Abstract
Variations in the TP53 gene have been suggested to play a role in many cancers, including breast. We previously observed an association between TP53 haplotypes based on four polymorphisms (rs17878362, rs1042522, rs12947788, and rs17884306) and the risk of colorectal and pancreatic cancer. Based on these results, in the present study, we have investigated the same polymorphisms and their haplotypes in 705 breast cancer cases and 611 healthy controls in relation to the disease risk, histopathological features of the tumor and clinical outcomes. In comparison to the most common haplotype A1-G-C-G, all the other identified haplotypes were globally associated with a significantly decreased breast cancer risk (P = 0.006). In particular, the A2-G-C-G haplotype was associated with a marked decreased risk of breast cancer when compared with the common haplotype (P = 0.0001). Moreover, rs1042522 in patients carrying the GC genotype and receiving only the anthracycline-based chemotherapy was associated with both overall and disease-free survival (recessive model for overall survival HR = 0.30 95% CI 0.11–0.80, P = 0.02 and for disease-free survival HR = 0.42 95% CI 0.21–0.84, P = 0.01). Present results suggest common genetic features in the susceptibility to breast and gastrointestinal cancers in respect to TP53 variations. In fact, similar haplotype distributions were observed for breast, colorectal, and pancreatic patients in associations with cancer risk. Rs1042522 polymorphism (even after applying the Dunn-Bonferroni correction for multiple testing) appears to be an independent prognostic marker in breast cancer patients.
Introduction
Breast cancer (BC) is the third most common tumor worldwide (and the most frequent in women), representing 9% of the global cancer burden. Epidemiologic studies have suggested a number of risk factors including personal, family, and gynecological history, as well as an interplay between genetic and environmental factors [1]. Mutations in the rare high penetrance BC predisposing genes BRCA1 and BRCA2 account for 16–25% of the inherited component of this cancer [2]. To date, GWAS conducted for BC have identified more than 80 breast cancer susceptibility loci (all summarized in [3]). Although, several candidate gene association studies for BC have been conducted during the last decade [4–10], many of them were underpowered due to small sample size, resulting in inconsistent and not reproducible findings [11,12]. To date, only one polymorphism located in the coding region of CASP8 (rs1045485) has shown promise as a breast cancer predisposition risk factor [3,13].
The prognosis of breast carcinoma is mostly influenced by tumor stage, grade, overexpression of v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2), and hormonal receptors. These factors are routinely considered in selecting the treatment regimen for each individual [14]. Anthracycline combination chemotherapies are the most effective and widely used regimens for the treatment of BC [15]. Radiotherapy, on the other hand, improves BC survival by reducing the risk of local recurrence, whereas it has no direct impact on metastasis [16]. Besides, still novel biomarkers are needed to identify individuals who would benefit most from a given treatment regimen. Such markers might arise from inherited genetic variation, which—apart from BC susceptibility—may affect disease progression, response to treatment and outcome [17].
p53 protein (encoded by TP53 gene) is a key player in stress responses that preserve genomic stability responding to a variety of insults including DNA damage, hypoxia, metabolite stress and oncogene activation [18]. In addition, p53 interacts with numerous cellular proteins including those controlling the programmed cell death. This tumor suppressor gene is frequently mutated in various solid tumors, including colorectum, pancreas, and breast. These mutations result in the absence or dysfunction of the corresponding protein [19]. TP53 is also polymorphic: so far, 547 single nucleotide polymorphisms (SNPs) have been identified (http://genecards.org). Owing to the importance of p53 in tumor suppression, SNPs that may alter its function might affect not only cancer risk but also its progression and/or response to the treatment [18]. TP53 rs1042522 is one of the most commonly investigated variants in cancer genetic epidemiology. However, two recent reviews [20,21] suggested that this polymorphism is unlikely to be a risk factor for BC. Instead, a recent meta-analysis on rs17878362 (PIN3, Ins11951_11966, allele A2 carries the 16-bp insertion within intron 3) has found an increased risk of BC in A2A2 carriers [22]. Previous studies have suggested rs1042522 as a prognostic marker for BC [23–26]. On the contrary, the role of TP53 haplotypes has not yet been fully investigated for BC susceptibility and its clinical outcome.
Previously, we have studied the role of four TP53 polymorphisms on colorectal cancer (CRC) and pancreatic cancer risk. In both studies, we have observed a consistent differential distribution of the major haplotypes built on the four variants between cases and controls, suggesting that prevalent haplotypes within the TP53 gene may modulate both CRC and pancreatic cancer risk [27,28]. In the present investigation, we evaluated the association of the same variations within TP53 gene and resulting haplotypes (loci in the order: rs17878362, rs1042522, rs12947788, and rs17884306) with the risk of BC in a hospital-based case-control study from the Czech Republic. In this country, around 6,000 women every year are diagnosed with BC, and about 2,000 women die because of this cancer (incidence rank: 30th worldwide and 18th in Europe; mortality rates rank: 117th worldwide and 36th in Europe; www.mamo.cz). In a set of the BC patients with complete follow-up, we also investigated the prognostic relevance of TP53 gene variability.
Material and Methods
Study population
The study population comprised 705 BC patients and 611 healthy controls. Blood samples were obtained from women with incident BC consecutively diagnosed in three hospitals in Prague between February 2002 and December 2010. Sample collection from patients was described elsewhere [29–31]. All subjects were informed and provided written consent to participate in the study and to use their biological samples for genetic analyses, according to the Helsinki declaration. The design of the study was approved by the Ethical Committee of the Institute of Experimental Medicine, Prague, Czech Republic.
The control group consisted of healthy women whose information were previously described in other association studies [27,32–34]. Two control groups were included in the study. The first group consisted of 268 hospital-based individuals admitted to gastroenterological departments that had negative colonoscopy results for malignancy or idiopathic bowel diseases. The reasons for undergoing the colonoscopy were: (i) positive fecal occult blood test, (ii) hemorrhoids, (iii) abdominal pain of unknown origin, and (iv) macroscopic bleeding. They did not have any malignancy at the time of the sampling. The second group of controls consisted of 343 healthy blood donor volunteers collected from a blood donor center in Prague. All individuals were subjected to standard examinations to verify the health status for blood donation and were cancer-free at the time of the sampling.
The following data on patients were retrieved from medical records: date of cancer diagnosis, age, menopausal status, family history of cancer (number of relatives affected by BC, ovarian cancer or other malignant diseases), tumor size, UICC (International Union Against Cancer) tumor-node-metastasis (TNM) system classification, histological type and grade of tumor, expression of estrogen receptor (ER), progesterone receptor (PR) and ERBB2, expression of the Ki-67 protein, chemotherapy and hormonal regimen. ERBB2 status was defined as positive in samples with an immunohistochemical score of 2+ or 3+ [35]. Due to different treatment of patients with metastasis, those women with metastasis were excluded from the analysis.
For all cases, detailed information on radiotherapy, first line chemotherapy, and hormonal therapy was available. Patients were treated with chemotherapy regimens containing 5-fluorouracil, anthracycline, cyclophosphamide, and/or taxanes (for all treatments see S1 File). Information about distant metastasis, relapse and date of death was also collected, with a follow-up until December 31, 2013. In this study, the outcome variables measured were overall survival (OS) and disease-free survival (DFS). The same variables were also investigated after stratification according to anthracycline-, taxane-based chemotherapy, or hormonal-based therapy,
Selection of polymorphisms
The TP53 SNPs analysed were the same as those previously investigated for CRC [27] and pancreatic cancer [28] studies and represent part of the genetic variation in TP53 gene with MAF≥5%. In particular, three tag SNPs were selected (rs1042522:G>C (Ex4+119 G>C, Arg72Pro), rs12947788:C>T (IVS7+72 C>T) and rs17884306:G>A (Ex11-363 G>A)). In the study, we also included the 16 bp insertion/deletion polymorphism within the intron 3 (rs17878362:A1>.A2, PIN3, Ins11951_11966, in which the A2 allele carries the 16 bp insertion within the intron 3).
SNP Genotyping
Genomic DNA was isolated from peripheral blood lymphocytes using standard procedures as described in [36]. DNA samples from cases and controls were randomly placed on plates where an equal number of cases and controls were run simultaneously. Genotyping of the selected SNPs was carried out by using the KASPar chemistry of LGC Genomics (http://www.lgcgenomics.com/genotyping/kasp-genotyping-reagents/kasp-technical-resources/), as described in [37]. Duplicate samples (5%) and non-template controls in each plate were used as quality control tests.
Rs17878362 in TP53 gene was genotyped by PCR-RFLP as previously described in [27] and [28].
Statistical analyses
Chi-square test (1 degree of freedom), with a type-I error threshold set at α = 0.05, was used to verify whether the genotypes were in Hardy-Weinberg equilibrium in controls. The haplotype frequencies in cases and controls were estimated with the SAS/Genetics software module (SAS Institute, Cary, NC, USA). The analysis was carried out to examine the phase of TP53 SNPs using the expectation–maximization algorithm to generate maximum likelihood estimates of haplotype frequencies.
The association between SNPs and BC risk was calculated by estimating the odds ratios (ORs) and their 95% confidence intervals (CI) adjusted for age. For all SNPs, the co-dominant, the dominant or the recessive models were calculated.
OS was defined as the time from the surgery to the date of death or the date of the end of the study (December 31, 2013). DFS was defined as the time from surgery/end of therapy to the occurrence of distant metastasis, local recurrence or death, whichever came first. The relative risk of death and recurrence was estimated as hazard ratio (HR) using Univariate Cox regression, Multivariate Cox regression for significant results and Long-rank test. The survival curves for overall and disease-free survival were derived by the Kaplan–Meier method. Multivariate survival analyses were adjusted for age and therapy. Statistical analyses were performed using SAS software (SAS Institute, Cary, NC, USA).
Results
Case-control study
The study included 705 cases and 611 controls (detailed information in S1 File). The median age (range) of patients at diagnosis was 59 (65–92) years while the median age of controls at recruitment was 49 (57–85) years. There was a statistically significant difference in the age distribution between BC patients and controls (P<0.0001). Due to differences in age among cases and controls, we have considered this factor as a confounding variable and used in our analysis adjusting OR for it.
The distribution of genotypes within the four selected TP53 SNPs in controls was in agreement with the Hardy-Weinberg equilibrium (Table 1). No significant differences were found among cases and controls in the genotype frequencies for any of the polymorphisms when analyzed individually (Table 1). Rs17884306 resulted monomorphic (only GG genotype in all BC subjects). Additionally, no clinicopathological parameters were associated with the risk of BC after both univariate and multivariate analysis.
Table 1. Genotype distribution of investigated TP53 polymorphisms in BC patients and controls.
| Genotypes | Controls a | Cases a | OR b | 95% CI | P | HWE c |
|---|---|---|---|---|---|---|
| (n = 611) | (n = 705) | Χ2, P | ||||
| rs17878362 d | 0.01, 0.99 | |||||
| A 1 A 1 | 421 | 474 | REF | |||
| A 1 A 2 | 172 | 164 | 0.88 | 0.67–1.16 | 0.36 | |
| A 2 A 2 | 18 | 24 | 1.13 | 0.57–2.24 | 0.71 | |
| A 1 A 2 + A 2 A 2 | 190 | 188 | 0.91 | 0.70–1.18 | 0.46 | |
| rs1042522 | 0.35, 0.84 | |||||
| GG | 301 | 370 | REF | |||
| GC | 260 | 275 | 0.89 | 0.70–1.14 | 0.35 | |
| CC | 50 | 55 | 0.93 | 0.61–1.43 | 0.74 | |
| GC+CC | 310 | 330 | 0.90 | 0.71–1.14 | 0.37 | |
| rs12947788 | 0.24, 0.89 | |||||
| CC | 530 | 594 | REF | |||
| CT | 78 | 102 | 1.26 | 0.90–1.77 | 0.19 | |
| TT | 2 | 5 | 1.60 | 0.30–8.53 | 0.58 | |
| CT+ TT | 80 | 107 | 1.27 | 0.91–1.77 | 0.16 | |
| rs17884306 | 0.48, 0.79 | |||||
| GG | 540 | 702 | ||||
| GA | 66 | 0 | - | - | - | |
| AA | 1 | 0 | - | - | - | |
| GA+AA | 67 | 0 | - | - | - |
aNumbers may not add up to 100% of subjects due to genotyping failure. All samples that did not give a reliable result in the first round of genotyping were resubmitted to up to two additional rounds. Data points that were still not filled after this procedure had been left blank.
bLogistic regression analysis values are adjusted for age.
cX2 and P-values for the deviation of observed and the numbers expected from the Hardy-Weinberg equilibrium (HWE) in the controls.
dAllele A2 carries the 16-bp insertion within intron 3.
OR, odds ratio; CI, confidence interval.
In the present study, the two most frequent haplotypes A1-G-C-G and A2-C-C-G comprised the 83% of cases and only the 77% of controls. The A2-G-C-G haplotype was associated with a significantly decreased risk of BC when compared with the reference haplotype A1-G-C-G, comprising only the common alleles (OR = 0.36, 95%CI 0.21–0.61, P = 0.0001; Table 2). When the reference haplotype A1-G-C-G was compared to all other haplotypes, it resulted in association with an increased BC risk (OR = 1.29, 95%CI 1.08–1.54, P = 0.006).
Table 2. TP53 haplotype distribution between BC patients and controls.
| Haplotypes a | Controls b | Cases b | OR c | 95% CI | P |
|---|---|---|---|---|---|
| n = 1222 | n = 1410 | ||||
| d A 1 -G-C-G | 775 | 898 | REF | ||
| All others | 437 | 394 | 0.78 | 0.65–0.93 | 0.006* |
| A 2 -C-C-G | 153 | 179 | 1.06 | 0.82–1.37 | 0.67 |
| A 1 -C-C-G | 84 | 85 | 0.81 | 0.58–1.13 | 0.21 |
| A 1 -C-T-G | 65 | 95 | 1.36 | 0.96–1.93 | 0.09 |
| A 1 -C-C-A | 55 | 0 | - | - | - |
| A 2 -G-C-G | 50 | 24 | 0.36 | 0.21–0.61 | 0.0001* |
| A 1 -G-T-G | 17 | 8 | - | - | - |
| A 1 -G-C-A | 9 | 0 | - | - | - |
| A 2 -G-C-A | 3 | 0 | - | - | - |
| A 2 -C-C-A | 1 | 0 | - | - | - |
| A 2 -C-T-G | 0 | 2 | - | - | - |
| A 2 -G-T-G | 0 | 1 | - | - | - |
aLoci rs17878362, rs1042522, rs12947788, rs17884306.
bNumber of alleles are reported. Because each individual has two alleles, the total number of alleles will be twice the total number of individuals. Individuals with missing haplotype data were not included in the analyses.
cAdjusted for age.
dAllele A2 carries the 16-bp insertion within intron 3
OR, odds ratio; CI, confidence interval. Significant P-values are in bold.
During the follow-up of BC cases, overall 77 patients died, with 39 of them due to cancer progression. In addition, 78 subjects had recurrence (S1 File). Forty four patients were not retrievable during follow-up, therefore, they were excluded from the analysis. The median OS and DFS for the studied population were 54.3 and 52.2 months, respectively.
One hundred forty-four (20.4%) patients received neoadjuvant chemotherapy before surgery. Concerning the adjuvant chemotherapy, 192 (51.9%) patients were administered with antracyclines, 93 (25.1%) with taxanes, 34 (9.2%) with both agents in combination and 44 (11.9%) with others agents as first-line postoperative therapy (for more detail see S1 File). Two hundred ninety-one (41.3%) subjects did not receive any adjuvant chemotherapy after surgery, while 218 (30.9%) women received the hormonal therapy as a first line postoperative therapy instead of adjuvant therapy. Finally, 318 (45.1%) patients were administrated with both adjuvant and hormonal therapy, comprising mainly tamoxifen or inhibitors of aromatases. Patients not receiving any hormonal therapy or radiotherapy or with advanced age (>60 years) resulted to have a shorter OS (log-rank P = 0.04; P = 0.006 and P = 0.01, respectively) and DFS (log-rank P = 0.04; P = 0.002 and P = 0.02, respectively) when compared with those receiving hormonal or radiotherapy or with age lower than 60, respectively. No other clinicopathological parameters were associated with survival (after both univariate and multivariate analysis).
Patients with homozygous variant genotype A2A2 of rs17878362 polymorphism were at higher risk of relapse or metastasis (HR = 2.15, 95%CI 1.04–4.44, P = 0.04; Table 3). On the other hand, patients carrying GC genotype of rs1042522 were at lower risk of relapse or metastasis (HR = 0.66, 95%CI 0.44–1.00, P = 0.05). Concerning haplotype analyses, the haplotype A1-C-C-G was associated with lower risk of relapse or metastasis (HR = 0.34, 95%CI 0.14–0.84, P = 0.02; Table 4).
Table 3. Overall (OS) and disease-free (DFS) survival in relation to SNP distributions (Cox regression).
| Genotype | OS | DFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Events | HR | 95% CI | P | Events | HR | 95%CI | P | |
| rs17878362 | |||||||||
| A 1 A 1 | 470 | 52 | REF | - | - | 78 | REF | - | - |
| A 1 A 2 | 163 | 16 | 0.87 | 0.50–1.52 | 0.63 | 24 | 0.88 | 0.56–1.39 | 0.58 |
| A 2 A 2 | 24 | 3 | 1.07 | 0.33–3.43 | 0.91 | 8 | 2.15 | 1.04–4.44 | 0.04 |
| A 1 A 2 + A 2 A 2 | 187 | 19 | 0.90 | 0.53–1.52 | 0.69 | 32 | 1.03 | 0.68–1.56 | 0.88 |
| rs1042522 | |||||||||
| GG | 369 | 44 | REF | - | - | 71 | REF | - | - |
| GC | 271 | 25 | 0.82 | 0.50–1.34 | 0.42 | 33 | 0.66 | 0.44–1.00 | 0.05 |
| CC | 55 | 4 | 0.62 | 0.22–1.71 | 0.35 | 9 | 0.88 | 0.44–1.75 | 0.71 |
| GC+CC | 326 | 29 | 0.78 | 0.49–1.25 | 0.30 | 42 | 0.70 | 0.48–1.02 | 0.06 |
| rs12947788 | |||||||||
| CC | 592 | 63 | REF | - | - | 97 | REF | - | - |
| CT | 99 | 11 | 1.06 | 0.56–2.00 | 0.87 | 16 | 1.00 | 0.59–1.70 | 0.99 |
| TT | 5 | 0 | - | - | - | 0 | - | - | - |
| CT+TT | 104 | 11 | 1.00 | 0.53–1.90 | 0.99 | 16 | 0.95 | 0.56–1.61 | 0.85 |
The SNP rs17884306 was monomorphic in cases, thus not presented.
HR, hazard ratio; 95% CI, confidence interval. Significant results in bold.
Table 4. OS and DFS in relation to haplotype distributions (Cox regression).
| Haplotype a | OS | DFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases b | Events b | HR | 95%CI | P | Events | HR | 95%CI | P | |
| c A 1 -G-C-G | 896 | 104 | REF | 157 | REF | ||||
| All others | 390 | 36 | 0.81 | 0.55–1.18 | 0.26 | 59 | 0.88 | 0.65–1.19 | 0.40 |
| A 2 -C-C-G | 178 | 16 | 0.78 | 0.46–1.32 | 0.35 | 29 | 0.95 | 0.64–1.42 | 0.81 |
| A 1 -C-T-G | 92 | 10 | 0.97 | 0.50–1.85 | 0.91 | 15 | 0.96 | 0.57–1.64 | 0.89 |
| A 1 -C-C-G | 85 | 5 | 0.54 | 0.22–1.31 | 0.17 | 5 | 0.34 | 0.14–0.84 | 0.02* |
aLoci rs17878362, rs1042522, rs12947788, rs17884306.
b Number of alleles are reported. Because each individual has two alleles, the total number of alleles is twice the total number of individuals. Individuals with missing haplotype data were not included in the analyses.
cAllele A2 carries the 16-bp insertion within intron 3
HR, hazard ratio; 95% CI, confidence interval. Significant results in bold.
For significant associations, we have also applied the multivariate Cox regression analysis. Patients with advanced age (>60 years old) and those not receiving any hormonal or radiotherapy resulted in association with a shortened OS and DFS (S1 File). In the recessive model, rs1042522 was still associated with lower risk of relapse or metastasis (HR = 0.65, 95%CI 0.44–0.96, P = 0.03).
Adjuvant chemotherapy
In patients receiving only adjuvant chemotherapy, rs1042522 was significantly associated with OS (log-rank for recessive model P = 0.02; Fig 1). In particular, when patients with the homozygous GG genotype were compared to C allele carriers, they showed longer survival (HR = 0.21, 95%CI 0.05–0.94, P = 0.04).
Fig 1. Kaplan-Meier overall survival curves for TP53 rs1042522 polymorphism in patients receiving only adjuvant chemotherapy (log-rank for recessive model P = 0.02).
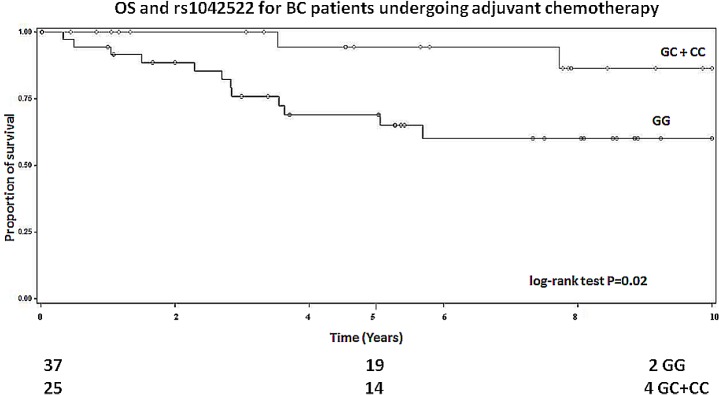
Numbers of patients at risk are indicated in the lower part of the plot.
After stratification for anthracycline-based chemotherapy, rs17878362 was associated with OS (log-rank P = 0.02). In particular, a better survival was observed for those patients carrying the A1A2 genotype and undergoing anthracycline-based therapy (HR = 0.14, 95%CI 0.02–1.00, P = 0.05; Table 5). In the same group of treated patients, rs1042522 polymorphism was also associated with both OS and DFS (log-rank P = 0.02 and 0.007, respectively; Table 5). Namely, we have observed a better survival for those patients carrying the GC genotype and undergoing anthracycline-based chemotherapy (OS: HR = 0.31, 95%CI 0.11–0.90, P = 0.03; DFS: HR = 0.34, 95%CI 0.15–0.77, P = 0.01 respectively). In a recessive model, the associations remained the same (log-rank P = 0.01 and 0.01, respectively; Table 5).
Table 5. OS and DFS in relation to SNP distributions in patients treated with anthracycline-based chemotherapy (Cox regression).
| Genotype | OS | DFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Events | HR | 95% CI | P | Events | HR | 95%CI | P | |
| rs17878362 | |||||||||
| A 1 A 1 | 159 | 22 | REF | - | - | 35 | REF | - | - |
| A 1 A 2 | 53 | 1 | 0.14 | 0.02–1.00 | 0.05 | 5 | 0.42 | 0.16–1.06 | 0.07 |
| A 2 A 2 | 7 | 2 | 1.86 | 0.44–7.93 | 0.40 | 3 | 2.03 | 0.62–6.61 | 0.24 |
| A 1 A 2 + A 2 A 2 | 60 | 3 | 0.35 | 0.11–1.18 | 0.09 | 8 | 0.60 | 0.28–1.28 | 0.19 |
| rs1042522 | |||||||||
| GG | 120 | 21 | REF | - | - | 33 | REF | - | - |
| GC | 82 | 4 | 0.31 | 0.11–0.90 | 0.03 | 7 | 0.34 | 0.15–0.77 | 0.01* |
| CC | 20 | 1 | 0.29 | 0.04–2.15 | 0.23 | 4 | 0.73 | 0.26–2.07 | 0.56 |
| GC+CC | 102 | 5 | 0.30 | 0.11–0.80 | 0.02* | 11 | 0.42 | 0.21–0.84 | 0.01* |
| rs12947788 | |||||||||
| CC | 189 | 21 | REF | - | - | 34 | REF | - | - |
| CT | 33 | 5 | 1.44 | 0.54–3.82 | 0.47 | 10 | 2.00 | 0.98–4.00 | 0.06 |
| TT | 2 | 0 | - | - | - | 0 | - | - | - |
| CT+TT | 35 | 5 | 1.33 | 0.50–3.53 | 0.57 | 10 | 1.80 | 0.89–3.65 | 0.10 |
The SNP rs17884306 was monomorphic in cases, thus not presented.
HR, hazard ratio; 95% CI, confidence interval. Significant results in bold; significant differences after Dunn–Bonferroni correction (P<0.02) are marked with an asterisk.
The stratification for anthracycline-based chemotherapy or for taxane-based chemotherapy did not show any significant associations of TP53 haplotype distribution with the OS or DFS (data not shown).
Hormonal therapy
Among patients receiving only the hormonal therapy, rs17878362 polymorphism was significantly associated with DFS (log-rank for recessive model P = 0.04; Fig 2). In particular, we have observed a worse survival for those patients carrying the variant A2A2 genotype (HR = 2.66, 95%CI 1.02–6.94, P = 0.05).
Fig 2. Kaplan-Meier disease-free survival curves for TP53 rs17878362 polymorphism in patients receiving only the hormonal therapy (log-rank for recessive model).
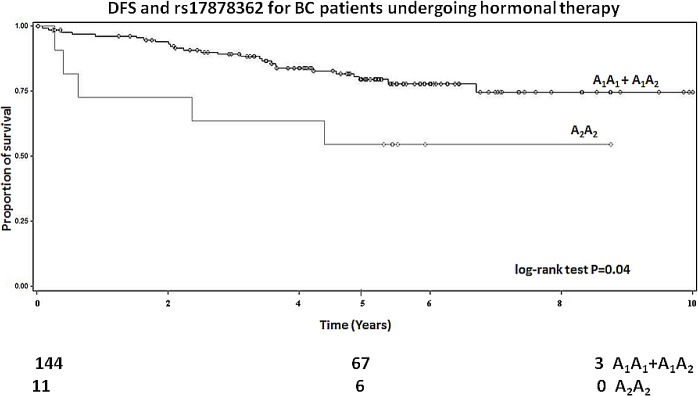
Numbers of patients at risk are indicated in the lower part of the plot.
Further stratification for hormonal-based therapy and the regime containing either tamoxifen or inhibitors of aromatases agents did not show any significant associations with BC prognosis (data not shown).
Discussion
In the present study, we investigated the role of genetic variations within TP53 gene on BC risk and its clinical outcomes in a population from the Czech Republic. Recent meta-analyses suggested that polymorphisms in TP53 taken individually are not presumably risk factors for BC [20,21]. Our data on single TP53 polymorphisms are in good agreement with the above postulation. In contrast, there are also meta-analyses reporting that rs17878362 may contribute to susceptibility of BC [22,38]. On the other hand, TP53 haplotypes have not been comprehensively explored regarding the BC risk and prognosis.
Interestingly, in the case-control study we found that the TP53 A2-G-C-G haplotype was associated with a decreased risk of BC when compared with the most common haplotype A1-G-C-G. Globally, all the detected haplotypes were associated with decreased risk of the disease when compared with A1-G-C-G.
Although some studies found an association between sporadic BC and rs1042522, rs17878362 or their respective haplotypes [39–41], the results are rather inconclusive due to limited sample size (108, 159, 122 patients, respectively), differences in allele frequencies between ethnic groups or staging of the malignancy.
In previous reports, we have observed a differential distribution of specific haplotypes between cancer patients and controls, suggesting that prevalent haplotypes within TP53 may modulate both CRC and pancreatic cancer risk [27,28]. Interestingly, we have observed a similar pattern also for BC patients. The haplotypes within TP53 along with SNPs in other genes in its pathway may impact the onset of pancreatic cancer and CRC, as well as BC. Thus, our results seem to point to general mechanisms based on common genetic background involved in the development of solid tumors. As previously observed for colorectal and pancreatic malignancies, the different distribution of TP53 haplotypes also in BC cases and controls may be due to a linkage of the disease to yet unknown functional polymorphism(s) within TP53 or in some neighboring genes on 17p that could carry putative functional variant(s) linked to the detected haplotypes. This argument is supported by predictions of such associations and the existence of large haplotype blocks within the human genome [42]. On the other hand, there are several studies in which deletion of 17p have been frequently observed in BC patients [43–45]. The reversal of the modulation effect of TP53 polymorphisms within haplotypes may also point to additional polymorphism(s) that cause differential cancer risk, either directly or through interaction with environmental factors [27]. p53 interacts with numerous cellular proteins, including those in the control of programmed cell death. Above molecular interactions might contribute to its inhibitory role in the tumorigenesis. TP53 is frequently mutated in various solid tumors, including colorectal, pancreatic and breast cancers, and these mutations result in the absence or dysfunction of the p53 protein [46].
Another novel finding emerging from this study was that two individual SNPs in TP53 (rs1042522 and rs17878362) were associated with patient´s survival in a set of 705 BC cases. In particular, patients with variant genotype A2A2 in rs17878362 were at higher risk of relapse or metastasis. After stratification for anthracycline-based chemotherapy, rs17878362 was also associated with OS. The association of SNPs with longer OS and DFS, particularly in relation to the anthracycline–based chemotherapy, is of interest since it is in contrast to mutations in this gene that are generally associated with a poor prognosis in BC patients [47,48]. Apart from few studies [23,49–51], the effect of TP53 polymorphisms on patients survival has not been studied in detail. The role of TP53 as a predictive biomarker for treatment response still remains a matter of debate. Interestingly, [49] observed on a relatively large set of Finnish samples that patients carrying the homozygous variant CC genotype of rs1042522 had significantly poorer survival than BC patients with other genotypes. A similar pattern of association was also observed in the study of Toyama et al. (2007) on Japanese BC women, in the one on a Chinese population [24] and more recently in the work of [26] on a Spanish group of BC. Interestingly, patients with rs1042522 GC genotype were at lower risk of relapse or metastasis, as well as those with the less frequent haplotype A1-C-C-G.
The association of rs17878362 to patient’s survival is of particular interest since the functional relevance of rs17878362 is unknown. Intronic sequences in this gene have been implicated in the regulation of gene expression and in DNA–protein interactions [52]. Rs17878362 may also be in linkage disequilibrium with other yet unidentified genes, thus explaining its association with a distinctive phenotype.
The common G allele of rs1042522 is associated with a form of the p53 protein that is a more potent inducer of apoptosis than the one containing the C allele [53]. It has been suggested that patients carrying the GG genotype may respond more favorably to radiation or chemotherapy [54]. For example, the higher efficiency of GG genotype in inducing apoptosis is reflected in vivo in a better outcome in carriers of this genotype with advanced squamous head and neck carcinoma, and receiving chemo-radiotherapy [55]. However, these effects of the G allele may be reversed by a somatic p53 mutation on this allele [56]. Retention of the G allele with loss of the C allele in the tumor tissue has been associated with reduced survival in GC heterozygous BC patients [57].
It has been shown in various in vitro and mouse experiments that cell cycle arrest or apoptosis induced by radiotherapy and various chemotherapeutic drugs depends on an intact TP53 pathway [58,59]. Preclinical studies suggested p53-dependent anthracycline-induced apoptosis and p53-independent taxane activity [58,60,61]. Other investigations have presented data in support of reduced anthracycline activity in TP53 mutated tumors [62,63]. Thus, the presence of variations in the TP53 gene could be one of the underlying causes of drug resistance, the major cause of treatment failure and cancer death.
We are aware of certain limitations of this study, such as: i) the different age distribution among cases and controls, ii) the non-homogenous proportion of patients with neoadjuvant and adjuvant treatment, iii) the wide range of therapeutic agents used. BC risk is also tightly linked to non-genetic factors such as pregnancy and hormonal history. Unfortunately, we were not able to assemble this information. Other variables such as age at menarche, oophorectomy, pregnancy and lactation history were also not included for comparison between cases and controls, since they were not available for both groups. These limitations may be counterbalanced by the homogeneity of the population and the detailed follow-up. To overcome the possible effect of age, we have repeated the analyses selecting patients and controls by matching for ±5 years. After matching, 570 patients (median age 56 years, range 27–71) and 575 controls (median age 51 years; range 33–91) were included in the analyses. For all the analyses performed, we obtained similar results for both study groups (whole unmatched and matched case-control group) (S1 File).
For the three studied polymorphisms, we also applied the Dunn-Bonferroni correction for multiple comparisons. After correction, the new adjusted threshold of P-value significance is 0.02 for analyses of individual SNPs. The rs1042522 polymorphism still appeared to be an independent prognostic marker in breast cancer patients.
In conclusion, in our study conducted on a Czech population of Caucasian origin, we reported for the first time that the most frequent TP53 haplotype, previously associated with an increased CRC and pancreatic cancer risks, was also associated with an increased BC susceptibility. These results suggest that a particular genetic background based on certain haplotypes in relevant genes such as TP53 may play a role in the susceptibility of solid cancers, as demonstrated on gastrointestinal cancers and BC. Importantly, we have also observed that both individual polymorphisms or specific haplotypes of the TP53 gene were associated with BC clinical outcome.
Further studies are needed to replicate these findings in independent and larger populations to support the hypothesis that SNPs and haplotypes in TP53 may be reliable predictive biomarkers in solid tumors and may help in directing to a better pre-selection of patients according to administered chemotherapeutic agents. Moreover, it is important to characterize functional relevance of the identified genetic variants and to elucidate their biologic associations.
Supporting Information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Grant agency of the Czech Republic (GACR) [CZ:GACR: P304/12/1585] and by Prvouk-P27/LF1/1 from Ministry of Education, Youth and Sport, Czech Republic (First Medical Faculty, Charles University, Prague, Czech Republic as a recipient) and by Internal Grant Agency Ministry of Health: NT 13424 and NT 14055-3.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2. Beggs AD, Hodgson SV (2009) Genomics and breast cancer: the different levels of inherited susceptibility. Eur J Hum Genet 17: 855–856. 10.1038/ejhg.2008.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sapkota Y (2014) Germline DNA variations in breast cancer predisposition and prognosis: a systematic review of the literature. Cytogenet Genome Res 144: 77–91. 10.1159/000369045 [DOI] [PubMed] [Google Scholar]
- 4. Allen-Brady K, Cannon-Albright LA, Neuhausen SL, Camp NJ (2006) A role for XRCC4 in age at diagnosis and breast cancer risk. Cancer Epidemiol Biomarkers Prev 15: 1306–1310. [DOI] [PubMed] [Google Scholar]
- 5. Bewick MA, Conlon MS, Lafrenie RM (2006) Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol 24: 5645–5651. [DOI] [PubMed] [Google Scholar]
- 6. Haiman CA, Hsu C, de Bakker PI, Frasco M, Sheng X, Van Den Berg D, et al. (2008) Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet 17: 825–834. [DOI] [PubMed] [Google Scholar]
- 7. Pooley KA, Baynes C, Driver KE, Tyrer J, Azzato EM, Pharoah PD, et al. (2008) Common single-nucleotide polymorphisms in DNA double-strand break repair genes and breast cancer risk. Cancer Epidemiol Biomarkers Prev 17: 3482–3489. 10.1158/1055-9965.EPI-08-0594 [DOI] [PubMed] [Google Scholar]
- 8. Sehl ME, Langer LR, Papp JC, Kwan L, Seldon JL, Arellano G, et al. (2009) Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clin Cancer Res 15: 2192–2203. 10.1158/1078-0432.CCR-08-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin WY, Camp NJ, Cannon-Albright LA, Allen-Brady K, Balasubramanian S, Reed MW, et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J Med Genet 48: 477–484. 10.1136/jmedgenet-2011-100018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, et al. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys 81: 52–58. 10.1016/j.ijrobp.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 11. Dunning AM, Healey CS, Pharoah PD, Teare MD, Ponder BA, Easton DF. (1999) A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 8: 843–854. [PubMed] [Google Scholar]
- 12. Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. (2002) Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet 31: 33–36. [DOI] [PubMed] [Google Scholar]
- 13. Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. (2007) A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet 39: 352–358. [DOI] [PubMed] [Google Scholar]
- 14. Goldhirsch A, Gelber RD, Yothers G, Gray RJ, Green S, Bryant J, et al. (2001) Adjuvant therapy for very young women with breast cancer: need for tailored treatments. J Natl Cancer Inst Monogr: 44–51. [DOI] [PubMed] [Google Scholar]
- 15. Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjäkoski K, et al. (2008) NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet 40: 844–853. 10.1038/ng.155 [DOI] [PubMed] [Google Scholar]
- 16. Budach W, Kammers K, Boelke E, Matuschek C (2013) Adjuvant radiotherapy of regional lymph nodes in breast cancer—a meta-analysis of randomized trials. Radiat Oncol 8: 267 10.1186/1748-717X-8-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9: 95–107. 10.1038/nrc2584 [DOI] [PubMed] [Google Scholar]
- 19. Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. (2014) Analysis of TP53 Mutation Status in Human Cancer CellLines: A Reassessment. Hum Mutat. 35:756–65. 10.1002/humu.22556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dahabreh IJ, Schmid CH, Lau J, Varvarigou V, Murray S, Trikalinos TA. (2013) Genotype misclassification in genetic association studies of the rs1042522 TP53 (Arg72Pro) polymorphism: a systematic review of studies of breast, lung, colorectal, ovarian, and endometrial cancer. Am J Epidemiol 177: 1317–1325. 10.1093/aje/kws394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng H, Ma B, Jiang R, Wang W, Guo H, Shen N, et al. (2012) Individual and combined effects of MDM2 SNP309 and TP53 Arg72Pro on breast cancer risk: an updated meta-analysis. Mol Biol Rep 39: 9265–9274. 10.1007/s11033-012-1800-z [DOI] [PubMed] [Google Scholar]
- 22. Sagne C, Marcel V, Amadou A, Hainaut P, Olivier M, Hall J. (2013) A meta-analysis of cancer risk associated with the TP53 intron 3 duplication polymorphism (rs17878362): geographic and tumor-specific effects. Cell Death Dis 4: e492 10.1038/cddis.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toyama T, Zhang Z, Nishio M, Hamaguchi M, Kondo N, Iwase H, et al. (2007) Association of TP53 codon 72 polymorphism and the outcome of adjuvant therapy in breast cancer patients. Breast Cancer Res 9: R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, et al. (2005) p53 Codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res 11: 7328–7333. [DOI] [PubMed] [Google Scholar]
- 25. Szkandera J, Absenger G, Dandachi N, Regitnig P, Lax S, Stotz M, et al. Analysis of functional germline polymorphisms for prediction of response to anthracycline-based neoadjuvant chemotherapy in breast cancer. Mol Genet Genomics 287: 755–764. 10.1007/s00438-012-0715-7 [DOI] [PubMed] [Google Scholar]
- 26. Rodrigues P, Furriol J, Tormo E, Ballester S, Lluch A, Eroles P. (2013) Epistatic interaction of Arg72Pro TP53 and -710 C/T VEGFR1 polymorphisms in breast cancer: predisposition and survival. Mol Cell Biochem 379: 181–190. 10.1007/s11010-013-1640-8 [DOI] [PubMed] [Google Scholar]
- 27. Polakova V, Pardini B, Naccarati A, Landi S, Slyskova J, Novotny J, et al. (2009) Genotype and haplotype analysis of cell cycle genes in sporadic colorectal cancer in the Czech Republic. Hum Mutat 30: 661–668. 10.1002/humu.20931 [DOI] [PubMed] [Google Scholar]
- 28. Naccarati A, Pardini B, Polakova V, Smerhovsky Z, Vodickova L, Soucek P, et al. (2010) Genotype and haplotype analysis of TP53 gene and the risk of pancreatic cancer: an association study in the Czech Republic. Carcinogenesis 31: 666–670. 10.1093/carcin/bgq032 [DOI] [PubMed] [Google Scholar]
- 29. Hubackova M, Vaclavikova R, Ehrlichova M, Mrhalova M, Kodet R, Kubackova K, et al. (2012) Association of superoxide dismutases and NAD(P)H quinone oxidoreductases with prognosis of patients with breast carcinomas. Int J Cancer 130: 338–348. 10.1002/ijc.26006 [DOI] [PubMed] [Google Scholar]
- 30. Vaclavikova R, Ehrlichova M, Hlavata I, Pecha V, Kozevnikovova R, Trnkova M, et al. (2012) Detection of frequent ABCB1 polymorphisms by high-resolution melting curve analysis and their effect on breast carcinoma prognosis. Clin Chem Lab Med 50: 1999–2007. 10.1515/cclm-2012-0103 [DOI] [PubMed] [Google Scholar]
- 31. Kunicka T, Soucek P (2014) Importance of ABCC1 for cancer therapy and prognosis. Drug Metab Rev. [DOI] [PubMed] [Google Scholar]
- 32. Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, et al. (2012) Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis 33: 1346–1351. 10.1093/carcin/bgs172 [DOI] [PubMed] [Google Scholar]
- 33. Naccarati A, Polakova V, Pardini B, Vodickova L, Hemminki K, Kumar R, et al. (2012) Mutations and polymorphisms in TP53 gene—an overview on the role in colorectal cancer. Mutagenesis 27: 211–218. 10.1093/mutage/ger067 [DOI] [PubMed] [Google Scholar]
- 34. Pardini B, Kumar R, Naccarati A, Prasad RB, Forsti A, Polakova V, et al. (2011) MTHFR and MTRR genotype and haplotype analysis and colorectal cancer susceptibility in a case-control study from the Czech Republic. Mutat Res 721: 74–80. 10.1016/j.mrgentox.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 35. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344: 783–792. [DOI] [PubMed] [Google Scholar]
- 36. Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, et al. (2008) DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res 638: 146–153. [DOI] [PubMed] [Google Scholar]
- 37. Vymetalkova V, Pardini B, Rosa F, Di Gaetano C, Novotny J, Levy M, et al. (2014) Variations in mismatch repair genes and colorectal cancer risk and clinical outcome. Mutagenesis 29: 259–265. 10.1093/mutage/geu014 [DOI] [PubMed] [Google Scholar]
- 38. Wu D, Zhang Z, Chu H, Xu M, Xue Y, Zhu H, et al. (2013) Intron 3 sixteen base pairs duplication polymorphism of p53 contributes to breast cancer susceptibility: evidence from meta-analysis. PLoS One 8: e61662 10.1371/journal.pone.0061662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bisof V, Salihovic MP, Narancic NS, Skaric-Juric T, Jakic-Razumovic J, Janićijević B, et al. (2010) TP53 gene polymorphisms and breast cancer in Croatian women: a pilot study. Eur J Gynaecol Oncol 31: 539–544. [PubMed] [Google Scholar]
- 40. Trifa F, Karray-Chouayekh S, Mabrouk I, Baccouche S, Khabir A, Sellami-Boudawara T, et al. (2010) Haplotype analysis of p53 polymorphisms: Arg72Pro, Ins16bp and G13964C in Tunisian patients with familial or sporadic breast cancer. Cancer Epidemiol 34: 184–188. 10.1016/j.canep.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 41. Lajin B, Alhaj Sakur A, Alachkar A (2013) Association between polymorphisms in apoptotic genes and susceptibility for developing breast cancer in Syrian women. Breast Cancer Res Treat 138: 611–619. 10.1007/s10549-013-2467-4 [DOI] [PubMed] [Google Scholar]
- 42. Crawford DC, Nickerson DA (2005) Definition and clinical importance of haplotypes. Annu Rev Med 56: 303–320. [DOI] [PubMed] [Google Scholar]
- 43. Baudis M (2007) Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. BMC Cancer 7: 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghazani AA, Arneson N, Warren K, Pintilie M, Bayani J, Squire JA, et al. (2007) Genomic alterations in sporadic synchronous primary breast cancer using array and metaphase comparative genomic hybridization. Neoplasia 9: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao Y, Niu Y, Wang X, Wei L, Zhang R, Lv S, et al. Chromosome aberrations associated with centrosome defects: a study of comparative genomic hybridization in breast cancer. Hum Pathol 42: 1693–1701. 10.1016/j.humpath.2010.12.027 [DOI] [PubMed] [Google Scholar]
- 46. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M (2007) TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 26: 2157–2165. [DOI] [PubMed] [Google Scholar]
- 47. Pharoah PD, Day NE, Caldas C (1999) Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 80: 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167. [DOI] [PubMed] [Google Scholar]
- 49. Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, et al. (2005) Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res 11: 5098–5103. [DOI] [PubMed] [Google Scholar]
- 50. Bisof V, Salihovic MP, Narancic NS, Skaric-Juric T, Jakic-Razumovic J, Janićijević B, et al. (2012) The TP53 gene polymorphisms and survival of sporadic breast cancer patients. Med Oncol 29: 472–478. 10.1007/s12032-011-9875-2 [DOI] [PubMed] [Google Scholar]
- 51. Szkandera J, Absenger G, Dandachi N, Regitnig P, Lax S, Stotz M, et al. (2012) Analysis of functional germline polymorphisms for prediction of response to anthracycline-based neoadjuvant chemotherapy in breast cancer. Mol Genet Genomics 287: 755–764. 10.1007/s00438-012-0715-7 [DOI] [PubMed] [Google Scholar]
- 52. Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutiérrez-Enríquez S, et al. (2004) A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene 23: 1954–1956. [DOI] [PubMed] [Google Scholar]
- 53. Pim D, Banks L (2004) p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer 108: 196–199. [DOI] [PubMed] [Google Scholar]
- 54. Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- 55. Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, et al. (2004) Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene 23: 3328–3337. [DOI] [PubMed] [Google Scholar]
- 56. Schneider-Stock R, Boltze C, Peters B, Szibor R, Landt O, Meyer F, et al. (2004) Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia 6: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, et al. (2003) Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res 9: 4860–4864. [PubMed] [Google Scholar]
- 58. O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, et al. (1997) Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res 57: 4285–4300. [PubMed] [Google Scholar]
- 59. Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, et al. (1994) p53 status and the efficacy of cancer therapy in vivo. Science 266: 807–810. [DOI] [PubMed] [Google Scholar]
- 60. Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, et al. (1996) Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat Med 2: 72–79. [DOI] [PubMed] [Google Scholar]
- 61. Fernandez-Cuesta L, Oakman C, Falagan-Lotsch P, Smoth KS, Quinaux E, Buyse M, et al. (2012) Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02–98 phase III trial. Breast Cancer Res 14: R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Geisler S, Lonning PE, Aas T, Johnsen H, Fluge O, Haugen DF, et al. (2001) Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res 61: 2505–2512. [PubMed] [Google Scholar]
- 63. Di Leo A, Tanner M, Desmedt C, Paesmans M, Cardoso F, Durbecq V, et al. (2007) p-53 gene mutations as a predictive marker in a population of advanced breast cancer patients randomly treated with doxorubicin or docetaxel in the context of a phase III clinical trial. Ann Oncol 18: 997–1003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


