Abstract
Plant and soil properties cooperatively structure soil microbial communities, with implications for ecosystem functioning. However, the extent to which each factor contributes to community structuring is not fully understood. To quantify the influence of plants and soil properties on microbial diversity and composition in an agricultural context, we conducted an experiment within a corn-based annual cropping system and a perennial switchgrass cropping system across three topographic positions. We sequenced barcoded 16S ribosomal RNA genes from whole soil three times throughout a single growing season and across two years in July. To target the belowground effects of plants, we also sampled rhizosphere soil in July. We hypothesized that microbial community α-diversity and composition (β-diversity) would be more sensitive to cropping system effects (annual vs. perennial inputs) than edaphic differences among topographic positions, with greater differences occurring in the rhizosphere compared to whole soil. We found that microbial community composition consistently varied with topographic position, and cropping system and the rhizosphere influenced α-diversity. In July, cropping system and rhizosphere structured a small but specific group of microbes implying a subset of microbial taxa, rather than broad shifts in community composition, may explain previously observed differences in resource cycling between treatments. Using rank abundance analysis, we detected enrichment of Saprospirales and Actinomycetales, including cellulose and chitin degraders, in the rhizosphere soil and enrichment of Nitrospirales, Syntrophobacterales, and MND1 in the whole soil. Overall, these findings support environmental filtering for the soil microbial community first by soil and second by the rhizosphere. Across cropping systems, plants selected for a general rhizosphere community with evidence for plant-specific effects related to time of sampling.
Introduction
Soil microorganisms mediate biogeochemical transformations that underpin important ecosystem functions. Bacteria and fungi decompose organic matter and mineralize it to plant available forms [1], retain nutrients [2], and influence atmospheric concentrations of greenhouse gases [3]. Thus, for agricultural ecosystems, the response of soil microorganisms to environmental parameters has important implications for crop productivity and the long-term suitability of a soil to agriculture. There is evidence that soil microbial community function can be influenced by community structure [4–7], and that both are cooperatively shaped by plants and soil properties [8,9]. There are contrasting reports in the literature, however, as to the relative contribution of plant and soil properties as factors structuring microbial community composition and diversity.
Edaphic factors are thought to be an important environmental filter shaping soil microbial communities, including the microbial seed back, at a range of scales [8]. For example, bacterial community composition and diversity are shaped largely by soil pH at relatively coarse scales [10,11]. At finer scales, pH along with soil texture and nutrient status can shape soil microbiota [12]. Within an individual soil, the quantity and chemistry of root exudates and plant residues can stimulate or inhibit different microbial taxa, resulting in distinct microbial communities associated with specific plants [13–15]. Differences in root production and phenology can alter resource availability in time and space, and thus also structure microbial communities [16]. In addition to patches of plant residues [17], a hotspot for a plant’s influence on microbial communities is the rhizosphere, the narrow zone of soil that surrounds and is influenced by plant roots [8,18]. Here, microbial communities are enriched from the whole soil in response to the availability of low-molecular-mass compounds and polymerized sugar from root cells. Additionally, plant secondary metabolites are involved in establishing symbioses and repelling pests and pathogens, and root water and nutrient uptake alter pH and resource availability for microbes [8]. These influences illustrate the role that the rhizosphere plays as a second environmental filter on soil microbiota, with cascading effects to biogeochemical cycling of carbon (C) and nitrogen (N) [19].
In agricultural ecosystems, management practices add another level of complexity to the edaphic forces structuring soil microbial communities. Management influences microbial communities directly by altering soil properties, and indirectly through changes in plant nutrient and water requirements. One management action common across diverse types of agriculture (e.g. pasture, row-crop) and cropping systems (e.g. annual- and perennial-based bioenergy crops) is N enrichment from fertilization. Nitrogen fertilization, especially from inorganic sources, can affect soil microbiota directly by increasing N availability for microbes [20], and indirectly by altering plant-derived C inputs to soils [21,22] and increasing soil acidity [23,24]. These microbial shifts, in turn, may alter rates of decomposition [20] and weaken plant-microbe linkages critical to ecosystem nutrient retention [25].
In a previous study contrasting cropping system and topographic position as a proxy for edaphic factors, cropping system had a stronger influence on the functional capacity of the microbial community (potential enzyme activity, net N mineralization and respiration) than edaphic factors associated with topographic position [26]. We did not observe changes in microbial activity concomitant to changes in microbial biomass, suggesting the physiological response of the microbial community influenced differences in C and N cycling. At the microbial community scale, we hypothesized that the physiological response of microbial activity to plants could be driven by a shift in community structure, i.e. diversity and composition [5]. Alternatively, if topographic position or soil edaphic characteristics drive microbial community composition and structure, then responses of microbial activity to cropping system are likely due to localized differences in the physiological response of individual species to plant inputs (i.e. physiology or plasticity; [27,28]) rather than community scale changes in composition.
To test the influences of topography, cropping system, and rhizosphere on soil microbial (bacteria, archaea) community structure, we investigated community responses to a fertilized perennial cropping system compared to an annual-based system across a range of edaphic conditions associated with three topographic positions at the Landscape Biomass Project. We used topographic position as a master variable for changes in soil properties because topography affects factors such as soil pH, N availability, moisture regime and plant productivity [29,30]. Given the temporal dynamics of plant effects [16], we sampled whole soil at three times throughout a single growing season and mid-summer in July, around peak aboveground biomass, over two growing seasons. We expected plant effects to be most pronounced mid-summer and in the rhizosphere. Thus, we sampled both whole and rhizosphere soil during mid-summer. Specifically, we hypothesized that 1) α-diversity and community composition (β-diversity, defined here as membership and relative abundance of specific OTUs) of the whole soil would respond more to cropping system than to topographic position, both (a) within a growing season and (b) between years. We predicted greater microbial α-diversity in the perennial cropping system due to increased overall resource supply [31] and greater niche dimensionality as a result of year-round interactions with plants. We also specifically hypothesized that 2) differences in microbial community composition between the annual and perennial cropping systems would be greater in the rhizosphere soil compared to the whole soil. If greater α-diversity (species richness) is due to direct effects of plant inputs, we predicted unique communities would be present in the rhizosphere of specific crop plants (i.e. greater β-diversity or compositional dissimilarity among samples).
Materials and Methods
Study site
We conducted this study as part of the Landscape Biomass Project at the Uthe Research & Demonstration Farm in Boone County, Iowa, USA (41°55'N, 93°45'W), where plots of approximately 0.05 ha were established in fall 2008 and first planted in spring 2009. The Uthe Research and Demonstration Farm is owned by the Council for Agricultural Development and managed by Iowa State University, who granted permission for field access and sampling. The field studies did not involve endangered or protected species. Soils at the site are classified as fine-loamy Hapludoll Mollisols and follow a topographic gradient with a slope of approximately 0.5% on the summit and approximately 2.5% on the side slope (back slope and toe slope), as described in detail in Wilson et al. [32]. Soil organic C averaged 17.2 g kg-1, total soil N averaged 1.42 g kg-1, bulk density averaged 1.57 g cm-3, and soil texture averaged 49.4% silt + clay [33]. Notwithstanding similarities in chemistry and root inputs among summit, back slope and toe slope positions, soils from the back slope have significantly lower concentrations of potassium and phosphorus, and soils on the toe slope have significantly greater aggregate geometric mean diameter [33]. Salt-extractable organic C, nitrate, ammonium, pH and water content did not vary with topographic position [26]. Prior to study initiation in 2008, summit and side slopes were managed under a corn-soybean rotation with corn in rotation in 2008. This history of annual-based agriculture may leave a legacy on the microbial community such that changes between microbial communities in the whole soil of annual and perennial cropping systems may be relatively slow (i.e. decades) [34]. Mean monthly air temperatures in 2011 and 2012 did not differ from the long-term mean, but rainfall was extremely variable. Below average rainfall was recorded at the end of summer 2011, followed by a record drought in 2012 [32].
Experimental design
To assess the impact of cropping system and soil properties on microbial communities, we sampled from a subset of the Landscape Biomass experimental plots, including a no-till annual monoculture (continuous corn, Zea mays L.; “annual”) and a perennial monoculture (switchgrass, Panicum virgatum L., cv: ‘Cave-In-Rock’; “perennial”), replicated three times at three topographic positions (summit, back slope, and toe slope) following a randomized complete block design (n = 3; S1 Fig). Corn was fertilized with N at a rate of 168 kg urea-N ha-1 in 2011 and 175 kg urea-N ha-1 in 2012, and switchgrass was fertilized at a rate 134 kg urea-N ha-1 in 2011 and 135 kg urea-N ha-1 in 2012. Both cropping systems received 56 kg P2O5 ha-1 and 112 kg KCl ha-1 in 2011. At harvest, maximum aboveground biomass was removed, leaving ~10% of the aboveground biomass from all cropping systems. Refer to Wilson et al. [32] for complete management details.
Soil sampling
We sampled whole soil, defined as root-free soil collected randomly within plots, and rhizosphere soil, defined as soil adhering to roots from samples collected below a plant. We sampled whole soil by collecting and compositing ten randomly distributed soil cores (2.2 cm in diameter x 15 cm depth) within each plot. We then sieved the soil to 4-mm in the laboratory and subsampled for gravimetric water content and DNA extraction, which we froze at -80°C. To test the effect of soil origin with regard to comparisons between whole and rhizosphere soil, we sampled soil under five randomly chosen plants by taking one core directly under a plant, angled towards the plant stalk. We placed the five cores per plot in sterile bags and transported to the laboratory on ice. Using aseptic technique in the laboratory, we placed the soil on sterilized bench paper and isolated roots from the sample using sterilized tweezers, and removed excess soil from root surfaces with gentle shaking. We sampled the remaining soil that was directly adhered to the root (“rhizosphere soil”) for DNA extraction [9].
We collected whole soil samples in spring after corn emergence and new switchgrass growth (June), mid-summer around peak aboveground biomass (July), and late summer at the beginning of plant senescence (August) in 2011 for a total of 54 composited samples collected in 2011 (3 topographic positions x 2 cropping systems x 3 plots x 3 dates). Based on previous work showing greater microbial activity in the perennial cropping system around peak aboveground biomass in July 2011 [26], we focused sampling efforts to July in 2012 and sampled both whole soil and rhizosphere soil from summit and toe slope positions, for an additional 24 composited samples collected in 2012 (2 topographic positions x 2 cropping systems x 2 soil origins x 3 plots) (S1 Table). To compare extremes of the topographic gradient on rhizosphere communities, we sampled only on the summit and toe slope.
DNA extraction and 16S rRNA gene sequencing
We extracted DNA from a 0.25 g sub-sample using the PowerSoil-htp 96 Well Soil DNA Isolation Kit (MoBio, Carlsbad, CA, USA), with modifications following the Earth Microbiome Project (EMP; www.earthmicrobiome.org/emp-standard-protocols). We quantified DNA via PicoGreen fluorometry. To assess the α-diversity and β-diversity of the microbial communities, we obtained 16S rRNA gene sequences following EMP standard protocols [35]. Briefly, we used the 515f/806r primer set to amplify the V4/V5 region of the 16S rRNA gene and obtained overlapping paired-end 150 base reads using an Illumina MiSeq sequencing system at the Next Generation Sequencing Core (Argonne National Laboratory). We deposited all sequences in the National Center for Biotechnology Information database (BioProject ID: PRJNA248482).
We first merged the raw 150-bp sequence reads using EA-Utils fastq-join [36] and obtained a median merged sequence length of 253-bp. We then processed merged sequences with QIIME v.1.7.0 [37] using default parameters for quality filtering (Phred quality score, Q20) and demultiplexing. We assigned taxonomy by using closed reference UCLUST clustering [38] against the May 2013 release of the Greengenes database filtered at 97% sequence identity [39]. Before downstream analysis, we required that all operational taxonomy units (OTUs) have a count of at least two reads across all samples and rarefied to 6678 sequences per sample to correct for differences in reads across samples. Library size ranged from 6678 to 23,359 sequences per sample with an average of 13,378 sequences per sample.
Statistical analyses
We quantified microbial communities in the whole soil and rhizosphere in four ways, including three α-diversity measurements (richness, evenness, Shannon’s H’) and the Bray-Curtis dissimilarly index to test for differences in community composition, or β-diversity. For each α-diversity measurement (richness, evenness, Shannon’s H’), we conducted a full-factorial analysis of variance (ANOVA) on normalized data. We found no difference between results with block as a random effect and with no random effects and present results of the simpler model with no random effects, which allowed us to include Tukey’s HSD to test for multiple comparisons of means. We tested full models with interaction between all main effects and removed non-significant interaction terms from final models. To assess β-diversity, we tested for differences between the Bray-Curtis dissimilarity index using PERMANOVAs with 9999 permutations [40]. Given the large number of OTUs used in multivariate analyses of community composition, we chose PERMANOVAs over parametric tests, such as MANOVA, in order to not violate the assumption of normality. We visualized changes in composition using non-metric multidimensional scaling (NMDS).
For hypothesis 1, we tested for full-factorial differences in α-diversity (ANOVAs on richness, evenness and Shannon’s H’) and community composition (PERMANOVAs on Bray-Curtis dissimilarity) in whole soil for samples collected on the three dates in 2011 and between samples collected in July 2011 and 2012. For hypothesis 2, we tested for full-factorial differences in α-diversity and β-diversity between whole and rhizosphere soil (soil origin) collected in July 2012. We also performed ANOVAs and PERMANOVAs with un-rarefied data but found no difference in results compared to rarefied data.
In addition, we summarized our OTUs at the order level and tested for taxa that significantly increased or decreased in rank within communities between whole and rhizosphere soils. We chose to analyze based on rank because this analysis is sensitive to how changes in relative abundance translate to changes in dominance of specific microbial taxa. In this way, changes in rank can catch differences in community structure that are not captured by measurements of community composition. We generated rank-abundance curves for whole and rhizosphere samples within each plot, then calculated differences in rank (Δrank) for microbial orders between whole and rhizosphere soil. We then bootstrapped 95% confidence intervals for Δrank by subsampling changes in rank position of a specific order within each cropping system and topographic position. When we found significant relationships for a particular microbial order (not containing 0 within the confidence interval), we extended the Δrank analysis to families and genera within that order.
We performed all analyses in R v.3.0.2 [41] and deposited all R scripts, including libraries used, in an online repository (https://github.com/chnops/Landscape_Biomass_16S; PERMANOVAs, “multivariate_tests”; ANOVAs, “diversity_tests”; delta rank, “delta_rank”).
Results
Response in the whole soil
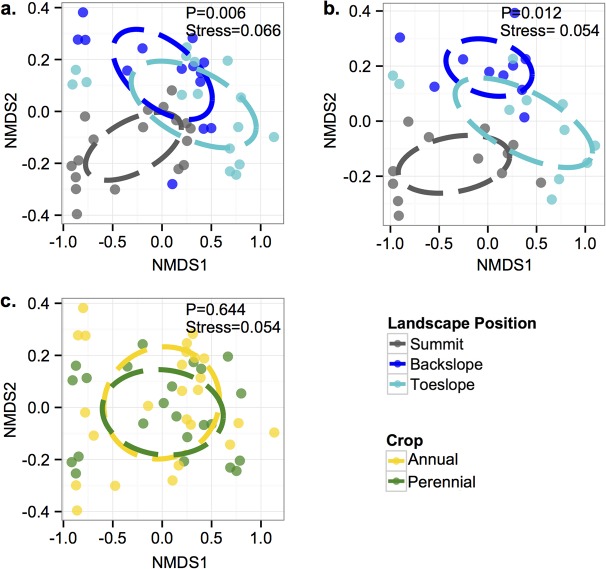
Within whole soil (hypothesis 1), topography influenced microbial community composition (Bray-Curtis dissimilarity of OTUs) for all analyses. The microbial communities as measured by soil from the summit differed from the back slope and toe slope for all time points in 2011 (Fig 1A; Table 1, hypothesis 1a) and in July 2011 and 2012 (Fig 1B; Table 1, hypothesis 1b). Cropping system did not influence microbial community composition between any of the comparisons tested (Fig 1C; Table 1). There was no detectable effect of time on whole soil microbial community composition, neither within 2011, nor between years in July (Table 1).
Fig 1. Differences in microbial community composition in the whole soil using the Bray-Curtis dissimilarity index for β-diversity using NMDS.
Community composition (a) within the 2011 growing season for each topographic position (3 months x 3 plots x 2 cropping systems), (b) in July 2011 and 2012 for each topographic position (2 years x 3 plots x 2 cropping systems) and (c) between the annual (corn) and perennial (switchgrass) cropping systems (3 months x 3 plots x 3 topographic positions). Using PERMANOVAs, we detected only effects of topographic position for all comparisons. Ellipses represent a centroid of one standard deviation of points.
Table 1. P-values for ANOVA for microbial richness, Shannon’s H’ and evenness and PERMANOVA for Bray-Curtis dissimilarity.
| Hypothesis | Source of variation | Richness | Shannon’s H’ | Evenness | Community Composition* |
|---|---|---|---|---|---|
| 1a: Whole soil within year, June, July, August 2011 | CS | 0.839 | 0.753 | 0.800 | 0.644 |
| LP | <0.001 | <0.001 | <0.001 | 0.006 | |
| Month | 0.029 | 0.136 | 0.240 | 0.836 | |
| LP x CS | NS | NS | NS | NS | |
| LP x Month | NS | NS | NS | NS | |
| CS x Month | NS | NS | NS | NS | |
| LP x CS x Month | NS | NS | NS | NS | |
| 1b: Whole soil between years, July 2011 & 2012 | CS | 0.014 | 0.073 | 0.177 | 0.592 |
| LP | <0.001 | <0.001 | 0.001 | 0.012 | |
| Year | 0.206 | 0.514 | 0.752 | 0.663 | |
| LP x CS | NS | NS | NS | NS | |
| LP x Year | NS | NS | NS | NS | |
| CS x Year | NS | NS | NS | NS | |
| LP x CS x Year | NS | NS | NS | NS | |
| 2: Whole or rhizosphere soil, July 2012 | CS | 0.007 | <0.001 | <0.001 | 0.268 |
| LP | <0.001 | <0.001 | <0.001 | 0.011 | |
| Soil origin | 0.002 | 0.070 | 0.620 | 0.184 | |
| LP x CS | NS | NS | NS | NS | |
| LP x Soil origin | NS | NS | NS | NS | |
| CS x Soil origin | NS | NS | NS | NS | |
| LP x CS x Soil origin | NS | NS | NS | NS |
LP, topographic position; CS,cropping System; NS, non-signification interaction terms at α > 0.1 and terms were removed from the model
*Community composition evaluated from Bray-Curtis dissimilarity matrices based on OTU abundances.
Microbial α-diversity in the whole soil consistently changed with topographic position. In 2011 and 2012, richness, Shannon’s H’ and evenness decreased from summit to toe slope (Fig 2; Table 1, hypothesis 1a). We observed greater richness in the whole soil in June compared to July and August in 2011 (Fig 2). There was no difference in microbial richness between years (2011, 2012) for samples taken in July (Table 1, hypothesis 1b).
Fig 2. Changes in microbial richness, Shannon’s H’ and evenness at three topographic positions throughout the 2011 growing season on the summit, back slope and toe slope.
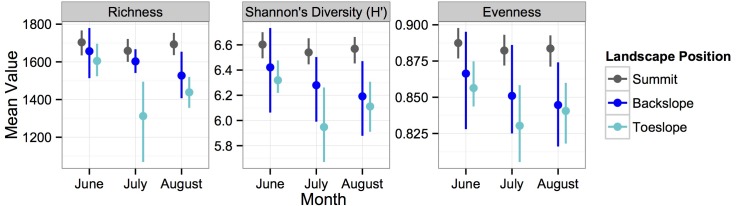
These variables were not affected by cropping system or sampling date so values represent means and 95% CI’s of combined annual and perennial treatments at each sampling time.
We did not detect cropping system effects in the whole soil among the three sampling dates in 2011 or within individual sampling dates (Table 1, hypothesis 1a). However, when we compared measurements between years (July 2011, 2012), we detected consistent and significant cropping system effects (Table 1, hypothesis 1b). Our results revealed that cropping system effects on microbial communities were only detectable in July, when plants are fully grown. Specifically, we observed greater Shannon’s H’ and evenness in the perennial than annual system (Fig 3; Table 1). Similarly, we observed greater microbial richness (P = 0.014) and Shannon’s H’ (P = 0.073) in the perennial compared to annual cropping system in July 2011 and 2012 (Fig 3).
Fig 3. Changes in α-diversity of the microbial communities in July in the whole soil and rhizosphere soil from annual (corn) and perennial (switchgrass) cropping systems at two topographic positions at the Landscape Biomass Project.
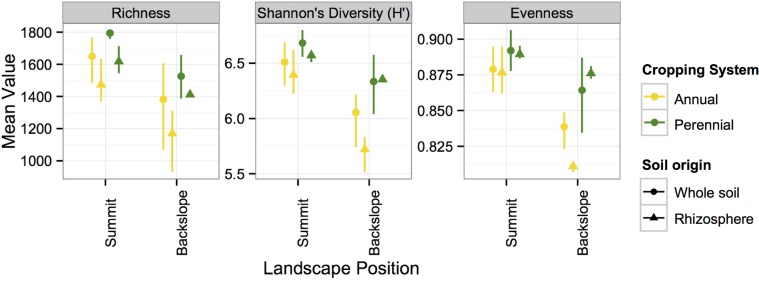
Means and 95% CI’s are shown.
Response in the rhizosphere
With respect to differences between whole and rhizosphere soil (hypothesis 2), topographic position also influenced differences in microbial community composition in 2012 (Table 1). Visualization of microbial community composition via NDMS shows that rhizosphere and whole soil differentiated, though this separation was not statistically significant (P = 0.184; Fig 4). When we compared whole and rhizosphere soils in July 2012, we observed greater microbial richness, Shannon’s H’ and evenness on the summit than on the toe slope (Fig 3; Table 1). Overall, the rhizosphere was less rich (P = 0.002) and less diverse (Shannon’s H’; P = 0.07) than the whole soil with no detectable changes in evenness (P = 0.620).
Fig 4. Differences in microbial community composition between whole soil and rhizosphere soil using the Bray-Curtis dissimilarity index for β-diversity (1 date x 3 plots x 2 cropping systems x 2 topographic positions).
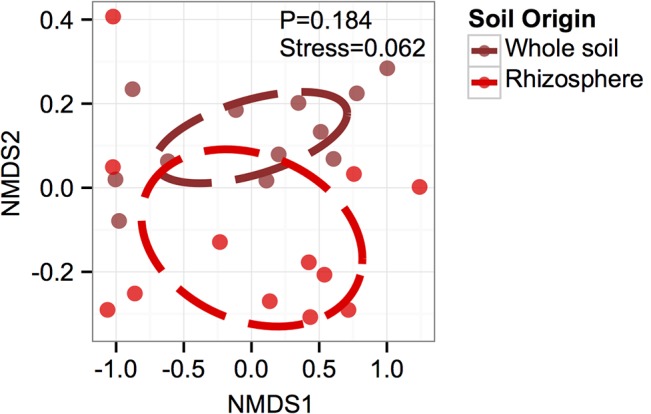
We detected only effects of topographic position for all comparisons. Ellipses represent a centroid of one standard deviation of points.
Of the 243 orders captured in this study, we used differences in rank (Δrank) to detect shifts in the relative abundance of 16 bacterial orders between whole and rhizosphere soil (S2 Table). Of these, we observed consistent enrichment of Actinomycetales and Saprospirales in the rhizosphere soil on both summit and toe slope and of Nitrospirales, Syntrophobacterales and MND1 in the whole soil on both summit and toe slope (Fig 5). For two orders enriched in the rhizosphere, Actinomycetales and Saprospirales, we also observed greater abundance of specific families and genera in the rhizosphere. Within the Actinomycetales, we observed a greater abundance of Actinospicaceae, Frankiaceae, and Cellulomonadaceae, especially genera Cellulomonas, in the rhizosphere. Within the Saprospirales, we observed a greater abundance in the rhizosphere of bacteria within the family Chitinophagaceae. In contrast, we observed equal enrichment of families and genera within Nitrospirales, Syntrophobacterales and MND1 in the rhizosphere and whole soil.
Fig 5. Change in rank abundance from whole to rhizosphere soil for Actinomycetales, Saprospirales, Syntrophobacterales, Nitrospirales, and MND1 bacterial orders.
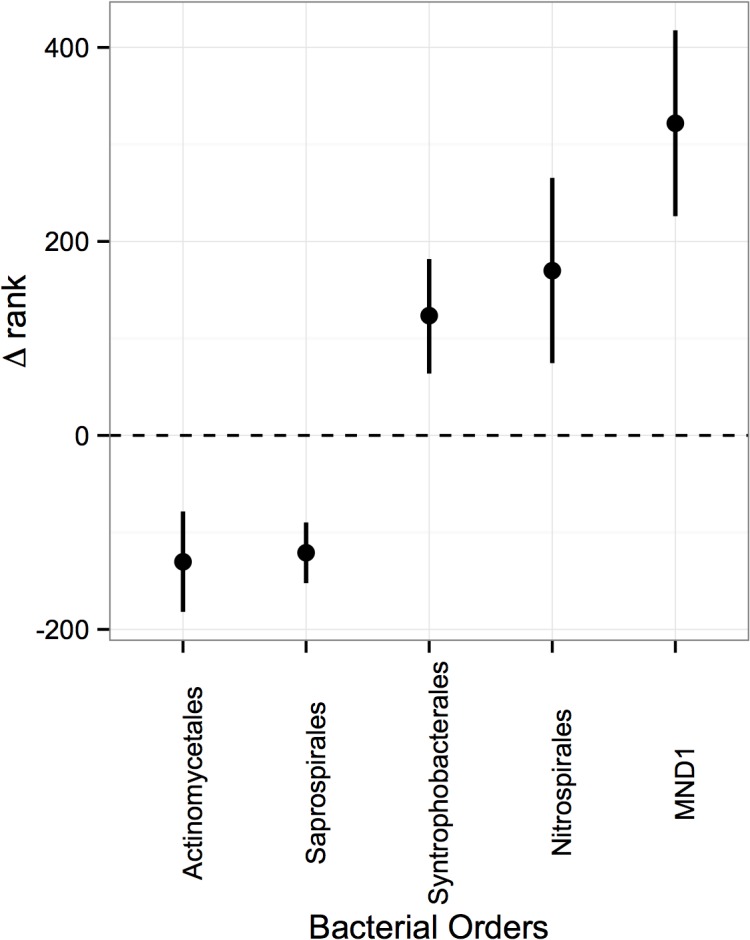
Positive values indicate the order has a higher rank in the whole soil and negative values indicate the order has a higher rank in the rhizosphere soil. Error bars represent 95% CI’s.
Discussion
We sought to investigate the role of cropping system, rhizosphere effects and topographic position in shaping microbial communities under field conditions. Based on previous results demonstrating differences in microbial activity between cropping systems but not topographic positions [26], we hypothesized that microbial α-diversity and β-diversity in the whole soil would be primarily shaped by cropping system (hypothesis 1). Consistent with previously published work on microbial activity at this site [26], we only detected cropping system effects on the microbial α-diversity mid summer in July, coincident with peak aboveground biomass. Our finding of greater α-diversity without a change in composition indicates that the dominant taxa are similar between cropping systems, but microbial communities under perennials contain more less-abundant taxa relative to the annual cropping system. Further, because we only detected cropping system effects around peak aboveground biomass, our data suggest that plant phenological traits, such as timing of rhizodeposition and fine root inputs, stimulated these shifts in less abundant taxa [16,42–44]. Considering the remarkable stability of microbial composition across time, even with a drought in 2012, these shifts in α-diversity (richness, Shannon’s H’, evenness) may account for previously observed temporal changes in microbial enzyme activity, respiration and mineralization [26].
In contrast to hypothesis 1, microbial community composition was most strongly and consistently affected by topographic position, despite no differences in pH. Generally, we observed distinct microbial communities on the summit with greater α-diversity than those on the back slope and top slope positions. While most soil physiochemical characteristics do not differ among the three topographic positions, previous research at this site has shown that the summit has higher concentrations of potassium and phosphorus than the back slope and toe slope [33]. Although not often considered in studies of microbial biogeography, phosphorus and potassium concentrations correlate with the abundance of microbial phyla in agricultural fields [45] and, thus, may be important drivers of microbial communities when N-limitation is relieved through the addition of fertilizer. We recognize that edaphic factors often co-vary such that topographic position results in uncharacterized effects; additional work is needed to test the effects of individual or suites of edaphic factors on microbial community structure.
Contrary to hypothesis 2, the rhizosphere of the perennial and annual plant did not uniquely shape microbial communities from the whole soil (i.e. no soil origin x cropping system interaction). Differences between rhizosphere and whole soil were not cropping system-specific. Rhizosphere communities were less rich than whole soil communities, indicating a general enrichment of root-associated taxa in rhizosphere soil. Together with less rich microbial communities, we observed consistent changes in relative abundance of a small number of the most dominant microbial orders, irrespective of cropping system and topographic gradient. In rhizosphere soils, we observed a greater abundance of the ubiquitous Actinomycetales, a filamentous group of bacteria that can promote plant growth [46,47], and Saprospirales, previously shown in high alpine systems to be predictive of plant cover, β-Glucosidase activity, soil water, dissolved organic C, and pH [48]. Enrichment of specific families and genera within these orders suggests an importance of plant cell wall material (i.e. cellulose degraders Cellulomonadaceae) and fungal cell wall material,(i.e. chitin degraders Chitinophagaceae) for the selection of organisms to the rhizosphere. The fact that shifts in these taxa were not cropping system-specific suggests that they may be more general rhizosphere colonizers.
Unlike previous findings suggesting Nitrospira are also rhizosphere generalists in unmanaged systems [49,50], we observed Nitrospirales in less abundance in the rhizosphere compared to whole agricultural soil. Given findings by Turner et al. [27], who found depletion of Nitrospira in the rhizosphere of a legume (pea) relative to the whole soil, the depletion of Nitrospirales in the rhizosphere that we observed might be attributable to N fertilization. Under N limiting conditions, N-rich microsites can select for both roots and nitrifiers [51]. When N is not limiting for plants and/or nitrifiers, however, other selective forces may take precedent over their distribution and co-occurrence, implying that resource needs by plants can lead to the enrichment or depletion of some taxa in the rhizosphere [52]. Selective conditions in the whole soil, such as compaction from lack of root penetration, may also lead to taxon enrichment or depletion, as demonstrated by the greater abundance of strict anaerobes like Syntrophobacterales in whole soil compared to the rhizosphere soil.
Our results support environmental filtering for the microbial community by the soil conditions, including general plant effects related to time of sampling but not plant-specific effects within the rhizosphere. The lack of observed effect of cropping system or soil origin (rhizosphere vs. whole soil) on overall microbial community composition stands in contrast to the current paradigm that plant-specific traits influence environmental filtering of rhizosphere communities [27,52,53]. Specific to our system, previous field studies that have found differences in microbial community composition in the whole soil of perennial bioenergy cropping systems [54,55]. Although the legacy of conventional, annual-based agriculture may have limited cropping system effects to overall community composition [56–58], it has not prevented changes in microbial communities in response to perennial bioenergy cropping systems in similar soils in the Corn Belt of the United States [54,55,59]. The most unique attribute of the perennial cropping system in this study is N fertilization, which is not part of perennial cropping systems in other studies. Indeed, recent work suggests plant N status drives microbial community structure in the rhizosphere [52]. Therefore, while other factors can not be ruled out, N fertilization at this site may have masked plant-specific effects on microbial community composition [20,21,60], suggesting soil nutrient amendments hinder crop plants’ ability to directly influence rhizosphere communities.
Conclusions
Topographic position was the strongest and most consistent driver of microbial community composition (β-diversity) in our system, both within whole and rhizosphere soils. Soil properties associated with topography, therefore, had precedent over plant effects in shaping microbial community composition in these soils. Given that both cropping systems were fertilized, N fertilization may also preclude cropping system-specific microbial response as demonstrated by similar microbial communities in annual and perennial systems.
Results presented here also provide evidence that cropping system shape a small subset of organisms in the whole soil at specific times in the growing season. Given that previously documented changes in microbial activity [26] coincided with our observed shifts in diversity, we hypothesize that the activity of less abundance organisms may provide novel insights into composition-function relationships of soil microbial communities. Further, factors shaping rhizosphere communities in this system are related to general plant effects, but not plant-specific effects. We identified lifestyles (i.e. filamentous) and metabolic capabilities (i.e. cellulose and chitin degradation, but not nitrate-oxidation) associated with the rhizosphere of these fertilized crops. Overall, this work demonstrates the overarching importance of edaphic factors for shaping microbial community composition and the importance of plant effects and phenology for determining changes in relative abundance of specific microbial taxa.
Supporting Information
Whole soil samples were taken only from the perennial switchgrass and annual corn cropping systems on the summit, back slope and toe slope, denoted in bold and by asterisks. Rhizosphere soil was sampled only from summit and toe slope positions. Modified from Wilson et al. 2014.
(DOCX)
(DOCX)
Soil samples were taken mid-summer, at peak biomass in 2012.
(DOCX)
Acknowledgments
We thank Theo Gunther and Nicholas Boersma for field maintenance, Eric Asbe, Megan Bartholomew, and Austin Putz for field and laboratory assistance, and members of the Landscape Biomass Project for logistical support. Fan Yang provided support with sequence processing in QIIME and Monika Maier Shea produced the map of the experimental site.
Data Availability
The Illumina MiSeq 16S rRNA sequencing data are available in the National Center for Biotechnology Information database (BioProject ID: PRJNA248482).
Funding Statement
Funding for this research was provided by the Agriculture and Food Research Initiative Competitive Grant No. 2010-85101-20471 from the USDA National Institute of Food and Agriculture (http://www.nifa.usda.gov/) to KSH and a National Science Foundation (http://www.nsf.gov/) Doctoral Dissertation Improve Grant (DEB-1210742) to SKH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Paul EA. Soil Microbiology, Ecology and Biochemistry. Academic Press, Burlingont, MA; 2006. [Google Scholar]
- 2. de Vries FT, Bardgett RD. Plant–microbial linkages and ecosystem nitrogen retention: lessons for sustainable agriculture. Frontiers in Ecology and the Environment. 2012;10(8):425–32. [Google Scholar]
- 3. Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol Rev. 1996;60(4):609–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90(2):441–51. [DOI] [PubMed] [Google Scholar]
- 5. Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458(7238):623–6. 10.1038/nature07840 [DOI] [PubMed] [Google Scholar]
- 6. Zhao M, Xue K, Wang F, Liu S, Bai S, Sun B, et al. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014;8(10):2045–55. 10.1038/ismej.2014.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martínez A, Ventouras L-A, Wilson ST, Karl DM, DeLong EF. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front Microbio. 2013;4; 10.3389/fmicb.2013.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Micro. 2013; 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- 9. Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology. 2009;68(1):1–13. 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- 10. Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103(3):626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biology and Biochemistry. 2008;40(9):2407–15. [Google Scholar]
- 12. Wessén E, Söderström M, Stenberg M, Bru D, Hellman M, Welsh A, et al. Spatial distribution of ammonia-oxidizing bacteria and archaea across a 44-hectare farm related to ecosystem functioning. ISME J. 2011;5(7):1213–25. 10.1038/ismej.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wardle DA. Ecological Linkages Between Aboveground and Belowground Biota. Science. 2004;304(5677):1629–33. [DOI] [PubMed] [Google Scholar]
- 14. Somers E, Vanderleyden J, Srinivasan M. Rhizosphere Bacterial Signalling: A Love Parade Beneath Our Feet. Critical Reviews in Microbiology. 2004;30(4):205–40. [DOI] [PubMed] [Google Scholar]
- 15. Ladygina N, Hedlund K. Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biology and Biochemistry. 2010;42(2):162–8. [Google Scholar]
- 16. Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE. 2013;8(2):e55731 10.1371/journal.pone.0055731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parkin TB. Soil microsites as a source of denitrification variability. Soil Science Society of America Journal. Soil Science Society of America; 1987;51(5):1194–9. [Google Scholar]
- 18. Berendsen RL, Pieterse CMJ, Bakker Pahm. The rhizosphere microbiome and plant health. Trends in Plant Science. 2012;17(8):478–86. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 19. Philippot L, Hallin S, Börjesson G, Baggs EM. Biochemical cycling in the rhizosphere having an impact on global change. Plant Soil. 2008;321(1–2):61–81. [Google Scholar]
- 20. Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Global Change Biology. 2012;18(6):1918–27. [Google Scholar]
- 21. Ramirez KS, Lauber CL, Knight R, Bradford MA, Fierer N. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology. 2010;91(12):3463–70. [DOI] [PubMed] [Google Scholar]
- 22. Waldrop M. Microbial community response to nitrogen deposition in northern forest ecosystems. Soil Biology and Biochemistry. 2004;36(9):1443–51. [Google Scholar]
- 23. Wei C, Yu Q, Bai E, Lü X, Li Q, Xia J, et al. Nitrogen deposition weakens plant-microbe interactions in grassland ecosystems. Global Change Biology. 2013;19(12):3688–97. 10.1111/gcb.12348 [DOI] [PubMed] [Google Scholar]
- 24. Enwall K, Nyberg K, Bertilsson S, Cederlund H, Stenström J, Hallin S. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biology and Biochemistry. 2007;39(1):106–15. [Google Scholar]
- 25. de Vries FT, Shade A. Controls on soil microbial community stability under climate change. Front Microbio. 2013;4:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hargreaves SK, Hofmockel KS. Physiological shifts in the microbial community drive changes in enzyme activity in a perennial agroecosystem. Biogeochemistry. 2014;117(1):67–79. [Google Scholar]
- 27. Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, et al. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013;7: 2248–2258. 10.1038/ismej.2013.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ofek M, Voronov-Goldman M, Hadar Y, Minz D. Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities. Environmental Microbiology. 2013;16(7):2157–2167. 10.1111/1462-2920.12228 [DOI] [PubMed] [Google Scholar]
- 29. Tscherko D, Hammesfahr U, Marx M-C, Kandeler E. Shifts in rhizosphere microbial communities and enzyme activity of Poa alpina across an alpine chronosequence. Soil Biology and Biochemistry. 2004;36(10):1685–98. [Google Scholar]
- 30. Högberg MN, Högberg P, Myrold DD. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia. 2006;150(4):590–601. [DOI] [PubMed] [Google Scholar]
- 31. Hurlbert AH, Stegen JC. When should species richness be energy limited, and how would we know? Ecology Letters. 2014;17(4):401–13. 10.1111/ele.12240 [DOI] [PubMed] [Google Scholar]
- 32. Wilson DM, Heaton EA, Schulte LA, Gunther TP, Shea ME, Hall RB, et al. Establishment and Short-term Productivity of Annual and Perennial Bioenergy Crops Across a Landscape Gradient. Bioenerg Res. 2014;7(3):885–898. [Google Scholar]
- 33. Ontl TA, Hofmockel KS, Cambardella CA, Schulte LA, Kolka RK. Topographic and soil influences on root productivity of three bioenergy cropping systems. New Phytol. 2013;199(3):727–37. 10.1111/nph.12302 [DOI] [PubMed] [Google Scholar]
- 34. Jangid K, Williams MA, Franzluebbers AJ, Schmidt TM, Coleman DC, Whitman WB. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biology and Biochemistry. 2011;43(10):2184–93. [Google Scholar]
- 35. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aronesty E. Comparison of Sequencing Utility Programs. Open Bioinformatics Journal. 2013. [Google Scholar]
- 37. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 39. McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011;6(3):610–8. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral Ecology. 2001;26(1):32–46. [Google Scholar]
- 41.Team RC. R: A Language for Environment and Statistical Computing. R Foundation for Statistical Computing [Internet]. Available from: http://www.R-project.org
- 42. Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiology Ecology. 2008;65(2):193–201. 10.1111/j.1574-6941.2008.00535.x [DOI] [PubMed] [Google Scholar]
- 43. Ziegler M, Engel M, Welzl G, Schloter M. Development of a simple root model to study the effects of single exudates on the development of bacterial community structure. Journal of Microbiological Methods. 2013;94(1):30–6. 10.1016/j.mimet.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 44. Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2013;8(4):790–803. 10.1038/ismej.2013.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao J, Ni T, Li Y, Xiong W, Ran W, Shen B, et al. Responses of Bacterial Communities in Arable Soils in a Rice-Wheat Cropping System to Different Fertilizer Regimes and Sampling Times. PLoS ONE. 2014;9(1):e85301 10.1371/journal.pone.0085301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Franco-Correa M, Quintana A, Duque C, Suarez C, Rodríguez MX, Barea J-M. Evaluation of actinomycete strains for key traits related with plant growth promotion and mycorrhiza helping activities. Applied Soil Ecology. 2010;45(3):209–17. [Google Scholar]
- 47. Palaniyandi SA, Yang SH, Zhang L, Suh J-W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl Microbiol Biotechnol. 2013;97(22):9621–36. 10.1007/s00253-013-5206-1 [DOI] [PubMed] [Google Scholar]
- 48. King AJ, Freeman KR, McCormick KF, Lynch RC, Lozupone C, Knight R, et al. Biogeography and habitat modelling of high-alpine bacteria. Nat Comms. 2010;1(5):1–6. [DOI] [PubMed] [Google Scholar]
- 49. DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2008;3(2):168–78. 10.1038/ismej.2008.103 [DOI] [PubMed] [Google Scholar]
- 50. Rosenzweig N, Bradeen JM, Tu ZJ, McKay SJ, Kinkel LL. Rhizosphere bacterial communities associated with long-lived perennial prairie plants vary in diversity, composition, and structure. Can J Microbiol. 2013;59(7):494–502. 10.1139/cjm-2012-0661 [DOI] [PubMed] [Google Scholar]
- 51. Schimel JP, Bennett J. Nitrogen mineralization: challenges of a changing paradigm. Ecology. 2004;85(3):591–602. [Google Scholar]
- 52. Bell CW, Asao S, Calderón F, Wolk B, Wallenstein MD. Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biology and Biochemistry. 2015;85:170–82. [Google Scholar]
- 53. Kourtev PS, Ehrenfeld JG, Häggblom M. Exotic plant species alter the microbial community structure and function in the soil. Ecology. 2002;83(11):3152–66. [Google Scholar]
- 54. Mao Y, Yannarell AC, Mackie RI. Changes in N-Transforming Archaea and Bacteria in Soil during the Establishment of Bioenergy Crops. PLoS ONE. 2011;6(9):e24750 10.1371/journal.pone.0024750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mao Y, Yannarell AC, Davis SC, Mackie RI. Impact of different bioenergy crops on N-cycling bacterial and archaeal communities in soil. Environmental Microbiology. 2012;15(3):928–42. 10.1111/j.1462-2920.2012.02844.x [DOI] [PubMed] [Google Scholar]
- 56. Salles JF, Van Veen JA, van Elsas JD. Multivariate Analyses of Burkholderia Species in Soil: Effect of Crop and Land Use History. Applied and Environmental Microbiology. 2004;70(7):4012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garbeva P, Elsas JD, Veen JA. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil. 2007;302(1–2):19–32. [Google Scholar]
- 58. Buckley DH, Schmidt TM. The Structure of Microbial Communities in Soil and the Lasting Impact of Cultivation. Microb Ecol. 2001;42(1):11–21. [DOI] [PubMed] [Google Scholar]
- 59. Anderson-Teixeira KJ, Masters MD, Black CK, Zeri M, Hussain MZ, Bernacchi CJ, et al. Altered Belowground Carbon Cycling Following Land-Use Change to Perennial Bioenergy Crops. Ecosystems. 2013;16(3):508–520. [Google Scholar]
- 60. Marschner P, Yang C-H, Lieberei R, Crowley DE. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biology and Biochemistry. 2001;33(11):1437–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole soil samples were taken only from the perennial switchgrass and annual corn cropping systems on the summit, back slope and toe slope, denoted in bold and by asterisks. Rhizosphere soil was sampled only from summit and toe slope positions. Modified from Wilson et al. 2014.
(DOCX)
(DOCX)
Soil samples were taken mid-summer, at peak biomass in 2012.
(DOCX)
Data Availability Statement
The Illumina MiSeq 16S rRNA sequencing data are available in the National Center for Biotechnology Information database (BioProject ID: PRJNA248482).