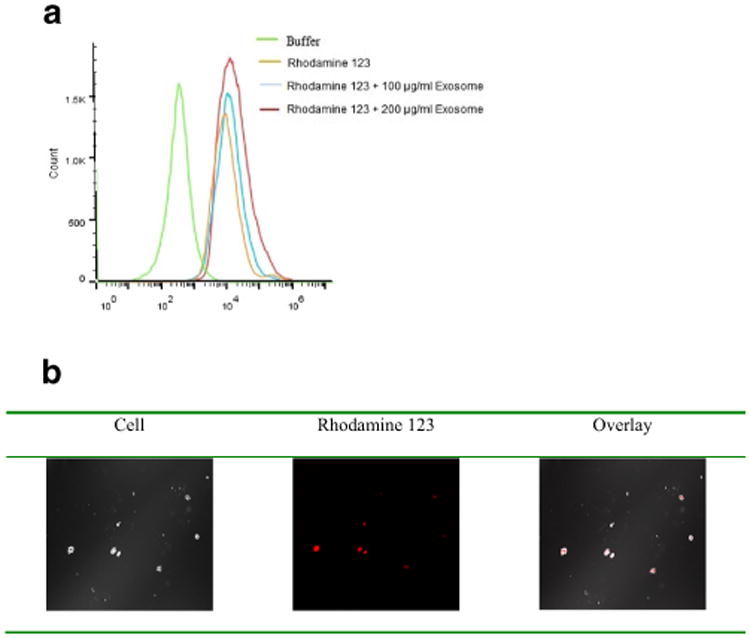
Uptake (a) and confocal imaging (b) of bEND.3 exosome delivered fluorescent marker in U-87 MG cells.
Cytotoxicity of Exosome Delivered Anticancer Drugs
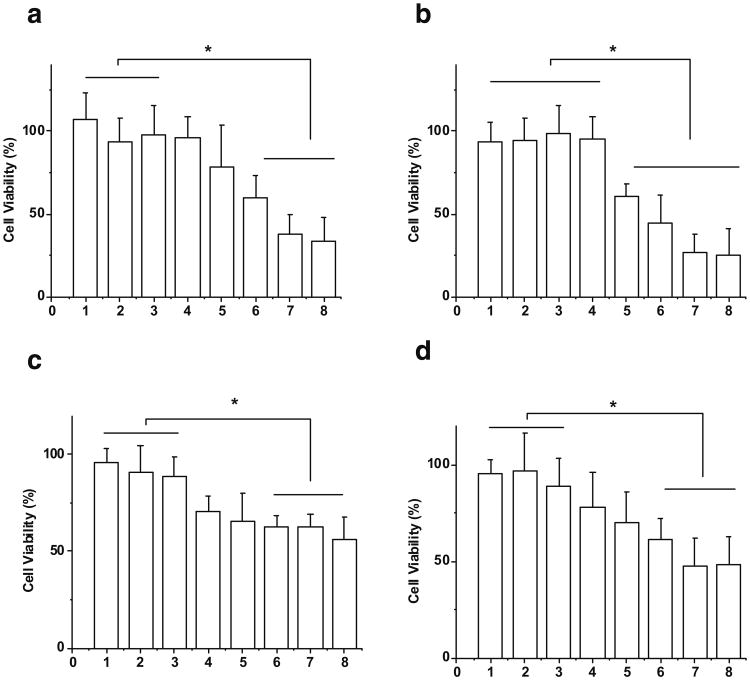
Cytotoxic effects of optimized bEND.3 and U-87 MG exosome delivered anticancer drugs doxorubicin and paclitaxel were examined on U-87 MG cells (Fig. 5). Doxorubicin (0.2 mg/ml) alone did not cause more cell death compared to medium control. Similarly, bEND.3 and U-87 MG cell released exosomes alone (100 or 200 μg/ml quantified by the amount of total protein) had no effect on cell viability compared to medium control. Both bEND3 and U-87 MG exosomes delivered doxorubicin significantly decreased cell viability in a dose-dependent fashion, with the highest concentration of exosomes (200 μg/ml) and doxorubicin decreasing cell viability by about 70% (Fig. 5a and b). Similar cytotoxic effects were found with paclitaxel. BEND.3 and U-87 MG exosomes delivered paclitaxel decreased cell viability (>50%) with the highest concentration of exosomes (200 μg/mL) (Fig. 5c and d).
Fig. 5.
Cytotoxicity on U-87 MG cells treated by (a) doxorubicin with U-87 MG cell derived exosome, (b) doxorubicin with bEND.3 cell derived exosome, (c) paclitaxel with U-87 MG cell derived exosome, and (d) paclitaxel with bEND.3 cell derived exosome (1: 25 μM drug; 2. 100 μg/ml exosome; 3. 200 μg/ml exosome; 4. drug + 20 μg/ml exosome; 5. drug + 40 μg/ml exosome; 6. drug + 80 μg/ml exosome; 7. drug + 100 μg/ml exosome; 8. drug + 200 μg/ml exosome, * Results are significantly different, p<0.05).
Exosome Mediated Delivery Across the BBB in Zebrafish
To determine the ability of exosomes to deliver drugs across the BBB in vivo, rhodamine 123 loaded exosomes were injected into the circulation in zebrafish embryos and fluorescence of rhodamine 123 was examined in the brain tissue. After 18 h post injection into the embryo's circulating system via the cardinal vein, rhodamine 123 was not observed in the brain tissue but remained stayed in the vessels (Fig. 6a). The images show only green vascularization and no red rhodamine 123 in the brain. The red rhodamine 123 was confined to vascular system (green) in the transgenic fish. Similarly, rhodamine 123 delivered by exosomes from U-87 MG, PFSK-1, and A-172 cells (Fig. 6b–d) also did not penetrate the brain in treated embryos. However, rhodamine 123 delivered by bEND.3 exosomes crossed the BBB and entered the brain. Seen in the overlay image, red rhodamine 123 was distributed into the brain area outside the green vessels (Fig. 6e). Since only the bEND3 derived exosomes were able to deliver rhodamine 123 across the BBB into the brain, this exosome was further tested for the delivery of anticancer drugs in vivo.
Fig. 6.
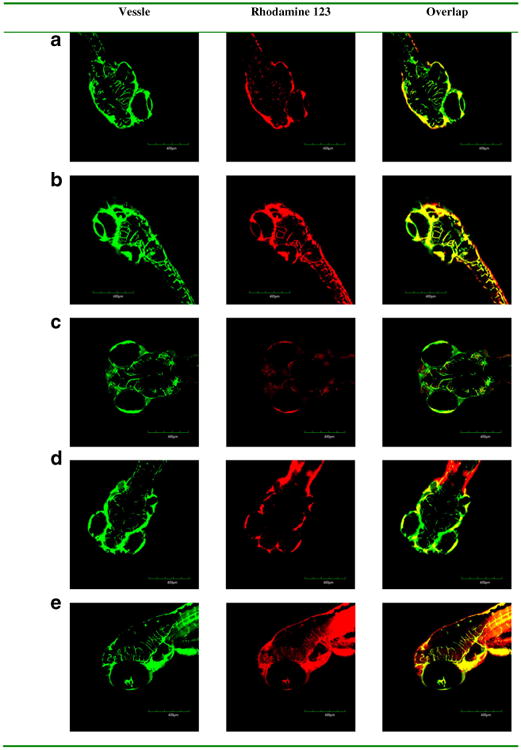
In vivo brain imaging of exosome delivered rhodamine 123 in Tg (fli1:GFP) embryonic zebrafish. Rhodamine 123 (red) retained within vessels (green) after the injected formulations without exosome (a) and with exosomes isolated from (b) neuroectodermal tumor PFSK-1, (c) glioblastoma A-172, and (d) glioblastoma-astrocytoma U-87 MG. Rhodamine 123 (red) crossed out of vessels (green) after the injected formulation with exosomes isolated from (e) brain endothelial bEND.3 cells.
Fluorescent doxorubicin and paclitaxel alone and formulated with bEND3 exosomes were injected into the cardinal vein in zebrafish embryos and penetration into brain tissue was evaluated. When given alone, the images showed no distribution of fluorescent drug out of the vasculature and into the brain (Fig. 7a and b). However, when formulated with exosomes both drugs distributed into the brain region of the zebrafish embryos (Fig. 7c and d). These results indicate that bEND.3 derived exosomes can effectively deliver the anticancer drugs across the BBB into the brain in vivo.
Fig. 7.
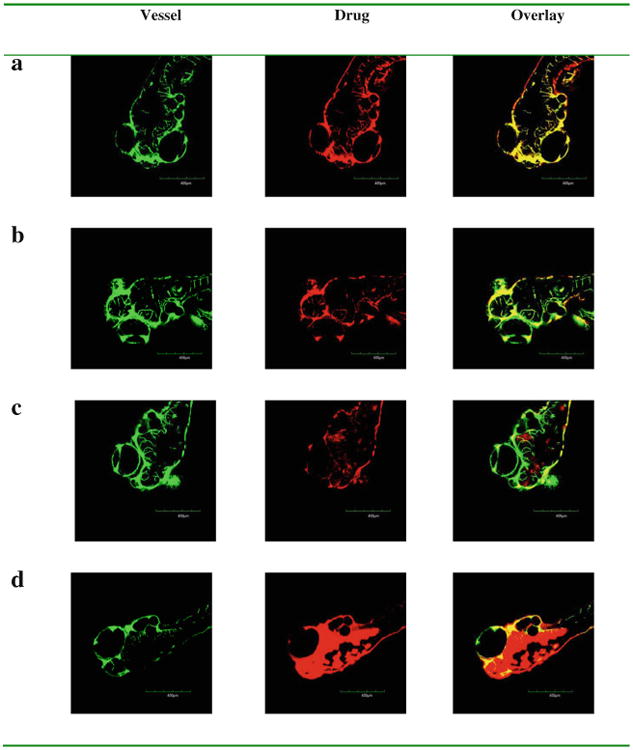
In vivo brain imaging of exosome delivered anticancer drugs in Tg (fli1:GFP) embryos. Doxorubicin (a) and fluorescence labelled paclitaxel (b) retained within vessels after the injected formulations without exosomes. Doxorubicin (c) and fluorescence labelled paclitaxel (d) (red) crossed out of vessels (green) after the injected formulations with brain endothelial bEND.3 cell derived exosomes.
Brain Cancer Treatment by Exosome Delivered Anticancer Drug in Zebrafish Model
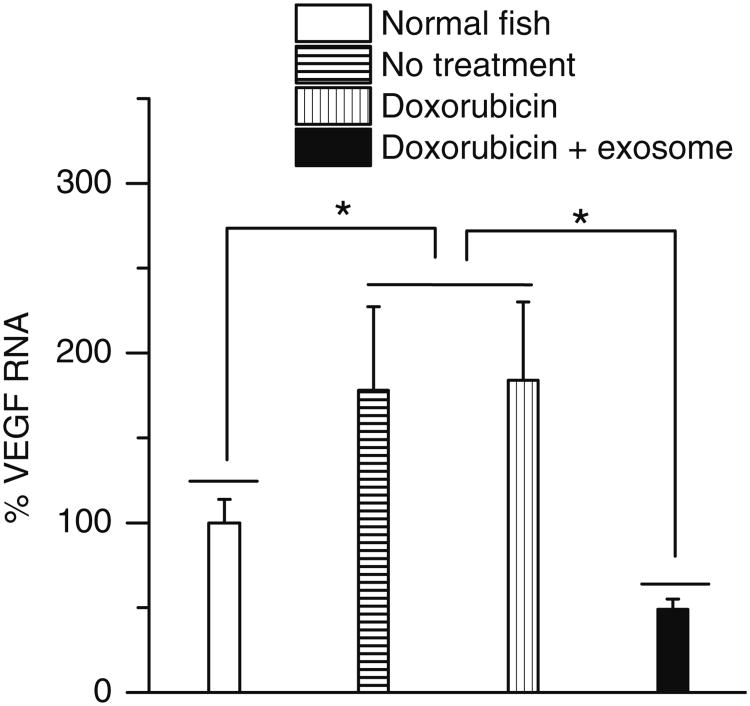
A model of primary brain cancer was developed by injecting DiD labelled U-87 MG cells into the zebrafish brain ventricle. At 2 h and 2 days post injection, the cells were visible in the brain area, with no migration into other areas (Data not shown). Similarly at 5 days post injection, the U-87 MG cells were located in the same region of the brain, with no migration (Fig. 8a). To test in vivo effects of exosome delivered doxorubicin, at 2 days post injection of U-87 MG cells into the brain ventricle, zebrafish embryos were treated with 4 nl of 0.2 mg/ml doxorubicin alone or delivered in 200 μg/ml of bEND.3 exosomes. Zebrafish treated with exosome delivered doxorubicin had a significantly smaller area of the U-87 MG cancer cells compared to embryos treated with drug alone or buffer control (Figs. 8b–d). In exosome delivered doxorubicin treated embryos, very few labelled cancer cells remained in the brain (Fig. 8d). Furthermore exosome delivered doxorubicin significantly suppressed the RNAs of VEGF in brain tumor model in zebrafish (Fig. 9). The data show doxorubicin delivered in 200 μg/ml of bEND.3 exosomes has significant therapeutic efficacy in this zebrafish model of brain cancer.
Fig. 8.
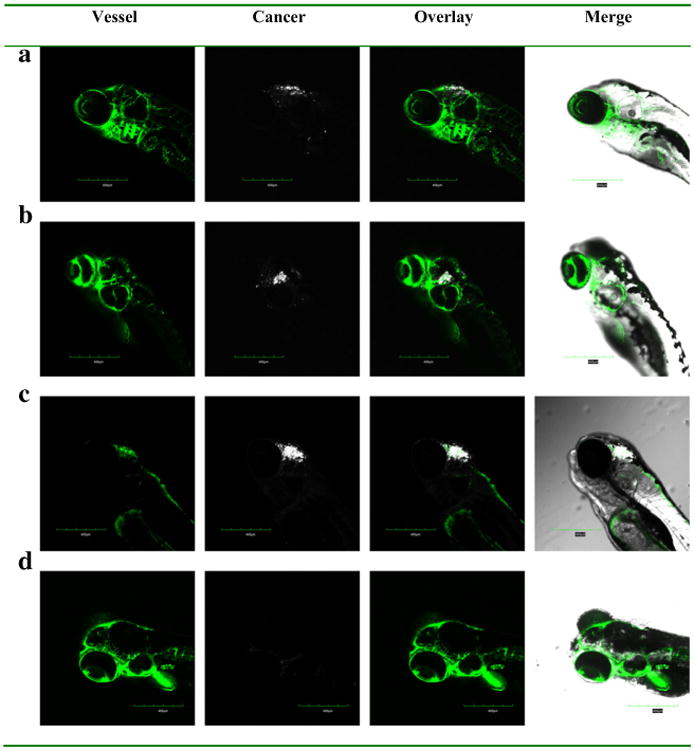
Efficacy of bEND.3 exosome delivered doxorubicin on zebrafish cancer model. DiD labeled cancer cells were injected into the hindbrain ventricle of 2 dpf zebrafish and further visualized at (a) without treatment, (b) treated by PBS buffer, (c) doxorubicin, (d) doxorubicin loaded exosome at 7 dpf.
Fig. 9.
Inhibitory of VEGF in vivo using bEND.3 exosome delivered doxorubicin on zebrafish cancer model (* Results are significantly different, p<0.05).
Discussion
Penetrating the blood–brain barrier (BBB) and delivering anti-cancer drugs to the tumor at therapeutic levels is a major challenge in the treatment of brain cancers (14,26). Finding a delivery approach that is minimally invasive but allows for maximum effectiveness is crucial for successful treatment of brain cancers. Currently, the main approach to deliver drugs across the BBB is to utilize endogenous transport mechanisms (27).
Exosomes are the most extensively characterized endogenous carriers, which transport information between cells by receptor mediated endocytosis (28). A breakthrough in understanding of the significance of exosomes was the finding that exosomes express a selected number of parent cell-derived markers (29–31), some of which are tissue-specific relating to the cell of origin (32). In this current study, we prepared and studied exosomes from brain neuronal glioblastoma-astrocytoma U-87 MG, brain endothelial bEND.3, neuroectodermal tumor PFSK-1, and glioblastoma A-172 cells. The hypothesis was that exosomes derived from brain cells would more likely display brain-specific biomarkers for delivery of molecules across the BBB.
Although the amount of exosome produced by brain cells was slightly variable, the size (30–100 nm) and appearance of all exosome types were consistent with previous studies (30,31,33). With a role in cell-cell communication, the surface of exosomes is typically enriched in cell-targeting adhesion molecules (tetraspanins and integrins), membrane trafficking proteins, antigen-presenting molecules (major histocompatibility complex class I and class II), and receptors/transmembrane proteins (32). For example, tetraspanin proteins CD9, CD63, and CD81 isolated from brain endothelial HCMEC/D3 cells played a very important role in communication between primary astrocytes and cortical neurons (32). In this study, levels of CD9 and CD81 were similar in all four types of exosomes. However, brain endothelial bEND.3 exosomes presented significantly higher CD63 levels compared to the other exosomes. This is the first documented identification of the molecular signature for brain endothelial cell-derived exosomes.
Exosomes can carry cell-type-specific proteins found in the membrane of the parent cell, such as myelin proteins in exosomes derived from oligodendrocytes, with the unique property of homing selectivity (7). Specific homing biomarkers (e.g., CD63) of exosomes derived from endothelial cells of the BBB would more likely provide transport across the BBB than exosomes derived from neuronal glioblastoma and neuroectodermal cells. Although CD63 may not be the only biomarker for the homing selectivity to the BBB, the bEND.3 exosomes derived from cells originally found in the BBB was expected to be able to cross the BBB through cell-type-specific proteins via receptor mediated endocytosis. Our results demonstrated that bEND.3 derived exosomes had greatly enhance internalization of the fluorescent marker in bEND.3 cells via an energy-dependent internalization process. The active process is assumed to be receptor-mediated endocytosis. This assumption has been supported by the observation that brain endothelial cell extracellular vesicles contained several receptors previously shown to carry macromolecules across the BBB, including transferrin, LDL, and insulin receptors (32).
BEND.3 exosomes were able to significantly enhance cell uptake of a fluorescent marker (rhodamine 123), and to significantly increase the cytotoxic effect of anticancer drugs doxorubicin and paclitaxel in U-87 MG cells brain cancer cells. In all cases, more exosomes produced higher cell uptake and cytotoxicity. Since exosomes resemble liposomes, with a bi-lipid membrane and an aqueous core, potentially they could load both hydrophilic and lipophilic drugs (28). As a nanosize carrier, the loading efficiency of nanoparticle is limited by their interior space (34), thus to load more drug would require more exosomes. Doxorubicin and paclitaxel are anti-cancer drugs commonly used to treat breast cancer and solid tumors. They are not used to treat brain tumors because they are unable to pass the BBB (16). When given alone, neither drug caused significant cytotoxicity in the U-87 MG cells. The significant cytotoxicity (up to 70%) seen when delivered in bEND3 exosomes most likely resulted from exosome-enhanced intracellular penetration of the two anticancer drugs. This exosome-induced penetration of doxorubicin or paclitaxel into cancer cells is consistent with results from previous cell uptake studies. It is also supported by the intracellular actions of the two drugs of intercalating and inhibiting DNA (doxorubicin) and inhibiting microtubules (paclitaxel). Interestingly, exosomes from brain endothelial bEND.3 cells and brain neuronal glioblastoma U-87 MG cells showed effective delivery of anticancer drugs. This may be because both types of exosomes promote cell uptake by endocytosis as do other polymeric nanoparticles such as liposomes (34).
The in vivo efficacy of exosome delivery of anticancer drug into the brain was studied in zebrafish embryos. Due to size, ease of care, rapid development, and highly conserved nature of both genetics and cell biology with higher vertebrates, the zebrafish is a suitable organism in which to model human diseases (35). Importantly, by 3 days post fertilization (dpf) the zebrafish embryos have a fully developed and functioning BBB that has similar properties to the human BBB (24), making them a suitable animal for brain drug delivery studies.
Previous research has shown that exosomes were viable carriers for drugs, such as curcumin, and able to deliver drugs to the treatment site (10). Moreover, exosomes were capable of delivering siRNA to the brain, across the BBB in mice (7). Our results are consistent with other studies of exosome facilitated drug delivery to the brain and to other regions of the body.
When given alone, rhodamine 123, doxorubicin and paclitaxel remained localized within the vasculature and did not penetrate the BBB in zebrafish. This finding also confirms that zebrafish embryos do form the BBB by 3 dpf. When delivered in bEND.3 exosomes, there was significant penetration of the BBB of the fluorescent marker and of the two anticancer drugs. In addition, preliminary data from the embryos with xenotransplanted U-87 MG cells support that the exosomes delivered cytotoxic levels of doxorubicin intracellularly to the brain cancer cells. This was seen as smaller U-87 MG cell mass in the doxorubicin treated embryos.
Zebrafish have been found to develop many tumor types seen in humans, with similar morphology, gene expression, and signaling pathways (23). Transplantation studies have been specifically effective in the study of vasculature remodeling, cancer invasion, and metastasis. The rapid development of zebrafish allows dynamic processes to be followed in real time, such as evaluating therapeutic efficacy of optimized formulations in transplanted animals (36). Ours is the first report of a xenotransplant brain cancer model in zebrafish, however, our approach was supported by a previous study on an orthotropic transplant in zebrafish. That study showed that human retinoblastoma cells could be injected into the vitreous cavity about 2 days post fertilization and maintain stability and size for about 4 days post injection (25). For the brain cancer model, we injected fluorescent labeled human glioblastoma-astrocytoma U-87 MG cells into the brain ventricles at 2 days post fertilization. From two to 5 days post injection, the cells were present as masses seen via confocal microscopy through the transparent head area of the zebrafish. Zebrafish survived through 7 days post fertilization, providing an adequate time window for drug screening. BEND.3 exosome-delivered doxorubicin significantly decreased the U-87 MG cell signal in xenotransplanted zebrafish, compared with vehicle treated controls.
This study evaluated the potential for exosomes to be used to deliver drugs across the BBB and into brain. Based on the presence of surface markers likely to aid BBB penetration, we optimized the exosomes isolated from brain endothelial bEND.3 cells. Delivery in these exosomes significantly increased the intracellular uptake and cytotoxicity of anticancer drugs doxorubicin and paclitaxel in human glioblastoma-astrocytoma U-87 MG cells in vitro. In zebrafish in vivo, exosome delivery allowed doxorubicin and paclitaxel to cross the BBB, whereas when given alone neither drug showed brain uptake. Preliminary data from zebrafish support that exosome-induced brain delivery of doxorubicin resulted in cytotoxicity against xenotransplanted U-87 MG cells. These data show the potential for use of exosomes with specific properties to deliver drugs across the BBB. This provides another avenue for pharmaceutical treatments for brain cancers and other neurological disorders that are currently untreatable via pharmaceuticals.
To create exosome-based drug delivery system is superior to synthetic drug carriers. However, some shortcomings and obstacles do exist that need to be overcome to reach maximum potential in the clinic. Important issues that still need to be addressed include the choice of exosome donor cell, type of loading procedure, and use of targeting peptides on the exosome surface. The solutions to these questions are awaited with great interest. We are cautious in interpreting in vitro and in vivo animal data, and in extrapolating to the clinical setting. Data on the stability, drug loading, pharmacokinetics properties, and safety of the carrier remains to be established. Further studies are planned to optimize exosome formulations and investigate long-term therapeutic efficacy in models of brain cancer.
Acknowledgments
Research reported in this project was supported by the Maine Cancer Foundation Pilot Research Grant 2013, the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423, and the Salisbury Cove Research Fund. The authors thank Drs. Michelle Goody and Carol Kim at the University of Maine for kind help on the zebrafish study.
Contributor Information
Tianzhi Yang, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Paige Martin, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Brittany Fogarty, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Alison Brown, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Kayla Schurman, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Roger Phipps, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA.
Viravuth P. Yin, Davis Center for Regenerative Biology and Medicine, Mount Desert Island Biological Laboratory, Salisbury Cove, Maine 04672, USA
Paul Lockman, Department of Basic Pharmaceutical Sciences, School of Pharmacy West Virginia University, Morgantown, West Virginia 26506, USA.
Shuhua Bai, Email: bais@husson.edu, Department of Basic Pharmaceutical Sciences, School of Pharmacy, Husson University, Bangor, Maine 04401, USA,; Davis Center for Regenerative Biology and Medicine, Mount Desert Island Biological Laboratory, Salisbury Cove, Maine 04672, USA.
References
- 1.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–41. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery—a novel application for the mesenchymal stem cell. Biotechnol Adv. 2012 doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 3.van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G. Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev. 2013;65:331–5. doi: 10.1016/j.addr.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Lakhaland S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. BioEssays News Rev Mol Cell Dev Biol. 2011;33:737–41. doi: 10.1002/bies.201100076. [DOI] [PubMed] [Google Scholar]
- 5.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery—a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543–51. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol JASN. 2009;20:363–79. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 8.Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, et al. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7:7698–710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 9.Yangand J, Aschner M. Developmental aspects of blood-brain barrier (BBB) and rat brain endothelial (RBE4) cells as in vitro model for studies on chlorpyrifos transport. Neurotoxicology. 2003;24:741–5. doi: 10.1016/S0161-813X(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 10.Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther J Am Soc Gene Ther. 2010;18:1606–14. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther J Am Soc Gene Ther. 2011;19:1769–79. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chenand Y, Liu L. Modern methods for delivery of drugs across the blood-brain barrier. Adv Drug Deliv Rev. 2012;64:640–65. doi: 10.1016/j.addr.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Morikawa A, Peereboom DM, Thorsheim HR, Samala R, Balyan R, Murphy CG et al. Capecitabine and lapatinib uptake in surgically resected brain metastases from metastatic breast cancer patients: a prospective study. Neuro-Oncol. 2014 doi: 10.1093/neuonc/nou141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn KA, et al. Heterogeneous blood-tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16:5664–78. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muldoon LL, Soussain C, Jahnke K, Johanson C, Siegal T, Smith QR, et al. Chemotherapy delivery issues in central nervous system malignancy: a reality check. J Clin Oncol Off J Am Soc Clin Oncol. 2007;25:2295–305. doi: 10.1200/JCO.2006.09.9861. [DOI] [PubMed] [Google Scholar]
- 16.Sathornsumeteeand S, Rich JN. New approaches to primary brain tumor treatment. Anti-Cancer Drugs. 2006;17:1003–16. doi: 10.1097/01.cad.0000231473.00030.1f. [DOI] [PubMed] [Google Scholar]
- 17.Yang T, Nyiawung D, Silber A, Hao J, Lai L, Bai S. Comparative studies on chitosan and polylactic-co-glycolic acid incorporated nanoparticles of low molecular weight heparin. AAPS PharmSciTech. 2012;13:1309–18. doi: 10.1208/s12249-012-9854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai S, Yang T, Abbruscato TJ, Ahsan F. Evaluation of human nasal RPMI 2650 cells grown at an air-liquid interface as a model for nasal drug transport studies. J Pharm Sci. 2008;97:1165–78. doi: 10.1002/jps.21031. [DOI] [PubMed] [Google Scholar]
- 19.Mittal A, Chitkara D, Kumar N. HPLC method for the determination of carboplatin and paclitaxel with cremophorEL in an amphiphilic polymer matrix. J Chromatogr B Anal Technol Biomed Life Sci. 2007;855:211–9. doi: 10.1016/j.jchromb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Iqbal T, Kinjo M, Dowling TC. Determination of Rhodamine 123 in cell lysate by HPLC with visible wavelength detection. J Chromatogr B Anal Technol Biomed Life Sci. 2005;814:259–62. doi: 10.1016/j.jchromb.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Al-Abd AM, Kim NH, Song SC, Lee SJ, Kuh HJ. A simple HPLC method for doxorubicin in plasma and tissues of nude mice. Arch Pharm Res. 2009;32:605–11. doi: 10.1007/s12272-009-1417-5. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Bantegui T, Pike K, Bloom R, Phipps R, Bai S. In vitro evaluation of optimized liposomes for delivery of small interfering RNA. J Liposome Res. 2014 doi: 10.3109/08982104.2014.907306. [DOI] [PubMed] [Google Scholar]
- 23.Westerfield M. The Zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene. Or: 1993. [Google Scholar]
- 24.Jeong JY, Kwon HB, Ahn JC, Kang D, Kwon SH, Park JA, et al. Functional and developmental analysis of the blood-brain barrier in zebrafish. Brain Res Bull. 2008;75:619–28. doi: 10.1016/j.brainresbull.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 25.Yang XJ, Cui W, Gu A, Xu C, Yu SC, Li TT, et al. A novel zebrafish xenotransplantation model for study of glioma stem cell invasion. PLoS One. 2013;8:e61801. doi: 10.1371/journal.pone.0061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardridge WM. Blood-brain barrier delivery. Drug Discov Today. 2007;12:54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Pardridge WM. The blood-brain barrier: bottleneck in brain drug development. NeuroRx J Am Soc Exp Neurother. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Record M, Subra C, Silvente-Poirot S, Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81:1171–82. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Schneiderand A, Simons M. Exosomes: vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013;352:33–47. doi: 10.1007/s00441-012-1428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonsand M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Haqqani AS, Delaney CE, Tremblay TL, Sodja C, Sandhu JK, Stanimirovic DB. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10:4. doi: 10.1186/2045-8118-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylorand DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33:441–54. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 34.De Rosa G, Salzano G, Caraglia M, Abbruzzese A. Nanotechnologies: a strategy to overcome blood-brain barrier. Curr Drug Metab. 2012;13:61–9. doi: 10.2174/138920012798356943. [DOI] [PubMed] [Google Scholar]
- 35.Umansand RA, Taylor MR. Zebrafish as a model to study drug transporters at the blood-brain barrier. Clin Pharmacol Ther. 2012;92:567–70. doi: 10.1038/clpt.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie J, Farage E, Sugimoto M, Anand-Apte B. A novel transgenic zebrafish model for blood-brain and blood-retinal barrier development. BMC Dev Biol. 2010;10:76. doi: 10.1186/1471-213X-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]