Introduction
There is growing evidence that the sugar N-acetylglucosamine (GlcNAc) plays diverse roles in cell signaling pathways that impact the virulence properties of microbes and host cells. GlcNAc is already well known as a ubiquitous structural component at the cell surface that forms part of bacterial cell wall peptidoglycan, cell wall chitin in fungi and parasites, and extracellular matrix glycosaminoglycans of animal cells. Chitin and peptidoglycan have been previously linked to cell signaling as they can stimulate responses in plant and animal host cells [1–3]. Recent studies now indicate that GlcNAc released from these polymers can also activate cell signaling via several different mechanisms [4–6]. The role of these new GlcNAc signaling pathways in the regulation of virulence factors will be the focus of this review.
GlcNAc Induces Morphogenesis and Virulence Pathways in Fungi
GlcNAc first attracted attention as a signaling molecule for fungi over 40 years ago, when it was discovered to induce a remarkable switch from budding to hyphal growth in the human pathogen Candida albicans (Fig 1A) [7]. GlcNAc was subsequently shown to induce filamentous growth in a diverse group of fungi [5]. Switching to filamentous hyphal morphology contributes to invasive growth of C. albicans in the host and influences the interaction with leukocytes [8]. GlcNAc also stimulates the expression of virulence genes, such as the adhesins that promote adherence to host cells and biofilm formation [5,8]. Although it is not clear whether GlcNAc plays a role in systemic candidiasis, it has been implicated in commensal growth in the mucosa of the GI tract [9]. Consistent with this, GlcNAc promotes an epigenetic switch in morphology from the “White Phase” to the “Opaque Phase,” which is better adapted to mucosal growth [10].
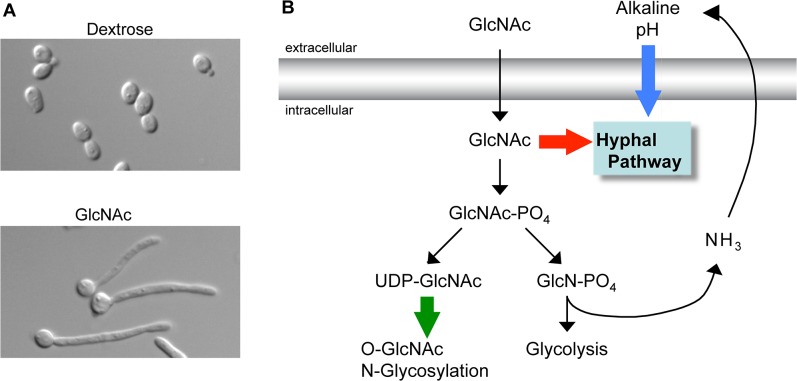
Fig 1. GlcNAc signaling pathways.
(A) C. albicans grown in dextrose form budding cells (top) whereas growth in GlcNAc induces them to switch to the filamentous hyphal form (bottom). (B) Summary of three types of GlcNAc-regulated pathways. GlcNAc itself can transduce a signal to induce hyphal growth in C. albicans (red arrow). Catabolism of GlcNAc releases excess ammonia whose export alkalinizes the extracellular pH and can synergize with GlcNAc to induce hyphal growth and gene expression (blue arrow). In mammals and some microbes conversion of GlcNAc to the building block UDP-GlcNAc promotes changes in O-GlcNAc modification of intracellular proteins and N-linked glycosylation of cell surface proteins (green arrow).
Identification of a GlcNAc transporter (Ngt1) in the C. albicans plasma membrane helped to resolve earlier controversies as to whether GlcNAc had to be imported into the cell to induce signaling [11]. An ngt1Δ mutant was defective in inducing hyphae, indicating that intracellular GlcNAc activates signaling. Since Ngt1 was the first eukaryotic GlcNAc transporter to be identified, its discovery has also helped to define the role of GlcNAc transport in other species. An interesting example of this is that Ngt1 orthologs were shown to mediate the ability of GlcNAc to induce hyphal growth in the dimorphic fungal pathogen Histoplasma capsulatum [12].
The ability of intracellular GlcNAc to transduce a signal raised the question of whether it had to be metabolized to induce signaling. Analysis of a C. albicans mutant lacking all three enzymes needed for GlcNAc catabolism (hxk1Δ nag1Δ dac1Δ) showed that the breakdown of this sugar was not needed for it to promote hyphal growth [13]. Furthermore, analysis of this mutant also indicated that GlcNAc did not have to be converted to the important building block UDP-GlcNAc. The hxk1Δ mutation blocks conversion of GlcNAc to GlcNAc-6-PO4, which is required for it to be subsequently processed into UDP-GlcNAc. These results indicate C. albicans uses a novel GlcNAc pathway (red arrow in Fig 1B) that is distinct from the major known signaling pathway in mammalian cells that requires conversion of GlcNAc into UDP-GlcNAc for use in O-GlcNAc modification of intracellular proteins (green arrow) [14]. The search for components in C. albicans that transduce the GlcNAc signal indicates that multiple pathways are activated. For example, the cAMP pathway is needed for GlcNAc to induce hyphal morphogenesis and virulence genes, but is not needed to induce the genes needed to catabolize GlcNAc [15].
Catabolism of GlcNAc Raises the Ambient pH: Synergy between GlcNAc and pH
Although GlcNAc catabolism is not required to induce hyphae, it can indirectly stimulate responses in C. albicans by raising the pH of the extracellular medium (blue arrow in Fig 1B) [16]. In contrast to acidification of the environment that occurs for cells grown in glucose, growth in GlcNAc raises the pH since cells export excess nitrogen as ammonia [17]. Studies with a mutant that lacks the GlcNAc metabolic genes (hxk1Δ nag1Δ dac1Δ) revealed an interesting synergy between GlcNAc and pH [16] as the mutant cells were able to induce hyphal morphology without the induction of hyphal-specific genes at low pH. However, the mutant cells could induce hyphal-specific genes when buffered to a higher pH (>5) that mimicked the effects of GlcNAc catabolism. Although alkaline pH can induce hyphal responses [8], the observed effects occurred at pH levels that were well below the levels required to induce hyphae, indicating that there is synergy between these pathways. These results are significant because they indicate that GlcNAc can stimulate hyphal morphogenesis independently of the induction of hyphal-specific genes, which had been linked to promoting the transition to filamentous growth [8]. This type of synergy between GlcNAc and pH likely occurs with other species, as cells from bacteria to humans export excess nitrogen as ammonia [18]. Thus, future studies must take care to distinguish between a direct role of GlcNAc in cell signaling and an indirect effect on the ambient pH.
GlcNAc Regulation of Virulence Factors in Bacteria
An important source of GlcNAc for cell signaling in many environments is due to release of this sugar during bacterial growth due to remodeling of cell wall peptidoglycan, which consists of alternating GlcNAc and N-acetylmuramic acid residues [19]. Approximately 50% of the sidewall peptidoglycan is broken down during each generation to accommodate the stepwise enlargement of the cell wall [19]. The presence of exogenous GlcNAc can therefore signal that cells nearby are dividing. One metabolic decision regulated by exogenous GlcNAc is to determine whether cells synthesize new GlcNAc, recycle exogenous GlcNAc back into peptidoglycan, or catabolize it for nutrition. Escherichia coli has streamlined this decision by placing the genes needed for GlcNAc synthesis and catabolism on opposite sides of a divergent operon that is regulated by the NagC transcription factor that responds to GlcNAc-6-PO4 [20]. Proper regulation of GlcNAc metabolism genes is significant, as it is important for colonization of the host by E. coli [21] and Vibrio cholera [22], and for production of virulence factors and biofilms by the cariogenic bacterium Streptococcus mutans [23].
GlcNAc has diverse effects in different bacteria by up-regulating or down-regulating virulence factors. In soil bacteria, it stimulates antibiotic production [24]. In polymicrobial infections, GlcNAc released from Gram-positive bacteria makes the infection more severe by stimulating Pseudomonas aeruginosa to produce toxins and virulence factors [6,25]. In contrast, GlcNAc down-regulates two extracellular adhesion factors in E. coli. GlcNAc inhibits production of type 1 fimbrial adhesins that promote urinary tract infections by mediating attachment to host cells [4]. GlcNAc also diminishes the production of the extracellular Curli fibers that play a role in biofilm formation, adhesion, and the internalization of E. coli by epithelial cells [26]. It has been suggested that rising GlcNAc levels during inflammation could signal to bacteria that host defenses are activated [4]. Inhibiting the expression of fimbriae and Curli fibers would therefore have two advantages for the bacteria: it would decrease the levels of these pro-inflammatory surface structures and the decreased levels of these adhesins would promote dissemination within the host. In this regard, it is interesting that many bacteria adhere via biofilms formed with extracellular poly-β,1–6 GlcNAc (distinct from β,1–4 linked chitin). Degradation of poly-β,1–6 GlcNAc disperses cells from biofilms and would also likely activate GlcNAc signaling that could affect adhesin production to further promote dissemination.
GlcNAc has additional roles in bacterial pathogenesis that depend on its metabolism and conversion to UDP-GlcNAc. For example, O-GlcNAc modification of proteins regulates cell motility in the pathogen Listeria monocytogenes [27]. In addition, exported toxins in other bacteria promote an unusual O-GlcNAc modification on arginine residues in cell death receptors and tyrosine residues in Rho that inactivates these host functions [28,29].
How Do Cells Distinguish Exogenous Versus Endogenous GlcNAc?
A key question is how do cells sense exogenous GlcNAc when they actively synthesize high levels of this sugar to create UDP-GlcNAc, a building block for glycosylation, GPI anchors, and the cell wall. Fungi and bacteria appear to distinguish exogenous GlcNAc because its phosphorylation status is distinct from endogenously synthesized GlcNAc [5]. For example, fungi take up unmodified GlcNAc, whereas they synthesize a phosphorylated form (GlcNAc-6-PO4) [30]. A variation of this occurs in bacteria that take up GlcNAc using a phosphotransferase system that converts it to GlcNAc-6-PO4, a form that is not synthesized in bacteria. Bacteria differ from fungi in that they avoid making GlcNAc-6-PO4 because the precursor sugar, glucosamine-6-PO4, is converted directly to GlcNAc-1-PO4, which is then further modified to create UDP-GlcNAc [30].
GlcNAc Signaling in Mammalian Cells
GlcNAc is known to induce responses in mammalian cells following its conversion to UDP-GlcNAc (green arrow in Fig 1B). Elevated UDP-GlcNAc increases O-GlcNAc modification of proteins and also increases N-GlcNAc branching on cell surface proteins, which changes cell signaling properties by altering the stability of receptors on the cell surface [14,31]. It is not clear whether GlcNAc itself can induce signaling in mammals. However, it is noteworthy that after infection with the fungus Cryptococcus neoformans, Th2 cell induction depended on cleavage of chitin by the mammalian chitinase, chitotriosidase, indicating that chitin fragments and perhaps GlcNAc are involved [32]. Although GlcNAc has also been reported to inhibit Th1 and Th17 cells, which play key roles in antifungal defense [33], further studies will be needed to determine how GlcNAc influences the immune system.
Concluding Comments
Emerging data indicate that the ubiquitous sugar GlcNAc is sensed by a broad range of organisms as a way to detect growth of neighboring cells or pathogenic attack. The ability of fungal and bacterial pathogens to regulate virulence functions in response to GlcNAc suggests that parasites will too, a possibility supported by the important role for GlcNAc metabolism in Leishmania [34]. The widespread presence of GlcNAc also suggests that is well suited to mediate interspecies communication with the host or between microorganisms to promote either symbiotic relationships or pathogenic interactions. In this way, GlcNAc is similar to many different chemical messengers, including quorum sensing factors, that are also used to communicate both intra- and interspecies [35]. Thus, it will be important to define the roles for GlcNAc signaling in complex environments, such as the human gut or in polymicrobial infections that contain a diverse array of bacteria, fungi, and human cells [36].
Acknowledgments
We thank Nick Carpino and the members of our lab for their helpful comments on the manuscript.
Funding Statement
Research in our lab is supported by grants from the National Institutes of Health awarded to James B. Konopka. http://www.nih.gov R21DE025200 RO1AI47837 The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kombrink A, Thomma BP. LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 2013;9(12):e1003769 10.1371/journal.ppat.1003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bueter CL, Specht CA, Levitz SM. Innate sensing of chitin and chitosan. PLoS Pathog. 2013;9(1):e1003080 10.1371/journal.ppat.1003080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wheeler R, Chevalier G, Eberl G, Gomperts Boneca I. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell Microbiol. 2014;16(7):1014–23. 10.1111/cmi.12304 [DOI] [PubMed] [Google Scholar]
- 4. Sohanpal BK, El-Labany S, Lahooti M, Plumbridge JA, Blomfield IC. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci U S A. 2004;101(46):16322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica. 2012;2012:489208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110(3):1059–64. 10.1073/pnas.1214550110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans . Nature. 1974;250(464):344–6. [DOI] [PubMed] [Google Scholar]
- 8. Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9(10):737–48. Epub 2011/08/17. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 9. Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45(9):1088–91. 10.1038/ng.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans . PLoS Pathog. 2010;6(3):e1000806 Epub 2010/03/20. 10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans . Mol Biol Cell. 2007;18:965–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gilmore SA, Naseem S, Konopka JB, Sil A. N-acetylglucosamine (GlcNAc) Triggers a Rapid, Temperature-Responsive Morphogenetic Program in Thermally Dimorphic Fungi. PLoS Genet. 2013;9(9):e1003799 10.1371/journal.pgen.1003799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naseem S, Gunasekera A, Araya E, Konopka JB. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J Biol Chem. 2011;286(33):28671–80. 10.1074/jbc.M111.249854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–58. Epub 2011/03/12. 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunasekera A, Alvarez FJ, Douglas LM, Wang HX, Rosebrock AP, Konopka JB. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot Cell. 2010;9(10):1476–83. Epub 2010/08/03. 10.1128/EC.00178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naseem S, Araya E, Konopka JB. Hyphal growth in Candida albicans does not require induction of hyphal-specific gene expression. Mol Biol Cell. 2015;26(6):1174–87. 10.1091/mbc.E14-08-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2(3):e00055–11. Epub 2011/05/19. 10.1128/mBio.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moye ZD, Burne RA, Zeng L. Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans . Appl Environ Microbiol. 2014;80(16):5053–67. 10.1128/AEM.00820-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev. 2008;72(2):211–27. Epub 2008/06/07. 10.1128/MMBR.00027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14(16):3958–65. Epub 1995/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101(19):7427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghosh S, Rao KH, Sengupta M, Bhattacharya SK, Datta A. Two gene clusters co-ordinate for a functional N-acetylglucosamine catabolic pathway in Vibrio cholerae. Mol Microbiol. 2011;80(6):1549–60. 10.1111/j.1365-2958.2011.07664.x [DOI] [PubMed] [Google Scholar]
- 23. Kawada-Matsuo M, Mazda Y, Oogai Y, Kajiya M, Kawai T, Yamada S, et al. GlmS and NagB regulate amino sugar metabolism in opposing directions and affect Streptococcus mutans virulence. PloS one. 2012;7(3):e33382 10.1371/journal.pone.0033382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, et al. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol. 2010;75(5):1133–44. Epub 2010/05/22. 10.1111/j.1365-2958.2009.07020.x [DOI] [PubMed] [Google Scholar]
- 25. Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193(4):909–17. Epub 2010/12/21. 10.1128/JB.01175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnhart MM, Lynem J, Chapman MR. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol. 2006;188(14):5212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen A, Kamp HD, Grundling A, Higgins DE. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 2006;20(23):3283–95. Epub 2006/12/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S, Zhang L, Yao Q, Li L, Dong N, Rong J, et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501(7466):242–6. 10.1038/nature12436 [DOI] [PubMed] [Google Scholar]
- 29. Jank T, Bogdanovic X, Wirth C, Haaf E, Spoerner M, Bohmer KE, et al. A bacterial toxin catalyzing tyrosine glycosylation of Rho and deamidation of Gq and Gi proteins. Nat Struct Mol Biol. 2013;20(11):1273–80. 10.1038/nsmb.2688 [DOI] [PubMed] [Google Scholar]
- 30. Milewski S, Gabriel I, Olchowy J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast. 2006;23(1):1–14. [DOI] [PubMed] [Google Scholar]
- 31. Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139(7):1229–41. Epub 2010/01/13. 10.1016/j.cell.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, et al. Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection. PLoS Pathog. 2015;11(3):e1004701 10.1371/journal.ppat.1004701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grigorian A, Araujo L, Naidu NN, Place DJ, Choudhury B, Demetriou M. N-acetylglucosamine inhibits T-helper 1 (Th1)/T-helper 17 (Th17) cell responses and treats experimental autoimmune encephalomyelitis. J Biol Chem. 2011;286(46):40133–41. Epub 2011/10/04. 10.1074/jbc.M111.277814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Naderer T, Heng J, McConville MJ. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathog. 2010;6(12):e1001245 Epub 2011/01/05. 10.1371/journal.ppat.1001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12(2):205–14. Epub 2009/03/03. 10.1016/j.mib.2009.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–7. Epub 2012/06/08. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]